Abstract
The C1q-tumor necrosis factor 5 (C1QTNF5) protein plays a significant role in retinal pigmented epithelium (RPE) cellular adhesion. The C1QTNF5 gene is co-transcribed with the frizzled-related protein (MFRP) gene. A Ser-to-Arg mutation at site 163 (S163R) in C1QTNF5 is known to cause late-onset retinal macular degeneration (L-ORMD). Here we also found that C1QTNF5 monomers can multimerize into a bouquet-like octadecamer. We found that a novel intermolecular hydrogen-bond network of S163 that glues adjacent globular heads of C1QTNF5 together and was weakened or abolished by the R163 pathogenic mutation. These findings could underlie the structural basis of this protein’s adhesive function and relate to the pathogenesis of its S163R mutation. Additionally, the fact that C1QTNF5 immobilized to a resin selectively enriched detergent extracted membrane-bound MFRP, further confirming their interaction and implying functions other than cellular adhesion for C1QTNF5.
Keywords: L-ORMD, L-ORD, age-related macular degeneration, cellular adhesion, CTRP5, C1QTNF5
Introduction
L-ORMD is a fully penetrant autosomal dominant eye disorder caused by a single mutation c.686C>G (p.S163R) in the C1QTNF5 protein (Ayyagari et al., 2005; Hayward et al., 2003). L-ORMD is a progressive eye disorder. Affected patients start to experience visual difficulties in their 40s and eventually lose their sight after their 60s. The exact prevalence of this condition is unknown, in part because it is easily misdiagnosed, mostly as AMD in its early stage and as retinitis pigmentosa subsequently (Borooah et al., 2009a). L-ORMD consistently exhibits thick extracellular deposits of lipid-rich material between the retinal pigmented epithelium (RPE) and Bruch’s membrane (Borooah et al., 2009b; Shu et al., 2006a). Understanding the molecular pathogenesis of L-ORMD is important for the design of appropriate therapeutics. However, it is still unclear how C1QTNF5 functions in RPE cellular adhesion and how the pathogenic S163R mutation modifies its molecular function.
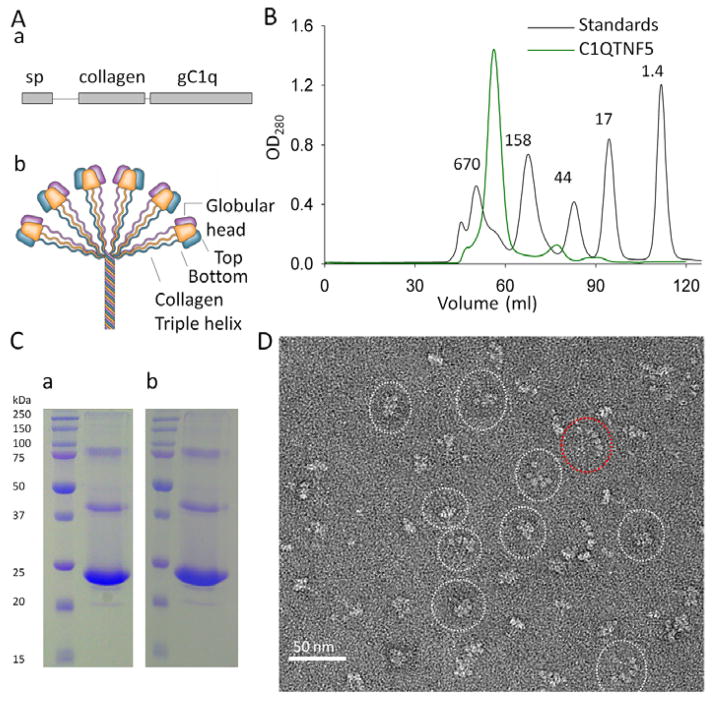
The human C1QTNF5 gene encodes a 25 kDa secretory and membrane-associated protein containing three domains: a signal peptide (sp domain, residues 1–15), a collagen domain (residues 30–98 composed of 23 uninterrupted Gly-X-Y repeats) and a globular C1q domain (gC1q domain, residues 103–243) (Hayward et al., 2003; Mandal et al., 2006b; Tu and Palczewski, 2012) (Fig. 1A). C1QTNF5 is a novel member of the C1q family that includes C1q, adiponectin, collagens, and other structurally related proteins. C1q family proteins share the common feature of trimerization (Shapiro and Scherer, 1998). Their collagen domains intertwine into a stalk-like triple helix and their gC1q domains trimerize into flower-like globular heads (Fig. 1A) (Tu and Palczewski, 2012). Although studies of complement protein C1q and adiponectin have shown that six trimers can further multimerize into a bouquet-like octadecameric assembly through their collagen triple helices (Fig. 1A), there is no direct evidence that C1QTNF5 also adopts similar architecture.
Fig. 1.
C1QTNF5 adopts a bouquet-like assembly without N-terminal disulfide bonding. (A) Primary structural features of the gC1q domain (a) and a bouquet-like assembly of C1q family proteins (b). sp, signal peptide; gC1q, globular C1q domain. (B) Appearance of C1QTNF5 after gel filtration chromatography. The column was calibrated by gel filtration molecular weight standards, namely bovine thyroglobulin (670 kDa), bovine γ-globulin (158 kDa), chicken ovalbumin (44 kDa), horse myoglobin (17 kDa) and vitamin B12 (1.4 kDa). (C) Non-reducing SDS-PAGE (a) and reducing SDS-PAGE (b) of the predominant peak fraction (~450 kDa) of C1QTNF5 obtained by gel filtration chromatography. (D) Appearance of C1QTNF5 expressed in bacteria after negative staining electron microscopy. Shapes of particles can be divided into two major categories: bouquet-shaped and fan-shaped. Representative images are outlined with white and red dotted ovals, respectively.
The S163R pathogenic mutation is located within the gC1q domain of C1QTNF5. This mutant is not secreted and displays a significant reduction in implementing cellular adhesion (Mandal et al., 2006b; Shu et al., 2006a). It also is unstable and prone to aggregation in vitro (Hayward et al., 2003; Shu et al., 2006b). Our previous study has revealed the structural mechanism underlying this instability resulted from aggregation of the S163R mutant (Tu and Palczewski, 2012) but it is still unclear how C1QTNF5 functions in RPE adhesion. Moreover, it is notable that all examined patients with this mutation are heterozygous, meaning that wild-type C1QTNF5 and its S163R mutant co-exist within secreted oligomer(s) of this protein. Thus, the structural effects of the S163R mutation which cause reduction or loss of function of the heterozygous C1QTNF5 oligomers leading to L-ORMD need to be investigated.
The C1QTNF5 gene is located entirely within the 3′-UTR of the MFRP gene and is co-transcribed with MFRP (Hayward et al., 2003). C1QTNF5 can co-immunoprecipitate with MFRP, and vice versa (Mandal et al., 2006a). Moreover, a study has shown that C1QTNF5 can pull down GST-tagged CUB domains of MFRP expressed in bacteria (Shu et al., 2006a), indicating that they directly interact with each other. Though native CUB domains contain two disulfide bonds, heterologously expressed CUB domains lack disulfide bonds so their interactions with C1QTNF5 could be artificial and caused by the partially folded CUB domain. Thus, this pull-down study needs to be further confirmed with MFRP expressed in mammalian cells that allow formation of disulfide bonds. Their interaction would imply that C1QTNF5 could have functions other than cellular adhesion.
Thus, in this study, we aimed to examine whether C1QTNF5 could form a bouquet-like octadecameric assembly, understand how C1QTNF5 expedites RPE cellular adhesion, gain insights into the molecular pathogenesis of the S163R mutant in attenuating RPE cell adhesion and provide further proof that C1QTNF5 can interact with MFRP.
Results and Discussion
Expression, purification and electron microscopic (EM) imaging of C1QTNF5
His-tagged C1QTNF5 was expressed in the cytosol of E.coli Rossetta (DE3) and purified by metal affinity in tandem with gel filtration chromatography. C1QTNF5 eluted as a predominant peak with an apparent mass of ~450 kDa on gel filtration chromatography (Fig. 1B). There were two minor peaks with apparent masses corresponding to the trimer (72 kDa) and monomer (24 kDa). One explanation for this phenomenon is that overexpression of C1QTNF5 in bacteria results in a small fraction of incompletely folded protein that could not be assembled into the octadecamer. Another possibility is that there exists an equilibrium among octadecamer, trimer and monomer. We then performed non-reducing sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on the purified ~450 kDa peak fraction and found that it migrated as a major band with an estimated molecular weight of ~24 kDa, close to the theoretical molecular weight (23.5 kDa) of a single C1QTNF5 molecule (Fig. 1Ca). This result indicates that monomeric C1QTNF5 can form a high-molecular-weight multimer. All C1q family protein multimers have a basic oligomerization unit of a trimer. A trimer of C1QTNF5 has a mass of ~72 kDa, so an apparent mass of ~450 kDa indicates that this multimer is possibly composed of 6 trimers, making it an octadecamer.
C1QTNF5 contains three Cys residues; two (C28 and C98) are localized in the N-terminal region and one (C145) is in the gC1q domain (Wong et al., 2008). With respect to the two visible minor oligomeric bands noted on non-reducing SDS-PAGE, it is known that Rossetta (DE3) bacteria contain a highly reduced cytosolic environment which prevents formation of disulfide bonds. We thus performed reducing SDS-PAGE by adding 10 mM fresh DTT to the sample loading buffer to rule out possible disulfide-bond formation during the purification process. However, the reducing SDS-PAGE did not provide any observable difference from non-reducing SDS-PAGE (Fig. 1C). Therefore, the minor C1QTNF5 oligomeric bands visualized on SDS-PAGE were not caused by intermolecular disulfide bonds. Previous studies have shown that the C1QTNF5 gC1q domain can migrate as oligomers on non-reducing or reducing SDS-PAGE (Shu et al., 2006a; Shu et al., 2006b), but it is also possible that the collagen super helices contribute to this appearance of oligomeric bands on SDS-PAGE. Furthermore, we performed EM imaging of the predominant ~450 kDa peak fraction, and the negatively stained EM images showed that C1QTNF5 multimerized into bouquet-like super structures. Each C1QTNF5 trimer has one globular head, so an octadecamer has six globular heads. Some particles clearly revealed six globular heads, demonstrating that C1QTNF5 can form an octadecameric assembly (Fig. 1D and Supplementary Fig. 1). Notably, the negatively stained particles of C1QTNF5 also displayed structural polymorphism indicated by globular heads of the C1QTNF5 multimer assuming various spatial positions relative to each other (Fig. 1D and Supplementary Fig. 1). A similar structural diversity also was observed for adiponectin (Radjainia et al., 2008). Such flexibility could allow C1QTNF5 to achieve the best binding geometry by adjusting the position of each globular head and therefore enhance its multivalent interactions. This could be an inherent feature of these multivalent C1q family proteins.
For two reasons, our EM images were not subjected to quantifying the distribution of the number of globular heads per multimer. First, the images that we have obtained are projections to a CCD camera. Projections of C1QTNF5 multimers to the camera could have overlapped and therefore obstructed portions of the proteins, so that six globular heads are not visible for all multimers. However, this does not mean that they do not have six globular heads. Some of the particles were in an orientation relative to the camera that resulted in projections with six visible globular heads (Fig. 1D, highlighted with dotted cycles, and Supplementary Fig. 1). Second, as mentioned previously, C1QTNF5 appeared to possess structural heterogeneity as indicated by various spatial layouts of its globular heads. We therefore could not construct three-dimensional structures for each particle, which prevented us from accurately counting globular heads for all particles.
Studies have shown that octadecameric assembly of C1q or adiponectin is dependent on N-terminal intermolecular disulfide bonds (Kishore et al., 2004; Tsao et al., 2003; Wong et al., 2008). When the N-terminal Cys residues (C28 and C98) of C1QTNF5 were mutated to Ala residues, the resulting mutant could only form trimers (Wong et al., 2008). These results prompted a proposal that C1QTNF5 also requires N-terminal intermolecular disulfide bonds to assemble into an octadecamer (Wong et al., 2008). Interestingly, our data show that C1QTNF5 also can adopt the same bouquet-like octadecameric assembly, like C1q and adiponectin, that is independent of N-terminal disulfide bonding. Indeed, there still is no direct evidence indicating that N-terminal Cys residues of C1QTNF5 form intermolecular disulfide bonds in vivo. Inability of the C1QTNF5 Cys-to-Ala mutant to form an octadecamer could result from the Ala residues inducing local changes that prevented the trimers from forming higher-order oligomers. The structural role(s) of the Cys side chain and whether potential N-terminal intermolecular disulfide bonds can stabilize the formed C1QTNF5 octadecamer require further investigation.
The C1QTNF5 gC1q domain structure crystallized in space group P63 reveals novel interactions of the pathogenic S163 mutation site
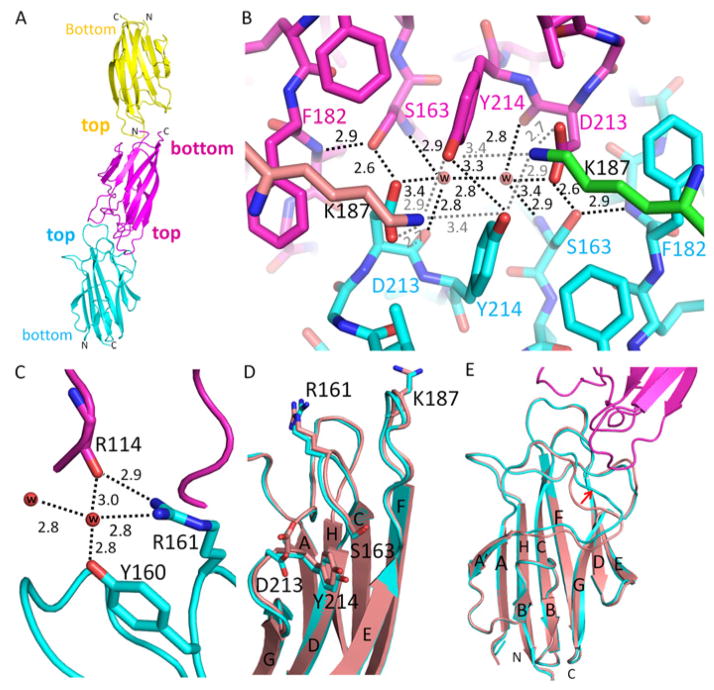
The previously reported structure of the C1QTNF5 gC1q domain was crystallized in space group H3 and solved to 1.34 Å (Tu and Palczewski, 2012). Here, we report a structure of the C1QTNF5 gC1q domain crystallized in a new space group, namely P63, and solved to a comparable resolution of 1.42 Å. Statistics of data processing and structural refinement are summarized in Table 1. The C1QTNF5 gC1q domain in this new space group reveals several novel molecular packing features. The asymmetric unit contains three molecules giving the Matthews’s coefficient of 2.02 A3/Da with 39.1% solvent content. It is known that the gC1q domain trimerizes into a stable globular head (Tu and Palczewski, 2012). However, because each of the three molecules in the asymmetric unit is derived from a different globular head, they are independent of each other with respect to trimerization. These three molecules stacked on each other in two different ways: top-to-bottom and top-to-top (Fig. 2A). The top-to-bottom packing has a negligible interface for interactions. Furthermore, considering the collagen domain stems from the bottom of the gC1q domain, this top-to-bottom intermolecular interaction is unlikely to occur in C1QTNF5 because the collagen domain would be spatially excluded. In contrast, top-to-top packing involves tight interactions with 759 Å2 of buried surface area, which is comparable to the trimeric interface area (1086 Å2) of each protomer of a C1QTNF5 globular head (Tu and Palczewski, 2012). Top-to-top packing thus permits intermolecular interactions between adjacent globular heads, which has implications for mediating RPE cellular adhesion. Notably, the pathogenic S163 mutation site is located at the interface and plays a key role in creating this top-to-top packing. Compared with the previously reported structure, S163 forms not only a common intramolecular hydrogen bond with the main chain nitrogen of F182 but also a novel intermolecular hydrogen-bond network together with D213, two water molecules and Y214 (Fig. 2B). The hydroxyl group of S163 interacts with the side chain of D213 from its top-to-top packing partner at a distance of ~2.6 Å (Fig. 2B), providing the strongest intermolecular hydrogen bond involved in this top-to-top packing. Two water molecules also play critical roles in forming this interaction network. One water molecule connects the main-chain nitrogen of S163 from the first partner with the main-chain oxygen and side chain of D213 from the second partner by hydrogen bonds, and a second water molecule forms the same interaction between the second and first partner, creating a symmetric intermolecular interaction network that holds these two gC1q domains together (Fig. 2B). Y214 interacts with Y214 from its top-to-top interacting partner by side-chain hydrogen bonding, which strengthens and stabilizes this S163-involved hydrogen-bonding network (Fig. 2B). Surprisingly, this S163 interaction network is not limited to these two interacting gC1q molecules, but further extends to their respective trimerization partners by their D213 and Y214 residues. D213 of one molecule forms a salt bridge with K187 from the molecule that trimerizes with its top-to-top interacting partner at a distance of 2.7 Å, and the Y214 residues of these two partners further interact with this K187 through their side chains at distances of 2.9 Å and 3.4 Å, respectively. All residues involved in this S163 hydrogen-bond network are highly evolutionally conserved in C1QTNF5 from fish to humans (Supplementary Fig. 2). In sharp contrast, S163 is not involved in any intermolecular interactions in the previously reported C1QTNF5 structure (Tu and Palczewski, 2012), nor does this hydrogen-bond network exist. Moreover, the C1QTNF5 gC1q domain crystallized in this reported H3 space group possessed only one gC1q molecule in the asymmetric unit (Tu and Palczewski, 2012).
Table 1.
Statistics of Data Collection and Refinement
| Data collection | |
| Space group | P63 |
| Unit cell dimensions | |
| a = b (Å) | 50.60 |
| c (Å) | 268.53 |
| Resolution range (Å) | 134.27 – 1.42 (1.50 – 1.42) |
| Unique reflections | 72,440 |
| Rmergeb | 0.041 (0.122) |
| <I>/<σI>c | 30.3 (3.0) |
| Redundancy | 4.8 (8.4) |
| Completeness (%) | 99.1 (93.8) |
| Refinement Statistics | |
| Resolution range (Å) | 46.28–1.42 |
| Number of atoms | |
| Protein | 3,202 |
| Water | 339 |
| Rmsd | |
| Bond length (Å) | 0.012 |
| Bond angle (°) | 1.645 |
| Rwork/Rfreed (%) | 10.4/14.3 |
| Ramachandran analysis (%)e | |
| Most favored | 85.5 |
| Additionally allowed | 12.7 |
| Generously allowed | 1.7 |
| Disallowed | 0.0 |
Selected statistics from data collection and refinement are shown. Values in parentheses are data from the highest resolution shell.
Rmerge is ΣhklΣj|Ij - 〈I〉 |/ΣhklΣj Ij. 〈I〉 is the mean intensity of j observations of reflection hkl and its symmetry equivalents.
<I>/<σI> is the mean intensity divided by the mean error.
Rwork is Σ||Fo|- |Fc||/Σ|Fo| where Fo is an observed amplitude and Fc is a calculated amplitude; Rfree is the same statistic calculated over a subset of the data that was not used for refinement.
Ramachandran analysis from PROCHECK (Laskowski et al., 1993).
Fig. 2.
Intermolecular interactions of CIQTNF5 globular heads. (A) Colored yellow, magenta and cyan, three gC1q molecules in the asymmetric unit of space group P63 are stacked on each other in two ways: top-to-bottom and top-to-top. (B) An intermolecular hydrogen bond network facilitates top-to-top packing of two adjacent gC1q molecules. This intermolecular hydrogen bond network involves the pathogenic mutation residue S163, D213, two water molecules and Y214. This intermolecular hydrogen bond network is further extended to other molecules by K187. The two K187 residues are not from these two top-to-top packed gC1q molecules but rather from their respective trimerization partners, so their carbon atoms are colored in green and tint, respectively. Water molecules are represented with red balls. Hydrogen-bonds are shown with black or gray dotted lines. Distances are given in Å. (C) Water molecules connect R114 of one gC1q molecule with Y160 and R161 of another gC1q molecule by hydrogen bonds. (D) Structural comparison of gC1q domains involved (cyan) and uninvolved (tint, PDB: 4F3J) in top-to-top packing. Side chains of D213 and R161 are flipped in top-to-top packing. Y214 shows a slight conformational shift but S163 retains a nearly identical conformation. (E) Top-to-top intermolecular interaction features a conformational shift of the β-sheet A|A′ loop indicated by the red arrow. Structures of gC1q domains involved in top-to-top packing are colored cyan and magenta. The gC1q domain structure uninvolved in this type of packing is colored with tint (PDB: 4F3J).
In addition to the above hydrogen network, other interactions facilitate this top-to-top packing. The R161 side chain forms a hydrogen bond with the main-chain oxygen of R114 from its top-to-top interacting partner at a distance of 2.9 Å (Fig. 2C), and vice versa. Moreover, water molecules other than those involved in the S163 hydrogen-bond network create additional intermolecular hydrogen-bond networks linking R161, Y160 and R114 of the two top-to-top packed partners (Fig. 2C). These residues are also highly evolutionally conserved in C1QTNF5 (Supplementary Fig. 2). Another water molecule connects the main-chain oxygens of G184 and P116 of these two partners at distances of ~2.8 Å and ~2.8 Å, respectively (Supplementary Fig. 3).
Although the three molecules in the asymmetric unit of the P63 space group shared the same overall conformation, they were not ordered to the same extent because of differences in their packing environments. The two molecules packed top-to-top were better ordered with similar average main-chain B factors of ~10 A2. These two molecules also had nearly identical main-chain and side-chain conformations. However, the third molecule forming the head-to-bottom packing configuration was significantly less ordered according to its electron density and an average main-chain B factor of ~39 A2. Thus, we used those molecules forming top-to-top packing for structural comparisons by default. C1QTNF5 adopts a 10-strand jelly-roll fold arranged in two anti-parallel, 5-stranded β-sheets (Tu and Palczewski, 2012). We did not observe any significant conformational shifts of these β-sheets after superposing this new structure on the previously reported structure of the gC1 domain (Fig. 2D and 2E), supported by the average coordinate root-mean-square deviation of ~0.13 Å. Although S163 formed the described novel intermolecular hydrogen-bonding network, it was still tightly surrounded by hydrophobic residues A162, F179, F181, F182 and Y214, just as observed in the reported structures crystallized in the H3 space group (Tu and Palczewski, 2012). Conformations of S163 and its surrounding residues in the P63 space group are nearly identical to those seen in the H3 space group (Fig. 2D). However, the side chains of D213 and K187 were flipped relative to their conformations in the H3 space group structure, a feature caused by the hydrogen bonding network (Fig. 2D). The R161 side chain also was flipped relative to the H3 space group structure and interacted with the main-chain oxygen of R114 from its packing partner. We also observed a significant main-chain conformation shift of residues 115–125, a loop of β-sheet A|A′ (Fig. 2E). This was caused primarily by top-to-top intermolecular packing, because the loop is located at the intermolecular interface (Fig. 2E).
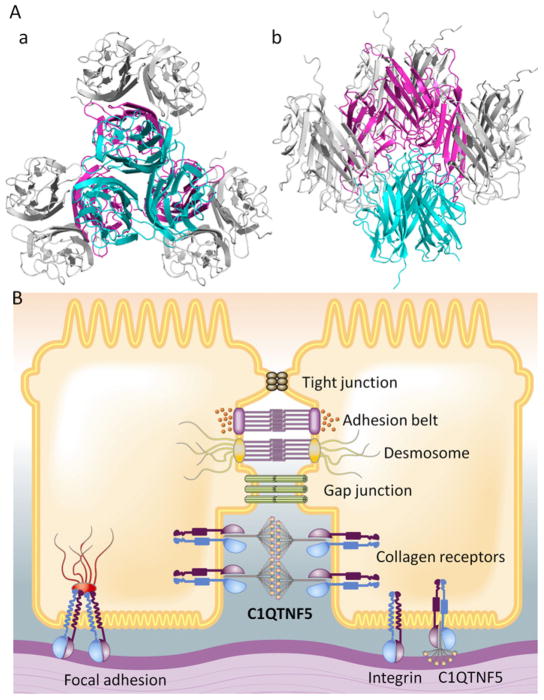
The gC1q domain was packed into the crystal lattice as a trimer that constitutes the globular head of C1QTNF5. Therefore, the top-to-top interaction of gC1q domains is occurs between adjacent C1QTNF5 globular heads (Fig. 3A). This self-interaction reveals that two adjacent C1QTNF5 molecules can interact with each other through their globular heads in a crowded environment, a feature with implications for its function at the lateral and basal membranes of RPE cells. C1QTNF5 is a secreted and membrane-associated protein of the RPE. One possible scenario is that the interaction of C1QTNF5 globular heads facilitates binding of C1QTNF5 short-chain collagen to collagen receptors on adjacent RPE cell membranes (Fig. 3B). This binding not only would enhance cellular adhesion but also fill gaps between adjacent RPE cells or RPE cells and Bruch’s membrane. Our structure thus provides a plausible hypothesis for the function of C1QTNF5 in cellular adhesion. Mutation of S163 to Arg not only causes exposure of hydrophobic residues in apex loops resulting in aggregation of C1QTNF5 (Tu and Palczewski, 2012), but also can abolish potential intermolecular hydrogen bonding with D213. With a significantly enlarged side-chain volume in the tightly packed interface, the S163R mutation may further prevent the top-to-top interaction of gC1q domains because of steric exclusion. All studied L-ORMD patients to date possess a heterozygous gene for C1QTNF5, so both wild-type and S163R mutants co-exist in the globular heads of this protein’s octadecameric assembly. In this scenario, the S163R mutation could destabilize the heterozygous globular heads and reduce the head-to-head interactions of the C1QTNF5 octadecamer, thereby compromising its ability to facilitate the adhesion of RPE cells. Consequently, affected individuals would accumulate lipids in their sub-RPE space with age.
Fig. 3.
C1QTNF5 functions in RPE cellular adhesion through its globular heads. (A) Head-to-head interactions of C1QTNF5 present in space group P63: perpendicular view (a) and side view (b). Globular heads were generated by applying crystallographic 3-fold axes to the top-to-top interacting partner. Three gC1q monomers of one globular head respectively interact with other three globular heads. These interwoven interactions can assemble all neighboring C1QTNF5 multimers on adjacent RPE cell membranes together. (B) Globular heads of two bouquet-like C1QTNF5 multimers can interact with each other, whereas their collagen domains bind to collagen receptors on membranes of two adjacent RPE cells, thus filling the sub-retinal space and gluing the two RPE cells together.
Pull-down of MFRP by immobilized C1QTNF5
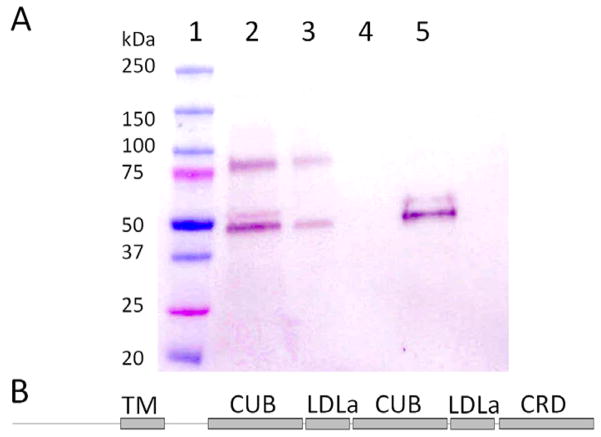
After immobilizing C1QTNF5 onto CNBr-activated agarose, we then pulled down MFRP from a DDM-solubilized cell membrane preparation of CHO-K1 cells. An immunoblot indicated that a significant portion of MFRP remained bound to the resin (Fig. 4A). MFRP was not washed out by a buffer containing 400 mM NaCl but eluted in 2% SDS (Fig. 4A). This result demonstrates that C1QTNF5 tightly and specifically interacted with MFRP. Previous studies have shown that C1QTNF5 can be co-immunoprecipitated with MFRP and that the C1QTNF5 gC1q domain can pull down bacteria-expressed MFRP domains (Mandal et al., 2006; Shu et al., 2006). We here present an analysis of mammalian cell-derived MFRP pulled down with C1QTNF5 covalently coupled to agarose beads. Our findings complemented those previously reported and provide further evidence that C1QTNF5 can indeed interact with MFRP.
Fig. 4.
C1QTNF5 specifically interacts with MFRP. (A) Immunoblot of 1D4-tagged MFRP pulled down by immobilized C1QTNF5. The complex was detected with alkaline phosphatase--conjugated 1D4 antibody: Lane 1. Protein standards. 2. DDM-solubilized CHO cell membranes expressing MFRP. 3. Flow-through. 4. Resin washed with 400 mM NaCl. 5. Elution of MFRP with 2% SDS. (B) Primary structural features of MFRP: TM, transmembrane domain; CUB, cubilin domain; LDLa, low density lipoprotein receptor class A domain; CRD, Cys-rich domain.
The genomic organization, co-transcription and interaction of C1QTNF5 and MFRP strongly suggest a functional collaboration of these two genes. However, the phenotypes of diseases associated with these two genes appear to differ. Although mutations in both can cause retinitis pigmentosa-like symptoms, those in MFRP are also associated with nanophthalmos and posterior microphthalmia, indicating that MFRP is involved in ocular development. In contrast, the S163R mutation in C1QTNF5 causes late-onset retinal degeneration wherein symptoms are first detected after the age of 40. These differences suggest that C1QTNF5 and MFRP are not completely dicistronic. MFRP is a type II membrane protein with a large C-terminal portion exposed to the extracellular space. This portion contains two cubilin (CUB) domains, two low density lipoprotein receptor class A (LDLa) domains and one frizzled-like Cys-rich domain (CRD). The CUB and LDLa domains are arranged in an alternating pattern (Fig. 4B). The CUB domain is a structural motif found almost exclusively in extracellular and plasma membrane-associated proteins. Notably, this domain participates in receptor-mediated endocytosis (Verroust, 2002). LDLa is a structural motif that binds low density lipoproteins which then are taken up by endocytosis to play a central role in mammalian cholesterol metabolism. CRD may be involved in signaling, as the CRDs of frizzled receptors bind Wingless/Ints (Wnts). Together, these analyses suggest that MFRP may be involved in lipid transport or serve as a sensor for lipids excreted by exocytosis into the sub-retinal space (the space between the RPE and Bruch’s membrane). Because the RPE routinely phagocytoses rod outer segment discs shed from photoreceptor cells, some lipids inevitably are excreted into the sub-retinal space. Thus, to avoid lipid accumulation in the sub-retinal space, there must be mechanisms to either return lipids to the RPE or transport them across Bruch’s membrane into blood stream. There is no evidence yet that MFRP and C1QTNF5 participate in such functions, but their interaction at least indicates that C1QTNF5 has functions other than cellular adhesion because mutations in MFRP are not associated with L-ORMD. Both proteins may have multiple actions, as their associated diseases display a variety of complex phenotypes.
Sequence comparisons of C1QTNF5 with its paralogues
Sequences of C1QTNF5 and its newly identified paralogues, termed C1q/TNF-related proteins (CTRP) (Schaffler and Buechler, 2012; Wong et al., 2008), have been aligned and compared. We previously identified a characteristic hydrophobic segment, FFGGWP (residues 181–186), within the apical loop of C1QTNF5 through structural comparison and sequence alignment with other well-established members including C1q, adiponectin and collagens (Tu and Palczewski, 2012). This hydrophobic segment is located close to residue S163, and its conformation is affected by the pathogenic mutation S163R. We found that this segment is unique, and S163 is not evolutionarily conserved among these newly identified paralogues (Fig. 5). CTRP1, 6, and 8 lack the structural elements for coordinating calcium ions, similar to C1QTNF5 (Fig. 5, highlighted by underscores). However, CTRP2, 3, 4, 7, 9, and 10 have these structural elements. Calcium ions are essential to the functions of C1q, adiponectin and collagens. Previous structural studies have shown that Y152, F230 and V232 form a hydrophobic core critical for trimerization of C1QTNF5 (Tu and Palczewski, 2012). These residues are conserved among C1q, adiponectin and collagens (Tu and Palczewski, 2012) but appear relatively more variable among the newly identified paralogues (Fig. 5, highlighted with star symbol). For the CTRP1, 6 and 8 group that lacks potential calcium binding sites, it is notable that both CTRP6 and CTRP8 have a His residue, and CTRP1 has a tryptophan at the site equivalent to F230 of C1QTNF5 (Fig. 5). Although CTRP6 and CTRP8 can still form trimers (Peterson et al., 2009; Wong et al., 2008), their trimerization could differ from that of C1QTNF5 as His residues are much less hydrophobic than Phe. The aromatic motif, identified by comparing the sequences of collagens X, VIII, C1q and fibrillar collagens (Brass et al., 1992; Tu and Palczewski, 2012), remains conserved among these paralogues (Fig. 5, highlighted with red box).
Fig. 5.
Sequence comparisons of human C1QTNF5 with its paralogues. Conserved residues of the gC1q domain are highlighted with white or black letters on a blue background according to their conservation. Gly residues in the collagen domain are highlighted with white font on a red background. The conserved hydrophobic motif (residues 143–153) and the characteristic hydrophobic segment FFGGWP (residues 181–186) of human C1QTNF5 are outlined with red rectangles. Residues forming the hydrophobic core at the trimerization interface are indicated with stars. The pathogenic mutation site of C1QTNF5 is indicated with a red arrow. Structural elements coordinating with Ca2+ ions are highlighted with red underlining.
Relative to their sequence variations in the gC1q domain, the sequences of collagen domains are even more variable among these paralogues (Fig. 5). Notably, CTRP4 does not even exhibit a typical primary collagen structure (Gly-X-Y repeats), so its N-terminal domain may not adopt a collagen-like structure. Moreover, CTRP9 has a significantly longer collagen domain than the other paralogues. Co-expression studies showed that CTRP1 could form a heterotrimer with CTRP6, and CTRP2 could form a heterotrimer with CTRP7 (Peterson et al., 2009; Wong et al., 2008). CTRP1 and CTRP6 have almost the same lengths and similar Gly-X-Y repeats, as do CTRP2 and CTRP7, so this may constitute the structural basis underlying the formation of CTRP1/6 and CTRP2/7 heterotrimers.
Conclusions
Our new C1QTNF5 structure reveals novel intermolecular interactions of S163 that could glue the globular heads of adjacent C1QTNF5 octadecamers. These interactions suggest a scenario wherein C1QTNF5 globular heads interact with each other, whereas their collagen domains bind to collagen receptors on the membranes of adjacent RPE cells. Accordingly, C1QTNF5 would fill in the sub-retinal space and enhance intercellular adhesion of the RPE. Although wild-type C1QTNF5 and its S163R mutant co-exist in affected humans, the pathogenic S163R mutation causes a reduction of C1QTNF5 functionality, as it destabilizes the protein’s globular heads and weakens its intermolecular interactions. As a consequence, affected individuals accumulate sub-RPE deposits with age. Thus, our structures provide insights into pathogenic mechanisms of L-ORMD at the molecular level.
C1QTNF5 can form bouquet-like octadecamers without N-terminal intermolecular disulfide bonds. In addition, C1QTNF5 displays structural heterogeneity evidenced by various spatial layouts of its globular heads. This flexibility in the spatial arrangement of C1QTNF5 globular heads could affect their self-interaction and/or potential ligand binding. Also, C1QTNF5 can interact with MFRP. Considering the differences in phenotypes of diseases associated with C1QTNF5 and MFRP, C1QTNF5 should have actions other than just enhancing the adhesion of RPE cells.
Materials and Methods
1. Expression and purification of human C1QTNF5 and its gC1q domain in Rosetta™(DE3) E. coli
Cloning, expression and purification of human C1QTNF5 were carried out according to the procedure used for its gC1q domain (Tu and Palczewski, 2012). Briefly, C1QTNF5 lacking the signal peptide (residues 18–243) was amplified from a synthesized human C1QTNF5 gene with natural NcoI restriction sites removed. The forward PCR primer introduced a NcoI restriction site and a Gly residue at the 5′-end, and the reverse primer introduced six His residues and an XhoI restriction site at the 3′-end. The PCR product and pETduet-1 vector were ligated together after digestion with the restriction enzymes NcoI/XhoI. The recombinant plasmid construct was sequenced and then transformed into Rosetta™(DE3) competent cells (Novagen). Expression of C1QTNF5 was induced overnight at 16 °C with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). Cell pellets were re-suspended in buffer A (40 mM Tris–HCl, pH 8.0, 300 mM NaCl, 0.5% Triton X-100, 0.5 mM β-mercaptoethanol, 5 mM imidazole and 10% glycerol) supplemented with protease inhibitors (cOmplete EDTA-free, Roche), disrupted on ice by sonication, and cleared by ultracentrifugation at 4 °C. The supernatant was applied to 4 ml of TALON–NTA affinity resin in a 1.5 × 6.3 cm column. This column was first washed with 400 ml of buffer B (20 mM Tris–HCl, pH 8.0, 300 mM NaCl, 0.5% Triton X-100, 10% glycerol, 0.5 mM β-mercaptoethanol and 10 mM imidazole) followed by 200 ml of buffer C (20 mM Tris–HCl, pH 8.0, 0.5 mM β-mercaptoethanol and 300 mM NaCl). Protein then was eluted with buffer D (20 mM Tris–HCl, pH 8.0, 300 mM NaCl, 0.5 mM β-mercaptoethanol and 300 mM imidazole). The eluted protein peak was immediately further purified by gel filtration chromatography on a Superdex 200 10/300 GL column (GE Healthcare) equilibrated and eluted with buffer E (20 mM Tris-HCl, pH 8.0, 100 mM NaCl). The gel filtration column was calibrated with Bio-Rad gel filtration standards containing bovine thyroglobulin (670 kDa), bovine gamma globulin (158 kDa), chicken ovalbumin (44 kDa), horse myoglobin (17 kDa) and vitamin B12 (1.4 kDa). Expression and purification of the C1QTNF5 gC1q domain from E. coli was described previously (Tu and Palczewski, 2012).
2. Negative staining of human C1QTNF5
Freshly isolated C1QTNF5 from the predominant ~450 kDa peak of the Superdex GL300 size exclusion chromatography was diluted to ~20 μg/ml and adsorbed onto glow-discharged 400 mesh carbon-coated grids for 1 min. Grids then were washed with 6–8 drops of distilled water and negatively stained with 0.75% (w/v) uranyl formate. Samples were imaged with a FEI Tecnai T12 microscope (FEI, Eindhoven, The Netherlands) operated at 100 kV.
3. Crystallization and diffraction data collection
Crystallization was performed at room temperature by a traditional hanging-drop method. Two microliters of protein sample were mixed with an equal volume of reservoir solution (2 M ammonium sulfate, 0.1 M sodium acetate, pH 5.0) and equilibrated against 500 μl of reservoir solution. Crystals grew to sizes suitable for X-ray diffraction in one week. These were soaked in cryoprotectant solution (reservoir solution with ethylene glycol concentration elevated to 25%) for ~30 s, and then immediately flash-cooled in liquid nitrogen. Diffraction data were collected at beamline 24-ID-C of the Advanced Photon Source at Argonne National Laboratory.
4. Structural determination and analyses
Diffraction data were processed, reduced and scaled with software XDS (Kabsch, 2010). The initial phase was derived from the previously solved structure of C1QTNF5 gC1q (PDB: 4F3J) by the molecular replacement method implemented in the program Phaser (McCoy et al., 2007). Iterative model building and refinement were carried out with Coot (Emsley and Cowtan, 2004), Phenix (Adams et al., 2010) and Refmac (Murshudov et al., 1997). The anisotropic B factor was refined in the final stage of refinement. Composite omit maps and simulated annealing omit maps were calculated with CNS (Brunger et al., 1998). The final models and structural factors were deposited in the Protein Data Bank (www.pdb.org) under accession code 4NN0. The quality of the final model was analyzed by PROCHECK (Laskowski et al., 1993). Structural alignment was achieved with secondary-structure matching (Krissinel and Henrick, 2004) implemented in the Coot software. Figures were constructed with PyMOL (Schrodinger, 2010) and Jalview software (Waterhouse et al., 2009).
5. Stable expression of recombinant human MFRP in mammalian CHO-K1 cells
Human MFRP cDNA was purchased from GeneCopoeia™. MFRP with a 1D4 tag at the C-terminus was cloned into mammalian expression vector pcDNA™3.1(−) between the NheI and BamH1 sites. CHO-K1 cells were cultured in F-12 medium supplemented with 10% bovine serum albumin. The recombinant construct was transfected into CHO-K1 cells with the X-tremeGENE HP transfection reagent according to the manufacturer’s instructions. To obtain stable transfectants, transfected CHO-K1 cells were cultured in 750 μg/ml of Geneticin® for 3 weeks. The generated stable cell line expressing MFRP was expanded and washed with buffer (20 mM HEPES, pH 7.4, and 40 mM NaCl) four times before harvesting. Cells were harvested into lysis buffer (25 mM HEPES, pH 7.4, 40 mM NaCl, DNase I and Roche cOmplete protease inhibitor), lysed with a glass Dounce homogenizer and pelleted by centrifugation at 100,000×g and 4°C for 1 h. Membrane pellets were resuspended into a buffer containing 25 mM HEPES, pH 7.4, 100 mM NaCl and Roche cOmplete protease inhibitor and later solubilized by addition of 3% (w/v) of DDM for 3 h at 4°C before being cleared by centrifugation at 150,000×g for 1 h.
6. Pull-down of MFRP by immobilized C1QTNF5
Purified C1QTNF5 was immobilized on CNBr-activated agarose according to the manufacturer’s protocol (SIGMA-ALDRICH®). A DDM-solubilized membrane fraction of CHO-K1 cells expressing MFRP was loaded onto a C1QTNF5-loaded CNBr-activated agarose column equilibrated with 20 mM HEPES, pH 7.4, 75 mM NaCl, 0.05% DDM and 10% glycerol at 4°C. The loaded column was placed on a shaker for 3 h before it was drained and washed extensively (100 × resin volume) with a buffer containing 20 mM HEPES, pH 7.4, 400 mM NaCl, 0.05% DDM and 10% glycerol. Protein was finally eluted from the column with 2% SDS solution at room temperature. All collected fractions were assayed by immunoblotting with an alkaline phosphatase-conjugated 1D4 antibody specific for MFRP detection.
Supplementary Material
Acknowledgments
We thank Dr. Leslie T. Webster Jr. and members of Palczewski’s laboratory (Case Western Reserve University) for valuable comments on the manuscript. This research was supported in part by Grants EY008061 and P30EY11373 from the National Institutes of Health. The work also is based upon research conducted at the Advanced Photon Source on the Northeastern Collaborative Access Team beamlines supported by Grants from the National Center for Research Resources (5P41RR015301-10) and the National Institute of General Medical Sciences (8P41GM103403-10) from the National Institutes of Health. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under Contract No. DE-AC02-06CH11357.
Footnotes
Conflict of interest:
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyagari R, Mandal MN, Karoukis AJ, Chen L, McLaren NC, Lichter M, Wong DT, Hitchcock PF, Caruso RC, Moroi SE, Maumenee IH, Sieving PA. Late-onset macular degeneration and long anterior lens zonules result from a CTRP5 gene mutation. Invest Ophthalmol Vis Sci. 2005;46:3363–3371. doi: 10.1167/iovs.05-0159. [DOI] [PubMed] [Google Scholar]
- Borooah S, Collins C, Wright A, Dhillon B. Late-onset retinal macular degeneration: clinical insights into an inherited retinal degeneration. Br J Ophthalmol. 2009a;93:284–289. doi: 10.1136/bjo.2008.150151. [DOI] [PubMed] [Google Scholar]
- Borooah S, Collins C, Wright A, Dhillon B. Republished review. [corrected] Late-onset retinal macular degeneration: clinical insights into an inherited retinal degeneration. Postgrad Med J. 2009b;85:495–500. doi: 10.1136/bjo.2008.150151. [DOI] [PubMed] [Google Scholar]
- Brass A, Kadler KE, Thomas JT, Grant ME, Boot-Handford RP. The fibrillar collagens, collagen VIII, collagen X and the C1q complement proteins share a similar domain in their C-terminal non-collagenous regions. FEBS Lett. 1992;303:126–128. doi: 10.1016/0014-5793(92)80503-9. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Hayward C, Shu X, Cideciyan AV, Lennon A, Barran P, Zareparsi S, Sawyer L, Hendry G, Dhillon B, Milam AH, Luthert PJ, Swaroop A, Hastie ND, Jacobson SG, Wright AF. Mutation in a short-chain collagen gene, CTRP5, results in extracellular deposit formation in late-onset retinal degeneration: a genetic model for age-related macular degeneration. Hum Mol Genet. 2003;12:2657–2667. doi: 10.1093/hmg/ddg289. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore U, Gaboriaud C, Waters P, Shrive AK, Greenhough TJ, Reid KB, Sim RB, Arlaud GJ. C1q and tumor necrosis factor superfamily: modularity and versatility. Trends Immunol. 2004;25:551–561. doi: 10.1016/j.it.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- Mandal MN, Vasireddy V, Jablonski MM, Wang X, Heckenlively JR, Hughes BA, Reddy GB, Ayyagari R. Spatial and temporal expression of MFRP and its interaction with CTRP5. Invest Ophthalmol Vis Sci. 2006a;47:5514–5521. doi: 10.1167/iovs.06-0449. [DOI] [PubMed] [Google Scholar]
- Mandal MN, Vasireddy V, Reddy GB, Wang X, Moroi SE, Pattnaik BR, Hughes BA, Heckenlively JR, Hitchcock PF, Jablonski MM, Ayyagari R. CTRP5 is a membrane-associated and secretory protein in the RPE and ciliary body and the S163R mutation of CTRP5 impairs its secretion. Invest Ophthalmol Vis Sci. 2006b;47:5505–5513. doi: 10.1167/iovs.06-0312. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Peterson JM, Wei Z, Wong GW. CTRP8 and CTRP9B are novel proteins that hetero-oligomerize with C1q/TNF family members. Biochem Biophys Res Commun. 2009;388:360–365. doi: 10.1016/j.bbrc.2009.08.014. [DOI] [PubMed] [Google Scholar]
- Radjainia M, Wang Y, Mitra AK. Structural polymorphism of oligomeric adiponectin visualized by electron microscopy. J Mol Biol. 2008;381:419–430. doi: 10.1016/j.jmb.2008.06.015. [DOI] [PubMed] [Google Scholar]
- Schaffler A, Buechler C. CTRP family: linking immunity to metabolism. Trends Endocrinol Metab. 2012;23:194–204. doi: 10.1016/j.tem.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Schrodinger, LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010. [Google Scholar]
- Shapiro L, Scherer PE. The crystal structure of a complement-1q family protein suggests an evolutionary link to tumor necrosis factor. Curr Biol. 1998;8:335–338. doi: 10.1016/s0960-9822(98)70133-2. [DOI] [PubMed] [Google Scholar]
- Shu X, Tulloch B, Lennon A, Vlachantoni D, Zhou X, Hayward C, Wright AF. Disease mechanisms in late-onset retinal macular degeneration associated with mutation in C1QTNF5. Hum Mol Genet. 2006a;15:1680–1689. doi: 10.1093/hmg/ddl091. [DOI] [PubMed] [Google Scholar]
- Shu X, Tulloch B, Lennon A, Hayward C, O’Connell M, Cideciyan AV, Jacobson SG, Wright AF. Biochemical characterisation of the C1QTNF5 gene associated with late-onset retinal degeneration. A genetic model of age-related macular degeneration. Adv Exp Med Biol. 2006b;572:41–48. doi: 10.1007/0-387-32442-9_7. [DOI] [PubMed] [Google Scholar]
- Tsao TS, Tomas E, Murrey HE, Hug C, Lee DH, Ruderman NB, Heuser JE, Lodish HF. Role of disulfide bonds in Acrp30/adiponectin structure and signaling specificity. Different oligomers activate different signal transduction pathways. J Biol Chem. 2003;278:50810–50817. doi: 10.1074/jbc.M309469200. [DOI] [PubMed] [Google Scholar]
- Tu X, Palczewski K. Crystal structure of the globular domain of C1QTNF5: Implications for late-onset retinal macular degeneration. J Struct Biol. 2012;180:439–446. doi: 10.1016/j.jsb.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong GW, Krawczyk SA, Kitidis-Mitrokostas C, Revett T, Gimeno R, Lodish HF. Molecular, biochemical and functional characterizations of C1q/TNF family members: adipose-tissue-selective expression patterns, regulation by PPAR-gamma agonist, cysteine-mediated oligomerizations, combinatorial associations and metabolic functions. Biochem J. 2008;416:161–177. doi: 10.1042/BJ20081240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.