Abstract
Ubiquitin is a tag that often initiates degradation of proteins by the proteasome in the ubiquitin proteasome system. Targeted expression of K6W mutant ubiquitin (K6W-Ub) in the lens results in defects in lens development and cataract formation, suggesting critical functions for ubiquitin in lens. To study the developmental processes that require intact ubiquitin, we executed the most extensive characterization of the lens proteome to date. We quantified lens protein expression changes in multiple replicate pools of P1 wild-type and K6W-Ub-expressing mouse lenses. Lens proteins were digested with trypsin, peptides were separated using strong cation exchange and reversed-phase liquid chromatography, and tandem mass (MS/MS) spectra were collected with a linear ion trap. Transgenic mice that expressed low levels of K6W-Ub (low expressers) had normal, clear lenses at birth, whereas the lenses that expressed high levels of K6W-Ub (higher expressers) had abnormal lenses and cataracts at birth. A total of 2052 proteins were identified, of which 996 were reliably quantified and compared between wild-type and K6W-Ub transgenic mice. Consistent with a delayed developmental program, fiber-cell-specific proteins, such as γ-crystallins (γA, γB, γC, and γE), were down-regulated in K6W-Ub higher expressers. Up-regulated proteins were involved in energy metabolism, signal transduction, and proteolysis. The K6W-Ub low expressers exhibited delayed onset and milder cataract consistent with smaller changes in protein expression. Because lens protein expression changes occurred prior to lens morphological abnormalities and cataract formation in K6W-Ub low expressers, it appears that expression of K6W-Ub sets in motion a process of altered protein expression that results in developmental defects and cataract.
Keywords: proteomics, mass spectrometry, mouse lens, nuclear cataract, ubiquitin, proteasome
Introduction
The ubiquitin proteasome system (UPS) is a highly selective intracellular protein degradation system.1,2 In addition to selective degradation of various forms of aberrant proteins, the UPS plays an important role in regulating many facets of cellular functions, such as signal transduction, cell cycle control, and differentiation.3−12 In the UPS, proteins to be degraded are first tagged by the covalent attachment of multiple ubiquitin molecules. Then, the ubiquitin-tagged proteins are recognized and degraded by the 26S proteasome with the release of free and reusable ubiquitin.13
Ubiquitin is a highly conserved 76 amino acid polypeptide. Covalent attachment of ubiquitin to the protein substrate proceeds via a three-step cascade mechanism.14 Ubiquitin is most often conjugated to ε-amino groups of an internal lysine residue of the target protein. For efficient targeting of substrate to the proteasome for degradation, multiple ubiquitins are often conjugated to the initial substrate-bound ubiquitin moiety to form polyubiquitin chains that are linked via internal lysine residues or the N-terminus of the previously conjugated ubiquitin molecule.15−17 The structures of polyubiquitin chains in cells can be highly diverse, and different linkages of the polyubiquitin chains may have specific functions,18−21 For example, K48-linked ubiquitin chains are often involved in targeting substrate for proteasomal degradation18,19 whereas K63-linked polyubiquitin chains are often involved in signal transduction and DNA repair.21,22
Few substrates that require K6 on ubiquitin for their conjugation have been identified, and the functions of K6-linked polyubiquitin chains remain largely unknown. However, our previous work demonstrated that modification or mutation of the K6 residue in ubiquitin does not affect its ability to form polyubiquitin chains, but ubiquitin conjugates formed with K6-modified or mutant ubiquitin are not recognized by the proteasome and are thus resistant to proteasomal degradation.20 Thus, K6-modified or mutant ubiquitin becomes a specific upstream dominant negative inhibitor of the UPS. In comparison, virtually all of the small molecule inhibitors of the UPS diminish proteasomal activity per se. Expression of K6W mutant ubiquitin (K6W-Ub) in cells or in lenses in vivo resulted in accumulation of ubiquitin conjugates, stabilization of many regulatory proteins, such as cyclins, p21, and p27, along with associated cell cycle arrest,23,24 as well as defects in development and cataract formation.25 However, the mechanism of the development defects and cataract caused by K6W-Ub remains to be fully elucidated.
To gain a broader understanding of the mechanism by which expression of K6W-Ub causes defects of lens development and cataractogenesis, we compared the proteomes of wild type (WT) and K6W-Ub expressing lenses from newborn mice (P1) using mass spectrometry (MS). A total of 2052 proteins were identified in these lenses, expanding the known mouse lens proteome26 over five-fold, and 996 of these were reliably quantified. The effects of K6W-Ub on the lens proteome were dose-dependent. In lenses that expressed low levels of K6W-Ub, the protein profiles were similar to those of WT lenses. However, in lenses that expressed high levels of K6W-ubiquitin, there were significant changes in levels of 32 proteins as compared with WT. Total crystallin expression was down-regulated, with the γ-crystallins (γ-A, γ-B, γ-C, and γ-E) showing the greatest expression declines in K6W-Ub high expressers. Proteins that were up-regulated were involved in energy metabolism, signal transduction, and proteolysis. These data significantly strengthen and expand the hypothesis that alteration in protein expression may be related to the defect in lens development and cataract formation in mice that express K6W-Ub in the lens and that ubiquitin with K6 intact is required for progress of a proper differentiation program.
Materials and Methods
Generation of Transgenic Mice
All animal experiments were approved by the Animal Care and Use Committee HNRCA at Tufts University and were performed according to NIH guidelines for using experimental animals. Transgenic mice were generated and maintained in FVB/N background. In these animals, the expression of MRGS (His)6-WT-Ub and MRGS(His)6-K6W-Ub is under the control of the αA-crystallin promoter with DCR1 enhancer as previously described.25 This assures that the expression of the transgene is lens-specific. Genotypes of the transgenic mice were determined by PCR-based assays using this pair of primers: GAGGGTGCCTCAGTACAATGTGG and TGCGATCCTCTCATATCGATTGG with DNA extracted from mouse tails as templates. Levels of expression of the transgene were determined by Western blotting using an antibody specific to the transgene product (MRGS-His4) from Qiagen.
Slit Lamp and Histologic Examination of WT and Transgenic Lens
The eyes of WT and transgene-positive mice were examined with a slit-lamp microscope monthly to monitor lens transparency. To study the morphology, the eyes from 3-month old mice were fixed in 4% paraformaldehyde. Paraffin sections were stained with H&E to examine the histology of the lenses.
Sample Preparation for Mass Spectrometry Assay
Twelve lenses from new born (P1) mice of each of the groups (WT, K6W-Ub low expressers, and K6W-Ub high expressers) were used for proteomic analysis. Three lenses from each group were pooled, so there were four biological replicates in each group and a total of 12 samples. The pooled lenses were suspended in 250 μL of 20 mM phosphate buffer containing 1 mM EGTA, pH 7.0, and sonicated for 15 s (3 × 5 s with cooling on ice for 30 s between treatments) using a Model 60 Sonic Dismembrator (Fisher Scientific). The protein concentrations of the lens homogenates were determined using a BCA assay (Thermo Scientific). One hundred μg protein aliquots from each pooled sample were dried by vacuum centrifugation. Proteins in the dried samples were dissolved in 20 μL of 8 M deionized urea, 1.0 M Tris-HCl, pH 8.5, 8 mM CaCl2, and 0.2 M methylamine. Disulfides in the samples were then reduced by the addition of 2 μL of 0.2 M dithioerythritol (DTT) at 50 °C for 15 min. Samples were then alkylated by the addition of 2 μL of 0.5 M iodoacetamide and incubated at room temperature for 15 min. An additional 4 μL of 0.2 M DTT was added to neutralize any excess iodoacetamide. Twelve μL of water was then added to each sample, followed by 40 μL of 0.1 μg/μL trypsin (Proteomics grade, Sigma). Following overnight digestion at 37 °C, 5 μL of formic acid was added to each sample to stop the reaction. The resulting peptides were desalted by solid-phase extraction using Sep Pak Light C18 cartridges (Waters Corporation). To enhance protein detection, the desalted peptides were then separated into eight fractions by loading onto a 2.1 × 5 mm Reliasil cation exchange cartridge (Opti-Lynx, 11-02870-ES, Optimize Technologies) and eluted by manual injection of 300 μL of 17, 21, 25, 37.5, 56.5, 75, 150, and 375 mM ammonium formate dissolved in 25% acetonitrile containing 0.4% formic acid. The nine fractions (flow through was also collected) from the digest of each pooled sample were then dried by vacuum concentration and redissolved in 42 μL of 5% formic acid, and 40 μL was analyzed by LC–MS/MS.
Two-Dimensional LC–MS/MS Analysis
Cation exchange fractions were analyzed by LC–MS/MS using an Agilent 1100 series capillary LC system (Agilent Technologies) and an LTQ linear ion trap mass spectrometer (Thermo Scientific). Electrospray ionization was performed with an ion max source fitted with a 34 gauge metal needle (Thermo Scientific cat. no. 97144-20040) at a potential of 2.4 kV. Samples were applied at 20 μL/min to a trap cartridge (Michrom BioResources) and then switched onto a 0.5 × 250 mm Zorbax SB-C18 column with 5 μM particles (Agilent Technologies) using a mobile phase containing 0.1% formic acid, 7–30% acetonitrile gradient over 200 min, and 10 μL/min flow rate. Data-dependent collection of MS/MS spectra used a collision energy of 35% with the dynamic exclusion feature of the instrument’s control software enabled (repeat count equal to 1, exclusion list size of 50, exclusion duration of 30 s, and exclusion mass width of −1 to +4) to obtain MS/MS spectra of the three most abundant parent ions following each survey scan from m/z 400–2000. The tune file was configured with no averaging of microscans, a maximum inject time of 200 ms, and automatic gain control (AGC) targets of 3 × 104 in MS mode and 1 × 104 in tandem mass spectrometry (MSn) mode.
Analysis of 2D LC–MS/MS Data
DTA file creation (peak lists) was done with BioWorks 3.3 (Thermo Scientific) using the charge-state algorithm (ZSA) to determine the charge state. A minimum ion count of 25 and absolute intensity of 500 were used with no grouping requirements or combining of similar mass spectra. To determine protein sequences, we created a UniProt Sprot mouse database (release 2011.2 with 16 339 mouse proteins) with common contaminants (179 sequences) and decoy sequences (16 518 reversed protein sequences). Peptides were identified using SEQUEST (version 28, rev 12, Thermo Scientific) with trypsin cleavage specificity. The parent ion mass tolerance was 2.5 Da (average), and the fragment ion tolerance was 1.0 Da (average). Other parameters include a static C+57 modification (alkylation), variable modifications of M+16 (oxidation), and K+114 (ubiquitination). A maximum of two missed cleavages and a maximum of three variable modifications per peptide were permitted.
SEQUEST results were filtered to strict peptide and protein false discovery rates (FDRs), estimated from decoy sequences using the PAW software27 with explicit error control for modified peptides. Peptides were filtered to FDRs of 1.5% independently for unmodified and modified peptides. A maximum of two homogeneous modifications per peptide was allowed. Confidently identified peptides were mapped to proteins using basic parsimony principles. To be included in the final analyses, proteins were required to have a minimum of two distinct mapped peptide sequences. Modified sequences or sequences in different charge states were not considered to be distinct. This criterion was applied to each biological sample independently.
Quantitative Analysis
Protein abundances are correlated with the number of MS/MS spectra observed (spectral counting) in bottom-up proteomics experiments and can be used to measure protein differential abundance. A complication in bottom-up proteomics experiments is that the same peptides can come from more than one protein (shared peptides). To quantitatively assign peptides that are found in multiple proteins, we split-shared peptide counts into fractions based on the relative abundance of the proteins that share the peptide and combined the fractional counts with the unique peptide counts to generate a “corrected total spectral count” for each protein as previously described.27 This method becomes less reliable when unique counts drop to small numbers. In such cases, an in-house algorithm was used to group together proteins having high proportions of shared counts (highly homologous proteins) into families, and quantification was done on the total family counts instead of the individual family member counts.
Because equal quantities of total protein were loaded for each sample and spectral counts are highly correlated with protein abundance,28 the total corrected spectral counts per sample should be equal. To correct for any possible protein loading and instrumental variations, the total number of confidently identified MS/MS spectra per sample (excluding matches to contaminants) was tabulated and averaged over the 12 samples. Global scaling factors were calculated for each sample to adjust that sample’s total spectral count to the average total spectral count. The corrected spectral counts for each protein in each sample were adjusted by the respective scaling factors to generate normalized corrected spectral count for each protein. The normalization adjustments were very small, with factors ranging from 0.98 to 1.02
Statistics
One-way ANOVA analysis was performed for each protein’s normalized, corrected total spectral counts using the f_oneway function from the stats module of the SciPy package (www.scipy.org). Proteins having average normalized, corrected total spectral counts of less than 2.5 (across the 12 samples) were excluded from quantification because the proteins having average spectral counts <2.5 had a missing data (the spectral count of a given protein in a given sample being zero) rate as high as 40%, and greatly reduced the reliability of spectral counts-based quantitative comparisons. Excluding the proteins with average spectral count <2.5 greatly increased the reliability of quantitative comparison. To estimate background of false-positive candidates arising from the tests by chance, we generated a density histogram of the ANOVA test p values for 996 quantifiable proteins.29 The p-value distribution is composed of a uniform (flat) density from 0 to 1 for the nondifferentially expressed proteins and a standout density of low p-value associated with the true differential expressed proteins (Supplemental Figure 1 in the Supporting Information). On the basis of the p-value distribution pattern, p < 0.01 cutoff was chosen to identify the differential proteins. The average number of proteins in each interval of 0.01 p-value was 9.86, suggesting that ∼10 proteins with p < 0.01 could be detected by chance. Thus, the estimated FDR was ∼25%. Application of the excel Benjamini–Hochberg multiple comparison analysis30 with 25% FDR identified the same set of differentially expressed proteins.
Identification of Proteins from SDS-Gel Bands
Total proteins from P1 lenses were resolved by 12% gel under reducing conditions. After staining with Coomassie blue, the bands of interest were excised. Excised bands from Coomassie Blue stained gels were reduced, alkylated, and subjected to in-gel digestion with trypsin, as previously described.31 Peptides were then filtered (Millipore, 0.45 μm Ultrafree-MC), dried by vacuum centrifugation, and dissolved in 40 μL of 5% formic acid, and 10 μL was analyzed by LC–MS/MS. Peptides were separated using a NanoAcquity UPLC system (Waters), then delivered to an LTQ Velos dual pressure linear ion trap mass spectrometer (Thermo Scientific) with a Captive Spray Source (Microm Biosciences) fitted with a 20 μm taper spray tip and 1.0 kV source voltage. Samples were applied at 15 μL/min to a Symmetry C18 trap cartridge (Waters) for 10 min, then switched onto a 75 μm × 250 mm NanoAcquity BEH 130 C18 column with 1.7 μm particles (Waters) using mobile phases water (A) and acetonitrile (B) containing 0.1% formic acid, 7.5–30% acetonitrile gradient over 60 min, and 300 nL/min flow rate. Data-dependent collection of MS/MS spectra used a collision energy of 30% with dynamic exclusion enabled (repeat count equal to 1, exclusion list size of 500, exclusion duration of 24 s, and exclusion mass width of −1 to +4) to obtain MS/MS spectra of the 10 most abundant parent ions (minimum signal of 5000) following each survey scan from m/z 400–1400. The tune file settings were as previously described for the LTQ instrument.
Creation of DTA files were done using the parameters previously listed for the 2D LC–MS/MS study. SEQUEST parameters were also similar except that K+114 was not specified as a variable modification, and the 2013.04 Sprot mouse database (16 604 sequences) was used. The target/decoy method was employed to filter peptides and keep the protein FDR <2% per gel band. We used MS/MS-intensity-weighted spectral counts and identified the most abundant proteins that constituted at least 90% of the total gel band abundance. Because of limited resolution of 1-D gel electrophoresis, each band contained multiple proteins. Only proteins that accounted for >10% of the total spectral counts of each band were indicated in Figure 5A.
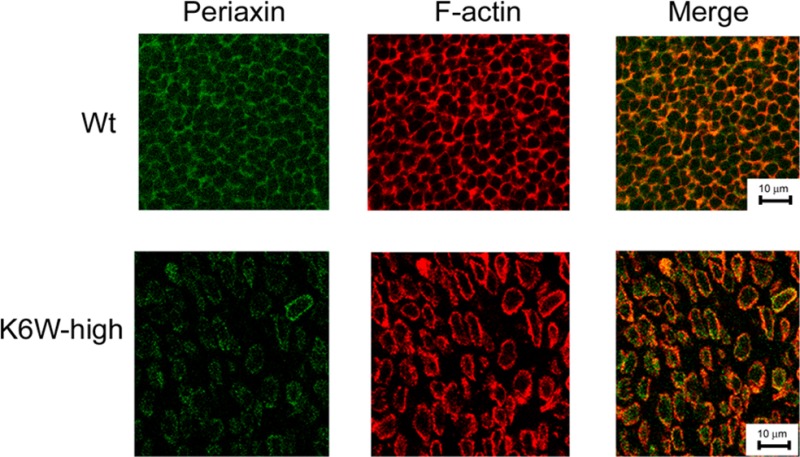
Figure 5.
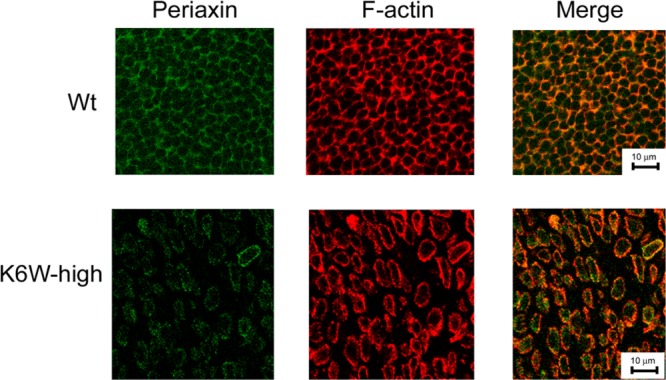
Expression of K6W-ubiquitin reduces the levels of periaxin and disrupts the fiber organization. Lenses from WT and high expressing K6W-Ub transgenic newborn (P1) mice were cryosectioned at the equatorial plane. The sections were immunostained with antibody to periaxin and FITC-labeled secondary antibody. F-actin was stained with rhodamine-labeled phalloidin. Confocal photomicrographs were taken using a Leica confocal system. These data indicate that periaxin is down-regulated in most lens fibers of K6W-Ub lenses and the diameter of lens fibers of K6W-ub lens is significantly larger than that of WT lens.
Western Blot Analysis
Lenses of P1 mice were dissected and homogenized directly with SDS-gel loading buffer. Protein concentrations were measured by densitometry analysis of Coomassie-stained gels. Total lens proteins were resolved by 10–15% SDS-PAGE and transferred to nitrocellulose membrane. Target proteins were probed with respective rabbit antibodies. Polyclonal antibody to γ-crystallins was a gift from Dr. Sam Zigler at Johns Hopkins University, polyclonal antibody to filensin was a gift from Dr. Paul FitzGerald at University of California (Davis, CA), and rabbit antibody to caprin2 was a gift from Dr. Christina E. Lorén at University of British Columbia (Vancouver, BC, Canada). Antibodies to periaxin, Hsp27, and GAPDH were purchased from Sigma-Aldrich. Antibody to PGK1 was purchased from Thermo Scientific. Antibody to annexin A was purchased from Cell Signaling. After incubation with horseradish peroxidase-conjugated antirabbit secondary antibodies (Jackson Immuno Research), the specifically bound antibody was visualized using a Super Signal chemiluminescent detection kit (Thermo Scientific).
Immunohistochemistry
Eyes were dissected from WT and transgenic P1 mice, fixed in 4% paraformaldehyde for 30 min, and then embedded in OCT and sectioned on equatorial plane. Cryosections were permeabilized with 0.05% Triton X-100 in PBS, blocked with 5% BSA in PBS, and incubated with primary antibody to periaxin (Sigma-Aldrich) and FITC-labeled secondary antibody using standard protocols. F-actin was stained with rhodamine-labeled phalloidin. Photomicrographs were taken using a Leica confocal system.
Results
Expression of K6W-Ub in Lens Caused Nuclear Cataract and Defects in Lens Development
Our previous work demonstrated that lens cells, including fibers in the nuclear region, have a functional UPS32,33 but that the activities and levels of the ubiquitin-conjugating machinery change during cell cycle and differentiation of lens cells.34,35 Furthermore, impairment of the UPS blocks lens cell proliferation and fiber differentiation in cell/tissue culture systems.24,36,37 To test the hypothesis that a fully functional UPS is essential for lens transparency, we generated transgenic mice in which K6W-Ub expression is targeted specifically in the lens starting at E10.5.
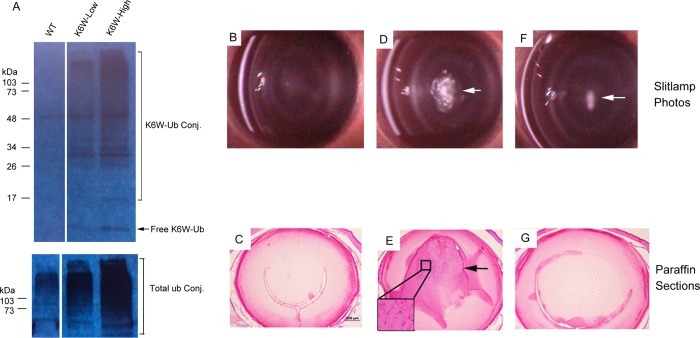
Two varieties of germ line-transmitted transgenic mice were generated. The two lines expressed different levels of K6W-Ub in the lenses. We designated the line that expresses higher levels of the transgene as “high expresser” and the line that expresses lower levels of the transgene as “low expresser”. Western blotting analysis using an antibody specific to the transgene product (anti-MRGS His4) indicates that lenses from the high expressers expressed ∼5-fold higher levels of K6W-Ub than the lenses from low expressers (Figure 1A). In lenses of the low expresser, levels of K6W-Ub were ∼50% the levels of endogenous WT-Ub, and in lenses of the high expresser, levels of K6W-Ub were about 2.5 times the levels of endogenous WT-Ub based on the increase in total ubiquitin in the lenses. As previously documented,25 the sizes and weights of lenses from K6W-Ub low expressers were comparable to those of WT mice, but the sizes and weights of lenses from K6W-Ub high expressers were ∼30% lower than those of WT mice at birth. Furthermore, lens-specific expression of K6W-Ub did not affect the size or body weight of the mice.
Figure 1.
Expression of K6W-ubiquitin in the lens causes nuclear cataract and defects in lens development in a dose-dependent manner. Two lines of transgenic mice were generated. Lenses from two mice of WT and each line of the transgenic mice were analyzed for expression of the transgenes. Western blot with antibody specific to the transgene showed that the two lines expressed different levels of K6W-Ub and ubiquitin conjugates in the lens (Panel A: lane 1, lenses from WT mice without expression of the transgene; lane 2, lenses from the transgenic line that expresses lower levels of transgene; lane 3, lenses from the transgenic line that express higher levels of transgene. Upper panel, probe with transgene specific antibody; Lower panel, probe with antibody to total ubiquitin). Slit-lamp microscope photographs of examination of mice at 3 months of age showed that K6W-Ub transgenic mice that expressed higher levels of K6W-Ub mice developed dense nuclear cataracts (panel D, white area), whereas mice that expressed lower levels of the transgene showed no or mild cataracts (panel F, white arrow). The WT mice have clear lenses (panel B). H&E staining of paraffin embedded sections of eyes from transgenic mice revealed that the high expresser had defects in lens development, including retention of nuclei in the primary fibers (panel E, indicated by arrow and insert). Similar to WT mice, the lenses of low expressers showed normal histology (panels C and G).
Whereas lenses from WT mice were clear, with normal histology (Figure 1B,C), all of the high expressers had cataracts at birth and developed dense nuclear cataracts by 3 months of age (Figure 1D). The lens outer cortex remained transparent and was not significantly affected by the expression of K6W-Ub. Histological examination showed that the high expresser had defects in maturation of primary fibers in the center of lenses (Figure 1E), including the persistence of nuclei in the normally organelle free zone (Figure 1E, insert). Similar to our previous observations of embryonic and neonatal lenses of these transgenic mice,25 the low expressers had lens histology that was similar to those of WT lens at 3 months of age (Figure 1G), and the lenses of low expressers had clear lenses at birth25 and retained clear lenses until 3 months of age, when a fraction (∼30%) of the lower expressers developed mild nuclear cataract (Figure 1F). These data corroborate prior indications that the effects of K6W-Ub on lens development and transparency were dose-dependent.
Expression of K6W-Ub in Lens Altered the Lens Proteome
To test the hypothesis that the morphological changes in lenses from high expressers of K6W-Ub were caused by changes in lens protein expression, we compared the proteomes between lenses from WT, K6W-Ub low expresser, and K6W-Ub high expresser mice using MS. A total of 2052 proteins were detected in the 12 pooled samples and are listed in Appendix A in the Supporting Information. This is a dramatic five-fold increase in mouse lens proteome relative to our prior report26 and is the largest lens proteome reported to date for any species. The estimated number of incorrect identifications was 46 proteins based on matches to reversed sequences, and the FDR was estimated to be ∼2.3%.
The major functional categories of proteins identified in the P1 WT lenses are summarized in Table 1. Spectral counts of all crystallins represented 28.7% of total spectral counts. Spectral counts for all energy metabolism enzymes, mainly carbohydrate metabolic enzymes, represented 6.2% of the total spectral counts, consistent with observations that most of the cells in lenses at this age are metabolically active. It is known that protein synthesis in lenses from P1 lenses is rapid, and we get the first corroboration of this phenomenon from findings that ribosomal proteins were the third largest family of proteins detected in the lens. This is supported by the observation of a high abundance of translation initiation factors and elongation factors. Cytoskeletal proteins, including tubulins, actins, filaments, and spectrin, accounted for a large portion of total spectral counts. The relatively high levels of proteasomal proteins, heat shock proteins (excluding αA-and αB-crystallins), and other protein-folding-related proteins, such as T-complex proteins and peptidyl-prolyl cis–trans isomerases, suggest that lenses in this stage have very active protein-quality control processes. 14–3–3 proteins, a family of proteins that are extensively involved in regulation of kinase, phosphatase, and receptor function, were also among the high abundant proteins in the newborn mouse lenses. Consistent with active iron metabolism in the lens and essential role of transferrin in maintaining lens transparency,38−40 a high content of transferrin was detected in the neonatal lenses.
Table 1. Major Functional Categories of Proteins in WT Newborn Mouse Lensesa.
| categories | average spectral counts | % of total spectral counts |
|---|---|---|
| crystallins | 7486 | 28.7 |
| energy metabolism enzymes | 1611 | 6.2 |
| ribosomal proteins | 1363 | 5.2 |
| tubulins | 644 | 2.5 |
| heat shock proteinsb | 579 | 2.2 |
| histones | 502 | 1.9 |
| actins | 470 | 1.8 |
| membrane proteins | 449 | 1.7 |
| spectrin | 435 | 1.7 |
| elongation factors | 375 | 1.4 |
| translation initiation factors | 318 | 1.2 |
| transferrin | 265 | 1.0 |
| filaments | 262 | 1.0 |
| proteasomal proteins | 242 | 0.9 |
| T-complex proteins | 227 | 0.9 |
| 14–3–3 proteins | 189 | 0.7 |
| peptidyl-prolyl cis–trans isomerases | 180 | 0.7 |
| calpains | 127 | 0.5 |
| othersc | 10808 | 41 |
| total | 26083 | 100 |
Major proteins or functional families of proteins with average spectral counts of >0.5% of total spectral counts (average of the 4 WT samples) are listed. Crystallins, the major lens-specific gene products, represented 28.7% of total spectral counts. Energy metabolism enzymes, mainly sugar metabolic enzymes, represented 6.2% of the total spectral counts. Protein synthesis machinery, including ribosomal proteins, translation initiation factors, and elongation factors, represented 7.8% of the total spectral counts. Cytoskeletal proteins, including tubulins, actins, filaments and spectrin, accounted for 7% of total spectral counts. Protein quality control machinery, including proteins involved in protein folding (heat shock proteins, T-complex proteins, peptidyl-prolyl cis–trans isomerases) and selective protein degradation (proteasomal proteins) accounted for 4.7% of the total spectral counts. Transferrin, 14–3–3 proteins (regulatory proteins), and calpains (calcium activated proteases) represented 1, 0.7, and 0.5% of the total spectral counts, respectively.
Heat shock proteins except for αA-and αB-crystallins.
All other detected proteins or protein families with average spectral counts <0.5% of total spectral counts in each sample.
To gain additional insights into alterations in composition, development, and function due to expressing K6W-Ub, we compared the expression levels of each protein in the three types of lenses using quantitative proteomic analysis. Because common statistical tests have difficulty with missing data points and fluctuations in small spectral count quantities, many low abundant proteins are not reliably quantifiable by spectral counting. To reduce the negative influence of missing data and small-count fluctuations, we used a minimum average spectral count of 2.5 across all samples as our threshold for quantitative analysis. With these criteria, we were able to reliably quantify roughly half (996) of the identified proteins using spectral counting. The quantitative analysis results, including the normalized spectral counts and one-way ANOVA test p-values, are also listed in Appendix A in the Supporting Information. The distribution of the p-values from the ANOVA test informed that 0.01 was an appropriate p-value threshold for differential expression candidates among the three groups of biological samples and to estimate the number of protein candidates that occurred by chance.
As shown in Figure 2, spectral counts for most lens proteins were not distinguishable between WT and K6W-Ub groups. There were only 40 candidate proteins having p-values <0.01 when comparing WT and K6W-Ub high expressers (Appendix 1 in the Supporting Information). On the basis of the average frequency of the distribution for p-values between 0.01 and 1.0 (assuming a flat, uniform distribution from nondifferential candidates) (Supplemental Figure 1 in the Supporting Information), 10 candidates would be expected to have p < 0.01 by chance. Most of the differentially expressed proteins had the same trend of changes between K6W-low expressers and high expressers. A few proteins showed changes in opposite directions. We assume that the levels of truly changed proteins in the low expressers should be between the levels of WT and high expressers. To reduce the number of candidates that might be detected by chance, we applied a simple trend criterion. We assumed that true differential candidates would have average spectral counts in the low expresser that should fall in between WT and high expresser values. Thirty two candidates, listed in Table 2, satisfied both the p-value and simple trend criteria. Their spectral count values and standard deviations are shown in Figure 3. For some proteins, the difference in expression was largely driven by K6W-Ub high expressers and the spectral counts for these proteins in low expressers were not significantly different from the WT lens (Table 2, indicated by f). The smaller changes of protein expression in low expresser in comparison with high expressers were consistent with the milder lens opacity in low expressers. A concern regarding application of the simple trend test to reduce FDRs is that it might also eliminate a few truly differentially expressed proteins. For example, levels of the ubiquitin-conjugating enzyme UbE2O and protein PARK7 were significantly decreased in the lenses of K6W-Ub high expressers, but there was no statistical difference between WT and low expressers, albeit the average spectral counts were slightly higher in K6W-Ub low expressers than in WT lenses (Appendix A in the Supporting Information). Conversely, levels of GRP78, MYH9, and PP2AA were significantly higher in lenses of K6W-Ub high expressers as compared with WT, but there was no real difference between WT and K6W-Ub low expresser (Appendix A in the Supporting Information). All of these proteins were excluded from Table 2 by the simple trend test.
Figure 2.
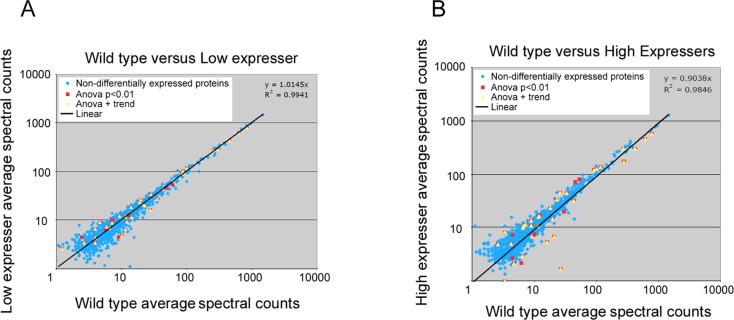
Comparison of spectral counts of lens proteins between WT and of K6W-ubiquitin expressing lenses. Panel A shows spectral counts of peptides in WT versus lower K6W-Ubiquitin expressers. Panel B shows spectral counts of peptides in WT versus K6W-Ub high expressers. The data indicate that the majority of the proteins have similar abundance in WT lens and K6W-Ub expressers (blue dots). However, 40 proteins showed expression difference between WT and K6W-Ub expressing lenses (red dots, ANOVA test p < 0.01). The yellow triangles indicates 32 proteins that were differentially expressed between WT and the higher and lower expressing K6W-Ub lenses with both criteria of ANOVA test <0.01 and passing the trend test. The extents of expression differences (in spectral counts) were more pronounced in K6W-Ub high expressers.
Table 2. Summary of Proteins That Were Differentially Expressed in K6W-Ub Expressing Lensesa.
| accessionb | descriptionc | WT aved | LE aved | HE aved | direction vs WTe |
|---|---|---|---|---|---|
| Structural Proteins | |||||
| CRGE_MOUSE_family | (gamma-crystallin E) | 835.17 | 785.93 | 571.32 | down |
| CRGA_MOUSE | gamma-crystallin A | 633.91 | 613.40 | 451.37 | down |
| CRGC_MOUSE | gamma-crystallin C | 450.12 | 420.63 | 303.46 | down |
| CRGB_MOUSE | gamma-crystallin B | 305.54 | 280.75 | 171.83 | down |
| BFSP1_MOUSE | filensin | 35.23 | 26.57 | 1.64 | down |
| CAPR2_MOUSE | caprin-2 | 27.78 | 16.09 | 5.30 | down |
| PRAX_MOUSE | periaxin | 10.18 | 4.70 | 2.11 | down |
| DBNL_MOUSE | drebrin-like protein | 22.83 | 19.52f | 14.33 | down |
| CLH_MOUSE | clathrin heavy chain 1 | 84.14 | 90.58 | 120.79 | up |
| Translation Factors | |||||
| PABP1_MOUSE | polyadenylate-binding protein 1 | 42.99 | 42.90 | 31.72 | down |
| IF2B_MOUSE | eukaryotic translation initiation factor 2 subunit 2 | 14.29 | 11.42 | 10.55 | down |
| IF4G1_MOUSE | eukaryotic translation initiation factor 4 gamma 1 | 12.07 | 9.07 | 6.73 | down |
| Sugar Metabolic Enzymes | |||||
| KPYM_MOUSE_family | (pyruvate kinase isozymes M1/M2) | 254.27 | 271.44 | 305.79 | up |
| PGK1_MOUSE | phosphoglycerate kinase 1 | 99.17 | 116.39 | 136.04 | up |
| ALDOA_MOUSE | fructose-bisphosphate aldolase A | 86.27 | 100.41 | 125.14 | up |
| K6PL_MOUSE | 6-phosphofructokinase, liver type | 18.81 | 23.17 | 32.94 | up |
| PGAM2_MOUSE | phosphoglycerate mutase 2 | 21.86 | 20.68f | 6.68 | down |
| Proteases and Related Proteins | |||||
| PSA2_MOUSE | proteasome subunit alpha type-2 | 9.35 | 11.99 | 16.13 | up |
| DNPEP_MOUSE | aspartyl aminopeptidase | 3.95 | 6.56 | 9.94 | up |
| CASP7_MOUSE | caspase-7 | 3.94 | 3.40f | 0.99 | down |
| RS27A_MOUSE_family | (ubiquitin-40S ribosomal protein S27a) | 20.01 | 25.99f | 44.83 | up |
| Basement Membrane and Related Proteins | |||||
| CO4A2_MOUSE | collagen alpha-2(IV) chain | 14.05 | 18.20 | 23.48 | up |
| P4HA1_MOUSE | prolyl 4-hydroxylase subunit alpha-1 | 7.14 | 8.84 | 10.95 | up |
| NID2_MOUSE | nidogen-2 | 3.94 | 4.26f | 4.97 | up |
| GRN_MOUSE | granulins | 2.66 | 3.38f | 8.12 | up |
| Carrier Proteins | |||||
| TTHY_MOUSE | transthyretin | 5.92 | 7.28 | 11.32 | up |
| MPCP_MOUSE | phosphate carrier protein, mitochondrial | 6.60 | 6.76f | 10.77 | up |
| Other Proteins | |||||
| SET_MOUSE | protein SET | 11.55 | 11.70f | 15.87 | up |
| ANXA1_MOUSE | annexin A1 | 28.01 | 32.97 | 44.35 | up |
| ATX10_MOUSE | ataxin-10 | 4.64 | 8.89 | 13.55 | up |
| HSPB1_MOUSE | heat shock protein beta-1 | 11.55 | 6.49 | 2.95 | down |
| PRPS1_MOUSE | ribose-phosphate pyrophosphokinase 1 | 1.23 | 2.59 | 4.86 | up |
Differentially expressed proteins that met both one-way ANOVA test p < 0.01 and trend criteria were grouped into seven different functional categories. The data showed that eight of nine structural proteins and all three translation factors were down-regulated in K6W-Ub lenses. In contrast, four of five sugar metabolic enzymes, all basement membrane-related proteins, and carrier proteins were up-regulated in K6W-Ub lens. Other proteins that were up-regulated in K6W-ub lenses include protein SET (a multitasking protein with antiapoptotic activity), annexin A (a calcium/phospholipid-binding protein that inhibits phospholipase A2 activity), ataxin-10 (without known function), and ribose-phosphate pyrophosphokinase (5-phosphoribose 1-diphosphate synthase involved in nucleotide, histidine, and tryptophan biosynthesis).
Sprot identifiers denoted with “family” indicate that the highly homologous proteins were grouped for quantification.
Descriptions in parentheses are the protein with the highest unique spectral count for each family group.
WT, wild type; HE, high expresser; LE, low expresser; Average, normalized total spectral count.
Direction of expression change relative to WT.
Proteins that have no significant difference between WT and LE.
Figure 3.
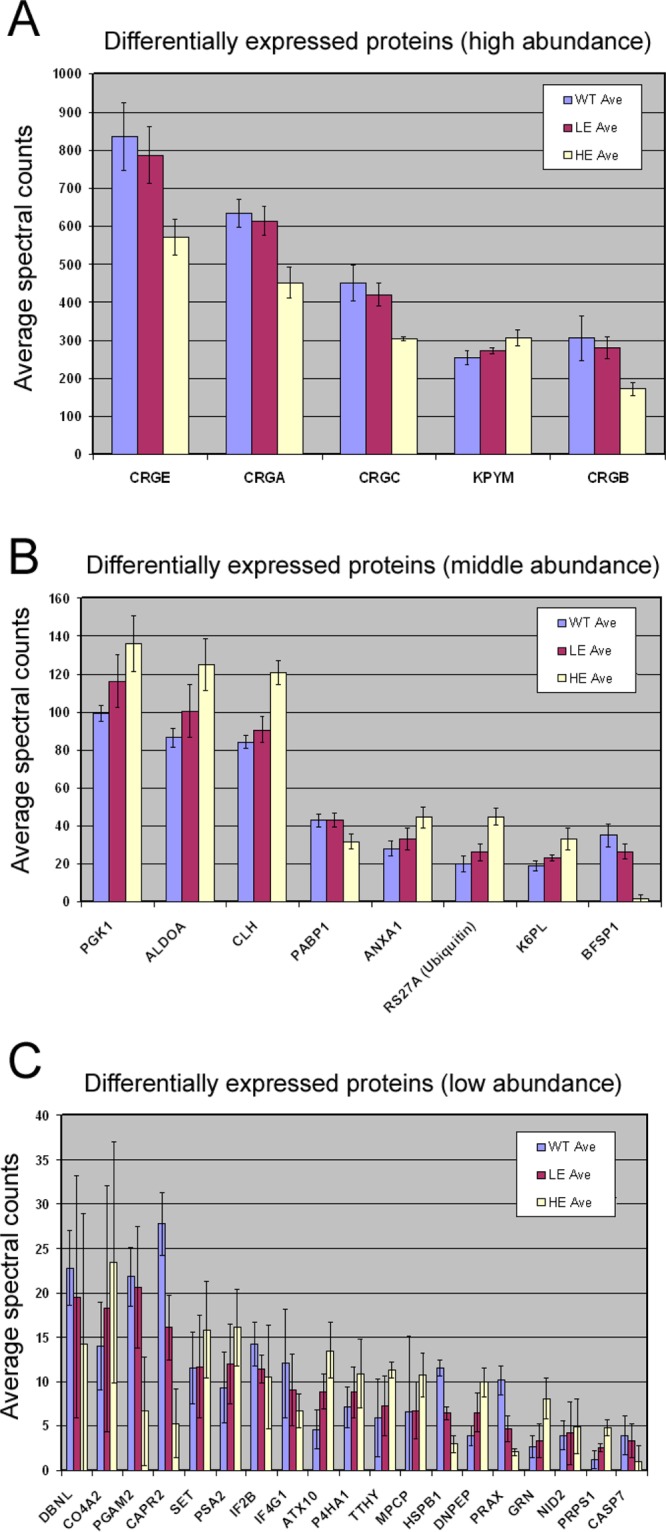
Average normalized, corrected spectral counts for the 32 differentially expressed protein candidates. The five proteins with the highest spectral counts are shown in panel A (top). Eight proteins with intermediate spectral counts are shown in panel B (middle). Differentially expressed proteins with low spectral counts are shown in panel C (bottom). Error bars correspond to one standard deviation of the averages. WT values are blue bars, low expressers are red bars, and high expressers are yellow bars. X-axis labels are Sprot protein identifiers for mouse, and the description of Sprot identifiers is listed in Table 2. These data indicate that the proteins that were down-regulated in K6W-Ub lenses were mainly lens-specific proteins, and the proteins that are up-regulated were mainly metabolic enzymes.
One of the differentially expressed proteins was RS27a. RS27a can represent both ribosomal protein RS27a and ubiquitin because of the shared identical peptide sequences. However, as reflected by the 2.5-fold increase in total ubiquitin expression in the lenses (Figure 1A), it appears that the majority of the identified peptides were derived from ubiquitin. This is corroborated by finding that low expressers showed a small increase in ubiquitin (RS27a) expression over WT, whereas high expressers showed an over two-fold increase in total ubiquitin counts (Figure 1).
Verification of Selected Protein Expression Levels
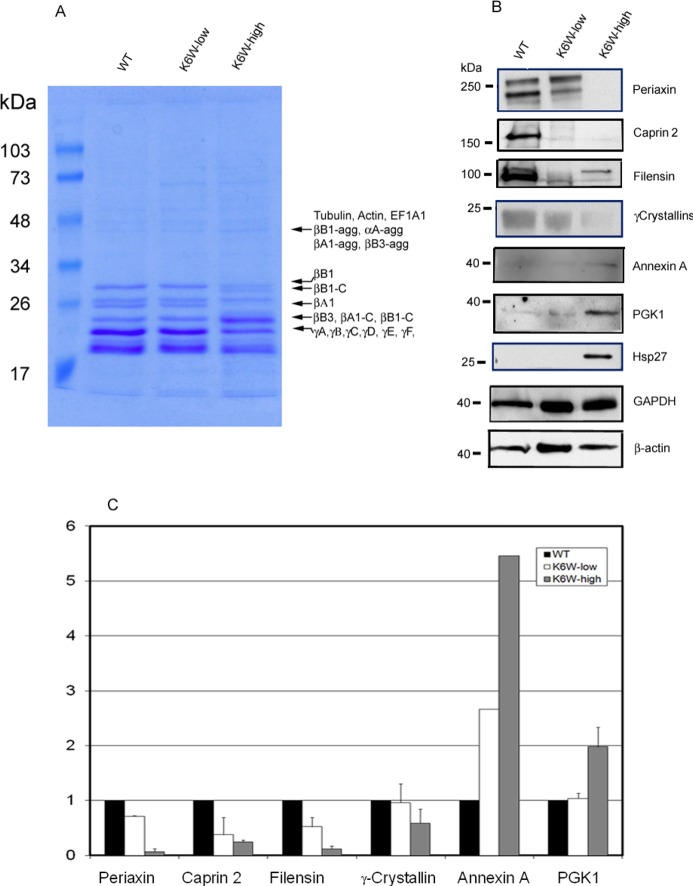
To further test the reliability of our MS methods, we selected some of the differentially expressed proteins for verification by secondary assays. As shown in Figure 4A, qualitative comparisons of Coomassie blue staining of SDS-PAGE gels also indicated that some proteins were down-regulated and others were up-regulated in K6W-Ub expressing lenses (Figure 4A). One of the decreased bands in the SDS-gel (Figure 4A) contained several γ-crystallins (γA, γB, γC, γD, and γF). The decrease in γ-crystallin band on the gel was consistent with the dramatic decrease in spectral counts of these proteins (Table 2). Similarly, levels of intact βB1-crystallin decreased and cleaved βB1-crystallin increased (Figure 4A, compare βB1 vs βB1C). Because the spectral-counting methods cannot distinguish cleaved protein from its intact form when using in-solution digestion methods, it is not surprising that there was a lack of significant changes in total spectral count of βB1-crystallin (Appendix A in the Supporting Information). The SDS-gel also showed that the level of intact βA1-crystallin was considerably decreased in lenses of high expresser (Figure 4A), but the total spectral counts for all forms of this protein did not change. Like βB1-crystallin, this discrepancy appears to be due to aggregation or fragmentation. Western blotting analysis with antibody to β-crystallins confirmed the presence of fragmented and aggregated forms of β-crystallins in lenses of K6W-Ub high expressers (Supplemental Figure 2 in the Supporting Information). Mass spectrometric analysis of the increased gel band ∼50 kDa also showed the presence of aggregated βA1, βB1, and βB3 in K6W-Ub lenses (Figure 4A).
Figure 4.
Western blotting verification of differentially expressed proteins among lenses from WT, K6W-ubiqutin low expresser, and K6W-ubiquitin high expresser. P1 lenses from WT, low expressers, and high expressers of K6W-Ub transgenic mice were collected and homogenized. Proteins were separated by SDS-PAGE and stained by Coomassie blue to visualize proteins profiles (A). The bands that had visibly different staining were excised and identified by mass spectrometry after trypsin in-gel-digestion. The protein identifications of these proteins are indicated on the right of the gel. βB1-agg, αA-agg, βA1-agg, and βB3-agg represent aggregated forms of βB1, αA-,βB1-, and βB3-crystallins, respectively; βB1–C, and βA1-C represent cleaved forms of βB1- and βA1-crystallins. Levels of periaxin, caprin 2, filensin, γ-crystallins, annexin A, PGK1, and Hsp27 were determined by Western blotting (B). GAPDH and β-actin were used as protein loading controls. All experiments were repeated two times using pooled lens samples (three to five lenses in each group). Because of the weak signal, the level of annexin A in the second experiment was not quantified. Panel C is the densitometry quantitation of panel B using GAPDH as a reference. The candidate protein/GAPDH ratios for the WT lenses were designated as 1.0 to present relative levels of proteins in low and high expressers.
Western blotting analysis confirmed the differential expression of six of the seven tested proteins that were detected by mass spectrometric analyses. Consistent with the MS data, levels of periaxin, caprin2, filensin, and γ-crystallins were significantly decreased in lenses in which K6W-Ub was expressed and the down regulation of these proteins depended on the levels of K6W-Ub in the lenses. Lenses from the K6W-Ub high expressers had much less periaxin, caprin-2, filensin, and γ-crystallins (Figure 4B,C) compared with lenses from low expressers. Probably due to posttranslational modifications, periaxin and filensin were detected by Western blotting as doublets. Similar results have also been reported by others.41 Also consistent with the MS data, levels of annexin A and PKG1 (Figure 4B,C) increased in K6W-Ub lenses, and the extent of the increase was also related to the levels of K6W-Ub. However, the Western blotting results for Hsp27 (also called HSPB1) were inconsistent with MS analysis. The MS analysis showed that spectral counts for HSPB1 decreased in K6W-Ub expressing lenses, but Hsp27 was only detected by Western blotting in the lenses of K6W-Ub high expressers (Figure 4B). The failure to detect Hsp27 in lenses of WT or K6W-Ub low expressers by Western blotting indicates that levels of Hsp27 in normal lenses may be below the detection limit by Western blotting. Consistently, spectral counts for this protein were also low, indicating relatively low abundance of Hsp27 in the newborn lens.
We further determined the levels and distribution of periaxin by immunohistochemistry. The neonatal lenses were cryosectioned on the equatorial plane. Sections near the equator region were immunostained with antibody against periaxin. As previously shown,41 periaxin was located on the fiber cell plasma membrane along the hexagonal fiber cells. The periaxin distribution patterns were similar to that of F-actin (Figure 5). Consistent with the results of MS and Western blot, the immunostaining signal for periaxin in the high K6W-Ub lens was much weaker than that in the WT lenses, particularly at the center of the lenses (Figure 5). In addition, expression of high levels of K6W-Ub resulted in disruption of the organization of lens fibers, particularly in the center cataractous region. In WT lenses, fibers were well-organized hexagons. In the lenses of K6W-Ub high expressers, the fibers were disorganized and detached from each other. Probably because of loss of gap junction coupling, the diameter of lens fibers in the center of K6W-Ub appears to be about two times bigger than that of WT lenses (Figure 5, compare lower panels with upper panels). Gaps between the lens fibers were found only at the center of equatorial sections of newborn K6W-Ub lenses, and they are not observed in sagittal sections of older lenses (Figure 1E) or sagittal sections of embryonic lenses.
To begin to test whether the alteration of proteome in K6W-Ub lenses was due to altered translation, we measured the mRNA levels of two lens fiber-specific proteins by real-time RT-PCR. When GAPDH mRNA was used as control, the mRNA levels for filensin and caprin in the lenses of newborn K6W-Ub high expressers were 60 and 80% of those found in newborn WT lenses. The trend of changes in mRNA levels of these gene was consistent with the changes in their protein levels, suggesting that alteration of transcription may be partially responsible for the altered protein levels revealed by the proteomic analysis.
Discussion
Using a deep proteomic analysis, together with Western blots and immunohistochemistry analysis, this work extended our understanding of how a functional UPS is critical in the lens for proper epithelial cell proliferation, fiber differentiation, and establishing a transparent lens.24,25,37,42 Significant changes in levels of many essential proteins (Table 2), abnormalities of lens differentiation and maturation, and nuclear cataract at birth were obvious when higher levels of K6W-Ub were expressed. The effects of expression of the transgene on the lens proteome and structure were dose-dependent. The findings in the low expressers of clear lenses, but with some alterations in expression of several critical lens fiber-specific proteins, suggest that proteomic changes in the K6W-Ub lens precede and are likely etiologically related to cataract formation. Taken together, these data suggest that proteomic changes may be the contributing factor for cataract formation rather than the consequence of cataract. These data also indicate that a functional UPS, with K6 on ubiquitin intact, is required for lens protein homeostasis, including transcription, translation, and selective protein degradation. It remains to be determined which of the altered proteins plays the key role in cataractogenesis.
More than 2000 proteins were identified in this proteomic study, and ∼50% of these proteins were quantitatively compared between WT and K6W-Ub expressing lenses. Among the 966 reliably quantified proteins, only 32 of the 966 quantified proteins in the lens showed significant changes in protein levels. Among these differentially expressed proteins, 17 were up-regulated and 15 were down-regulated (Table 2). The down-regulated proteins in K6W-Ub lenses were mainly lens-specific proteins, including γ-crystallins (γA, γB, γC, and γE), filensin, and periaxin. These proteins are involved in maintaining structural integrity. In addition, several translation factors were also down-regulated, suggesting that down-regulation of these translation factors might be responsible for the reduced levels of lens-specific proteins in K6W-Ub expressing lenses. The up-regulated proteins in K6W-Ub lenses are related to sugar metabolism, protein degradation, and basement membrane structures, suggesting that expression of K6W-Ub may also alter lens metabolism.
γ-Crystallins are a family of highly homologous lens fiber structural proteins. Many of the γ-crystallins are expressed in primary fibers during lens development, whereas γ-S-crystallin, like βB2-crystallin, is mainly expressed in secondary fibers during later stages of lens development. The high levels of these proteins in mature lens fibers play an important role in establishing and maintaining the proper optical properties of the lenses. Mutations of γ-crystallins are associated with congenital cataracts in humans and in mice.43−45 It was reported that the down-regulation of γ-S-crystallin expression was a potential mechanism for cataract formation in mice that lack functional Hsf4.46 Furthermore, loss of γ-D-crystallin due to mutation in start codon caused cataract in rat.47 The significant down-regulation of γ-crystallins (γA, γB, γC, and γE) in the K6W-Ub lenses might partially contribute to cataract formation. It is likely that both total crystallin levels and the relative stoichiometry of crystallins are important factors for lens transparency, albeit it is not yet clear whether the decrease in overall crystallin expression or the decrease in particular γ-crystallins relative to other crystallins (or both) has a more significant influence on cataractogenesis.
Filensin is one of the two members of the intermediate filament protein superfamily that are exclusively expressed in the lens. It is proposed that filensin plays an essential role in maintaining the architecture of lens fibers.48 Mutation of filensin causes recessive cataract,49 and it has been proposed that such mutations would also predispose heterozygote carriers to the early onset of age-related nuclear cataract.50 Filensin normally forms a complex with the other intermediate filament protein CP49 in lens, and the CP49/filensin complex appears to be important for the stability of the individual proteins.51 CP49 knockouts showed no obvious changes in lens transparency or lens architecture at the light microscope level.51,52 However, CP49 deletion sharply reduced the protein level of filensin but did not alter the mRNA levels, suggesting that loss of CP49 promotes the degradation of filensin.51 Genetic analysis showed that inbred mice of 129 genetic background, including FVB/N mice used for this study, carry a mutation that deleted the acceptor site of exon 2, resulting in erroneous splicing, a frameshift in the reading frame, and the loss of CP49 protein.53,54 Consistent with the observations in CP49 knock-out mice,51 all 129 mice have reduced levels of filensin in the lens as compared with mice C57 background.53,55 Despite the instability of filensin in the absence of CP49, filensin was readily detected by MS and Western blot in the lens of newborn WT FBV/N mice (Table 1, Figure 5). The further reduction of the filensin in K6W-Ub expressing lens may contribute to the disorganization of lens fibers and cataractogenesis (Figures 1E and 5).
Periaxin is a PDZ domain protein involved in myelin sheath stabilization, and it is also a component of adherens plaques in lens fiber cells. A recent study showed that periaxin is required for hexagonal geometry and membrane organization of mature lens fibers.41 Lenses from periaxin-null mice are softer and deformable, similar to lenses from K6W-Ub high expressers. Thus, down-regulation of this protein in K6W-Ub transgenic mice may precede and contribute to defects in fiber formation. Indeed, we found that the organization of lens fibers in the lenses of K6W-Ub high expressers was disrupted, resulting in disconnected and larger fibers (Figure 5). This work indicates that together with other membrane and cytoskeleton proteins, periaxin might be involved in establishing proper gap junctions. The loss of gap junction coupling in the nuclear region of K6W-Ub high expressers may result in fiber swelling and contribute to the increased diameter of lens fibers. Another possible mechanism for the observed histologic changes may be just due to altered cell adhesion in the K6W-expressing lens, which results in cell detachment during the mechanical strain of sectioning.
Caprin-2 is an FGF-induced protein and one of the early markers of lens fiber differentiation.56 Caprin-2 is also an RNA granule protein,57 and it is likely involved in mRNA stability and accessibility for translation. The down-regulation of caprin-2 in lens by expression of K6W-Ub could affect the translation of fiber-specific proteins, including γ-crystallins (γA, γB, γC, and γE).
This work demonstrated that disruption of function of the UPS by expressing K6W-ubiquitin altered the lens proteome, suggesting that a functional UPS is essential for establishing and maintaining a proper lens proteome. Although <10% of total ubiquitin conjugates involves K6-linkage,58−60 we found that expression of K6W-Ub results in accumulation of significant levels of ubiquitin conjugates (Figure 1A). The phenotypes due to expression of K6W-Ub imply that K6 in ubiquitin is of critical importance for the function of UPS. Expression of K6W-Ub may alter the lens development and lens proteome by limiting the formation of K6-linked ubiquitin conjugates or by its dominant negative effects through the formation of nondegradable conjugates.20 However, it still remains to be investigated how K6W-Ub specifically alters the lens proteome. The UPS is a proteolytic system that selectively degrades abnormal, obsolete, and regulatory proteins, including cell signaling and transcription factors.12,61,62 Impairment of the UPS by expressing K6W-Ub might directly block the degradation and account for the increased level of some of the up-regulated proteins. Disruption of the UPS also blocks the timely degradation of cell signaling and regulatory proteins and subsequently impedes the lens development process and contributes to the down-regulation of lens-fiber specific proteins, such as γ-crystallins (γA, γB, γC, and γE), filensin, and periaxin. The decrease in fiber mass associated with the impairment of lens fiber maturation in high expressers would increase the proportion of lens epithelium in the samples and might indirectly contribute to the increased abundance of epithelial-specific proteins, such as some of the metabolic enzymes. However, because of the sensitivity and dynamic range limitations in proteomic experiments, more than 50% of the identified proteins in the lens, most of which were low-abundance regulatory proteins, could not be reliably quantified. Future improvement of instrument sensitivity or analytic methods will help to identify and quantify low abundance signaling and regulatory proteins in the lens.
Another possibility for the down-regulation of lens fiber-specific proteins in K6W-Ub expressing lens is the increased protein degradation by other proteases, which might be up-regulated in the absence of functional UPS. We found that proteasome subunit alpha type-2 was up-regulated in K6W-Ub lens (Table 2). Another three proteasome subunits were also up-regulated in K6W-Ub lenses but to a less significant extent (with ANOVA test >0.01 but <0.05) (Appendix A in the Supporting Information). The up-regulation of the proteasome family in K6W-Ub expressing lenses may reflect the compensatory feedback of dysfunction of the UPS caused by the K6W-Ub expression. It was reported that impairment of the UPS function by proteasome inhibitors up-regulates proteasome levels.63 The up-regulation of the proteasome might not be able to improve the function of the UPS in K6W-Ub expressing lenses because the conjugates formed with K6W-Ub are resistant to proteasomal degradation.20 Furthermore, we found that calpain 3 was activated in K6W-Ub expressing lenses due to altered calcium homeostasis. Calpain activation may contribute to the cleavage of lens proteins (Supplemental Figure 2 in the Supporting Information) in K6W-Ub lens, but we do not expect that it will cause a reduction of spectral counts of specific proteins (Table 2).
In addition to demonstrating that expression of K6W-Ub alters the lens proteome, this work also greatly expanded the known lens proteome. In the past, the majority of global lens proteome analyses were performed by 2D gel electrophoresis separation coupled to mass spectrometric protein identification.64−68 Because of limited sensitivity of that method, less than one hundred proteins were identified in the lens, and the majority of the identified proteins were high abundance structural proteins, such as crystallins. The development of 2-D LC/mass spectrometric approaches significantly increased the sensitivity and expanded the known lens proteome to a few hundreds.26,27 With the improvement of instruments and analytic methods, the number of proteins identified in the lens has increased significantly. A recent proteomic work identified a total of 951 proteins in human lenses.69 Although previous lens proteomic studies focused on different fractions of lens proteins, such as membrane proteins, total proteins identified were also reported in those studies26,69 The current work reliably identified more than 2000 proteins in the newborn mouse lenses, doubling the known lens proteome in any species. A previous proteomic analysis of lenses from P18 to P28 mouse showed that crystallins account for 78% of the total protein spectral counts.26 In comparison, the current work indicates that crystallins accounted for only 28.7% of the total spectral counts of lens proteins in the WT newborn mouse lenses (Table 1). The smaller proportion of crystallin in the lenses of newborn mice is consistent with a previous study that showed that levels of βB2-, γS-, and αB-crystallins in lenses from 1.5 week old mice were much lower than those in lenses from 6 week old mice.65 In addition to exploiting modern techniques, the relatively lower abundance of crystallins in lenses of newborn mice offered the opportunity to detect other classes of proteins that are present in lower abundance in the lenses and helped to expand the known lens proteome reported in this study.
This study was focused on the quantitative analysis of lens proteins. We measured mRNA levels of two representative genes (filensin and caprin 2) by real-time PCR. The levels of mRNA levels of these two genes were consistent with their protein levels. However, it is not clear whether the alteration in other lens proteins occurred at the transcription levels or at posttranslational levels. Future comprehensive mRNA analysis by a high-throughput method, as mRNA microassay or RNA-seq, will help to elucidate the mechanism by which K6W-Ub alters lens proteome.
In conclusion, we demonstrate that this bottom-up proteomic method provides markedly enhanced sensitivity in comparison with previous lens proteomic work and affords a comprehensive approach to quantitatively analyze and compare steady-state protein levels between different groups of tissues. Using this approach, we found that the extent of alteration of the lens proteome is related to levels of K6W-ubiquitin and precedes alterations in lens morphology and cataract formation in low expressers. Secondary analysis by Western blot confirmed the reliability of the high-throughput MS method. These data corroborate our prior finding that the lysine in position 6 of ubiquitin has essential functions in maintaining proteomes and homeostasis and that these are essential for directing proper cell proliferation and differentiation as well as organ function. In turn, the data inform about the exquisite sensitivity of cells and tissues to perturbations in the ubiquitin pathway, specifically ubiquitin per se. Further investigation of these candidate proteins will help to elucidate their role in maintaining lens structure and lens transparency.
Acknowledgments
This work is supported by NIH grants EY 013250, EY 021212, EY 011717, EY 007755, and EY 010572 and USDA contract 1950-510000-060-01A.
Supporting Information Available
Quantitative protein results, one-way ANOVA product p-value distribution, ANOVA candidates (p < 0.01) with TREND requirement, and peptide identifications. Density histogram of ANOVA test p-value of the quantified 996 proteins. Expression of high levels of K6W-ubiquitin results in aggregation and cleavage of β-crystallins. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Ciechanover A.; Orian A.; Schwartz A. L. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays 2000, 225442–51. [DOI] [PubMed] [Google Scholar]
- Glickman M. H.; Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 2002, 822373–428. [DOI] [PubMed] [Google Scholar]
- Goldberg A. L. Protein degradation and protection against misfolded or damaged proteins. Nature 2003, 4266968895–9. [DOI] [PubMed] [Google Scholar]
- Shang F.; Taylor A. Ubiquitin-proteasome pathway and cellular responses to oxidative stress. Free Radic. Biol. Med. 2011, 5115–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr D. M.; Hohfeld J.; Patterson C. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem. Sci. 2002, 277368–75. [DOI] [PubMed] [Google Scholar]
- Dai Q.; Zhang C.; Wu Y.; McDonough H.; Whaley R. A.; Godfrey V.; Li H. H.; Madamanchi N.; Xu W.; Neckers L.; Cyr D.; Patterson C. CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 2003, 22205446–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frescas D.; Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat. Rev. Cancer 2008, 86438–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickliffe K.; Williamson A.; Jin L.; Rape M. The multiple layers of ubiquitin-dependent cell cycle control. Chem. Rev. 2009, 10941537–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. J. Ubiquitin signalling in the NF-kappaB pathway. Nat. Cell Biol. 2005, 78758–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes A. F.; Bian Q.; Jiang J. K.; Thomas C. J.; Taylor A.; Pereira P.; Shang F. Proteasome inactivation promotes p38 mitogen-activated protein kinase-dependent phosphatidylinositol 3-kinase activation and increases interleukin-8 production in retinal pigment epithelial cells. Mol. Biol. Cell 2009, 20163690–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Q.; Fernandes A. F.; Taylor A.; Wu M.; Pereira P.; Shang F. Expression of K6W-ubiquitin in lens epithelial cells leads to upregulation of a broad spectrum of molecular chaperones. Mol. Vision 2008, 14, 403–12. [PMC free article] [PubMed] [Google Scholar]
- Shang F.; Taylor A. Roles for the ubiquitin-proteasome pathway in protein quality control and signaling in the retina: Implications in the pathogenesis of age-related macular degeneration. Mol. Aspects Med. 2012, 334446–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A.; Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–79. [DOI] [PubMed] [Google Scholar]
- Hershko A.; Ciechanover A. The ubiquitin system for protein degradation. Annu. Rev. Biochem. 1992, 61, 761–807. [DOI] [PubMed] [Google Scholar]
- Beal R.; Deveraux Q.; Xia G.; Rechsteiner M.; Pickart C. Surface hydrophobic residues of multiubiquitin chains essential for proteolytic targeting. Proc. Natl. Acad. Sci. U.S.A. 1996, 932861–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam Y. A.; Xu W.; DeMartino G. N.; Cohen R. E. Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature 1997, 3856618737–40. [DOI] [PubMed] [Google Scholar]
- Lam Y. A.; Lawson T. G.; Velayutham M.; Zweier J. L.; Pickart C. M. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature 2002, 4166882763–7. [DOI] [PubMed] [Google Scholar]
- Chau V.; Tobias J. W.; Bachmair A.; Marriott D.; Ecker D. J.; Gonda D. K.; Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 1989, 243, 1576–1583. [DOI] [PubMed] [Google Scholar]
- Thrower J. S.; Hoffman L.; Rechsteiner M.; Pickart C. M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000, 19194–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang F.; Deng G.; Liu Q.; Guo W.; Haas A. L.; Crosas B.; Finley D.; Taylor A. Lys6-modified ubiquitin inhibits ubiquitin-dependent protein degradation. J. Biol. Chem. 2005, 2802120365–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L.; Wang C.; Spencer E.; Yang L.; Braun A.; You J.; Slaughter C.; Pickart C.; Chen Z. J. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 2000, 1032351–61. [DOI] [PubMed] [Google Scholar]
- Chiu R. K.; Brun J.; Ramaekers C.; Theys J.; Weng L.; Lambin P.; Gray D. A.; Wouters B. G. Lysine 63-polyubiquitination guards against translesion synthesis-induced mutations. PLoS Genet. 2006, 27e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek E. J.; Shang F.; Valverde P.; Liu Q.; Hobbs M.; Taylor A. Selectivity of the ubiquitin pathway for oxidatively modified proteins: relevance to protein precipitation diseases. FASEB J. 2005, 19121707–9. [DOI] [PubMed] [Google Scholar]
- Liu Q.; Shang F.; Zhang X.; Li W.; Taylor A. Expression of K6W-ubiquitin inhibits proliferation of human lens epithelial cells. Mol. Vision 2006, 12, 931–6. [PubMed] [Google Scholar]
- Caceres A.; Shang F.; Wawrousek E.; Liu Q.; Avidan O.; Cvekl A.; Yang Y.; Haririnia A.; Storaska A.; Fushman D.; Kuszak J.; Dudek E.; Smith D.; Taylor A. Perturbing the ubiquitin pathway reveals how mitosis is hijacked to denucleate and regulate cell proliferation and differentiation in vivo. PLoS One 2010, 510e13331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassnett S.; Wilmarth P. A.; David L. L. The membrane proteome of the mouse lens fiber cell. Mol. Vision 2009, 15, 2448–63. [PMC free article] [PubMed] [Google Scholar]
- Wilmarth P. A.; Riviere M. A.; David L. L. Techniques for accurate protein identification in shotgun proteomic studies of human, mouse, bovine, and chicken lenses. J. Ocul. Biol. Dis. Inf. 2009, 24223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.; Sadygov R. G.; Yates J. R. 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004, 76144193–201. [DOI] [PubMed] [Google Scholar]
- Storey J. D.; Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 2003, 100169440–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thissen D.; Steinberg L.; Kuang D. Quick and Easy Implementation of the Benjamini-Hochberg Procedure for Controlling the False Positive Rate in Multiple Comparisons. J. Educ. Behav. Stat. 2002, 27177–83. [Google Scholar]
- Shevchenko A.; Tomas H.; Havlis J.; Olsen J. V.; Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 162856–60. [DOI] [PubMed] [Google Scholar]
- Pereira P.; Shang F.; Girão H.; Taylor A. Lens fibers have a fully functional ubiquitin-proteasome pathway. Exp. Eye Res. 2003, 76, 623–631. [DOI] [PubMed] [Google Scholar]
- Girao H.; Pereira P.; Taylor A.; Shang F. Subcellular redistribution of components of the ubiquitin-proteasome pathway during lens differentiation and maturation. Invest. Ophthalmol. Visual Sci. 2005, 4641386–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q.; Shang F.; Guo W.; Hobbs M.; Valverde P.; Reddy V.; Taylor A. Regulation of the ubiquitin proteasome pathway in human lens epithelial cells during the cell cycle. Exp. Eye Res. 2004, 78, 197–205. [DOI] [PubMed] [Google Scholar]
- Guo W.; Shang F.; Liu Q.; Urim L.; West-Mays J.; Taylor A. Differential regulation of components of the ubiquitin-proteasome pathway during lens cell differentiation. Invest. Ophthalmol. Visual Sci. 2004, 45, 1194–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes A. F.; Guo W.; Zhang X.; Gallagher M.; Ivan M.; Taylor A.; Pereira P.; Shang F. Proteasome-dependent regulation of signal transduction in retinal pigment epithelial cells. Exp. Eye Res. 2006, 8361472–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q.; Shang F.; Whitcomb E.; Guo W.; Li W.; Taylor A. Ubiquitin-conjugating enzyme 3 delays human lens epithelial cells in metaphase. Invest. Ophthalmol. Visual Sci. 2006, 4741302–9. [DOI] [PubMed] [Google Scholar]
- Millonig G.; Muckenthaler M. U.; Mueller S. Hyperferritinaemia-cataract syndrome: worldwide mutations and phenotype of an increasingly diagnosed genetic disorder. Hum. Genomics 2010, 44250–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harned J.; Fleisher L. N.; McGahan M. C. Lens epithelial cells synthesize and secrete ceruloplasmin: effects of ceruloplasmin and transferrin on iron efflux and intracellular iron dynamics. Exp. Eye Res. 2006, 834721–7. [DOI] [PubMed] [Google Scholar]
- Harned J.; Ferrell J.; Nagar S.; Goralska M.; Fleisher L. N.; McGahan M. C. Ceruloplasmin alters intracellular iron regulated proteins and pathways: ferritin, transferrin receptor, glutamate and hypoxia-inducible factor-1alpha. Exp. Eye Res. 2012, 97190–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddala R.; Skiba N. P.; Lalane R. 3rd; Sherman D. L.; Brophy P. J.; Rao P. V. Periaxin is required for hexagonal geometry and membrane organization of mature lens fibers. Dev. Biol. 2011, 3571179–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W.; Shang F.; Liu Q.; Urim L.; Zhang M.; Taylor A. Ubiquitin-proteasome pathway function is required for lens cell proliferation and differentiation. Invest. Ophthalmol. Visual Sci. 2006, 4762569–75. [DOI] [PubMed] [Google Scholar]
- Talla V.; Narayanan C.; Srinivasan N.; Balasubramanian D. Mutation causing self-aggregation in human gammaC-Crystallin leading to congenital cataract. Invest. Ophthalmol. Visual Sci. 2006, 47125212–7. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Huerta L. M.; Messina-Baas O.; Urueta H.; Toral-Lopez J.; Cuevas-Covarrubias S. A. A CRYGC gene mutation associated with autosomal dominant pulverulent cataract. Gene 2013, 5291181–5. [DOI] [PubMed] [Google Scholar]
- Li L.; Chang B.; Cheng C.; Chang D.; Hawes N. L.; Xia C. H.; Gong X. Dense nuclear cataract caused by the gammaB-Crystallin S11R point mutation. Invest. Ophthalmol. Visual Sci. 2008, 491304–9. [DOI] [PubMed] [Google Scholar]
- Shi X.; Cui B.; Wang Z.; Weng L.; Xu Z.; Ma J.; Xu G.; Kong X.; Hu L. Removal of Hsf4 leads to cataract development in mice through down-regulation of gamma S-Crystallin and Bfsp expression. BMC Mol. Biol. 2009, 10, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. C.; Lee J. W.; Harmon A. C.; Morris Z.; Wang X.; Fratkin J.; Rapp J. P.; Gomez-Sanchez E.; Garrett M. R. A mutation in the start codon of gamma-Crystallin D leads to nuclear cataracts in the Dahl SS/Jr-Ctr strain. Mamm. Genome 2013, 243–495–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudge D. S.; McCuaig J. V.; Van Stralen S.; Hess J. F.; Wang H.; Mathias R. T.; FitzGerald P. G. Intermediate filaments regulate tissue size and stiffness in the murine lens. Invest. Ophthalmol. Visual Sci. 2011, 5263860–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R. D.; Perumalsamy V.; Hejtmancik J. F. Autosomal recessive juvenile onset cataract associated with mutation in BFSP1. Hum. Genet. 2007, 1213–4475–82. [DOI] [PubMed] [Google Scholar]
- Song S.; Landsbury A.; Dahm R.; Liu Y.; Zhang Q.; Quinlan R. A. Functions of the intermediate filament cytoskeleton in the eye lens. J. Clin. Invest. 2009, 11971837–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizadeh A.; Clark J. I.; Seeberger T.; Hess J.; Blankenship T.; Spicer A.; FitzGerald P. G. Targeted genomic deletion of the lens-specific intermediate filament protein CP49. Invest. Ophthalmol. Visual Sci. 2002, 43123722–7. [PubMed] [Google Scholar]
- Sandilands A.; Prescott A. R.; Wegener A.; Zoltoski R. K.; Hutcheson A. M.; Masaki S.; Kuszak J. R.; Quinlan R. A. Knockout of the intermediate filament protein CP49 destabilises the lens fibre cell cytoskeleton and decreases lens optical quality, but does not induce cataract. Exp. Eye Res. 2003, 763385–91. [DOI] [PubMed] [Google Scholar]
- Alizadeh A.; Clark J.; Seeberger T.; Hess J.; Blankenship T.; FitzGerald P. G. Characterization of a mutation in the lens-specific CP49 in the 129 strain of mouse. Invest. Ophthalmol. Visual Sci. 2004, 453884–91. [DOI] [PubMed] [Google Scholar]
- Sandilands A.; Wang X.; Hutcheson A. M.; James J.; Prescott A. R.; Wegener A.; Pekny M.; Gong X.; Quinlan R. A. Bfsp2 mutation found in mouse 129 strains causes the loss of CP49′ and induces vimentin-dependent changes in the lens fibre cell cytoskeleton. Exp. Eye Res. 2004, 784875–89. [DOI] [PubMed] [Google Scholar]
- Simirskii V. N.; Lee R. S.; Wawrousek E. F.; Duncan M. K. Inbred FVB/N mice are mutant at the cp49/Bfsp2 locus and lack beaded filament proteins in the lens. Invest. Ophthalmol. Visual Sci. 2006, 47114931–4. [DOI] [PubMed] [Google Scholar]
- Loren C. E.; Schrader J. W.; Ahlgren U.; Gunhaga L. FGF signals induce Caprin2 expression in the vertebrate lens. Differentiation 2009, 774386–94. [DOI] [PubMed] [Google Scholar]
- Shiina N.; Tokunaga M. RNA granule protein 140 (RNG140), a paralog of RNG105 localized to distinct RNA granules in neuronal dendrites in the adult vertebrate brain. J. Biol. Chem. 2010, 2853124260–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick D. S.; Hathaway N. A.; Hanna J.; Elsasser S.; Rush J.; Finley D.; King R. W.; Gygi S. P. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat. Cell Biol. 2006, 87700–10. [DOI] [PubMed] [Google Scholar]
- Xu P.; Duong D. M.; Seyfried N. T.; Cheng D.; Xie Y.; Robert J.; Rush J.; Hochstrasser M.; Finley D.; Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 2009, 1371133–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser S. E.; Riley B. E.; Shaler T. A.; Trevino R. S.; Becker C. H.; Schulman H.; Kopito R. R. Protein standard absolute quantification (PSAQ) method for the measurement of cellular ubiquitin pools. Nat. Methods 2011, 88691–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat K. P.; Greer S. F. Proteolytic and non-proteolytic roles of ubiquitin and the ubiquitin proteasome system in transcriptional regulation. Biochim. Biophys. Acta 2010, 18092150–5. [DOI] [PubMed] [Google Scholar]
- Geng F.; Wenzel S.; Tansey W. P. Ubiquitin and proteasomes in transcription. Annu. Rev. Biochem. 2012, 81, 177–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S.; Kanade S.; Han B.; Eckert R. L. A proteasome inhibitor-stimulated Nrf1 protein-dependent compensatory increase in proteasome subunit gene expression reduces polycomb group protein level. J. Biol. Chem. 2012, 2874336179–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampi K. J.; Shih M.; Ueda Y.; Shearer T. R.; David L. L. Lens proteomics: analysis of rat Crystallin sequences and two-dimensional electrophoresis map. Invest. Ophthalmol. Visual Sci. 2002, 431216–24. [PubMed] [Google Scholar]
- Ueda Y.; Duncan M. K.; David L. L. Lens proteomics: the accumulation of Crystallin modifications in the mouse lens with age. Invest. Ophthalmol. Visual Sci. 2002, 431205–15. [PubMed] [Google Scholar]
- Hoehenwarter W.; Kumar N. M.; Wacker M.; Zimny-Arndt U.; Klose J.; Jungblut P. R. Eye lens proteomics: from global approach to detailed information about phakinin and gamma E and F Crystallin genes. Proteomics 2005, 51245–57. [DOI] [PubMed] [Google Scholar]
- Su S.; Liu P.; Zhang H.; Li Z.; Song Z.; Zhang L.; Chen S. Proteomic analysis of human age-related nuclear cataracts and normal lens nuclei. Invest. Ophthalmol. Visual Sci. 2011, 5274182–91. [DOI] [PubMed] [Google Scholar]
- Andley U. P.; Malone J. P.; Hamilton P. D.; Ravi N.; Townsend R. R. Comparative proteomic analysis identifies age-dependent increases in the abundance of specific proteins after deletion of the small heat shock proteins alphaA- and alphaB-Crystallin. Biochemistry 2013, 52172933–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.; Han J.; David L. L.; Schey K. L. Proteomics and phosphoproteomics analysis of human lens fiber cell membranes. Invest. Ophthalmol. Visual Sci. 2013, 5421135–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.