Abstract
Purpose
Previously, we developed a radiosensitivity molecular signature (RSI) that was clinically-validated in three independent datasets (rectal, esophageal, head and neck) in 118 patients. Here, we test RSI in radiotherapy (RT) treated breast cancer patients.
Experimental Design
RSI was tested in two previously published breast cancer datasets. Patients were treated at the Karolinska University Hospital (n=159) and Erasmus Medical Center (n=344). RSI was applied as previously described.
Results
We tested RSI in RT-treated patients (Karolinska). Patients predicted to be radiosensitive (RS) had an improved 5 yr relapse-free survival when compared with radioresistant (RR) patients (95% vs. 75%, p=0.0212) but there was no difference between RS/RR patients treated without RT (71% vs. 77%, p=0.6744), consistent with RSI being RT-specific (interaction term RSIxRT, p=0.05). Similarly, in the Erasmus dataset RT-treated RS patients had an improved 5-year distant-metastasis-free survival over RR patients (77% vs. 64%, p=0.0409) but no difference was observed in patients treated without RT (RS vs. RR, 80% vs. 81%, p=0.9425). Multivariable analysis showed RSI is the strongest variable in RT-treated patients (Karolinska, HR=5.53, p=0.0987, Erasmus, HR=1.64, p=0.0758) and in backward selection (removal alpha of 0.10) RSI was the only variable remaining in the final model. Finally, RSI is an independent predictor of outcome in RT-treated ER+ patients (Erasmus, multivariable analysis, HR=2.64, p=0.0085).
Conclusions
RSI is validated in two independent breast cancer datasets totaling 503 patients. Including prior data, RSI is validated in five independent cohorts (621 patients) and represents, to our knowledge, the most extensively validated molecular signature in radiation oncology.
Keywords: radiosensitivity, predictive biomarkers, gene expression, molecular signature, breast cancer
Introduction
The development of a radiosensitivity predictive assay has been a central goal of radiation biology for several decades (1, 2). The clinical impact of a successful assay would be broad and significant since radiation therapy (RT) is the single most common therapeutic agent in clinical oncology. Approximately 60% of all cancer patients receive RT at some point during their treatment (3).
In the era of personalized medicine, there is significant emphasis on the development of companion diagnostics and/or molecular signatures to guide therapeutic decisions (4). For example, two recurrence risk signatures (Oncotype Dx and Mammaprint) are commonly used to guide chemotherapy in women with node negative breast cancer (5–7). In addition, K-ras mutation has been shown to be predictive of panitumimab and cetuximab non-benefit in colorectal cancer (8, 9). Furthermore, EGFR mutations have been shown to predict benefit from tyrosine kinase inhibitors (TKIs) and more recently ALK gene rearrangement has shown to be predictive for crizotinib benefit in non-small cell lung cancer (10–12). In contrast, clinical decision making in radiation oncology is still mainly based on clinico-pathological features. Thus, there is a great need to develop molecular diagnostics to more efficiently utilize RT.
A reasonable criticism of the biomarker development field in radiation oncology is the lack of a strategy for the discovery of RT-specific biomarkers. In general, biomarkers that have been evaluated have not been necessarily chosen based on their specificity for RT. Thus, most biomarkers that have been shown to correlate with outcome after RT are also prognostic in patients that do not receive RT. For example, Ki-67 has been shown to be prognostic in prostate cancer patients after prostatectomy (13, 14) and after definitive RT (15, 16). However in the personalized medicine era, biomarkers are needed that are clinically-useful and that can be linked to a specific therapeutic intervention.
To address this, our group has recently developed a radiosensitivity molecular signature (RSI) which was exclusively developed as a biomarker of cellular radiosensitivity. The signature is based on gene expression for 10 specific genes and a linear regression algorithm. RSI was developed in 48 cancer cell lines, using a systems-biology strategy focused on identifying biomarkers specific for cellular radiosensitivity. The survival fraction at 2 Gy (SF2), a measure of cellular radiosensitivity, was the main criteria utilized to identify the 10 genes in the signature (AR, cJun, STAT1, PKC, RelA, cABL, SUMO1, CDK1, HDAC1, IRF1) out of an original pool of over 7,000 genes. Biological pathways represented in the signature include: DNA damage response, histone deacetylation, cell cycle, apoptosis and proliferation. Finally, the locked-down linear algorithm was exclusively trained and developed to predict SF2 in the cell line database. Therefore cellular radiosensitivity (as defined by SF2) was the central criteria both in feature selection and final model training for RSI development. Importantly, RSI has been clinically-validated in three independent datasets (rectal, esophageal and head and neck cancer) in 118 patients. It has been shown to predict for pathological response to pre-operative chemoradiation in two independent cohorts of patients with esophageal and rectal cancer. Finally, RSI was shown to predict for locoregional recurrence in patients with locally-advanced head and neck cancer treated with definitive chemoradiation (17–19).
In this study we test whether RSI predicts for clinical outcome in RT-treated breast cancer patients. We tested the signature in two independent datasets totaling 503 patients. We show evidence that the molecular signature is RT-specific and propose that it may serve as a predictive biomarker of RT therapeutic benefit in breast cancer.
Patients and Methods
Patients
Two previously published clinical datasets were utilized to test the radiosensitivity signature (20–22). Clinical details for each cohort are presented in online table 1
Karolinska University Hospital, Radiumhemmet Prospective/Observational Cohort
The study subjects in this cohort are part of a prospective/observational cohort treated at the Karolinska Hospital between January 1 1994 and December 31st 1996 which has been previously described (20). The ethical committee at the Karolinska Institute approved the microarray expression project. Tumor material was frozen on dry ice or liquid nitrogen and stored at −70C. Reasons for exclusion have been previously described (20). The final study cohort includes 159 patients with tissue and gene expression data. Differences in the clinical characteristics between patients without tissue and the final study cohort have been described (20). Primary treatment was segmentectomy/mastectomy + RT in 77 patients (experimental group) and mastectomy /no RT in 82 patients (negative control). All patients underwent axillary dissection. Standard RT was 50 Gy in 25 fractions delivered to the conserved breast/chest wall with (locoregional) or without (local) regional nodes (axillary, supraclavicular). Adjuvant therapy included both endocrine therapy (tamoxifen and/or goserelin) in 104 patients and chemotherapy (most commonly intravenous cyclophosphamide, methotrexate and 5-fluorouracil (CMF) on days 1 and 8). High-risk patients were offered participation in the Scandinavian Breast Group 9401 study (23). Follow up was obtained from the Swedish Breast Cancer Registry and was supplemented with patient charts as previously described (20). Mean follow up was 72 months.
Erasmus Dataset
This dataset consists of a total of 344 lymph node negative breast cancer patients who did not receive any adjuvant systemic treatment (chemotherapy and/or endocrine therapy) (22). The dataset includes 286 patients used to generate and validate a 76 gene signature of early distant recurrence (21) supplemented with an additional 58 ER negative patients to perform reliable pathway analysis (24). The study was approved by the Medical Ethics Committee of the Erasmus Medical Center. Primary treatment was breast conserving therapy in 80% of patients (lumpectomy + RT). The remaining 20% of the dataset was treated with mastectomy alone. Reasons for exclusion, clinical characteristics follow-up and treatment details were previously described (21, 22). Early metastasis was defined as a distant recurrence in the first 5 years following completion of primary treatment.
RNA preparation and Gene Expression Profiling
This has been previously described (20–22). Raw gene expression data for both datasets are publicly-available in GEO (Karolinska - GSE1456, Erasmus – GSE2034, GSE5327). The robust multi-array (RMA) normalization method was applied to the Affymetrix U133A CEL files (25–27).
Radiosensitivity Signature: Radiosensitivity Index (RSI) determination
This was performed as previously described. Probesets utilized for each gene were the same as in previous studies (17, 18). Briefly, each of the ten genes in the assay was ranked according to gene expression (from the highest (10) to the lowest expressed gene (1)). RSI was determined using the previously published ranked-based linear algorithm:
Statistical Analyses
Each dataset was analyzed independently. The 25th percentile for RSI in the subset of patients that received RT was pre-defined as the cutpoint to dichotomize the patients into radiosensitive (RS) and radioresistant (RR) groups, as in a previous study (18). This RS/RR variable was compared to prognostic variables, as well as relapse-free survival (RFS, Karolinska) and distant-metastasis-free survival (DMFS, Karolinska and Erasmus). RFS (Karolinska) was defined as any relapse distant, regional or local from the end of primary treatment. DMFS (Erasmus) was defined as any distant recurrence in the first 5 years following completion of treatment. Exact Chi-square test using Monte Carlo estimation or Mann-Whitney-Wilcoxon test was used to study association between RR/RS variable and prognostic variables. Kaplan-Meier survival curves for RFS/DMFS were fit for the RS/RR groups, along with the Log-Rank test to determine difference between the curves. Cox proportional hazard models were also fit to obtain hazard ratios. Multivariable Cox proportional hazard models were used to select potential predictors for RFS/DFMS. The final model was fitted with backwards selection, at a 0.10 significance level for removal. An interaction model was fit for RFS/DFMS, with the covariates RR/RS, RT/no-RT, and the interaction between RR/RS and RT/no-RT, to determine if RR/RS is predictive of RT/no-RT benefit. The analysis was conducted for all patients in both datasets, including the ER subset analyses. All analyses were done with SAS (version 9.3), tests were two sided, and had a significance level of 0·05.
Results
The Radiosensitivity Molecular Signature Predicts Clinical Outcome Only in RT-Treated Patients
We first examined whether there was any association between radiophenotype (as predicted by the RSI) and clinical outcome in patients that underwent primary treatment with surgery and RT in the Karolinska dataset. As hypothesized, RS patients had an improved 5-yr RFS compared with RR patients (RS vs. RR, 95% vs. 75%, p=0.0212, Figure 1). In contrast, there was no difference in outcome between predicted RS and RR patients that did not receive RT (71% vs. 77%, p=0.6744) suggesting that RSI is RT-specific; i.e. a predictive biomarker. Importantly, the interaction term (RSIxRT) p value was 0.05, consistent with RSI being a predictive biomarker in this cohort (Figure 1).
Figure 1. Association of the Radiosensitivity Signature with Clinical Outcome (Karolinksa dataset).
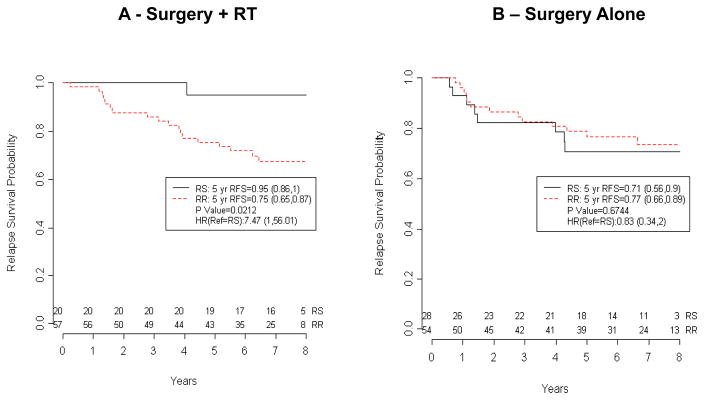
(A) RSI identifies a radiosensitive population (25th percentile) that has an improved 5-yr RFS in patients treated with surgery (lumpectomy/ segmentectomy) and RT. An interaction model between RSI and RT is consistent with RSI being RT-specific (p=0.05). (B) Kaplan-Meier curves of predicted radiosensitive and radioresistant patients treated with mastectomy and no RT. Patients at risk at different time points are indicated
A total of 40 relapse events were observed in the Karolinska cohort (n=159) and 75% of these events were distant relapses. Local/Regional relapses were rare in the RT group (n=2). Thus, an impact of RSI on local-regional recurrence could not be determined. A distribution of local-regional/distant events in each of the groups is shown in online table 2. The main impact observed in the RS/RT population when compared with the RR/RT group is a decrease in the development of distant metastasis (DM), consistent with the RSI being predictive of DM risk exclusively in RT-treated patients. When the same analysis was performed for the DMFS endpoint similar results were observed (RS vs. RR, 5-yr DMFS, 95% vs. 76.8%, p=0.0343, data not shown).
To confirm this observation in a second and more clinically homogenous dataset, we tested the RSI in the Erasmus dataset. This dataset, which was specifically developed to identify a molecular signature to predict early distant metastasis risk, includes only lymph node negative patients that received no adjuvant systemic treatment (chemotherapy and/or endocrine therapy), reducing the potential impact of treatment-related confounding variables in the analysis. As shown in figure 2, the radiosensitivity signature predicts for early DM in the cohort of patients treated with RT (n=282). Predicted RS patients had an improved 5-year DMFS when compared with RR patients (77% vs. 64%, p=0.0409). However, similar to the Karolinska dataset, the RSI did not show any statistical differences in patients treated without RT (RS vs. RR, 80% vs. 81%, p=0.9425), consistent with RSI being RT-specific, although the interaction term RSIxRT was not significant in this dataset (p=0.4506).
Figure 2. Association of the Radiosensitivity Signature with Distant Metastasis-Free Survival (Erasmus dataset).
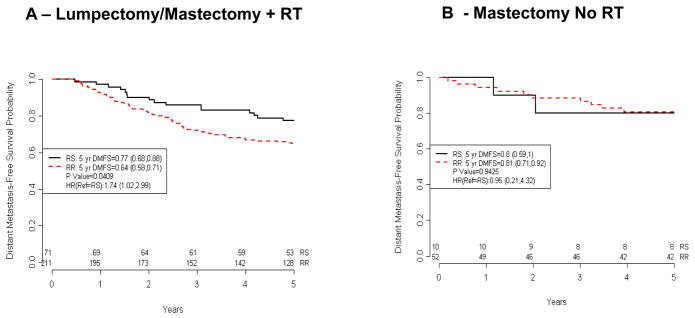
A) Kaplan-Meier curves of 282 patients treated with surgery + RT. (B) Kaplan-Meir curves of 62 patients treated with mastectomy alone. Patients at risk at different time points are indicated
Distribution of Clinical Characteristics between RS and RR patients
We determined whether there were any differences in the distribution of clinical and treatment variables between predicted RS and RR patients in both study datasets. As shown in table 1, there was no statistical difference in the distribution of any of the variables between RS and RR patients in the RT-treated cohorts. Similar results were observed in the patients that received no RT (data not shown). Of note, the Karolinska dataset was more heterogeneous as it included both lymph node negative and positive patients. In addition, treatment in the Karolinska dataset was more heterogeneous and included patients treated with adjuvant endocrine therapy and/or chemotherapy. This is reflective of this cohort being a prospective/observational cohort of patients treated following standard of care.
Table 1.
Clinical Characteristics of RT Patients
| ERASMUS (n=282) | N (%) Radiosensitive |
N(%) Radioresistant |
p value1 |
|---|---|---|---|
|
| |||
| n=71 | n=211 | ||
| ER/PR | |||
| ER+PR+ | 34 (51.5) | 98 (47.3) | |
| ER−PR+ or ER+PR− | 17 (25.8) | 47 (22.7) | 0.5579 |
| ER−PR− | 15 (22.7) | 62 (30) | |
| Size | |||
| T1 | 43 (60.6) | 103 (48.8) | 0.0980 |
| T2–4 | 28 (39.4) | 108 (51.2) | |
| Menopause | |||
| Premenopausal | 38 (53.5) | 121 (57.4) | 0.5783 |
| Postmenopausal | 33 (46.5) | 90 (42.7) | |
| Age | |||
| 26–40 | 9 (12.7) | 30 (14.2) | |
| 41–55 | 29 (40.9) | 97 (46.0) | 0.2296 |
| 56–70 | 29 (40.9) | 61 (28.9) | |
| 71–83 | 4 (5.6) | 23 (10.9) | |
| Surgery | |||
| Lumpectomy | 64 (90.1) | 184 (87.2) | 0.5415 |
| Mastectomy | 7 (9.9) | 27 (12.8) | |
| KAROLINSKA (n=77) | Radiosensitive | Radioresistant | p value1 |
|---|---|---|---|
|
| |||
| n=20 | n=57 | ||
| LN+ | 10(50) | 36(63.2) | 0.4329 |
| ER/PR | |||
| ER+PR+ | 13 (65.0) | 43 (75.4) | |
| ER−PR+ or ER+PR− | 3 (15.0) | 8 (14.0) | 0.5861 |
| ER−PR− | 4 (20.0) | 6 (10.5) | |
| Size | |||
| T1 | 14 (73.7) | 35 (62.5) | |
| T2–4 | 5 (26.3) | 21 (37.5) | 0.4200 |
| Grade‡: | |||
| elston 1 | 4 (23.5) | 10 (17.9) | 0.4827 |
| elston 2 | 5 (29.4) | 26 (46.4) | |
| elston 3 | 8 (47.1) | 20 (35.7) | |
| ET† | 18 (90.0) | 40 (70.2) | 0.1300 |
| CT† | 3 (15.0) | 15 (26.3) | 0.3703 |
| Surgery: | |||
| Segmentectomy | 12 (63.2) | 29 (51.8) | 0.4263 |
| Mastectomy | 7 (36.8) | 27 (48.2) | |
| RT | |||
| - Local | 11 (55.0) | 23 (40.3) | 0.3800 |
| Locoregional | 7 (35.0) | 31 (54.4) | |
| Unknown | 2 (10.0) | 3 (5.3) | |
P Values calculated using exact chi-squared test, with Monte Carlo estimation.
ET=endocrine therapy, CT=chemotherapy, RT=radiation therapy
Grade: 4 Missing data points
Multivariable and Backward Selection Models Identify RSI as the Strongest Variable in both Datasets
On multivariable analysis, RSI is the strongest variable in RT-treated patients in both datasets (Table 2, Erasmus, HR=1.64, p=0.0758, Karolinska, HR=5.53, p=0.0987). Of note, none of the variables in either dataset reached statistical significance, except for age category 71–83 in the Erasmus dataset (HR=0.31, p=0.0398). The importance of RSI was confirmed by the use of a backward selection model. Using a 0.10 significance level for removal, RSI was the only remaining significant variable in each final model.
Table 2.
Multivariate Cox Regression Analysis of RT-Treated Patients
| ERASMUS (n=273) (Ref vs. Level) | Hazard Ratio | p value |
|---|---|---|
| RSI (RS vs. RR) | 1.641 (0.950, 2.836) | 0.0758 |
| ER/PR | ||
| (ER+PR+ vs. ER−PR−) | 1.246 (0.747, 2.080) | 0.3995 |
| (ER+PR+ vs. ER+PR− or ER−PR+) | 1.134 ((0.685, 1.879) | 0.6249 |
| T-stage (T1 vs. T2,T3,T4) | 1.325 (0.853, 2.057) | 0.2106 |
| Age | ||
| (26–40 vs. 41–55) | 0.810 (0.445, 1.471) | 0.4882 |
| (26–40 vs. 56–70) | 0.891 (0.478, 1.659) | 0.7152 |
| (26–40 vs. 71–83) | 0.312 (0.103, 0.947) | 0.0398 |
| Surgery | ||
| (Mastectomy vs. Lumpectomy) | 1.467 (0.716, 3.006) | 0.2956 |
| KAROLINSKA (n=75) (Ref vs. Level) | Hazard Ratio | p value |
|---|---|---|
| RSI (RS vs. RR) | 5.533 (0.726,42.15) | 0.0987 |
| ER/PR | ||
| (ER+PR+ vs. ER−PR−) | 0.684 (0.122, 3.836) | 0.6662 |
| (ER+PR+ vs. ER+PR− or ER−PR+) | 2.321 (0.715, 7.541) | 0.1613 |
| T-stage (T1 vs. T2,T3,T4) | 1.452 (0.535, 3.937) | 0.464 |
| LN (No vs. Yes) | 1.395 (0.42, 4.629) | 0.5863 |
| ET (No vs. Yes) | 0.433 (0.123, 1.525) | 0.1926 |
| CT (No vs. Yes) | 0.83 (0.199, 3.465) | 0.7978 |
Subset Analysis Indicates RSI Is Predictive of Outcome in RT-treated ER+ Patients in the Erasmus Dataset
ER+ and ER− breast cancer patients have different biology. Therefore, we were interested in determining whether RSI is equally predictive of outcome in both ER+ and ER− patients. We conducted a subset analysis in the Erasmus dataset, which had enough DMFS events in both groups to warrant further study. As shown in figure 3, our analysis is consistent with RSI being predictive in the RT-treated ER+ patients. RS patients had a superior 5-yr DMFS compared to RR patients (figure 3A, RS vs. RR 81% vs. 61%, p=0.0124). In contrast no difference was seen between RS and RR patients in the RT-treated ER− subset (figure 3B, RS vs. RR 71% vs. 70%, p=0.952) or in either ER subset that did not receive RT (data not shown).
Figure 3. Association of the Radiosensitivity Signature with Distant Metastasis-Free Survival in in RT-treated ER+ Subset Patients in the Erasmus dataset.
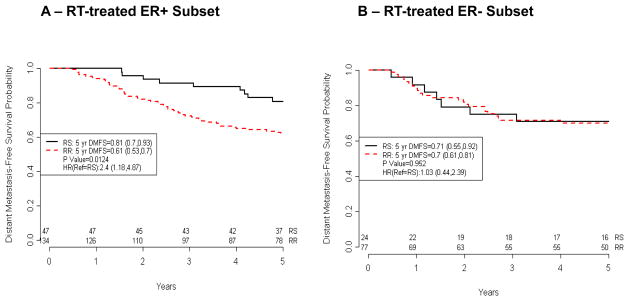
A) Kaplan-Meier curves of 181 RT-treated ER+ patients. B) Kaplan-Meier curves of 101 RT-treated ER− patients. Patients at risk at different time points are indicated
Interestingly, as reported above when the full Erasmus dataset was considered, the interaction term (RSIxRT) was not significant (p=0.4506). However when only the ER+ cohort was analyzed, the interaction between RSI and RT trended towards statistical significance (RSIxRT, p=0.0789), suggestive of RSI being a predictive biomarker in ER+ patients.
Importantly, as in the full datasets we confirmed no differences in clinical characteristics between RS and RR patients in each of the ER-subsets (data not shown). Finally, on multivariable analysis, RSI is an independent predictor of outcome in the RT-treated ER+ subset (Table 3, HR=2.64, p=0.0085), confirming the importance of RSI in this group of patients.
Table 3.
Multivariate Cox Regression Analysis of RT-Treated ER+ Patients
| ERASMUS (n=176) (Ref vs. Level) | Hazard Ratio | p value |
|---|---|---|
| RSI (RS vs. RR) | 2.640 (1.281, 5.438) | 0.0085 |
| PR (PR+ vs. PR−) | 1.746 (1.020, 2.988) | 0.0423 |
| T-stage (T1 vs. T2,T3,T4) | 1.525 (0.872, 2.666) | 0.1390 |
| Age | ||
| (26–40 vs. 41–55) | 0.491 (0.230, 1.052) | 0.0673 |
| (26–40 vs. 56–70) | 0.534 (0.241, 1.184) | 0.1226 |
| (26–40 vs. 71–83) | 0.225 (0.069, 0.732) | 0.0132 |
| Surgery | ||
| (Mastectomy vs. Lumpectomy) | 1.865 (0.702, 4.952 | 0.2112 |
Discussion
The development of biomarker-based models to guide therapeutic decisions is a central tenet of the personalized medicine era (28). In this paper, we tested a previously developed and validated (both biologically and clinically) 10-gene expression radiosensitivity signature, in two independent breast cancer datasets totaling 503 patients. Importantly, we minimized the potential biases associated with retrospective analyses by: 1. testing only one radiosensitivity signature, 2. testing a pre-determined cut-point, 3. including a negative control (surgery only patients) and 4. having a good understanding of patients within both datasets that were excluded from the tissue study. Including prior studies, the radiosensitivity signature is validated in five independent datasets in a total of 621 patients and represents to our knowledge the most clinically validated signature in radiation oncology. In a recent review article Simon and colleagues proposed a level of evidence (LOE) scale, to evaluate the clinical validity of prognostic and predictive biomarkers (29). Based on their criteria, the clinical validity of the radiosensitivity signature is supported by possibly level II scientific evidence based on the consistency of results in five independent datasets (combination of Category B and C studies).
Effective predictive biomarkers are a central requirement for the development of personalized treatment in clinical oncology. Unlike prognostic biomarkers which predict clinical outcome independent of treatment, predictive biomarkers are treatment specific and thus are critical for therapeutic decision-making (30). For example, several targeted drugs are now routinely offered to patients whose tumors harbor a specific marker for benefit or non-benefit (i.e Her-2/neu expression and trastuzumab benefit (31), K-ras mutation and panitumab non-benefit(8)). In contrast, radiation therapy is still recommended based on standard clinico-pathological features, which generally address tumor burden/aggressiveness and serve as prognostic biomarkers of outcome rather than a specific marker for RT therapeutic benefit.
Our data supports that the radiosensitivity signature may serve as a predictive marker of RT therapeutic benefit. We show that in both breast cancer datasets, the signature is RT-specific and only predicts outcome in RT-treated patients (Karolinska dataset, interaction term RSIxRT, p=0.05, Erasmus dataset ER+ subset, interaction term, RSIxRT, p=0.0789). In both datasets, predicted radiosensitive patients had a better outcome than radioresistant patients only when treated with RT. In patients treated without RT, patients predicted to be radiosensitive and radioresistant fared similarly. Furthermore, the effect size for the signature in each dataset is consistent with the expected therapeutic benefit for RT in lymph node positive/negative patients. The Oxford meta-analysis has shown that RT therapeutic benefit is proportional to the risk of recurrence in unirradiated women (32). Since 60% of the women in the Karolinska dataset had lymph node positive disease, a larger impact for RSI in this population would be expected. In addition, the Erasmus dataset only involved lymph node negative patients that received no adjuvant systemic hormonal or chemotherapy, making it unlikely that treatment factors other than RT are responsible for the differences in outcome observed. Moreover, on subset analysis we demonstrate RSI is an independent predictor of clinical outcome in RT-treated ER+ patients in the Erasmus dataset. In contrast RSI had no impact in ER− patients. Since ER− patients have a higher risk of distant micro-metastasis at diagnosis, it is reasonable to expect a lower impact for RSI in this subset, since the potential therapeutic impact of locoregional RT is outweighed by the absence of adjuvant systemic chemotherapy. Finally, the signature was developed specifically for cellular radiosensitivity and had been previously shown to predict for clinical response to pre-operative chemoradiation in two independent cohorts of rectal and esophageal cancer patients. Taken altogether, we think it is reasonable to propose that the radiosensitivity signature is a predictive biomarker of RT therapeutic benefit.
Prevention of locoregional recurrence has been long held as the most important therapeutic effect for RT in breast cancer and therefore the ideal endpoint for a radiosensitivity signature in breast cancer. Although the impact of RT in decreasing distant metastases is more controversial, data from randomized prospective clinical trials have shown that post-mastectomy RT to the chest wall and regional nodes decreases distant recurrences and improve OS after mastectomy in patients with positive axillary lymph nodes (33–35). In addition, the extensive Oxford meta-analysis has shown that RT reduces 15 year overall mortality presumably by preventing the development of distant metastasis (32). Finally, the Intergroup trial of Regional Nodal Irradiation in Early Breast Cancer (NCIC-CTG MA.20) which was recently reported at ASCO demonstrated that nodal irradiation resulted in a decrease in distant DFS (HR=0.64, p=0.002, 5 year risk, whole breast irradiation vs. whole breast + nodal irradiation, 92.4% vs. 87.0%) (36). Therefore we think this supports distant metastases risk as endpoint for our model. We have yet to test RSI as a predictor of local recurrence but we think this is a next logical step.
Recent findings have emphasized the difficulty of generating signatures to predict for local recurrence risk in breast cancer. Kreike and colleagues developed and validated a 111-gene classifier to predict local recurrence in breast cancer patients (37, 38). However in a subsequent study, Servant and colleagues were unable to validate the same signature in an independent dataset of 195 patients (39). In addition these investigators tested an additional 21 published signatures. None of the signatures achieved a higher accuracy than 59% in predicting local recurrence. Furthermore using the full dataset of 343 patients they were unable to generate a gene expression signature that outperformed standard clinical variables in predicting for local recurrence risk. These authors concluded that there are no significant differences in gene expression between patients with and without a local recurrence.
However a central difference between the approach discussed above and RSI is that we focused our model exclusively as a surrogate for cellular radiosensitivity. Loco-regional recurrence risk after BCT is related to a number of different factors some of which are non-biological (i.e. quality of surgery). Thus, there are potentially a number of significant confounding factors that can impact the robustness of a signature developed based on loco-regional recurrence. For example two similarly radioresistant patients might have different outcomes based on the quality of the surgery. However a model developed based on clinical outcome would be trained to call both of these samples different (i.e. one recurrent and one recurrence-free). This is a fundamental issue that is difficult to address and perhaps a central reason why many molecular signatures eventually fail. In contrast, we developed RSI exclusively on cellular radiosensitivity in cell lines, since this eliminates non-biological confounding factors that are an inherent part of clinical samples.
There is evidence to support that the factors that mediate locoregional failure risk in breast cancer are different between patients treated with and without RT. For example in a recent study, Oncotype DX recurrence score was shown to be predictive for risk of locoregional recurrence (40). In patients treated without RT (mastectomy alone), there was a clear association between the Oncotype DX-based recurrence score and locoregional recurrence risk. In contrast the association was significantly weaker for patients that received lumpectomy and RT (BCT). Multivariable analyses showed that the interaction between Oncotype-DX recurrence score and type of primary treatment was statistically significant. In addition, recent data shows that that in patients receiving BCT, locoregional recurrence risk was higher in HER2-enriched and basal subtypes whereas after mastectomy, luminal B, luminal-HER2, HER-2 enriched and basal subtypes were all associated with an increase risk of locoregional recurrences (41). A similar observation was made in triple negative breast cancer with a higher risk for locoregional recurrence in mastectomy patients when compared with BCT (42). One possible explanation for these findings is that RT effect is not equal across all molecular subtypes in breast cancer. Our data is consistent with RSI having a larger impact in ER+ patients (Luminal A, B and HER2 subtypes), at least as determined by the distant metastasis endpoint. Since ER status is one of the markers used to determine molecular subtype it is reasonable to hypothesize that the clinical impact of RSI may vary depending on molecular subtype.
The strategy to develop RSI was based on a systems-biology approach specific for the discovery of radiosensitivity biomarkers. In previous studies, we developed a linear regression algorithm to identify radiosensitivity biomarkers in 48 cancer cell lines. Gene expression was publicly-available and radiosensitivity was defined using the clonogenic survival of cell lines after 2 Gy of radiation (SF2) (17). As discussed above, the resulting radiosensitivity gene expression model to generate RSI has now been clinically validated in five independent datasets in a total of 621 patients. The successful translation from cell lines to patients in multiple disease sites argues that the biological basis of cellular radiosensitivity is conserved between cell lines and patients and across epithelial tumors.
There are several limitations to our study that are important to mention. First, treatment in both cohorts was not protocol-dictated and was based on established standard of care. Therefore, since this was not an experimental prospective trial, it is possible that there might be biases that we are unable to account for in our analysis. Second, in the Karolinska cohort (n=159) 70% of the original cohort (n=524) was excluded from the tissue/microarray study and excluded patients tended to have smaller tumors and a lower rate of events (20). Thus it is possible that this might have influenced the measured impact of RSI. Third, unlike the Karolinska dataset which is a populational-based cohort, the Erasmus cohort is a combination of a dataset of 286 patients specifically developed to identify a molecular signature to predict early distant metastatic risk supplemented with an additional 58 ER negative patients to increase the representation in this sub-group. Therefore, since this cohort is not exactly a random sample it may not accurately reflect risks and outcomes for the population. Finally, patients in both cohorts were generally undertreated compared to today standards. For example in the Karolinska cohort, 19% of patients received chemotherapy but 39% were lymph node positive. No patients in the Erasmus dataset received systemic chemotherapy and/or endocrine therapy but close to 50% were T2 tumors and 61% were ER positive. Therefore it is possible that the therapeutic benefit from RT was enhanced in both cohorts.
As discussed above RSI has been clinically-validated in 5 independent datasets in four disease sites. It should be noted that all clinical validation has been performed using publically-available datasets and further clinical utility demonstration and diagnostic platform developments are required before this technology can be applied in routine clinical care. A critical issue to address is standardizing the process for tissue acquisition, RNA isolation and gene expression measurement. A locked-down protocol for these steps as well as analytical validation of the process is required to meet regulatory body requirements and is currently under development. In addition, we think that transferring the diagnostic platform to work with formalin-fixed paraffin-embedded (FFPE) tissue will be critical to the routine use of this signature in the clinic. Our current strategy is to pursue additional validation in a more modern cohort of patients after these issues are addressed. An ideal validating cohort would be patients treated within a completed Phase 3 clinical trial that has addressed a controversial issue in radiation oncology. We think that samples from the NCIC-CTG MA.20 might be ideal since it would allow us to test whether the population benefiting from nodal irradiation is identified by RSI (36).
In conclusion we propose that RSI is a predictive biomarker of RT therapeutic benefit in breast cancer. This novel biomarker may provide an opportunity to integrate individual tumor biology with clinical decision-making in radiation oncology.
Supplementary Material
Translational Relevance.
Radiation Therapy (RT) is the single most commonly prescribed therapeutic agent in clinical oncology. Currently, clinical-decision making regarding RT is based on clinico-pathological features. Therefore, the development of molecular diagnostics to predict RT therapeutic benefit is of significant clinical importance. In this paper we test a previously developed and validated radiosensitivity molecular signature (RSI) in two independent breast cancer datasets (n=503). We show that in both datasets the signature predicts distant metastasis risk in RT-treated patients but not in patients treated without RT, suggesting that it may serve as a predictive biomarker of RT therapeutic benefit. Including prior data, RSI is validated in 5 independent datasets in a total of 621 patients. An accurate radiosensitivity molecular signature may lead to the refinement of treatment decision algorithms in breast cancer, to novel paradigms to test in signature-directed clinical trials and may open the door to biologically-guided radiation therapy.
Acknowledgments
Funding: National Institutes of Health (R21CA101355/R21CA135620), US Army Medical Research and Materiel Command, National Functional Genomics Center (170220051) and Bankhead-Coley Foundation (09BB-22). Karolinska Institutet/Karolinska University Hospital received unrestricted grants for pharmacogenetic studies from Merck and Bristol-Myers-Squibb. The research group by Jonas Bergh is supported by grants from grants from the Swedish Cancer Society, the Stockholm Cancer Society, the King Gustav V Jubilee Fund, the Swedish Research Council, the Stockholm City Council, Karolinska Institutet and Stockholm County Council Research Strategy Committee, The Swedish Breast Cancer Association (BRO), the Karolinska Institutet Research Funds, and Märit and Hans Rausing’s Initiative against Breast Cancer
Footnotes
Conflict of Interest: JTR and SAE are named as inventors in one awarded and two pending patent applications regarding the technology described. Both are c0-founders and officers of Cvergenx, Inc which holds an exclusive license for the commercialization of the technology
References
- 1.Peters LJ. The ESTRO Regaud lecture Inherent radio sensitivity of tumor and normal tissue cells as a predictor of human tumor response. Radiotherapy and Oncology 1990. 1990 Mar;17(3):177. doi: 10.1016/0167-8140(90)90202-8. [DOI] [PubMed] [Google Scholar]
- 2.Peters LJ, Brock WA, Chapman JD, Wilson G. Predictive assays of tumor radiocurability. Am J Clin Oncol 1988. 1988 Jun;11(3):275–87. doi: 10.1097/00000421-198806000-00005. [DOI] [PubMed] [Google Scholar]
- 3.Perez C. Principles and Management of Radiation Therapy. Philadelphia-NewYork: Lippincott-Raven; 1998. [Google Scholar]
- 4.Ramalingam SS, Owonikoko TK, Khuri FR. Lung cancer: New biological insights and recent therapeutic advances. CA Cancer J Clin. 2011 Mar-Apr;61(2):91–112. doi: 10.3322/caac.20102. [DOI] [PubMed] [Google Scholar]
- 5.van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002 Dec 19;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 6.van ‘t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002 Jan 31;415(6871):530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 7.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004 Dec 30;351(27):2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 8.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008 Apr 1;26(10):1626–34. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 9.Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008 Oct 23;359(17):1757–65. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 10.Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010 Feb;11(2):121–8. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 11.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010 Jun 24;362(25):2380–8. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 12.Shaw AT, Yeap BY, Solomon BJ, Riely GJ, Gainor J, Engelman JA, et al. Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol. 2011 Oct;12(11):1004–12. doi: 10.1016/S1470-2045(11)70232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laitinen S, Martikainen PM, Tolonen T, Isola J, Tammela TL, Visakorpi T. EZH2, Ki-67 and MCM7 are prognostic markers in prostatectomy treated patients. International journal of cancer. 2008 Feb 1;122(3):595–602. doi: 10.1002/ijc.23145. [DOI] [PubMed] [Google Scholar]
- 14.Rubio J, Ramos D, Lopez-Guerrero JA, Iborra I, Collado A, Solsona E, et al. Immunohistochemical expression of Ki-67 antigen, cox-2 and Bax/Bcl-2 in prostate cancer; prognostic value in biopsies and radical prostatectomy specimens. Eur Urol. 2005 Nov;48(5):745–51. doi: 10.1016/j.eururo.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 15.Li R, Heydon K, Hammond ME, Grignon DJ, Roach M, III, Wolkov HB, et al. Ki-67 Staining Index Predicts Distant Metastasis and Survival in Locally Advanced Prostate Cancer Treated With Radiotherapy: An Analysis of Patients in Radiation Therapy Oncology Group Protocol 86–10. Clin Cancer Res 2004. 2004 Jun 15;10(12):4118–24. doi: 10.1158/1078-0432.CCR-1052-03. [DOI] [PubMed] [Google Scholar]
- 16.Pollack A, DeSilvio M, Khor LY, Li R, Al-Saleem TI, Hammond ME, et al. Ki-67 Staining Is a Strong Predictor of Distant Metastasis and Mortality for Men With Prostate Cancer Treated With Radiotherapy Plus Androgen Deprivation: Radiation Therapy Oncology Group Trial 92–02. J Clin Oncol 2004. 2004 Jun 1;22(11):2133–40. doi: 10.1200/JCO.2004.09.150. [DOI] [PubMed] [Google Scholar]
- 17.Eschrich S, Zhang H, Zhao H, Boulware D, Lee JH, Bloom G, et al. Systems biology modeling of the radiation sensitivity network: a biomarker discovery platform. Int J Radiat Oncol Biol Phys. 2009 Oct 1;75(2):497–505. doi: 10.1016/j.ijrobp.2009.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eschrich SA, Pramana J, Zhang H, Zhao H, Boulware D, Lee JH, et al. A gene expression model of intrinsic tumor radiosensitivity: prediction of response and prognosis after chemoradiation. Int J Radiat Oncol Biol Phys. 2009 Oct 1;75(2):489–96. doi: 10.1016/j.ijrobp.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torres-Roca JF, Eschrich S, Zhao H, Bloom G, Sung J, McCarthy S, et al. Prediction of Radiation Sensitivity Using a Gene Expression Classifier. Cancer Res 2005. 2005 Aug 15;65(16):7169–76. doi: 10.1158/0008-5472.CAN-05-0656. [DOI] [PubMed] [Google Scholar]
- 20.Pawitan Y, Bjohle J, Amler L, Borg AL, Egyhazi S, Hall P, et al. Gene expression profiling spares early breast cancer patients from adjuvant therapy: derived and validated in two population-based cohorts. Breast Cancer Res. 2005;7(6):R953–64. doi: 10.1186/bcr1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005 Feb 19–25;365(9460):671–9. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 22.Smid M, Wang Y, Zhang Y, Sieuwerts AM, Yu J, Klijn JG, et al. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 2008 May 1;68(9):3108–14. doi: 10.1158/0008-5472.CAN-07-5644. [DOI] [PubMed] [Google Scholar]
- 23.Wilking N, Lidbrink E, Wiklund T, Erikstein B, Lindman H, Malmstrom P, et al. Long-term follow-up of the SBG 9401 study comparing tailored FEC-based therapy versus marrow-supported high-dose therapy. Ann Oncol. 2007 Apr;18(4):694–700. doi: 10.1093/annonc/mdl488. [DOI] [PubMed] [Google Scholar]
- 24.Yu JX, Sieuwerts AM, Zhang Y, Martens JW, Smid M, Klijn JG, et al. Pathway analysis of gene signatures predicting metastasis of node-negative primary breast cancer. BMC Cancer. 2007;7:182. doi: 10.1186/1471-2407-7-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003 Apr;4(2):249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 26.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003 Feb 15;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003 Jan 22;19(2):185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 28.Dalton WS, Friend SH. Cancer biomarkers--an invitation to the table. Science. 2006 May 26;312(5777):1165–8. doi: 10.1126/science.1125948. [DOI] [PubMed] [Google Scholar]
- 29.Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst. 2009 Nov 4;101(21):1446–52. doi: 10.1093/jnci/djp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark GM. Prognostic factors versus predictive factors: Examples from a clinical trial of erlotinib. Mol Oncol. 2008 Apr;1(4):406–12. doi: 10.1016/j.molonc.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baselga J. Treatment of HER2-overexpressing breast cancer. Ann Oncol. 2010 Oct;21(Suppl 7):vii36–40. doi: 10.1093/annonc/mdq421. [DOI] [PubMed] [Google Scholar]
- 32.Clarke M, Collins R, Darby S, Davies C, Elphinstone P, Evans E, et al. Effects of radiotherapy and of differences in the extent of surgery for early breast cancer on local recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005 Dec 17;366(9503):2087–106. doi: 10.1016/S0140-6736(05)67887-7. [DOI] [PubMed] [Google Scholar]
- 33.Overgaard M, Hansen PS, Overgaard J, Rose C, Andersson M, Bach F, et al. Postoperative radiotherapy in high-risk premenopausal women with breast cancer who receive adjuvant chemotherapy. Danish Breast Cancer Cooperative Group 82b Trial. N Engl J Med. 1997 Oct 2;337(14):949–55. doi: 10.1056/NEJM199710023371401. [DOI] [PubMed] [Google Scholar]
- 34.Overgaard M, Jensen MB, Overgaard J, Hansen PS, Rose C, Andersson M, et al. Postoperative radiotherapy in high-risk postmenopausal breast-cancer patients given adjuvant tamoxifen: Danish Breast Cancer Cooperative Group DBCG 82c randomised trial. Lancet. 1999 May 15;353(9165):1641–8. doi: 10.1016/S0140-6736(98)09201-0. [DOI] [PubMed] [Google Scholar]
- 35.Ragaz J, Olivotto IA, Spinelli JJ, Phillips N, Jackson SM, Wilson KS, et al. Locoregional radiation therapy in patients with high-risk breast cancer receiving adjuvant chemotherapy: 20-year results of the British Columbia randomized trial. J Natl Cancer Inst. 2005 Jan 19;97(2):116–26. doi: 10.1093/jnci/djh297. [DOI] [PubMed] [Google Scholar]
- 36.Whelan TJ, Olivotto I, Ackerman I, Chapman JW, Chua B, Nabid A, et al., editors. J Clin Oncol. ASCO; 2011. NCIC-CTG MA.20: An intergroup trial of regional nodal irradiation in early breast cancer. [Google Scholar]
- 37.Kreike B, Halfwerk H, Kristel P, Glas A, Peterse H, Bartelink H, et al. Gene expression profiles of primary breast carcinomas from patients at high risk for local recurrence after breast-conserving therapy. Clin Cancer Res. 2006 Oct 1;12(19):5705–12. doi: 10.1158/1078-0432.CCR-06-0805. [DOI] [PubMed] [Google Scholar]
- 38.Kreike B, Halfwerk H, Armstrong N, Bult P, Foekens JA, Veltkamp SC, et al. Local recurrence after breast-conserving therapy in relation to gene expression patterns in a large series of patients. Clin Cancer Res. 2009 Jun 15;15(12):4181–90. doi: 10.1158/1078-0432.CCR-08-2644. [DOI] [PubMed] [Google Scholar]
- 39.Servant N, Bollet MA, Halfwerk H, Bleakley K, Kreike B, Jacob L, et al. Search for a gene expression signature of breast cancer local recurrence in young women. Clin Cancer Res. 2012 Mar 15;18(6):1704–15. doi: 10.1158/1078-0432.CCR-11-1954. [DOI] [PubMed] [Google Scholar]
- 40.Mamounas EP, Tang G, Fisher B, Paik S, Shak S, Costantino JP, et al. Association between the 21-gene recurrence score assay and risk of locoregional recurrence in node-negative, estrogen receptor-positive breast cancer: results from NSABP B-14 and NSABP B-20. J Clin Oncol. 2010 Apr 1;28(10):1677–83. doi: 10.1200/JCO.2009.23.7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voduc KD, Cheang MC, Tyldesley S, Gelmon K, Nielsen TO, Kennecke H. Breast cancer subtypes and the risk of local and regional relapse. J Clin Oncol. 2010 Apr 1;28(10):1684–91. doi: 10.1200/JCO.2009.24.9284. [DOI] [PubMed] [Google Scholar]
- 42.Abdulkarim BS, Cuartero J, Hanson J, Deschenes J, Lesniak D, Sabri S. Increased risk of locoregional recurrence for women with T1-2N0 triple-negative breast cancer treated with modified radical mastectomy without adjuvant radiation therapy compared with breast-conserving therapy. J Clin Oncol. 2011 Jul 20;29(21):2852–8. doi: 10.1200/JCO.2010.33.4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


