Abstract
Fifty-two candidate DNA aptamer sequences were selected for binding to the cardiovascular biomarker B-type or brain natriuretic peptide (BNP). Candidate aptamers were screened to rank their relative affinities against BNP by an aptamer-based ELISA-like aptamer microplate assay (ELASA). The highest affinity aptamers from ELASA screening were also paired in all possible combinations and screened for electrochemiluminescence (ECL) assay potential in capture aptamer-magnetic bead and ruthenium trisbipyridine (Ru(bpy)32+)-reporter aptamer sandwich formats. The top ECL sandwich combinations utilized the same aptamer pair in either capture or reporting roles with nanogram to low picogram per mL levels of detection even in 50% human serum. ECL assay sensitivity and linearity even in 50% human serum suggest that the aptamer-based assay is at least comparable to other reported immunoassays for BNP.
Keywords: aptamer, BNP, cardiovascular, electrochemiluminescence, SELEX
Introduction
While numerous high quality immunoassays exist for many of the biomarkers associated with cardiovascular disease, these assays are dependent on animal-derived mono- or polyclonal antibodies. As such, they are subject to possible issues associated with antibodies such as lot-to-lot reproducibility, slower production and the cost of maintaining host animals or hybridomas and purification costs. Aptamers obviate problems and costs associated with antibody development and production by in vitro selection and PCR amplification of binding agents [1–5,15]. In the present work, we begin to explore the use of aptamers for potentially ultrasensitive and specific cardiovascular biomarker assays. In particular, we have developed DNA aptamers against B-type or brain natriuretic peptide (BNP) as an initial target. Despite some controversy over normal cut off and prognostic values of BNP and its related peptide assays, BNP is known to require relatively high sensitivity in the range of 25–100 pg/mL [6–8,10–16]. BNP is a cardiac biomarker of heart failure that is secreted into serum at minute levels when atrial and ventricular myocytes are stretched by hypertrophy or increased blood pressure from a variety of cardiac ailments [6,7,10].
The need for extreme sensitivity in BNP assays led Jang et al. [16] to develop a hybrid BNP aptamer plus antibody sandwich assay based on nanoparticle-enhanced surface plasmon resonance (SPR) with a BNP aptamer developed by Lin et al. [15]. The need for BNP detection sensitivity similarly led us to explore the possibility of maximizing sensitivity by coupling high affinity DNA aptamers to highly sensitive electrochemiluminescence (ECL) detection [4,5]. DNA aptamers have already been successfully used in sensitive ECL assay formats [17], especially in conjunction with magnetic microbead capture and analyte concentration [4,5,18].
Materials and Methods
Materials
Bovine serum albumin (BSA), human C-reactive protein (Cat. No. C4063), biologically active C-terminal BNP (32-amino acids, Cat. No. B5900), human interleukin-6 (IL-6; Cat. No. I3268-10KU), and bis(2,2′-bipyridine)-4′-methyl-4-carboxybipyridine-ruthenium N-succinimidyl ester-bis(hexafluorophosphate), hereby referred to as Ru(bpy)32+-succinimide, tripropylamine (TPA) and miscellaneous chemicals were purchased from Sigma-Aldrich Corp. (St. Louis, MO). An eight amino acid version of human C-telopeptide (CTx; EKAHDGGR) from human bone collagen I as well as natural pituitary human growth hormone (hGH) were obtained from GenWay Biotech, Inc. (San Diego, CA). Helical peptide (HP) of human α1(I) bone collagen from amino acids 620–633: GPPGPAGPAGERGE was purchased from Quidel Corp. (San Diego, CA.). N-telopeptide of human bone collagen I (NTx; DEKSTGG) was synthesized by GenScript, Inc. (Piscataway, NJ). Human D-dimer (DD; Cat. No. MBS173072), cardiac Troponin-T (Tpn-T; Cat. No. MBS142897), and human interleukin 18 (IL-18; Cat. No. MBS203090) were obtained from MyBioSource, LLC (San Diego, CA). Human serum was obtained from Lonza, Inc. (BioWhittaker brand; Walkersville, MD) and stored frozen at −20°C.
DNA aptamer development, cloning and sequencing
BNP was immobilizing on 2.8 micron Dynal (M280) tosyl coated-magnetic beads (MBs, Invitrogen Corp., Carlsbad, CA) for 2 hrs at 37°C. BNP-conjugated MBs were then collected using a Dynal MPC-S magnetic rack and washed three times in 1 mL of 1X binding buffer (1XBB; 0.5 M NaCl, 10 mM Tris-HCl, and 1 mM MgCl2, pH 7.5–7.6). BNP-MBs were next blocked for 2 hr at 37°C in 1XBB plus 2% ethanolamine and washed three times as before in 1XBB. DNA aptamers were developed against BNP-conjugated MBs through five rounds of selection and PCR amplification as previously described in the literature [1–3]. The presence of 72 bp aptamer PCR products were verified after each round of selection by ethidium bromide-stained 2% agarose gel electrophoresis against standard DNA ladders. BNP aptamers from the final round of selection and amplification were cloned into chemically competent E. coli using a Lucigen GC kit (Middleton, WI). All aptamers were sequenced by rolling circle amplification dideoxynucleotide methodology with proprietary treatment for high GC content DNA sequencing at Sequetech Corp. (Mountain View, CA).
ELASA screening and cross-reactivity assessments
One hundred ng of BNP or other targets for cross-reactivity studies were immobilized in flat-bottomed 96-well polystyrene plates in 100 μl of 0.1M sodium bicarbonate buffer (pH 8.5) overnight at 4°C. Plates were decanted and washed 3 times in 200 μl of Dulbecco’s phosphate buffered saline (PBS without calcium; pH 7.2). Plates were blocked with 150 μl of 2% ethanolamine in 0.1M sodium bicarbonate buffer for 1 hr at 37°C. The 52 unique 5′-biotinylated aptamer DNA sequences were purchased at 4.5 nanomoles each in separate wells of a 96-well microtiter plate from Integrated DNA Technologies (IDT; Coralville, IA). The biotin-DNA contents of each well were dissolved in 100 μl of PBS. All 100 μl of the biotinylated aptamers from each well were added to the corresponding wells of the BNP-coated polystyrene microtiter plate and gently mixed for 30 min at room temperature (RT). Wells were washed 3 times in 200 μl of PBS for 5 min and decanted. Each well then received 100 μl of 1:2,000 streptavidin-peroxidase (5 mg/mL stock from Southern Biotech, Birmingham, AL) and plates were mixed gently at RT for 30 mins. Plates were decanted and washed 3 times in 200 μL of PBS at 5 min per wash. Finally, 100 μl of onestep ABTS (Kirkegaard Perry Labs, KPL, pre-warmed to RT in the dark) were added per well and absorbance at 405 nm was determined at 5 min intervals over the next 15–20 minutes or until absorbance in the range of 1.5 to 2.0 was reached in some of the wells by use of a microplate reader.
Aptamer-magnetic bead ECL sandwich assays
ECL assay buffer consisting of 0.2M tripropylamine (TPA) in PBS plus 0.5% Triton X-100 was prepared in 18 MΩ deionized water. Cell cleaner buffer consisting of 0.71M KOH and 0.5% Triton X-100 in 18 MΩ deionized water (pH 13.85) was used to clean the ECL flow cell between readings. DNA aptamer-coated MBs were made by adding 100 μl of 1 to 1.5 mg/mL 5′-biotinylated aptamers in PBS (pH 7.2) to 100 μl of Dynal M280 streptavidin-MBs (~ 2 × 109 MBs/mL) and mixing with 1 mL of PBS for 1 hr at RT. Aptamer-biotin-streptavidin-MBs (or simply capture aptamer-MBs) were washed three times in 1 mL of filter-sterilized 1XBB per wash using a Dynal MPC-S magnetic rack. Capture aptamer-MBs were resuspended in 1 mL of 1XBB and stored at 4°C until needed for assays. Streptavidin-Ru(bpy)32+ was prepared by mixing 1 mg of streptavidin (Sigma-Aldrich Cat. No. S0677) with 1 mg of Ru(bpy)32+-succinimide (Sigma-Aldrich Cat. No. 96631) dissolved in 200 μl of methanol in 1 mL of PBS for 2 hr at 37°C. The streptavidin-Ru(bpy)32+ conjugate was purified through a PBS-equilibrated Sephadex G25 (PD-10, GE Healthcare) column. Fractions 4 and 5 were pooled (2 mL total volume), making the final concentration of the streptavidin component ~ 0.5 mg/mL. Since streptavidin is known to have four biotin-binding sites per molecule and weighs 52.8 kD, 100 μl of streptavidin-Ru(bpy)32+ was mixed with 50 μl of 5′-biotinylated aptamer to bind all of the available aptamer (i.e., both reagent concentrations were ~ 3.5 nanomoles). The streptavidin-Ru(bpy)32+ was then diluted to 1 mL with PBS to serve as the stock reporter reagent.
Serial two-fold dilutions of BNP were prepared in 1 mL of PBS in borosilicate glass 12 × 75 mm tubes. The final tube in each experiment received no BNP or other target and served as the background control blank. Forty μl of capture aptamer-MBs plus 40 μl of reporter aptamer stock reagent were added per tube with or without BNP or other target materials and tubes were vortexed on an Origen® ECL analyzer (previously marketed by the defunct Igen International or BioVeris corporations [4,5]) at 80 rpm for 30 minutes at RT. ECL values were obtained with an “assay gain” (PMT setting) of 900V using the “bead capture” mode with a ramp waveform.
Results
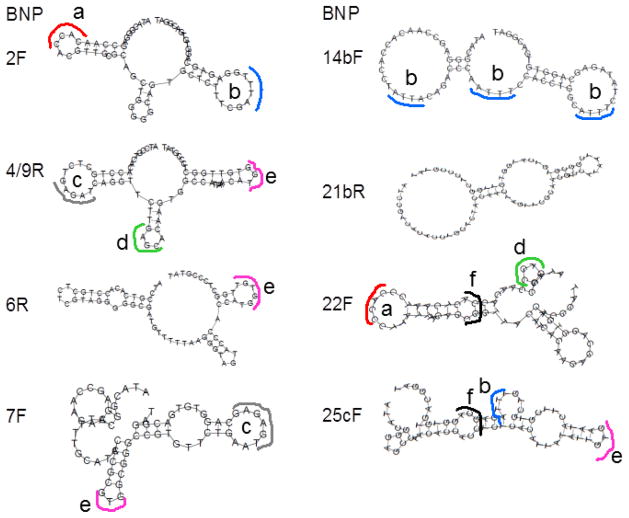
After sequencing of the cloned aptamers, 52 unique sequences emerged as shown in Table 1. Of these sequences, five forward and reverse sequences were identical across the full 72 base length of the aptamers. These five separately cloned sequences were then designated as one clone with each component clone number separated by slash marks (i.e., BNP - 5/11/15b/19/25bF or R). Numerous other shorter sequence segments which were identical or highly similar emerged from study of the highest affinity aptamer DNA sequences as determined by ELASA analysis (Table 2). Many of these shorter homologous sequence segments are bolded and italicized in Table 1. After aptamer secondary structures were obtained by use of Vienna RNA software (9) using DNA parameters and a temperature of 25°C, many of these common bolded and italicized sequence segments emerged in loop structures as shown by the highlighted and outlined loop regions identified by lower case letters (a–f) in Fig. 1. The discovery of common short sequence segments in naturally occurring loop structures suggests that these segments could be part of binding pockets for BNP epitopes.
Table 1.
Candidate DNA Aptamer Sequences Developed Against BNP
| BNP - 1F |
| ATACGGGAGCCAACACCATCACACACAATCCGTTTCTACGAAGGCATCCTGCATAGAGCAGGTGTGACGGAT |
| BNP - 1R |
| ATCCGTCACACCTGCTCTATGCAGGATGCCTTCGTAGAAACGGATTGTGTGTGATGGTGTTGGCTCCCGTAT |
| BNP - 2F |
| ATACGGGAGCCAACACCACGTTGCGCAGCTGGGGGCAGTGCTCTTTCGATTTGGAGAGCAGGTGTGACGGAT |
| BNP - 2R |
| ATCCGTCACACCTGCTCTCCAAATCGAAAGAGCACTGCCCCCAGCTGCGCAACGTGGTGTTGGCTCCCGTAT |
| BNP - 3F |
| ATACGGGAGCCAACACCATACTACCTGCTGCATTACTAAAGTAAGAGCGTATCTAGAGCAGGTGTGACGGAT |
| BNP - 3R |
| ATCCGTCACACCTGCTCTAGATACGCTCTTACTTTAGTAATGCAGCAGGTAGTATGGTGTTGGCTCCCGTAT |
| BNP - 4/9F |
| ATACGGGAGCCAACACCATGTTTAATGGCCACTTGTGCTCAAGAACCTGATCTCAGAGCAGGTGTGACGGAT |
| BNP - 4/9R |
| ATCCGTCACACCTGCTCTGAGATCAGGTTCTTGAGCACAAGTGGCCATTAAACATGGTGTTGGCTCCCGTAT |
| BNP - 5/11/15b/19/25bF |
| ATACGGGAGCCAACACCACCTTTTAAAACGCTAGCTAGCTTAGTCCATTCCACCAGAGCAGGTGTGACGGAT |
| BNP–5/11/15b/19/25bR |
| ATCCGTCACACCTGCTCTGGTGGAATGGACTAAGCTAGCTAGCGTTTTAAAAGGTGGTGTTGGCTCCCGTAT |
| BNP - 6F |
| ATACGGGAGCCAACACCATGGTGGGTACTACCCTTAAAAACATCGCCCCCTACGAGAGCAGGTGTGACGGAT |
| BNP - 6R |
| ATCCGTCACACCTGCTCTCGTAGGGGGCGATGTTTTTAAGGGTAGTACCCACCATGGTGTTGGCTCCCGTAT |
| BNP - 7F |
| ATACGGGAGCCAACACCATTTTGCATACCCGCGTGGCGGGCCGTGTTCTGAATGAGAGCAGGTGTGACGGAT |
| BNP - 7R |
| ATCCGTCACACCTGCTCTCATTCAGAACACGGCCCGCCACGCGGGTATGCAAAATGGTGTTGGCTCCCGTAT |
| BNP - 8F |
| ATACGGGAGCCAACACCAGTTAAGCACCCTCGTATCCCGCTATACTATGGAGTCAGAGCAGGTGTGACGGAT |
| BNP - 8R |
| ATCCGTCACACCTGCTCTGACTCCATAGTATAGCGGGATACGAGGGTGCTTAACTGGTGTTGGCTCCCGTAT |
| BNP - 10F |
| ATACGGGAGCCAACACCATTCCCGCATCGCGCGTTTTCAGCCTTTGACCGTTAGAGCAGGTGTGACGGAT |
| BNP - 10R |
| ATCCGTCACACCTGCTCTAACGGTCAAAGGCTGAAAACGCGCGATGCGGGAATGGTGTTGGCTCCCGTAT |
| BNP - 12F |
| ATACGGGAGCCAACACCAACGTGTGCTGTGTTACTGCCCTTCTCTGTAGCCGTGAGAGCAGGTGTGACGGAT |
| BNP - 12R |
| ATCCGTCACACCTGCTCTCACGGCTACAGAGAAGGGCAGTAACACAGCACACGTTGGTGTTGGCTCCCGTAT |
| BNP - 13F |
| ATACGGGAGCCAACACCACCTTTTAAAACGCTAGCCAGCTTAGTCCATTCCACCAGAGCAGGTGTGACGGAT |
| BNP - 13R |
| ATCCGTCACACCTGCTCTGGTGGAATGGACTAAGCTGGCTAGCGTTTTAAAAGGTGGTGTTGGCTCCCGTAT |
| BNP - 14aF |
| ATACGGGAGCCAACACCAATCTAACAGATTGCAGCTCGCCTGTCCCGGCGTACTAGAGCAGGTGTGACGGAT |
| BNP - 14aR |
| ATCCGTCACACCTGCTCTAGTACGCCGGGACAGGCGAGCTGCAATCTGTTAGATTGGTGTTGGCTCCCGTAT |
| BNP - 14bF |
| ATACGGGAGCCAACACCACCTATTACAGACCCAATTTCCACCTGGCATTTCTATAGAGCAGGTGTGACGGAT |
| BNP - 14bR |
| ATCCGTCACACCTGCTCTATAGAAATGCCAGGTGGAAATTGGGTCTGTAATAGGTGGTGTTGGCTCCCGTAT |
| BNP - 15aF |
| ATACGGGAGCCAACACCACATATCCTACACTCCCATACCCCACTGTAGACACGCAGAGCAGGTGTGACGGAT |
| BNP - 15aR |
| ATCCGTCACACCTGCTCTGCGTGTCTACAGTGGGGTATGGGAGTGTAGGATATGTGGTGTTGGCTCCCGTAT |
| BNP - 16F |
| ATACGGGAGCCAACACCAAACCGAGTGCTGGTGGCCCTCTCTGCCATATAAGTGAGAGCAGGTGTGACGGAT |
| BNP - 16R |
| ATCCGTCACACCTGCTCTCACTTATATGGCAGAGAGGGCCACCAGCACTCGGTTTGGTGTTGGCTCCCGTAT |
| BNP - 17F |
| ATACGGGAGCCAACACCACCTTTTAAAACGCTAGCTAGCTTAGTCCAATTCCACCAGAGCAGGTGTGACGGAT |
| BNP - 17R |
| ATCCGTCACACCTGCTCTGGTGGAATTGGACTAAGCTAGCTAGCGTTTTAAAAGGTGGTGTTGGCTCCCGTAT |
| BNP - 18F |
| ATACGGGAGCCAACACCATGACATTGCAACTATACGCTTACCCACGTCAGCTCCAGAGCAGGTGTGACGGAT |
| BNP - 18R |
| ATCCGTCACACCTGCTCTGGAGCTGACGTGGGTAAGCGTATAGTTGCAATGTCATGGTGTTGGCTCCCGTAT |
| BNP - 20F |
| ATACGGGAGCCAACACCATTAACCTGAAAGTACCAGTGTCAGTTTACCCTACCTAGAGCAGGTGTGACGGAT |
| BNP - 20R |
| ATCCGTCACACCTGCTCTAGGTAGGGTAAACTGACACTGGTACTTTCAGGTTAATGGTGTTGGCTCCCGTAT |
| BNP - 21aF |
| ATACGGGAGCCAACACCAGGCATTAGTGTAAAGCACTAAGAGTCAGGCTGTAGCAGAGCAGGTGTGACGGAT |
| BNP - 21aR |
| ATCCGTCACACCTGCTCTGCTACAGCCTGACTCTTAGTGCTTTACACTAATGCCTGGTGTTGGCTCCCGTAT |
| BNP - 21bF |
| ATACGGGAGCCAACACCATGACACGCCGATTATGGACGTTGCGAACTAGTTGGTAGAGCAGGTGTGACGGAT |
| BNP - 21bR |
| ATCCGTCACACCTGCTCTACCAACTAGTTCGCAACGTCCATAATCGGCGTGTCATGGTGTTGGCTCCCGTAT |
| BNP - 22F |
| ATACGGGAGCCAACACCACTCAATCCCACCCTTATTTAGAGCGGTTACATCACAAGAGCAGGTGTGACGGAT |
| BNP - 22R |
| ATCCGTCACACCTGCTCTTGTGATGTAACCGCTCTAAATAAGGGTGGGATTGAGTGGTGTTGGCTCCCGTAT |
| BNP - 23aF |
| ATACGGGAGCCAACACCAATTTGTGAAAATATTCCCGTGTTTTCCTTGAGCAGCAGAGCAGGTGTGACGGAT |
| BNP - 23aR |
| ATCCGTCACACCTGCTCTGCTGCTCAAGGAAAACACGGGAATATTTTCACAAATTGGTGTTGGCTCCCGTAT |
| BNP - 23bF |
| ATACGGGAGCCAACACCAGCGTAAAACCTTGTACCAATTGATGACACTAGCGGTAGAGCAGGTGTGACGGAT |
| BNP - 23bR |
| ATCCGTCACACCTGCTCTACCGCTAGTGTCATCAATTGGTACAAGGTTTTACGCTGGTGTTGGCTCCCGTAT |
| BNP - 24F |
| ATACGGGAGCCAACACCACAGCTCACCGCGCTTGCCGTGCCTTACGTCTGTCCAAGAGCAGGTGTGACGGAT |
| BNP - 24R |
| ATCCGTCACACCTGCTCTTGGACAGACGTAAGGCACGGCAAGCGCGGTGAGCTGTGGTGTTGGCTCCCGTAT |
| BNP-25aF |
| ATACGGGAGCCAACACCATCACCGTACTGGAGCCATCGTTCATCCAGCAATCTAGAGCAGGTGTGACGGAT |
| BNP - 25aR |
| ATCCGTCACACCTGCTCTAGATTGCTGGATGAACGATGGCTCCAGTACGGTGATGGTGTTGGCTCCCGTAT |
| BNP - 25cF |
| ATACGGGAGCCAACACCACCTCTCACATTATATTGTGAATACTTCGTGCTGTTTAGAGCAGGTGTGACGGAT |
| BNP - 25cR |
| ATCCGTCACACCTGCTCTAAACAGCACGAAGTATTCACAATATAATGTGAGAGGTGGTGTTGGCTCCCGTAT |
Notes: Identical sequence segments (forward or backward) are bolded and italicized. Many of these bolded an italicized sequences also appear in the loop structures shown in Fig. 1. All sequences are written 5′ → 3′ from left to right.
Table 2.
ELASA Rankings for BNP Aptamers
| Trial 1 | Trial 2 | Trial 3 | ||||
|---|---|---|---|---|---|---|
| Rank | Aptamer | A405nm | Aptamer | A405nm | Aptamer | A405nm |
| 1 | 6R | 2.766 | 6R | 2.171 | 6R | 2.285 |
| 2 | 14bF | 2.283 | 2F | 2.110 | 22F | 2.272 |
| 3 | 2F | 2.276 | 14bF | 2.089 | 25cF | 2.240 |
| 4 | 7F | 2.227 | 4/9R | 1.989 | 21bR | 2.212 |
| 5 | 25cF | 2.215 | 1F | 1.987 | 14bF | 2.206 |
| 6 | 1F | 2.184 | 13F | 1.986 | 8R | 2.197 |
| 7 | 8R | 2.176 | 5/11/15b/19/15bF | 1.977 | 5/11/15b/19/15bF | 2.196 |
| 8 | 4/9R | 2.163 | 14bR | 1.971 | 2F | 2.176 |
| 9 | 15aF | 2.162 | 25cF | 1.961 | 15aF | 2.165 |
| 10 | 21bR | 2.149 | 8R | 1.953 | 10F | 2.157 |
Notes: The consistent top four ranked and selected sequence designations are bolded.
Figure 1.
Secondary stem-loop structures of the top eight aptamers from ELASA rankings (Table 2) which also showed some commonality in the short sequences found in many of their loop regions suggesting common binding pockets. Similar loop sequence segments are similarly highlighted and coded by lower cases letters in the figure.
When all of the aptamer candidates were screened for affinity by several trials of ELASA microplate assay, several sequences consistently emerged among the top ten candidates as shown in Table 2. While the common BNP - 5/11/15b/19/15bF sequence emerged five times in the cloned and sequenced population, it only ranked seventh in two of the three trials and was not in the top ten for the other ELASA trial shown in Table 2. Therefore, BNP - 5/11/15b/19/15bF was not among the highest four affinities and ironically did not progress into actual aptamer-MB ECL sandwich assay development. When examined over three trials, the top four candidates from the ELASA screening were BNP – 2F, 6R, 14bF, and 25cR (bolded in Table 2). These four aptamer sequences proceeded into further sandwich ECL assay development. Table 3 summarizes the 4 × 4 matrix of 16 possible capture aptamer-MB paired with Ru(bpy)32+-reporter aptamer combinations. When all of these combinations were screened by ECL analysis three times against 1 ng/mL of BNP, the data in Fig. 2 resulted. It is of interest to note that combinations 4, 8, 13, and 15 gave the strongest ECL signals (indicated by asterisks in Fig. 2) and these combinations all shared the BNP 25cR aptamer either in a capture or reporter role (see Table 3).
Table 3.
BNP Sandwich Aptamer Combination Matrix
| Reporter Aptamer- Ru(bpy)32+ | Capture Aptamer-MBs | |||
|---|---|---|---|---|
| 2F | 6R | 14bF | 25cF | |
| 2F | Combination 1 | 2 | 3 | 4 |
| 6R | 5 | 6 | 7 | 8 |
| 14bF | 9 | 10 | 11 | 12 |
| 25cF | 13 | 14 | 15 | 16 |
Notes: The resulting top sandwich assay combinations are shaded.
Figure 2.
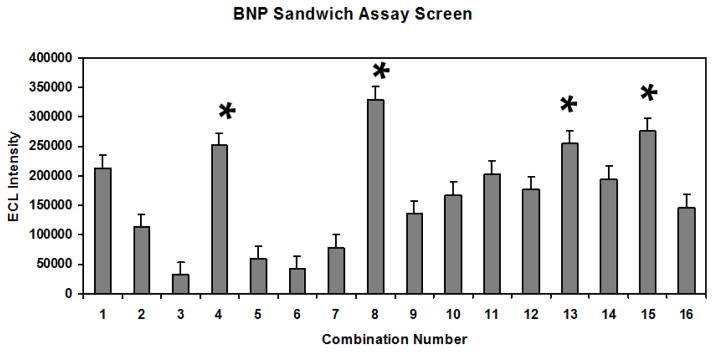
Results of BNP sandwich assay matrix screening in which capture aptamer-MBs were paired with different Ru(bpy)32+-reporter aptamer sequences as coded by combination number given in Table 3. Bar heights represent the mean ECL values of three independent measurements (N = 3) of 1 ng/mL BNP in 1XBB with standard deviation error bars.
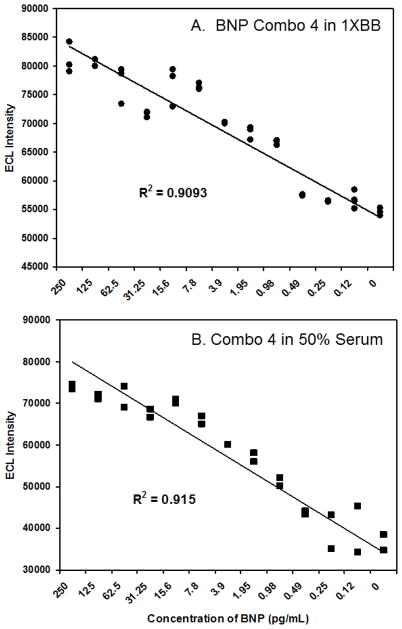
All four of the most intense ECL aptamer combinations from Fig. 2 were subjected to titration analyses with serial two-fold dilutions of BNP in 1XBB beginning with 250 pg/mL of BNP and ending with 120 fg/mL as well as a blank control with no BNP added. The initial titration results for the top four aptamer sandwich combinations are not shown for brevity, but combination 4 appeared marginally superior to combinations 8, 13 and 15 over this range in terms of sensitivity and linearity. Hence, combination 4 was pursued further with repeated titrations in 1XBB and 50% human serum (thawed commercial serum diluted 1:1 with 1XBB).
Fig. 3 demonstrates the relative linearity of the combination 4 (25cF capture aptamer paired with the 2F reporter aptamer) in 1XBB as well as 50% human serum. Dot plots of 3 trials in 1XBB are shown in Fig. 3A and two trials in 50% serum in Fig. 3B. The R2 correlation coefficients were greater than 0.9 in both 1XBB and 50% serum and both of these preliminary assays appear to exhibit sub-pg/mL sensitivity even in 50% human serum. Assays performed in 100% serum exhibited a tendency to clog or coat the ECL flow cell of the Origen® analyzer, leading to greater ECL variability and leading to extensive cleaning cycles and reconditioning of the flow cell between runs. Therefore, data presentation is limited to the 1XBB and 50% serum milieus.
Figure 3.
Combined dot plots of (A) three combination 4 BNP ECL assay titration trials in 1XBB and (B) two combination 4 assay trials in 50% human serum and 1XBB over the BNP concentration range shown in pg/mL.
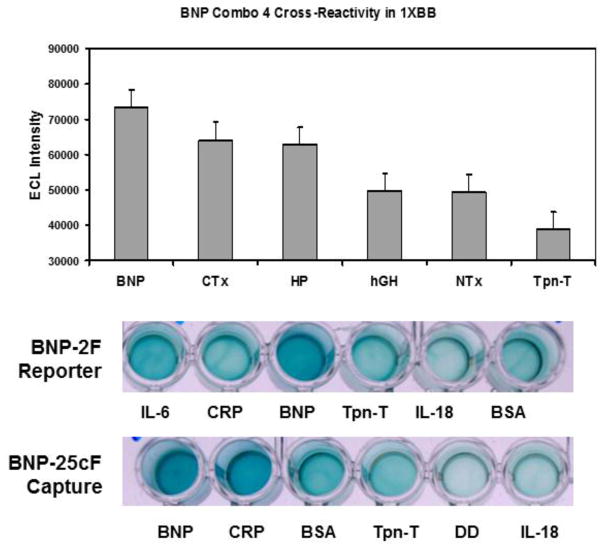
In an effort to evaluate specificity of the combination 4 BNP prototype ECL assay, we conducted a cross-reactivity study in 1XBB using 250 pg/mL of several peptides that might be found in human serum including several markers of osteoporosis (CTx, HP and NTx) and the cardiac infarction biomarker Tpn-T. While human growth hormone (hGH) is not expected to exist at high levels in middle-aged or elderly individuals expressing high levels of BNP, it was also added as a target analyte. ECL data presented in Fig. 4 (top) suggest significant cross-reactivity of the combination 4 BNP assay with CTx and HP collagen bone loss markers, although BNP still gave the strongest overall mean ECL response. Although CTx, HP, and NTx are not expected at high levels in serum unless a patient is suffering from osteoporosis [2], these levels of cross-reactivity by ECL assessment were not desirable. We therefore, decided to also evaluate the individual reporter and capture aptamers by ELASA (ELISA-like) plate assay. The results of the ELASA assessment are given in Fig. 4 (bottom) and again illustrate in general the strongest responses from aptamer interactions with BNP, but notable responses with BSA, CRP, and IL-6 in some cases.
Figure 4.
Top - Summary of BNP combination 4 assay cross-reactivity in 1XBB. Bar heights represent the mean ECL of five independent measurements (N = 5) with standard deviation error bars. Each analyte was measured at 250 pg/mL. Bottom – ELASA cross-reactivity data for each of the reporter (BNP 2F) and capture (BNP 25cF) aptamers used in the selected sandwich assay. Abbreviations: BSA; Bovine Serum Albumin, Brain Natriuretic Peptide; BNP, CRP; C-Reactive Protein, CTx; C-Telopeptide of Human Bone Collagen, DD; D-Dimer, HP; Helical Peptide of Human Bone Collagen, hGH; Human Growth Hormone, IL; Interleukin, NTx; N-Telopeptide of Human Bone Collagen, Tpn-T; Troponin-T.
Discussion
The present report summarizes an initial proof-of-concept study aimed at developing DNA aptamers and an ECL-based aptamer-magnetic bead sandwich assay for BNP in buffer and diluted serum. While preliminary, the work demonstrates the potential of aptamers to be employed in an ultrasensitive (low pg/mL detection) and linear assay of some potential clinical importance that could possibly be used in serum samples. The most promising capture and reporter aptamer combination (no. 4) demonstrated good linearity over a range useful for BNP assessment [10,12–16].
The reader is reminded that the ECL data were obtained with a relatively old ECL reader (Origen analyzer) which despite being maintained in good condition since the late 1990’s when its manufacturer (Igen International, Inc.) went out of business, may be exhibiting somewhat diminished performance at this point. In essence, this means that sensitivity could potentially be even better, if experiments are conducted with a more modern ECL instrument. Unfortunately, no newer ECL instruments were available for these studies.
While specificity of the chosen sandwich assay combination of aptamers was not ideal, some level of specificity was obtained. Future development in this area should focus on longer “multivalent” aptamers with multiple binding sites to improve specificity as other investigators have begun reporting [19,20]. Longer multivalent aptamers better emulate the six hypervariable region binding sites between heavy and light chains of antibody complementarity determining regions (CDRs), thereby leading to better selectivity [20]. In the past, we have been limited to 72 base aptamers because of the low yields of oligonucleotide synthesis beyond 72 bases, but recent advances in chemical oligonucleotide synthesis are enabling aptamers up to 200 bases which should significantly improve specificity for peptide or protein target detection [21].
Highlights.
52 DNA aptamers were developed and sequenced against BNP with some notable similarities.
Aptamers were attached to magnetic beads and paired in sandwich assay formats for electrochemiluminescence (ECL) assay potential assessment.
The optimal aptamer pairing led to low pg/mL detection of BNP even in serum with linearity across a physiologically useful range for detection of heart failure.
The assay maintained sensitivity even in 50% human serum.
Some cross-reactivity with osteoporosis biomarkers and other proteins was noted, but this may be overcome in the future by means of lengthier 200 base aptamers.
Acknowledgments
Work was funded by an SBIR Contract from the National Heart, Lung, and Blood Institute (NHLBI, No. HHSN268201000028C).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bruno JG, Carrillo MP, Phillips T, Edge A. Discrimination of recombinant from natural human growth hormone using DNA aptamers. J Biomolec Techn. 2011;22:27–36. [PMC free article] [PubMed] [Google Scholar]
- 2.Bruno JG, Carrillo MP, Phillips T, Hanson D, Bohmann JA. DNA aptamer beacon assay for C-telopeptide and handheld fluorometer to monitor bone resorption. J Fluoresc. 2011;21:2021–2033. doi: 10.1007/s10895-011-0903-6. [DOI] [PubMed] [Google Scholar]
- 3.Bruno JG, Carrillo MP, Richarte AM, Phillips T, Andrews C, Lee JS. Development, screening, and analysis of a small DNA aptamer library potentially useful for diagnosis and passive immunity of arboviruses. BMC Research Notes. 2012;5:633. doi: 10.1186/1756-0500-5-633. http://dx.doi.org/10.1186/1756-0500-5-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruno JG, Kiel JL. In vitro selection of DNA aptamers to anthrax spores with electrochemiluminescence detection. Biosens Bioelectron. 1999;14:457–464. doi: 10.1016/s0956-5663(99)00028-7. [DOI] [PubMed] [Google Scholar]
- 5.Bruno JG, Kiel JL. Use of magnetic beads in selection and detection of biotoxin aptamers by ECL and enzymatic methods. BioTechniques. 2002;32:178–183. doi: 10.2144/02321dd04. [DOI] [PubMed] [Google Scholar]
- 6.Casserly B, Klinger JR. Brain natriuretic peptide in pulmonary arterial hypertension: biomarker and potential therapeutic agent. Drug Design Develop Ther. 2009;3:269–287. doi: 10.2147/dddt.s4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dernellis J, Panaretou M. Assessment of cardiac risk before non-cardiac surgery: brain natriuretic peptide in 1590 patients. Heart. 2006;92:1645–1650. doi: 10.1136/hrt.2005.085530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Serio F, Ruggieri V, Varraso L, De Sario R, Mastrorilli A, Pansini N. Analytical evaluation of the Dade Behring Dimension RxL automated N-Terminal proBNP (NT-proBNP) method and comparison with the Roche Elecsys 2010. Clin Chem Lab Med. 2005;43:1263–1273. doi: 10.1515/CCLM.2005.217. [DOI] [PubMed] [Google Scholar]
- 9.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krishnaswami A. The role of B-type and other natriuretic peptides in health and disease. Permanante J. 2008;12:32–43. doi: 10.7812/tpp/08-019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noveanu M, Breidthardt T, Potocki M, Reichlin T, Twerenbold R, Uthoff H, Socrates T, Arenja N, Reiter M, Meissner J, Heinisch C, Stalder S, Mueller C. Direct comparison of serial B-type natriuretic peptide and NT-proBNP levels for prediction of short- and long-term outcome in acute decompensated heart failure. Crit Care. 2011;15:R1–R15. doi: 10.1186/cc9398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prontera C, Emdin M, Zucchelli GC, Ripoli A, Passino C, Clerico A. Analytical performance and diagnostic accuracy of a fully-automated electrochemiluminescent assay for the N-terminal fragment of the pro-peptide of brain natriuretic peptide in patients with cardiomyopathy: comparison with immunoradiometric assay methods for brain natriuretic peptide and atrial natriuretic peptide. Clin Chem Lab Med. 2004;42:37–44. doi: 10.1515/CCLM.2004.008. [DOI] [PubMed] [Google Scholar]
- 13.Rawlins ML, Owen WE, Roberst WL. Performance characteristics of four automated natriuretic peptide assays. Am J Clin Pathol. 2005;123:439–445. doi: 10.1309/PDJ2-RMM8-0FVR-DH7W. [DOI] [PubMed] [Google Scholar]
- 14.Sokoll LJ, Baum H, Collinson PO, Gurr E, Haass M, Luthe H, Morton JJ, Nowatzke W, Zingler C. Multicenter analytical performance evaluation of the Elecsys proBNP assay. Clin Chem Lab Med. 2004;42:965–972. doi: 10.1515/CCLM.2004.157. [DOI] [PubMed] [Google Scholar]
- 15.Lin MC, Nawarak J, Chen TY, Tsai HY, Hsieh JF, Sinchaikul S, Chen ST. Rapid detection of natriuretic peptides by a microfluidic LabChip analyzer with DNA aptamers: Application of natriuretic peptide detection. Biomicrofluidics. 2009;3:034101–1. doi: 10.1063/1.3194283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jang HR, Wark AW, Baek SH, Chung BH, Lee HJ. Ultrasensitive and ultrawide range detection of a cardiac biomarker on a SPR platform. Anal Chem. 2013 Dec 13; doi: 10.1021/ac4033565. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Chen P, Wu X, Chen J, Xu L, Chen G, Fu F. A signal-on electrochemiluminescence aptamer biosensor for the detection of ultratrace thrombin based on junction-probe. Biosens Bioelectron. 2011;26:2645–2650. doi: 10.1016/j.bios.2010.11.028. [DOI] [PubMed] [Google Scholar]
- 18.Zhu D, Zhou X, Xing D. A new kind of aptamer-based immunomagnetic electrochemiluminescence assay for quantitative detection of protein. Biosens Bioelectron. 2010;26:285–288. doi: 10.1016/j.bios.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 19.Mallikaratchy PR, Ruggiero A, Gardner JR, Kuryavyi V, Maguire WF, Heaney ML, McDevitt MR, Patel DJ, Scheinberg DA. A multivalent DNA aptamer specific for the B-cell receptor on human lymphoma and leukemia. Nucleic Acids Res. 2011;39:2458–2469. doi: 10.1093/nar/gkq996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McNamara JO, Kolonias D, Pastor F, Mittler RS, Chen L, Giangrande PH, Sullenger B, Gilboa E. Multivalent 4 1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice. J Clin Invest. 2008;118:376–386. doi: 10.1172/JCI33365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruno JG. A review of therapeutic aptamer conjugates with emphasis on new approaches. Pharmaceuticals. 2013;6:340–357. doi: 10.3390/ph6030340. http://dx.doi.org/doi:10.3390/ph6030340. [DOI] [PMC free article] [PubMed] [Google Scholar]