Abstract
Drug resistance in acute lymphoblastic leukemia (ALL) remains a major problem warranting new treatment strategies. Wnt/catenin signaling is critical for the self-renewal of normal hematopoietic progenitor cells. Deregulated Wnt signaling is evident in chronic and acute myeloid leukemia, however little is known about ALL. Differential interaction of catenin with either the Kat3 coactivator CREBBP (CBP) or the highly homologous EP300 (p300) is critical to determine divergent cellular responses and provides a rationale for the regulation of both proliferation and differentiation by the Wnt signaling pathway. Usage of the coactivator CBP by catenin leads to transcriptional activation of cassettes of genes that are involved in maintenance of progenitor cell self-renewal. However, the use of the coactivator p300, leads to activation of genes involved in the initiation of differentiation. ICG-001 is a novel small molecule modulator of Wnt/catenin signaling, which specifically binds to the N-terminus of CBP and not p300, within amino acids 1–110, thereby disrupting the interaction between CBP and catenin. Here, we report that selective disruption of the CBP/β- and γ-catenin interactions using ICG-001 leads to differentiation of pre-B ALL cells and loss of self-renewal capacity. Survivin, an inhibitor-of-apoptosis protein, was also downregulated in primary ALL after treatment with ICG-001. Using ChIP assay, we demonstrate occupancy by CBP of the survivin promoter, which is decreased by ICG-001 in primary ALL. CBP-mutations have been recently identified in a significant percentage of ALL patients, however, almost all of the identified mutations reported occur C-terminal to the binding site for ICG-001. Importantly, ICG-001, regardless of CBP mutational status and chromosomal aberration, leads to eradication of drug-resistant primary leukemia in combination with conventional therapy in vitro and significantly prolongs the survival of NOD/SCID mice engrafted with primary ALL. Therefore, specifically inhibiting CBP/catenin transcription represents a novel approach to overcome relapse in ALL.
Keywords: Acute lymphoblastic leukemia, drug resistance, small molecule inhibitor, CBP, p300, ICG-001
Introduction
Despite significant progress over the last decades, drug resistance remains a major problem in the treatment of acute lymphoblastic leukemia (ALL) (1). The potential for dose escalation of current chemotherapeutics is limited by acute and chronic toxicity, therefore new treatment modalities are required. The Wnt pathway has been implicated in the self-renewal and differentiation of normal hematopoietic stem/progenitor cells (2–4). Aberrant Wnt/catenin signaling has been described to play critical roles in acute myeloid leukemia (AML) cells (5), chronic myelogenous leukemia (CML) cells (6;7), where leukemic drug resistant clones have been associated with increased nuclear β-catenin levels (8). However, little is known about Wnt signaling in ALL. It has been described that Wnt-3a mediates proliferation of precursor B-ALL cell lines NALM6, REH and LK63 (9) and endogenous WNT16b expression has been found to be upregulated by the TCF3-PBX1 (E2A-PBX1) fusion gene (10). siRNA knockdown of WNT16b, decreasing canonical Wnt/β-catenin signaling, has been shown to initiate apoptosis and reduces the expression of the Wnt regulated inhibitor of apoptosis protein family member, survivin (BIRC5) (10), implicated in both the survival and drug resistance of leukemia cells (11–14). The Wnt pathway is classically mediated through the central signaling effector molecule β-catenin (15). Nuclear β-catenin recruits the Kat3 transcriptional co-activators, CREBBP (CBP) or its closely related homolog EP300 (p300), as well as other components of the basal transcriptional apparatus (16;17). The distinct roles of the co-activators, CBP in self-renewal and p300 in differentiation of normal hematopoietic cells has been previously described (18). The controversial dichotomous behavior of Wnt/catenin signaling in controlling both proliferation and differentiation can be readily explained by differential coactivator usage: -i.e. CBP/catenin-mediated transcription maintains cellular potency and that partnering with p300 is a first critical step to initiate differentiation (19–21). The usage of p300 as a coactivator can be pharmacologically induced by ICG-001, a novel low molecular weight small molecule modulator that binds specifically within amino acids 1–110 at the N-terminus of CBP (22). ICG-001 had been previously identified using a secondary structure-templated chemical library in a forward chemical genomic screen, looking for compounds that inhibited the β-catenin/T cell factor (TCF)-responsive reporter TOPFLASH in colorectal cancer cells (22). ICG-001, down-regulates β-catenin/TCF signaling by specifically binding to cyclic AMP response element-binding protein CREBBP (CBP) thereby disrupting the CBP/catenin interaction. We hypothesized that disrupting the CBP/catenin interaction could safely sensitize patient-derived (primary) pre-B ALL cells to conventional chemotherapy via forced differentiation associated with p300/catenin coactivator usage. Here, we report that drug resistant primary pre-B-ALL cells can be induced to differentiate using the novel CBP-specific small molecule inhibitor, ICG-001, thereby sensitizing them to either targeted or conventional chemotherapy, overcoming drug resistance.
Results
ICG-001 blocks the interaction of catenin with CBP in ALL cells
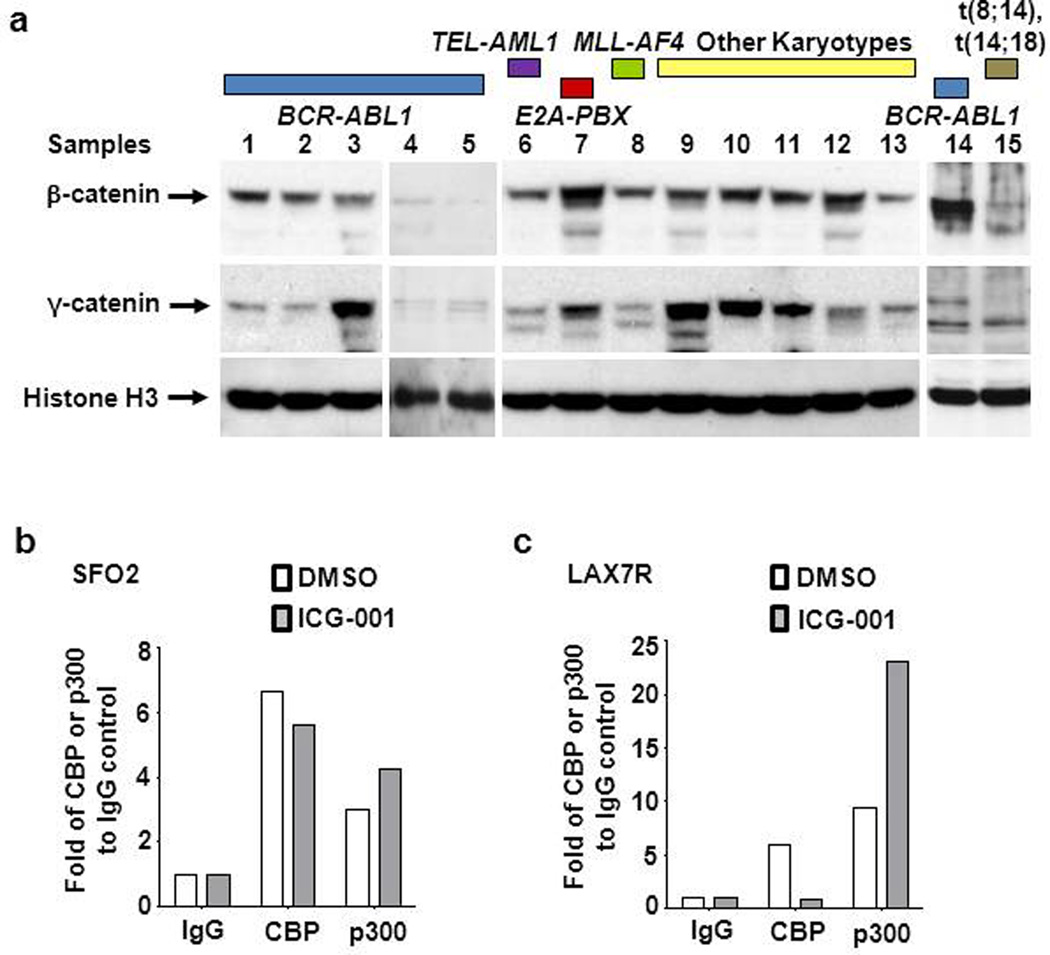
Initially, we examined nuclear β- and γ-catenin levels in 13 primary (pre-B) ALL cases encompassing various cytogenetic aberrations (BCR/ABL1, TEL-AML1, E2A-PBX1, MLL-AF4, and normal karyotype) and two pre-B ALL cell lines BV173 (BCR/ABL-1) and 380 (t(8;14), t(14;18)). Nuclear β and/or γ-catenin, albeit with variability in expression levels, were significantly increased compared with normal pre-B cells, as judged by immunoblot (Figure 1a and supplemental Table S1). As β- and γ-catenin have been implicated in self-renewal of leukemic cells, we evaluated a small molecule inhibitor of the CBP/catenin interaction, ICG-001. ICG-001 binds specifically to the N-terminus (amino acids 1–110) of the coactivator CBP, thereby disrupting CBP/β-catenin, as well as CBP/γ-catenin-mediated transcription (22). To confirm biochemically that ICG-001 also disrupts the CBP/catenin interaction in primary pre-B ALL cells, pre-B ALL cells SFO2 (BCR/ABL-1+) and LAX7R (normal karyotype) (Supplemental Table S1), who relapsed despite having received chemotherapy, were treated with ICG-001 (10µM) or vehicle control (DMSO). Isolated nuclear proteins were immunoprecipitated with CBP or p300 specific antibodies and probed for catenin. As anticipated, ICG-001 specifically decreased the binding of catenin, primarily γ-catenin in these cells, with CBP, with a concomitant increase in p300/γ-catenin association (Figure 1b, 1c; Supplementary Figure S2). As previously demonstrated in multiple cell types (21;22), ICG-001 also in primary ALL cells binds to CBP, sequesters the CBP N-terminus, making it less available for binding to catenin, and thereby enhancing the catenin/p300 interaction.
Figure 1.
ICG-001 blocks the CBP/γ-catenin interaction and enhances binding of p300/γ-catenin. (a) Nuclear β and γ-catenin protein expression levels of normal (CD19+) pre-B cells from healthy donors, 13 primary pre-B ALL (Samples: 1–13) and 2 pre-B ALL cell lines (Samples: 14 and 15) (Characteristics of pre-B cells was shown in Table S1) were determined by Western blot analysis. (b) Co-immunoprecipitation determines the interaction between endogenous γ-catenin and CBP or p300. SFO2 cells and LAX7R cells (c) were treated with ICG-001 at 10µM (ICG) or its vehicle control DMSO (D, 0.1%) for 48 hours. Nuclear proteins were subsequently incubated with CBP and p300 antibody. Western blot was performed using the antibodies against γ-catenin. Fold CBP or p300 compared to IgG control is shown.
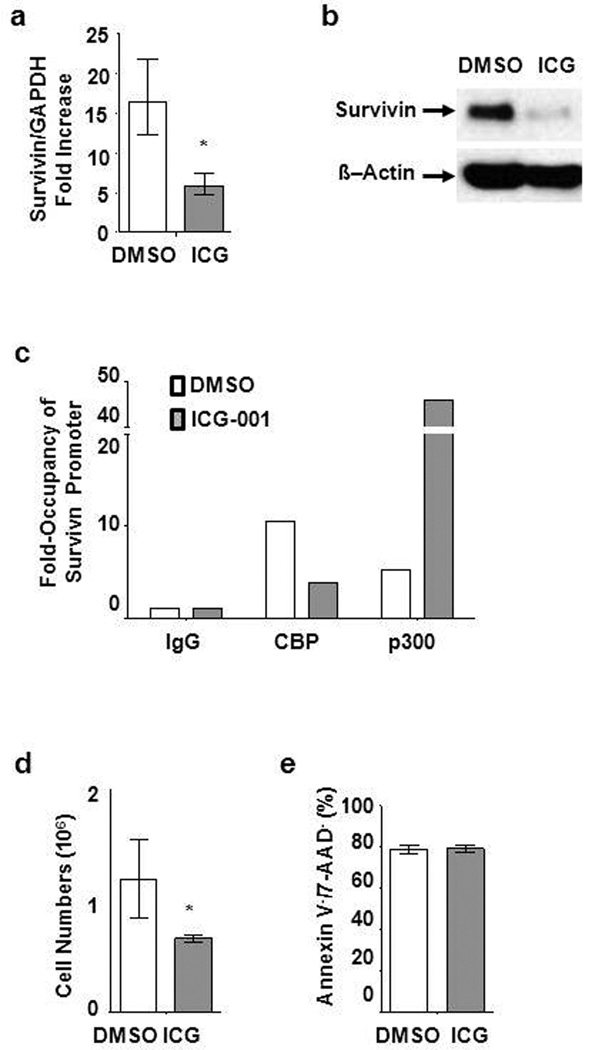
Survivin/BIRC5, is a member of the inhibitor of apoptosis protein family, which we have previously shown to be a CBP/catenin target gene in a number of cancer cell types (21;23). Survivin is highly expressed in primary ALL cells compared to CD19+ B cells from healthy donors (Supplementary Figure S3). Treatment with ICG-001 (10µM) demonstrated significant down-regulation of both survivin mRNA by qPCR and protein by Western blotting in primary LAX7R (Figure 2a, b; as well as other primary isolates, Supplementary Figure S4). Coactivator occupancy in LAX7R cells with or without ICG-001 treatment was assessed by chromatin immunoprecipitation (ChIP) assay. Without treatment, CBP is primarily associated with the survivin promoter (Figure 2c). After ICG-001 treatment, there is dramatically reduced occupancy of CBP at the survivin promoter, with a large concomitant increase in p300 occupancy. These data are consistent with our previous report of a repressive complex being assembled in conjunction with p300 recruitment to the survivin promoter (21). We also determined if a mutant CBP protein can bind to the survivin promoter utilizing ChIP assay with BV173 ALL cells (Supplementary Table S1). Similarly, in BV173 cells we found that CBP occupancy at the survivin promoter was decreased by ICG-001, with a concomitant increase in p300 occupancy (Supplementary Figure S5). We also observed decreased proliferation of primary pre-B ALL cells (LAX7R) (Figure 2d, Supplementary Figure S1). However, cell viability remained unchanged after treatment with ICG-001, as judged by Annexin V staining; demonstrating ICG-001 is not toxic to LAX7R cells (Figure 2e). Taken together, ICG-001 specifically binds to CBP and potently blocks catenin (both β and γ)-mediated expression of survivin and proliferation of ALL cells.
Figure 2.
ICG-001 downregulates survivin in ALL (LAX7R) cells and blocks occupancy of CBP at survivin promoter. (a) Real time PCR was applied to confirm downregulation of survivin gene expression by ICG-001 (10µM) on day 3 post-treatment in LAX7R cells. * p<0.05. (b) Western blot of survivin in ALL cells treated with DMSO (0.1%) or ICG-001 (10µM). β-Actin was used as protein loading control. (c) ChIP analysis of ICG-001 treated LAX7R cells in the presence or absence of ICG-001 for occupancy of CBP and p300 at survivin promoter. (d) Cell numbers were assessed by trypan blue exclusion of dead cells at 48 hours post-treatment of ALL cells (LAX7R) with either DMSO (0.1%) or ICG-001 at 10µM. *p<0.05 (e) Live cells (Annexin V−/7-AAD−) were measured by Annexin V and 7-AAD staining using flow cytometry.
ICG-001 decreases self-renewal capacity of ALL cells
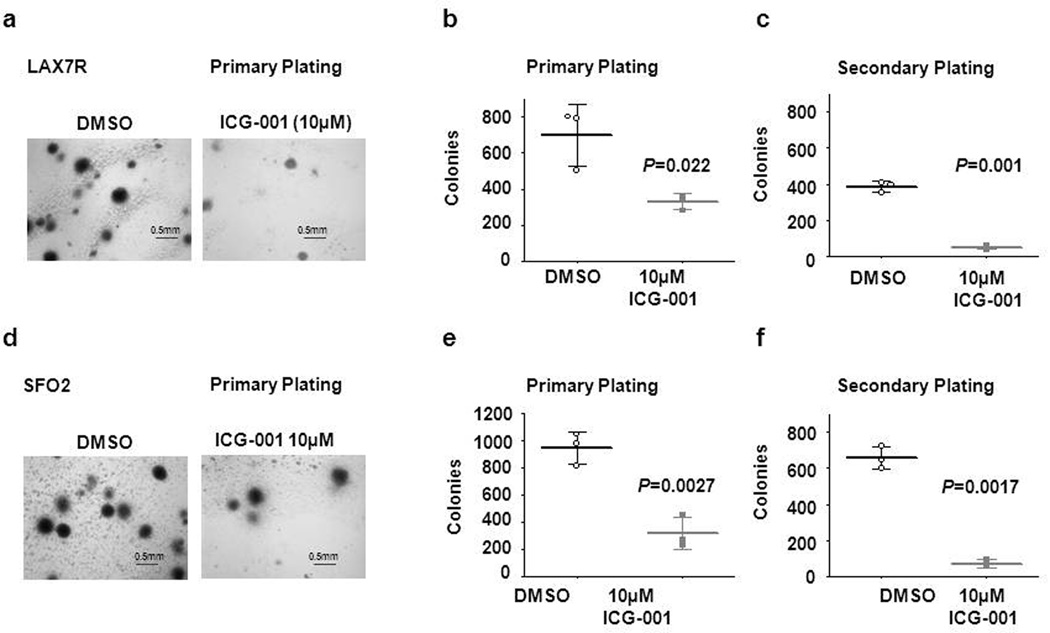
To determine the effect of ICG-001 on CBP/catenin-mediated self-renewal in ALL, we developed a colony forming unit (CFU) assay for primary ALL cells supplementing semisolid agar with an equal number of irradiated OP9 feeder layer cells at the bottom of each dish to enable the growth of primary ALL cells. Two primary ALL samples (LAX7R, SFO2) (Figure 3a–f), were treated with ICG-001 (10µM) or control, and plated in methocult. LAX7R cells showed a significant reduction of colony counts in primary platings after ICG-001 treatment (Figure 3a, b), as compared with the DMSO controls (p<0.05). Secondary colony formation was further significantly reduced (Figure 3c). As anticipated, the DMSO controls, remained re-platable. Similarly, with the second primary leukemia sample (SFO2), we observed a dramatic reduction in secondary colonies with ICG-001 treatment (10µM), whereas control treated cells continued to be serially re-platable (Figure 3f). Our data indicates that ICG-001 abrogates self-renewal of ALL cells.
Figure 3.
ICG-001 decreases self-renewal capacity of ALL cells in vitro. (a) Imaging of colonies of LAX7R cells from primary platings. Colony counts of primary (b) and secondary (c) platings of DMSO control and ICG-001-treated LAX7R cells (2.5× magnification). (d) Images of colonies from primary plating (2.5× magnification) of SFO2 cells. (e) and (f), colony counts of SFO2 cells after primary and secondary platings. Scale bar: 0.5mm
ICG-001 differentiates leukemia cells
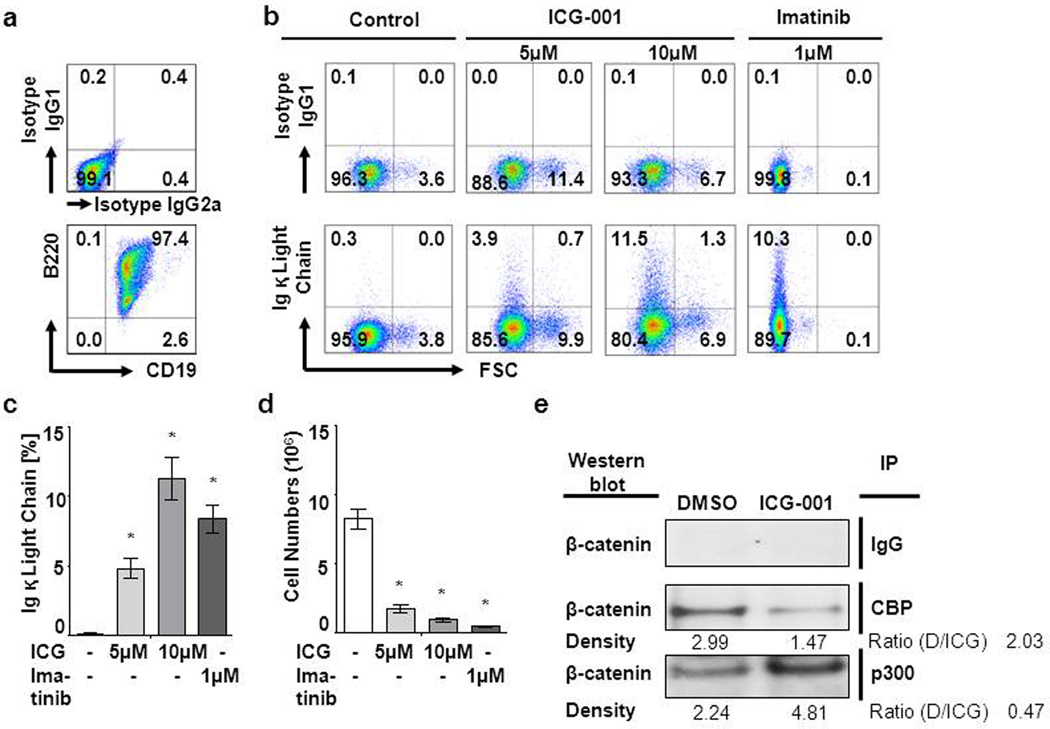
ICG-001 disrupts the interaction between CBP and catenins, resulting in the increased associations of catenins with p300 in ALL. Usage by catenin of p300 as its coactivator is the first critical step towards initiating the process of differentiation (19;20). To evaluate whether ICG-001 would induce differentiation of ALL leukemia cells, we utilized an in vitro system for inducible pre–B cell differentiation of Ig κ and λ light chain gene rearrangement (24;25). This system allows for a detailed analysis of the differentiation of large cycling pre–BII cells (26) to small resting pre–BII cells and ultimately to the immature B cell stage (27). SLP65−/− bone marrow cells were transduced using a murine stem cell (MSCV) retrovirus encoding BCR-ABL1 as a positive control (28). BCR-ABL1–transduced CD19+ B220+ pre–B cells (Figure 4a) were treated with ICG-001 (5µM and 10µM) for 2 days (Figure 4a). Murine leukemia cells differentiated as determined by flow cytometric analysis of increased κ light chain surface expression (Figure 4b and c) and decreased proliferation (Figure 4d). As a positive control, BCR-ABL1–transduced pre–B cells (CD19+ B220+) were induced to differentiate by inhibition of BCR-ABL1 kinase activity using Imatinib (1µM). To address whether this effect of ICG-001 was principally mediated through ICG-001 reduction of survivin, we examined ICG-001-induced differentiation in vector control cells or survivin-depleted murine BCR-ABL1+ ALL cells (Supplementary Figure S6a–d). We observed that κ light chain expression was not affected by survivin depletion (Supplementary Figure S6c), although survivin-depleted cells showed reduced cell number compared to survivin-competent cells (p<0.05) (Supplementary Figure S6d). We also demonstrated by co-immunoprecipitation followed by Western blotting that as anticipated the binding of β-catenin to CBP was dramatically inhibited and concomitantly the binding of β-catenin to p300 was increased by ICG-001 in BCR-ABL1–transduced pre–B ALL cells (Figure 4e). These data demonstrate that ICG-001 dose dependently induced the differentiation of leukemia cells.
Figure 4.
ICG-001 differentiates leukemia cells. Murine B220+CD19+BCR-ABL1 p210+ ALL cells were treated with ICG-001 (5 or 10µM) or Imatinib (1µM) or Control (media only) for 72 hours. (a) B220, CD19 and (b) Ig κ light chain were detected by flow cytometry and (c) quantified. (d) Cell numbers were determined by trypan blue exclusion of dead cells. (e) Co-immunoprecipitation determines the interaction between endogenous β-catenin and CBP or p300. Murine BCR-ABL1 p210+ ALL cells were treated with ICG-001 at 10µM (ICG) or its vehicle control DMSO (D, 0.1%) for 48 hours. Nuclear proteins were subsequently incubated with CBP and p300 antibody. Western blot was performed using the antibodies against β-catenin.
ICG-001 synergizes with chemotherapy to eradicate ALL cellsin vitro
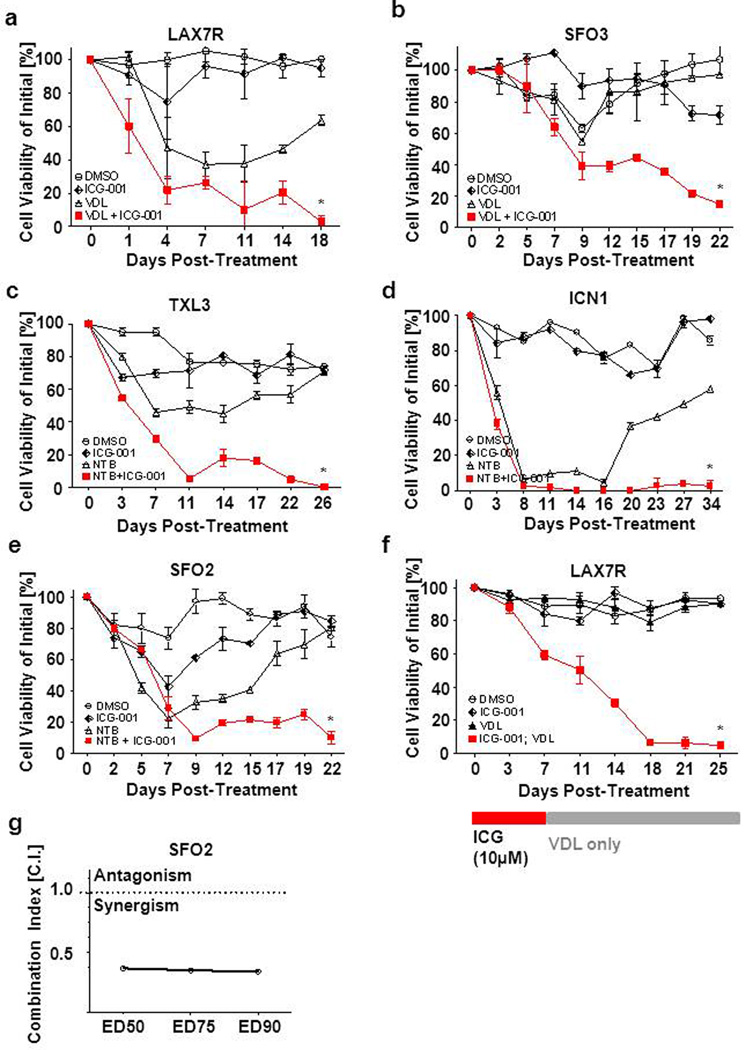
ICG-001 was not toxic to ALL cells but did induce their differentiation. Therefore, we examined if combination treatment of ICG-001 and chemotherapy could overcome drug resistance in primary leukemia cells in vitro. We have shown in Figure 1a, that nuclear β and/or γ-catenin are significantly increased, albeit at various levels, in both primary pre-B ALL and two pre-B ALL cell lines, compared to normal pre-B cells. We anticipated that ICG-001 would be broadly active in ALL regardless of the leukemia subtype. Therefore, we selected 5 ALL patient samples, from patients at diagnosis who relapsed after therapy, for treatment with chemotherapy for 19–34 days, with or without ICG-001. VDL was used to treat the Philadelphia chromosome negative (Ph−) ALL cells (LAX7R, SFO3). The tyrosine kinase inhibitor nilotinib, was used for the Philadelphia chromosome positive, Ph+, ALL cells (ICN1, TXL3, SFO2) (Supplementary Table S1). Persistent viability of all of the ALL samples after extended (2 – 4 weeks) treatment indicated that the ALL cells were resistant to conventional chemotherapy alone (Fig 5a–e). In marked contrast, within the same treatment period all five primary ALL cells that received a combination of chemotherapy plus ICG-001 had been eradicated as determined by decreased cell viability and cell number (Figure 5a–e, Figure S7). Not surprisingly, ICG-001 alone did not lead to elimination of ALL cells. However, importantly, when the primary pre B-ALL cells (LAX7R) were first treated with ICG-001 (10µM) as a single agent, followed by addition of VDL with no further addition of ICG-001 (Figure 5f), the primary ALL cells were also eradicated. The Combination Index (C.I.) for ED50 (median effective dose to inhibit 50% of cells) (29), ED75, and ED90 in SFO2 cells demonstrated synergism of ICG-001 with chemotherapy (Figure 5g). These results demonstrated that ICG-001 by specifically targeting the CBP/catenin interaction in ALL leads to the differentiation of ALL cells, thereby sensitizing drug resistant leukemia cells to chemotherapy. This was observed in five out of five cases examined, either when ICG-001 was given prior to or concomitantly with chemotherapy, leading to essentially complete ablation of the drug-resistant ALL cell population.
Figure 5.
ICG-001 combined with chemotherapy eradicates ALLs in vitro. (a) Cell viability of LAX7R treated continuously with ICG-001 and/or VDL (Vincristine 0.5nM, Dexamethasone 5pM, 0.0005 IU L-Asparaginase) for 31 days. (b) Cell viability of SFO3 cells treated with either ICG-001 or VDL or both for 22 days. Cell viability of ICN1 (c), TXL3 (d) and SFO2 (e) are shown, which were treated continuously with Nilotinib (1µM) and/or ICG-001 (10 µM) until the end of follow-up. (f) Pre B-ALL cells (LAX7R) were preincubated with ICG-001 (10µM) only for 7 days followed by addition of VDL (Vincristine 10nM, Dexamethasone 0.1nM and L-Asparaginase 0.01IU) to ALL cell culture media with no further addition of ICG-001. Cell viability of LAX7R was measured by trypan blue exclusion. (g) Combination index (C.I.) was determined in SFO2.
ICG-001 eradicates ALL cells bearing CBP mutations in vitro
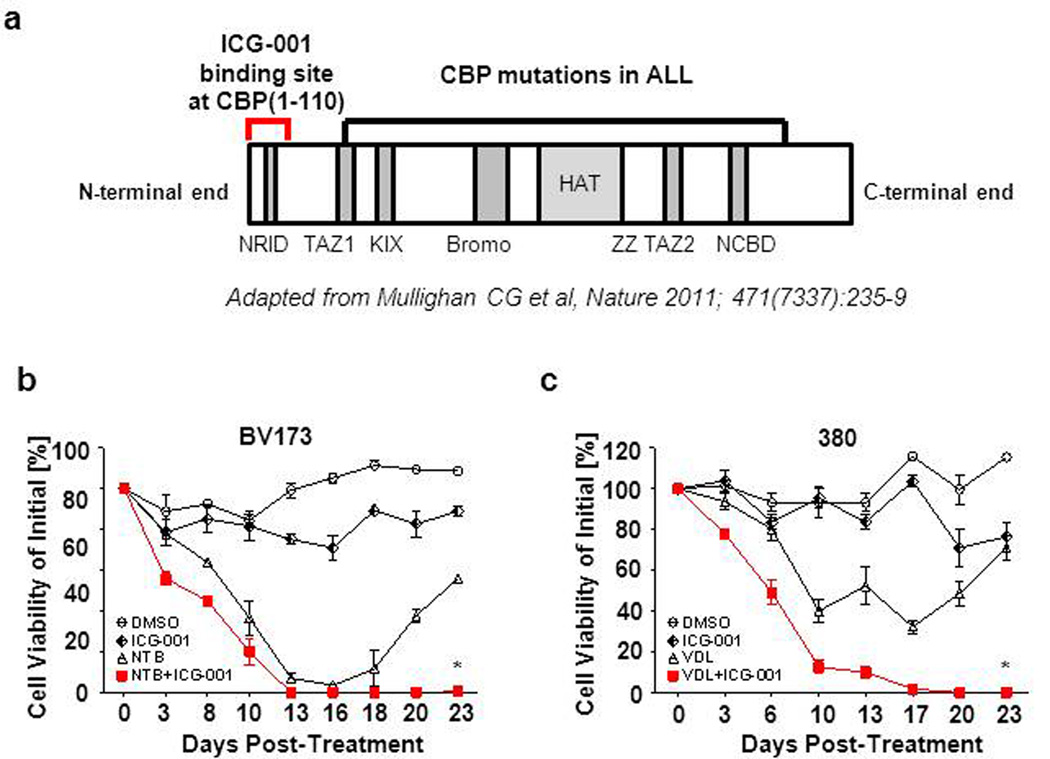
Recently, novel sequence or deletion mutations of CBP have been identified in ALL (30;31). Interestingly, none of these mutations has been found in the extreme N-terminus of CBP, the site at which both catenin and ICG-001 bind to the coactivator. The majority of mutations have been mapped to the HAT domain. Nevertheless, we evaluated the efficacy of ICG-001 on two ALL cell lines (380 and BV173) that harbor CBP mutations (Figure 6a) (30). These ALL cell lines were viable despite chemotherapy, reflecting their resistance to conventional chemotherapy alone (Figure 6b,c). Monotherapy with ICG-001 alone did not lead to elimination of these ALL cells. In marked contrast, chemotherapy in combination with ICG-001, within the same treatment period, completely eradicated both ALL cell lines. As anticipated, the presence of these mutations did not affect the molecular mechanism or efficacy of ICG-001, as the mutations are C-terminal to the ICG-001 binding site. Similar results were obtained for both the five primary ALL cases, for which the CBP mutational status are unknown (Figure 5), as well as both CBP mutated ALL cell lines, in that they were resistant to chemotherapy alone, yet eradicated at the end of the combination of chemotherapy plus ICG-001 (Figure 6 b,c). Our data demonstrates that using ICG-001 to specifically target the CBP/catenin in ALL cells synergizes with chemotherapy in vitro, leading to complete ablation of the drug-resistant cell population, even in ALL cells that harbor CBP mutations.
Figure 6.
ICG-001 eradicates ALL cells bearing CBP mutations in vitro. (a) N-terminal binding site of ICG-001 (1–110aa) at CBP and localization of identified patient mutations by Mullighan et al. (30) in the CBP domains (NRID: nuclear-receptor-interaction domain; TAZ1/2: transcriptional-adaptor zinc-finger ½; KIX, KID-binding domain; Bromo: bromodomain; HAT: histone acetyltransferase domain; ZZ: zinc-binding domain near the dystrophin WW domain; NCBD: nuclear-receptor coactivator-binding domain). (b) Cell viability of BV173 after continuous treatment with Nilotinib (5µM) and/or ICG-001 (10µM). Cell viabilities determined by trypan blue exclusion. *p<0.05. (c) Cell viability of 380 treated continuously with ICG-001(10µM) and/or VDL (Vincristine 2nM, Dexamethasone 20pM, 0.002 IU L-Asparaginase). Cell viabilities were determined by trypan blue exclusion of dead cells. *p<0.05.
Combination treatment of ICG-001 and chemotherapy prolongs survival of primary ALL recipient mice
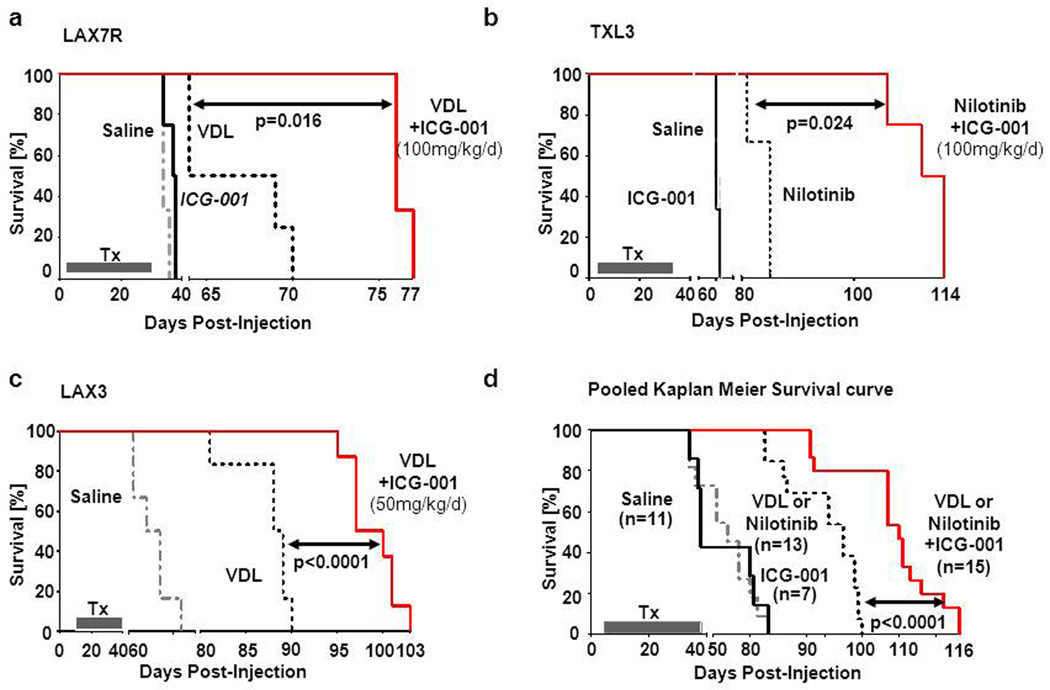
For preclinical in vivo evaluations of ICG-001, we used a xenograft model as described in (11;11) with three primary pre-B ALL cases – two Ph− ALLs (LAX7R, LAX3) with normal karyotype and one Ph+ ALL (TXL3). Cells were injected into sublethally irradiated NSG mice and treated with ICG-001 (50mg/kg/day for LAX3 or 100mg/kg/day for LAX7R and TXL3 via subcutaneous osmotic minipump), with or without respective chemotherapy; VDL for Ph− ALL (via i.p. injection) and Nilotinib (NTB p.o.) for Ph+ ALL. Pooled survival analysis of the three primary ALLs displayed markedly prolonged survival in the cohort that received chemotherapy plus ICG-001 (n=15), compared with the chemotherapy only treated mice (n=13) (Median Survival Time: MST=100 days vs. MST=85 days; p<0001) (Figure 7d). The saline-control treated (PBS) LAX7R mice died rapidly with a MST of 34 days (n=3). Similarly, ICG-only treated mice died with a MST of 37.5 days (n=4). Combined VDL and ICG-001 treatment (n=3) significantly prolonged survival, compared to the VDL only treated animals (n=4) (MST = 76 days vs. MST=66.5 days; p<0.016) (Figure 7a). In a pilot experiment lentiviral luciferase-labeled LAX7R cells were injected into NOD/SCID mice to determine reduction in leukemia burden by ICG-001 in combination with chemotherapy. Bioluminescent imaging demonstrated a significantly reduced leukemic load in the VDL+ICG-001-treated mice compared to the VDL only-treated mice (Figure S8a,b). As for the TXL3 (Ph+ ALL) mice, the saline treated group died rapidly (n=2) (MST=62.5 days), the Nilotinib (NTB) only group had a MST=85 days (n=3). Combined NTB+ICG-001 treated mice (n=4) lived significantly longer, as compared to the NTB only treated mice (MST=114 days vs. MST=85 days; p<0.024) (Figure 7b). Similar outcomes were observed in the Ph− ALLs LAX3 mice – VDL/ICG-001 treated group (n=8) has MST=98.5 days of vs. VDL-only treated mice (n=6) with MST=88.5 days; p<0.0001 (Figure 7c). Importantly, assessment of hematopoietic parameters after treatment of NSG mice with VDL with and without ICG-001 showed no significant changes (Supplementary Table S2). We interpret these results that ICG-001 has no deleterious effects on normal hematopoiesis.
Figure 7.
Combined ICG-001 and chemotherapy treatment prolongs survival in 3 primary ALL cases. Kaplan-Meier curves of NOD/SCID IL2Rγ−/− mice recipients of patient-derived LAX7R (a), TXL3 (b) and LAX3 (c) cells. (d) Pooled Kaplan-Meier curves of overall survival for 3 ALL xenografts (LAX7R, LAX3 and TXL3) in NOD/SCID IL2Rγ−/− (NSG) mice are shown. Mice were treated with Nilotinib (50mg/kg/day) or VDL (Vincristine 0.5mg/kg/day, Dexamethasone 10.5mg/kg/day and L-Asparaginase 1500IU/kg/day) combined with ICG-001 (50mg/kg/day for LAX3, while TXL3 and LAX7R received 100mg/kg/day) for 28 days. Pooled MST of the saline group (n=11) was 54 days, of the ICG-001 only group (n=7) was 38 days, of the chemotherapy (Nilotinib or VDL) only group (n=13) was 85 days. MST of chemotherapy/ICG-001 treated group (n=15) was 100 days (chemotherapy vs chemotherapy + ICG-001: p<0.0001).
In summary, our data clearly demonstrates that specifically targeting the CBP/catenin interaction using ICG-001 initiates the differentiation of ALL leukemia cells, thereby sensitizing them to chemotherapy, both in vitro and in vivo. These findings provide the basis for a novel therapeutic approach for ALL.
Discussion
Wnt signaling is a key signal transduction pathway in both normal and cancer stem cells (32) and β-catenin has been shown to be critical in the self-renewal of normal hematopoietic stem cells (2;3). Aberrant Wnt/β- and γ-catenin signaling has been described in the development of AML (5;33) and has also been identified as a critical pathway in the self-renewal of CML (6;7). Overexpression of γ-catenin has been observed in numerous cancers (23;33;34). Furthermore, transduction of γ -catenin into primitive hematopoietic progenitor cells preserved their immature phenotype during colony growth, which suggests enhanced self-renewal activity. Furthermore, γ -catenin transduced cells accelerated the development of leukemia in syngenic murine hosts (33). However, it should be noted that loss of both β- and γ-catenin leaves wnt signaling, hematopoiesis and lymphopoiesis intact (35), pointing to a yet uncharacterized catenin-like molecule that can compensate for the loss of both β- and γ-catenin. The role of Wnt/catenin signaling in ALL, however remains unclear. To generate a transcriptionally active complex, β-catenin recruits the coactivators CBP or its closely related homolog p300, as well as other members of the basal transcriptional apparatus (16;17). Wnt/β-catenin transcription activates a number of downstream target genes (e.g. c-myc, cyclin D1) including survivin (22). Survivin is a Wnt target gene whose expression has been shown to be CBP/catenin-mediated (21). Recently, it has been demonstrated that targeting survivin with shRNA or an antisense oligonucleotide increased primary ALL apoptosis, particularly in combination with chemotherapeutic agents (11;13;14). Targeting the CBP/catenin interaction with ICG-001, forcing differentiation and additionally down-regulating survivin expression, thereby sensitizing the cells to chemotherapy, would appear to be an attractive strategy to maximize chemotherapeutic potency without increasing toxicity to treat ALL.
The coactivators CREB-binding protein (CBP) and p300 comprise the unique KAT3 family of histone acetyltransferases (HATs) (36). A crucial role for both p300 and CBP in development was shown in mice where homozygous deletion of either gene (Ep300 and Crebbp for the proteins p300 and CBP respectively) resulted in embryonic lethality at a very early stage (37;38). Interestingly, double heterozygous Ep300+/−/Crebbp+/− mice also die in utero (37), indicating that a fine-tuned balance in the expression of both proteins is needed to ensure normal development. We previously demonstrated that CBP and p300 have distinct functions in the regulation of β-catenin-mediated gene transcription (22;39) including survivin (21). Rebel et al. (18), using a hematopoietic stem cell (HSC) model, concluded that CBP (and not p300) is essential for HSC self-renewal, whereas p300 is critical for proper hematopoietic differentiation. Kawasaki et al. (40) found that p300, but not CBP, is absolutely required for retinoic acid-induced F9 cell differentiation. Furthermore, both coactivators are critical in peripheral B cells (41). Despite their high degree of homology and similar ubiquitous patterns of expression, it is clear that CBP and p300 play unique and distinct roles in gene regulation, which has recently been confirmed at the genome wide level using ChIP-seq (42). The effects of CBP/catenin antagonism on wnt target genes are highly promoter-specific, as some wnt/catenin-regulated genes like survivin apparently utilize CBP as its co-activator for transcription, while others permissively use either CBP or p300, whereas others appear to utilize p300 almost exclusively (20;39). We have previously demonstrated reduction of survivin by ICG-001 in vivo in a xenograft model of colorectal cancer (22). Of note, even though survivin is involved in terminal differentiation in erythropoiesis (43), we demonstrate that ICG-001 induced ALL differentiation is independent of survivin reduction, as kappa light chain expression is similarly induced with or without survivin depletion in BCR-ABL1+ ALL cells (Supplementary Fig S6a–d). Therefore, survivin reduction is not sufficient to induce differentiation in ALL cells, demonstrating that decreased survivin is a not the primary driver for inducing differentiation. Upon inhibition of CBP/catenin binding, several potential promoter specific outcomes can occur including downregulation of CBP-only regulated genes, compensatory maintenance by p300 or even upregulation of genes using both CBP and p300, and finally activation of p300-dependent regulated genes (20;39). Utilizing chromatin immunoprecipitation assay (ChIP), we have previously demonstrated that coactivator switching from CBP to p300 at the survivin promoter is associated with the recruitment of transcriptionally repressive elements via p300, thereby decreasing gene expression (20;21).
A variety of ways to inhibit wnt signaling are currently under investigation for hematological malignancies (44–46). However, CBP/catenin antagonists such as ICG-001, and the second generation clinical compound PRI-724, are the only well characterized small molecule-inhibitors, that binds specifically to the N-terminal (a.a. 1–111) of CBP, thereby blocking the interaction with the C-terminal catenin trans activation domain (647–781), leaving the rest of the large CBP protein (300kDa) functionally intact (22). In addition, and of critical importance to its therapeutic utility, despite the close homology of CBP and p300, the specificity of ICG-001 avoids interference with p300/catenin dependent signaling (22), thereby allowing for cellular differentiation and maintenance of normal stem cell populations (20).
In humans, heterozygous point mutations and micro-deletions in CBP and rarely in p300, have been identified in Rubinstein-Taybi Syndrome patients. This rare disorder (~1 in 100,000 live births) is characterized by mental retardation, craniofacial malformations and increased occurrence of malignancies (47;48). Recently, sequence or deletion mutations of CBP have also been identified in ALL (30;31). Extensive analysis of an extended cohort of 71 diagnosis-ALL relapse cases and 270 acute leukemia cases that did not relapse found that 18.3% of relapse cases had sequence or deletion mutations of CBP. In addition, inactivating CBP mutations have been described as a common event in follicular lymphoma and diffuse large B-cell lymphoma (49) the two most frequent forms of B-cell Non-Hodgkin’s Lymphoma. Interestingly, of the hundreds of sequenced samples, most occurred within the HAT domain and none were described within the N-terminus of CBP, which constitutes both the catenin and ICG-001 binding site (Figure 6a). We propose that based upon the critical role for the CBP N-terminal/catenin interaction in maintaining the leukemia initiating cells that comprise the drug resistant ALL progenitor population, mutations in this region of CBP will generally not be selected for. Importantly from a therapeutic standpoint, we have demonstrated that as anticipated, ICG-001 can also sensitize B-ALL cell lines harboring CBP mutations to chemotherapy. These defective mutations are located in the HAT domain. HAT defective CBP may behave as a dominant negative, allowing binding of CBP to catenin without HAT enzymatic activity, which may be importantly involved in coactivator switching to p300 and subsequent p300 driven differentiation. ICG-001 does not work through the HAT domain, as it is located in the extreme N-terminus. ICG-001 by blocking the CBP/catenin interaction could thus remove the dominant negative HAT defective CBP from promoters, thereby allowing for enhanced p300 recruitment (Figure 1b).
Taken together, these recent findings emphasize an important role for the CBP/catenin interaction in maintaining drug resistant ALL leukemia initiating cells. The fundamental therapeutic concept outlined is that CBP/catenin antagonism can deplete drug resistant leukemia stem/progenitor cells by interruption of self-renewal and shift of catenin/coactivator function and selective induction of differentiation at the expense of self-renewal capacity (Figure 3) (20). Here, we demonstrate abrogation of self-renewal by inhibition of serial replatability of primary ALL cells. As analysis of differentiation of primary human pre-B ALL cells is difficult, we used an in vitro system for inducible pre–B cell differentiation and Ig κ and λ light chain gene rearrangement (24–27). ICG-001 induced dose-dependent differentiation of murine BCR-ABL1–transformed pre–B cells (CD19+B220+) as determined by analysis of κ light chain surface expression.
Taken together, we have shown that selective suppression of CBP/catenin signaling using the novel specific small-molecule inhibitor ICG-001 offers the opportunity to safely abrogate leukemia by disrupting the self-renewal of drug resistant leukemia initiating cells via forced differentiation, thereby sensitizing them to either cytotoxic or targeted chemotherapy to eradicate the leukemia. Therefore, inhibition of CBP/catenin driven self-renewal is a fundamental concept, and targeting it using specific CBP/catenin antagonists in combination with conventional therapy represents a promising therapeutic strategy to eradicate drug resistant leukemia initiating cells.
Materials and Methods
Patient samples
Bone marrow and peripheral blood samples from ALL patients (supplemental Table 1) were provided by USC (Los Angeles, USA), UCSF (San Francisco, USA) and Samsung Medical Center (Seoul, South-Korea), and the University Hospital Benjamin Franklin (Berlin, Germany) in compliance with the Institutional Review Board regulations of each institution. Informed consent was obtained from all human subjects.
Characterization of β- and γ-catenin expression in primary ALL
Patient xenograft nuclear protein samples were isolated using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific, Rockford, IL), with subsequent Western blotting.
Western blotting
Nuclear fractions isolated as described above or total cell lysate obtained from lysis in M-PER buffer (Thermo Scientific, Rockford, IL) supplemented with a 1% protease inhibitor cocktail (Pierce, Rockford, IL), were separated by 4–12% SDS-PAGE and electro-transferred to PVDF membrane (Invitrogen, Carlsbad, CA). The following antibodies were used: anti-survivin (D-8), anti-β-Actin (AC-15) (Santa Cruz Biotechnology, Santa Cruz, CA); anti-β-catenin (Clone 14) and anti-γ-catenin (Clone 15) (BD Biosciences, San Jose, CA).
Co-immunoprecipitation (Co-IP) assay
Nuclear proteins were incubated with primary antibodies (rabbit polyclonal CBP antibody, A-22; rabbit polyclonal p300 antibody, N-15; normal rabbit IgG) (Santa Cruz Biotechnology) overnight at 4°C. Protein-antibody aggregates were then incubated with Protein A-agarose (MP Biomedicals, Solon, OH) for 1 hour. The immunoprecipitated proteins were separated by 4–12% SDS-PAGE and analyzed by western blot using mouse monoclonal anti-β-catenin or γ-catenin (BD Biosciences). For quantification of Western blot band, we utilized the software Image J (java image processing program). Band densities were normalized to the corresponding IgG control.
ChIP Assay
ChIP assays were conducted essentially as described in Spencer et al. (2003) (50). Immunoprecipitation was performed with antibodies against CBP, p300 and control IgG (Santa Cruz Biotechnology). The captured immunocomplexes containing bound transcriptional DNA fragments were eluted, with recovered DNA fragments used for PCR amplification, using specific primers for the consensus human BIRC5 promoter, F: 5'-ggggcgctaggtgtggg-3' and R: 5'-ttcaaatctggcggttaatggc-3'.
CFU Assays
Primary ALL cells were plated with ICG-001 or DMSO vehicle control in triplicate on a murine OP-9 feeder layer in MethoCult GF+ H4435 (StemCell Tech, Vancouver, BC) and incubated at 37°C in humidified 5% CO2 for 14–21 days. Colonies were counted under phase contrast microscopy (Carl Zeiss Microimaging, Thornwood, NY) and Orca C4742-80-12AG camera (Hamamatsu Photonics, Bridgewater, NJ) at room temperature under 2.5×. The microscope and camera were controlled by Micro-Manager 1.4 software. 100× magnification photographs for colonies were acquired under light microscope using QCapture software 2.98 (Quantitative Imaging Corp; Surrey, BC Canada) via a QImaging QiCam, mounted to an Olympus IX71 microscope.
Flow cytometry
Anti-mouse Ig κ light chain -PE (Clone 187.1) as well as its isotype control Anti-mouse IgG1-PE (R3–34) antibodies (BD Biosciences) were used for phenotyping murine BCR-ABL1 p210 ALL cells treated with ICG-001 or Imatinib.
As a positive control for mouse Ig kappa light chain, bone marrow cells from SLP65−/− mice were harvested and cultured in the presence of 10ng/ml on retronectin (Takara Bio Inc.)-coated plates and retrovirally transformed by BCR-ABL1 as described earlier. 11 Ig kappa light chain was induced by either ICG-001 or imatinib treatment for 48hrs and determined by flow cytometry. Anti-mouse CD19 and B220 were purchased from eBioScience (San Diego, CA). PE-conjugated Annexin V and 7-AAD for apoptosis analyses were obtained from BD Biosciences (San Jose, CA).
In Vitro Drug Testing
Cells cultured on irradiated murine OP-9 were treated with VDL (Vincristine, Dexamethasone, and L-Aparaginase) or Nilotinib combined with ICG-001, at various concentrations. Cell viability was determined by either trypan blue exclusion or flow cytometry. The Combination Index (C.I.) for ED50 (median effective dose to inhibit 50% of cells), ED75, and ED90 were calculated using Chou and Talalay median effects analysis (CalcuSyn2.0 Software; Cambridge, United Kingdom) (29).
Xenograft model of primary leukemia
Primary ALL cells were transplanted into sublethally irradiated (250 cGy) NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) or NOD/SCID mice via intravenous injection as described previously (11).
In Vivo Drug Testing
Mice were treated with Nilotinib (p.o.) or VDL (Vincristine, Dexamethasone and L-Asparaginase) (i.p.) combined with ICG-001 for 28 days. ICG-001 was delivered via subcutaneous micro-osmotic pump (Model 1004; Alzet, Cupertino, CA) to ensure stable plasma dosing levels. Mice were under routine observation with individual weights tracked and used as criteria for sacrifice, upon 20% percent-loss from initial pre-treatment recordings. Animal care was in accordance with institutional guidelines.
In Vivo Imaging
Primary ALL cells were labeled with a lentiviral firefly luciferase vector by transduction with pCCL-MNDU3-LUC viral supernatant as described previously (11) and injected into sublethally irradiated NOD/SCID mice. Leukemia progression in mice at indicated time points was monitored using an in vivo IVIS 100 bioluminescence/optical imaging system (Xenogen). D-Luciferin (Promega) dissolved in PBS was injected intraperitoneally at a dose of 2.5mg per mouse, 15 minutes prior to measuring the luminescence signal. General anesthesia was induced with 5% isoflurane and continued during the procedure with 2% isoflurane introduced via a nose cone. Mice were monitored for weight loss and other leukemia symptoms. Moribund mice were sacrificed and tissues were analyzed for leukemia cell infiltration to confirm leukemia as the cause of death. All mouse experiments were subject to institutional approval by Children’s Hospital Los Angeles IACUC.
CBC Counts
Peripheral blood was withdrawn for CBC analysis via tail vein. Blood samples (~100µl/sample) were collected in BD microtainer tubes with EDTA (BD Biosciences) and analyzed by VetScan HM5 cell counter (Abaxis, Union City, CA).
Survivinfl/fl mice
Survivin-floxed mice were originally obtained from E.M. Conway (University of British Columbia, Vancouver, Canada). As previously described (11), survivinfl/fl bone marrow cells were retrovirally transduced with BCR-ABL1 p210. For competitive growth assays, oncogenically transformed cells were transduced with either retroviral pMSCV EmptyERt2 or pMSCV CreERt2 followed by puromycin selection (1µg/mL). Conditional deletion was achieved using tamoxifen (1µM) for 4 days. a. Survivin deleted (CreERt2) and undeleted (EmptyERt2) cells were then plated in triplicate and treated with either ICG-001 or imatinib or both for 48hrs prior to flow cytometric assay including apoptosis by Annexin V/7-AAD and Ig κ light chain expression by anti-mouse Ig κ light chain PE antibody (BD Bioscience).
Supplementary Material
Acknowledgement
This work was supported by grants from: William Lawrence and Blanche Hughes Foundation, Concern Foundation, Nautica Triathlon and the American Cancer Society and R01CA172896 (YMK); USC Norris Comprehensive Cancer Center Support Grant P30 CA014089 (MK); R01CA137060, R01CA139032, R01CA157644 (MM). EMC is supported by a CSL Behring Research Chair and a Canada Research Chair in Endothelial Cell Biology and by grants from the Canadian Institutes for Health Research (CIHR).
Footnotes
Conflict of Interest
MK owns stock and is a consultant for Prism Pharmnaceuticals.
Reference List
- 1.Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008 Mar 22;371(9617):1030–1043. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- 2.Reya T, Duncan AW, Ailles L, Domen J, Scherer DC, Willert K, et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003 May 22;423(6938):409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- 3.Luis TC, Naber BA, Roozen PP, Brugman MH, de Haas EF, Ghazvini M, et al. Canonical wnt signaling regulates hematopoiesis in a dosage-dependent fashion. Cell Stem Cell. 2011 Oct 4;9(4):345–356. doi: 10.1016/j.stem.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 4.Chan WI, Hannah RL, Dawson MA, Pridans C, Foster D, Joshi A, et al. The transcriptional coactivator Cbp regulates self-renewal and differentiation in adult hematopoietic stem cells. Mol Cell Biol. 2011 Dec;31(24):5046–5060. doi: 10.1128/MCB.05830-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Krivtsov AV, Sinha AU, North TE, Goessling W, Feng Z, et al. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010 Mar 26;327(5973):1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao C, Blum J, Chen A, Kwon HY, Jung SH, Cook JM, et al. Loss of beta-catenin impairs the renewal of normal and CML stem cells in vivo. Cancer Cell. 2007 Dec;12(6):528–541. doi: 10.1016/j.ccr.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Y, Chen Y, Douglas L, Li S. beta-Catenin is essential for survival of leukemic stem cells insensitive to kinase inhibition in mice with BCR-ABL-induced chronic myeloid leukemia. Leukemia. 2009 Jan;23(1):109–116. doi: 10.1038/leu.2008.262. [DOI] [PubMed] [Google Scholar]
- 8.Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004 Aug 12;351(7):657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 9.Khan NI, Bradstock KF, Bendall LJ. Activation of Wnt/beta-catenin pathway mediates growth and survival in B-cell progenitor acute lymphoblastic leukaemia. Br J Haematol. 2007 Aug;138(3):338–348. doi: 10.1111/j.1365-2141.2007.06667.x. [DOI] [PubMed] [Google Scholar]
- 10.Mazieres J, You L, He B, Xu Z, Lee AY, Mikami I, et al. Inhibition of Wnt16 in human acute lymphoblastoid leukemia cells containing the t(1;19) translocation induces apoptosis. Oncogene. 2005 Aug 11;24(34):5396–5400. doi: 10.1038/sj.onc.1208568. [DOI] [PubMed] [Google Scholar]
- 11.Park E, Gang EJ, Hsieh YT, Schaefer P, Chae S, Klemm L, et al. Targeting survivin overcomes drug resistance in acute lymphoblastic leukemia. Blood. 2011 Aug 25;118(8):2191–2199. doi: 10.1182/blood-2011-04-351239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008 Jan;8(1):61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- 13.Tyner JW, Jemal AM, Thayer M, Druker BJ, Chang BH. Targeting survivin and p53 in pediatric acute lymphoblastic leukemia. Leukemia. 2012 Apr;26(4):623–632. doi: 10.1038/leu.2011.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morrison DJ, Hogan LE, Condos G, Bhatla T, Germino N, Moskowitz NP, et al. Endogenous knockdown of survivin improves chemotherapeutic response in ALL models. Leukemia. 2012 Feb;26(2):271–279. doi: 10.1038/leu.2011.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005 Apr 14;434(7035):843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 16.Takemaru KI, Moon RT. The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J Cell Biol. 2000 Apr 17;149(2):249–254. doi: 10.1083/jcb.149.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hecht A, Vleminckx K, Stemmler MP, van RF, Kemler R. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 2000 Apr 17;19(8):1839–1850. doi: 10.1093/emboj/19.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rebel VI, Kung AL, Tanner EA, Yang H, Bronson RT, Livingston DM. Distinct roles for CREB-binding protein and p300 in hematopoietic stem cell self-renewal. Proc Natl Acad Sci U S A. 2002 Nov 12;99(23):14789–14794. doi: 10.1073/pnas.232568499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teo JL, Ma H, Nguyen C, Lam C, Kahn M. Specific inhibition of CBP/beta-catenin interaction rescues defects in neuronal differentiation caused by a presenilin-1 mutation. Proc Natl Acad Sci U S A. 2005 Aug 23;102(34):12171–12176. doi: 10.1073/pnas.0504600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kahn M. Symmetric division versus asymmetric division: a tale of two coactivators. Future Med Chem. 2011 Oct;3(14):1745–1763. doi: 10.4155/fmc.11.126. [DOI] [PubMed] [Google Scholar]
- 21.Ma H, Nguyen C, Lee KS, Kahn M. Differential roles for the coactivators CBP and p300 on TCF/beta-catenin-mediated survivin gene expression. Oncogene. 2005 May 19;24(22):3619–3631. doi: 10.1038/sj.onc.1208433. [DOI] [PubMed] [Google Scholar]
- 22.Emami KH, Nguyen C, Ma H, Kim DH, Jeong KW, Eguchi M, et al. A small molecule inhibitor of beta-catenin/CREB-binding protein transcription [corrected] [8-24-2004];Proc.Natl.Acad.Sci.U.S.A. 101(34):12682–12687. doi: 10.1073/pnas.0404875101. Ref Type: Magazine Article. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim YM, Ma H, Oehler VG, Gang EJ, Nguyen C, Masiello D, et al. The gamma catenin/CBP complex maintains survivin transcription in beta-catenin deficient/depleted cancer cells. Curr Cancer Drug Targets. 2011 Feb;11(2):213–225. doi: 10.2174/156800911794328420. [DOI] [PubMed] [Google Scholar]
- 24.Muljo SA, Schlissel MS. A small molecule Abl kinase inhibitor induces differentiation of Abelson virus-transformed pre-B cell lines. Nat Immunol. 2003 Jan;4(1):31–37. doi: 10.1038/ni870. [DOI] [PubMed] [Google Scholar]
- 25.Klein F, Feldhahn N, Mooster JL, Sprangers M, Hofmann WK, Wernet P, et al. Tracing the pre-B to immature B cell transition in human leukemia cells reveals a coordinated sequence of primary and secondary IGK gene rearrangement, IGK deletion, and IGL gene rearrangement. J Immunol. 2005 Jan 1;174(1):367–375. doi: 10.4049/jimmunol.174.1.367. [DOI] [PubMed] [Google Scholar]
- 26.Hardy RR, Hayakawa K. B cell development pathways. Annu Rev Immunol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- 27.Herzog S, Reth M, Jumaa H. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat Rev Immunol. 2009 Mar;9(3):195–205. doi: 10.1038/nri2491. [DOI] [PubMed] [Google Scholar]
- 28.Pear WS, Miller JP, Xu L, Pui JC, Soffer B, Quackenbush RC, et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998 Nov 15;92(10):3780–3792. [PubMed] [Google Scholar]
- 29.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 30.Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011 Mar 10;471(7337):235–239. doi: 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Mullighan CG, Harvey RC, Wu G, Chen X, Edmonson M, et al. Key pathways are frequently mutated in high-risk childhood acute lymphoblastic leukemia: a report from the Children's Oncology Group. Blood. 2011 Sep 15;118(11):3080–3087. doi: 10.1182/blood-2011-03-341412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 33.Zheng X, Beissert T, Kukoc-Zivojnov N, Puccetti E, Altschmied J, Strolz C, et al. Gamma-catenin contributes to leukemogenesis induced by AML-associated translocation products by increasing the self-renewal of very primitive progenitor cells. Blood. 2004 May 1;103(9):3535–3543. doi: 10.1182/blood-2003-09-3335. [DOI] [PubMed] [Google Scholar]
- 34.Gastaldi T, Bonvini P, Sartori F, Marrone A, Iolascon A, Rosolen A. Plakoglobin is differentially expressed in alveolar and embryonal rhabdomyosarcoma and is regulated by DNA methylation and histone acetylation. Carcinogenesis. 2006 Sep;27(9):1758–1767. doi: 10.1093/carcin/bgl008. [DOI] [PubMed] [Google Scholar]
- 35.Koch U, Wilson A, Cobas M, Kemler R, Macdonald HR, Radtke F. Simultaneous loss of beta- and gamma-catenin does not perturb hematopoiesis or lymphopoiesis. Blood. 2008 Jan 1;111(1):160–164. doi: 10.1182/blood-2007-07-099754. [DOI] [PubMed] [Google Scholar]
- 36.Marmorstein R. Structure and function of histone acetyltransferases. Cell Mol Life Sci. 2001 May;58(5–6):693–703. doi: 10.1007/PL00000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao TP, Oh SP, Fuchs M, Zhou ND, Ch'ng LE, Newsome D, et al. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998 May 1;93(3):361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka Y, Naruse I, Hongo T, Xu M, Nakahata T, Maekawa T, et al. Extensive brain hemorrhage and embryonic lethality in a mouse null mutant of CREB-binding protein. Mech Dev. 2000 Jul;95(1–2):133–145. doi: 10.1016/s0925-4773(00)00360-9. [DOI] [PubMed] [Google Scholar]
- 39.Kumar SR, Scehnet JS, Ley EJ, Singh J, Krasnoperov V, Liu R, et al. Preferential induction of EphB4 over EphB2 and its implication in colorectal cancer progression. Cancer Res. 2009 May 1;69(9):3736–3745. doi: 10.1158/0008-5472.CAN-08-3232. [DOI] [PubMed] [Google Scholar]
- 40.Kawasaki H, Taira K, Yokoyama KK. Functional analysis of the transcriptional coactivators p300 and CBP using ribozyme. Nucleic Acids Symp Ser. 1999;42:263–264. doi: 10.1093/nass/42.1.263. [DOI] [PubMed] [Google Scholar]
- 41.Xu W, Fukuyama T, Ney PA, Wang D, Rehg J, Boyd K, et al. Global transcriptional coactivators CREB-binding protein and p300 are highly essential collectively but not individually in peripheral B cells. Blood. 2006 Jun 1;107(11):4407–4416. doi: 10.1182/blood-2005-08-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramos YF, Hestand MS, Verlaan M, Krabbendam E, Ariyurek Y, van GM, et al. Genome-wide assessment of differential roles for p300 and CBP in transcription regulation. Nucleic Acids Res. 2010 Sep;38(16):5396–5408. doi: 10.1093/nar/gkq184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leung CG, Xu Y, Mularski B, Liu H, Gurbuxani S, Crispino JD. Requirements for survivin in terminal differentiation of erythroid cells and maintenance of hematopoietic stem and progenitor cells. J Exp Med. 2007 Jul 9;204(7):1603–1611. doi: 10.1084/jem.20062395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi-Yanaga F, Kahn M. Targeting Wnt signaling: can we safely eradicate cancer stem cells? Clin Cancer Res. 2010 Jun 15;16(12):3153–3162. doi: 10.1158/1078-0432.CCR-09-2943. [DOI] [PubMed] [Google Scholar]
- 45.Takebe N, Harris PJ, Warren RQ, Ivy SP. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat Rev Clin Oncol. 2011 Feb;8(2):97–106. doi: 10.1038/nrclinonc.2010.196. [DOI] [PubMed] [Google Scholar]
- 46.Verkaar F, van der SM, Blankesteijn WM, van der Doelen AA, Zaman GJ. Discovery of novel small molecule activators of beta-catenin signaling. PLoS One. 2011;6(4):e19185. doi: 10.1371/journal.pone.0019185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blough RI, Petrij F, Dauwerse JG, Milatovich-Cherry A, Weiss L, Saal HM, et al. Variation in microdeletions of the cyclic AMP-responsive element-binding protein gene at chromosome band 16p13.3 in the Rubinstein-Taybi syndrome. Am J Med Genet. 2000 Jan 3;90(1):29–34. doi: 10.1002/(sici)1096-8628(20000103)90:1<29::aid-ajmg6>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 48.Petrij F, Giles RH, Dauwerse HG, Saris JJ, Hennekam RC, Masuno M, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995 Jul 27;376(6538):348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 49.Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011 Sep;43(9):830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spencer VA, Sun JM, Li L, Davie JR. Chromatin immunoprecipitation: a tool for studying histone acetylation and transcription factor binding. Methods. 2003 Sep;31(1):67–75. doi: 10.1016/s1046-2023(03)00089-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.