Abstract
Heterozygous carriers of germ-line mutations in the BRCA2/FANCD1, PALB2/FANCN, and RAD51C/FANCO DNA repair genes have an increased life-time risk to develop breast, ovarian and other cancers; bi-allelic mutations in these genes clinically manifest as Fanconi anemia (FA). Here, we demonstrate that RAD51C is part of a novel protein complex that contains PALB2 and BRCA2. Further, the PALB2 WD40 domain can directly and independently bind RAD51C and BRCA2. To understand the role of these homologous recombination (HR) proteins in DNA repair, we functionally characterize effects of missense mutations of the PALB2 WD40 domain that have been reported in breast cancer patients. In contrast to large truncations of PALB2, which display a complete loss of interaction, the L939W, T1030I, and L1143P missense mutants/variants of PALB2 WD40 domain are associated with altered direct binding patterns to the RAD51C, RAD51 and BRCA2 HR proteins in biochemical assays. Further, the T1030I missense mutant is unstable, while the L939W and L1143P proteins are stable but partially disrupt the PALB2-RAD51C-BRCA2 complex in cells. Functionally, the L939W and L1143P mutants display a decreased capacity for DNA double-strand break-induced HR and an increased cellular sensitivity to ionizing radiation. As further evidence for the functional importance of the HR complex, RAD51C mutants that are associated with cancer susceptibility and FA also display decreased complex formation with PALB2. Together, our results suggest that three different cancer susceptibility and FA proteins function in a DNA repair pathway based upon the PALB2 WD40 domain binding to RAD51C and BRCA2.
Keywords: PALB2, RAD51C, BRCA2, homologous recombination, BRCA genes, DNA repair
INTRODUCTION
Inherited loss-of-function mutations in genes involved in the maintenance of genomic stability are associated with an increased cancer incidence in carriers of heterozygous mutations. BRCA1 and BRCA2, the two most prominent tumor suppressor genes linked to an inherited susceptibility to breast and ovarian cancer (reviewed in1–3), encode proteins which function in the homologous recombination (HR) DNA repair pathway4,5. BRCA2 regulates oligomerization of the RAD51 recombinase which is necessary for it to form a nucleoprotein filament with single-strand DNA6,7.
PALB2 and RAD51C are other DNA repair proteins linked to breast and/or ovarian cancer8–12. PALB2, the partner-and-localizer of BRCA2, interacts with both BRCA1 and BRCA2 through its N-terminal coiled-coil and C-terminal WD40 domains, respectively12–16. Notably, the recruitment of BRCA2 to sites of DNA damage, and its function in HR, critically depend on the presence of a normal PALB2 protein10,12,15,17.
WD40 domains are ring-like β-propeller structures with seven blades that mediate protein-protein interactions18. Truncation or frameshift mutants of PALB2 which perturb all, or a portion, of the WD40 domain abrogate the association of PALB2 with BRCA2, the assembly of RAD51 foci, and cellular resistance to DNA crosslinking agents such as mitomycin C12,13,15. The pathogenicity of these truncation mutants has generally been attributed to disruption of the interaction of PALB2 with BRCA2, but RAD51 also interacts with the WD40 domain of PALB219,20. Although missense mutants/variants of the PALB2 WD40 domain have also been identified in breast cancer patients21–31, their effect on the function of the PALB2 protein in DNA damage responses, and therefore their pathogeneticity, is still largely unknown.
RAD51C is one of five RAD51 paralogs which have been identified in somatic mammalian cells, along with RAD51B, RAD51D, XRCC2, and XRCC3. These proteins are of a similar size as RAD51, but have an overall low homology to RAD51 (20–30%)32. Like RAD51, each of the paralogs is required for DNA repair by HR33, but their specific roles in HR are not well characterized and seem to be distinct from the function of RAD51.
The RAD51 paralogs form at least two distinct protein complexes in cells: a RAD51B-RAD51C-RAD51D-XRCC2 complex and a RAD51C-XRCC3 complex. RAD51C is the only paralog present in both34. Aside of an apparent interaction with RAD1835, other RAD51C interactors have not been identified.
Here, we demonstrate that RAD51C has a direct protein-protein interaction with PALB2. In addition to their roles in breast/ovarian cancer, FANCD1/BRCA2, FANCN/PALB2, and FANCO/RAD51C are among the 16 currently identified Fanconi anemia (FA) genes36,37. The functional relationship of RAD51C to other FA proteins, including PALB2, was previously unknown. Our results suggest that the PALB2 WD40 domain may scaffold the RAD51C, RAD51, and BRCA2 HR proteins into a complex. Selected missense mutations of the PALB2 WD40 domain, found in breast cancer patients21, perturb this complex, but to a lesser degree than truncation of the WD40 domain. In addition, RAD51C mutants originally identified in individuals with breast/ovarian cancer or FA8,38 also diminish the complex of HR proteins. Together, our results suggest that PALB2, RAD51C, and BRCA2 directly cooperate in a network of proteins which mediate homologous recombination, and which may thereby maintain genomic stability. Interestingly, disruption of this network is genetically-linked to three distinct diseases: breast cancer, ovarian cancer, and Fanconi anemia.
RESULTS
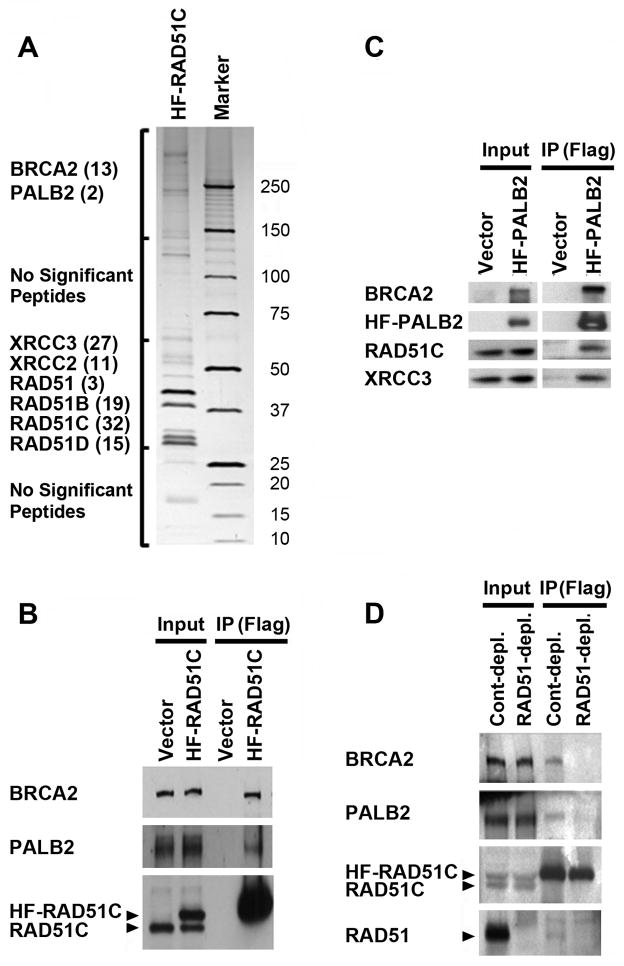
To better understand the function of RAD51C in DNA repair and the maintenance of genomic stability, we utilized mass spectrometry to identify proteins that co-purified with His6-FLAG-RAD51C (HF-RAD51C) from untreated cells (Figure 1A). As expected, peptides for each of the RAD51 paralogs and for RAD51 were detected by mass spectrometry. PALB2 and BRCA2 were the only other proteins detected which were not present in mock protein purifications. Further, these same proteins were also identified following treatment with MMC (data not shown). Interactions with PALB2 and BRCA2 were verified by co-immunoprecipitation with epitope-tagged RAD51C (Figure 1B). As confirmation, we also demonstrated the reciprocal co-immunoprecipitation of RAD51C with PALB2 (Figure 1C).
Figure 1. RAD51C interacts with PALB2 and BRCA2.
(A) Silver stained gel indicating components of a RAD51C complex identified by mass spectrometry following immunopurification from HeLa S3 cells. RAD51C was expressed with a N-terminal His6-Flag (HF) epitope tag. The gel was cut into four sections as indicated (left) and mass spectrometry was performed on each piece. The number of unique peptides identified, following subtraction of common peptides isolated from a mock-purified control, is shown in parentheses. (B) Immunoblot demonstrating that PALB2 and BRCA2 co-immunoprecipitate with HF-RAD51C. (C) Immunoblot demonstrating that RAD51C and XRCC3 co-immunoprecipitate with HF-PALB2. (B–C) Immunoprecipitates were prepared using M2 anti-Flag beads and extracts from HeLa (B) and EUFA1341 cells complemented with HF-PALB2 (C). (D) PALB2, BRCA2, RAD51C and RAD51 form a protein complex, as determined by depletion of a complex that could be immunoprecipitated with HF-RAD51C after prior immunodepletion of RAD51. HeLa cells were first incubated with anti-RAD51 or normal rabbit (Cont.) antisera. Supernatants were then incubated with anti-M2 agarose to immunoprecipitate HF-RAD51C and were immunoblotted with the indicated antibodies. (B–D) Immunoprecipitates represent 200-fold the levels loaded for Input lanes. The positions of HF-RAD51C and endogenous RAD51C, or RAD51, are indicated by arrowheads.
As evidence that PALB2, BRCA2, RAD51C and RAD51 form a protein complex together, we first immunodepleted RAD51 from extracts of HeLa cells which expressed HF-RAD51C (Figure 1D). While immunodepletion of RAD51 did not have a noticeable effect on the levels of BRCA2, PALB2 or RAD51C that remained in the extract, less PALB2, BRCA2 and RAD51 subsequently co-immunoprecipitated with HF-RAD51C from these extracts. These results suggest that RAD51 and RAD51C form a complex together with PALB2 and BRCA2.
While both RAD51 and RAD51C are minor components of the PALB2 complex(es), BRCA2 is a more abundant component (Supplemental Figure 1). Another RAD51 paralog, XRCC3, is also a minor component of the PALB2 complex (Supplemental Figure 1).
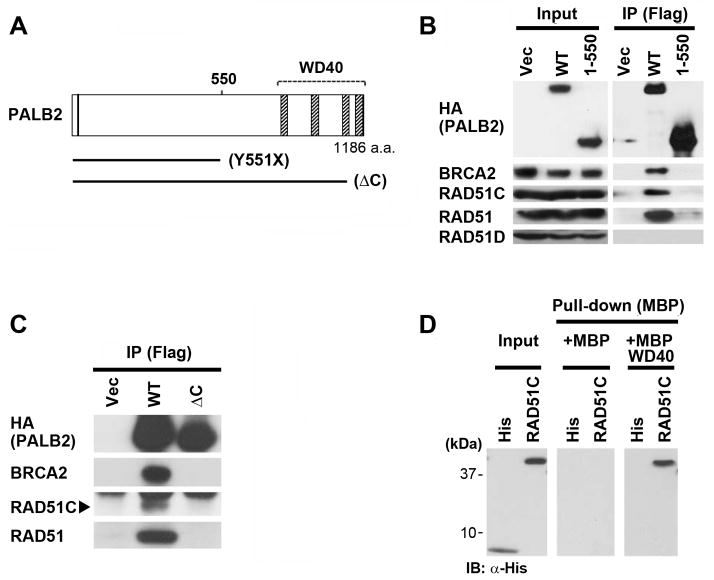
To better define the interaction of RAD51C with PALB2, we compared immunoprecipitates of full-length PALB2 to a version truncated in the middle of the protein (Y551X) (diagramed in Figure 2A). This mutant was first identified in EUFA1341 cells from a FA patient13. RAD51C, as well as BRCA2 and RAD51, co-immunoprecipitated with full-length Flag-HA-PALB2 from EUFA1341 cells (Figure 2B). Strikingly, however, neither RAD51C nor BRCA2 interacted with Flag-HA-PALB2-Y551X. This truncation mutant of PALB2 was also largely deficient for interaction with RAD51.
Figure 2. RAD51C directly binds to the WD40 domain of PALB2.
(A) Schematic diagram of PALB2 and selected mutants. Hatched boxes indicate WD40 repeats of PALB2. (B) Wild-type PALB2 and the Y551X truncation mutant containing N-terminal Flag and HA epitope tags were transiently transfected into 293T cells. Immunoprecipitates represent 500-fold the levels loaded for Input lanes. (C) Cell lysates were prepared from EUFA1341 cells reconstituted with wild-type PALB2 or the ΔC mutant truncated after P1097. The position of RAD51C is indicated by an arrowhead. (B–C) Cell lysates were immmunoprecipitated with α-Flag beads, followed by immunoblotting with the indicated antibodies. (D) Analysis of direct binding using bacterially-expressed maltose-binding protein (MBP) alone or MBP fused to the WD40 domain of PALB2 (amino acids 859–1186). Bacterially-expressed His6 alone, or fused to RAD51C, was incubated with MBP or MBP-PALB2 immobilized on maltose beads. Bound RAD51C was detected by immunoblotting with anti-His antibodies following elution.
RAD51D is a component of the RAD51B-RAD51C-RAD51D-XRCC2 complex of RAD51 paralogs34. RAD51D did not associate with the PALB2-BRCA2-RAD51C-RAD51 complex described above (Figure 2B). This demonstrates that these two protein complexes can be clearly distinguished.
The most prominent structural feature of the C-terminal half of PALB2 is a WD40 domain from amino acids 853–1186. We utilized PALB2ΔC (diagramed in Figure 2A), which we have described previously15, to determine whether RAD51C specifically interacts with the WD40 domain of PALB2. PALB2ΔC is truncated following amino acid P1097 and therefore lacks blades 5–7 of the PALB2 WD40 structure39. PALB2ΔC was completely deficient for interaction with RAD51C, RAD51, and BRCA2 (Figure 2C), suggesting that RAD51C and these other proteins interact with the WD40 domain of PALB2.
Next, we expressed and isolated fusion proteins containing RAD51C or the PALB2 WD40 domain from bacteria. Using this system, we demonstrate that RAD51C bound directly to the WD40 domain of PALB2 (Figure 2D).
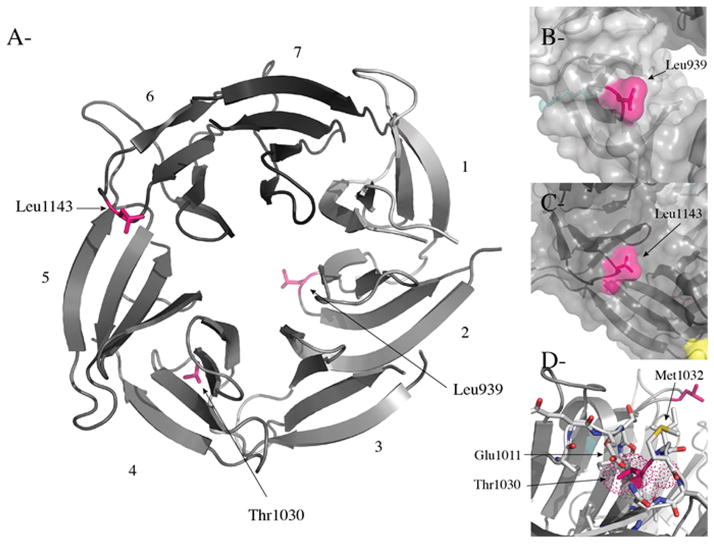
Truncation of the PALB2 WD40 domain completely abrogates interactions with BRCA2, RAD51C and RAD51 (Fig. 2C). Thus, as an alternative that might yield more insight into the interactions of BRCA2, RAD51C, and RAD51 with PALB2, we analyzed the effects of missense mutants/variants of PALB2 identified in a German population of breast cancer patients21. While missense mutants have been identified in other studies21–31, our study was not intended to be exhaustive but instead focused on selected mutants/variants of the PALB2 WD40 domain which were predicted to potentially be disease-causing. While the T1030I and L1143P mutants were only found in cancer patients, the L939W variant was found at a lower frequency in controls than in breast cancer families21 and has also been linked to breast cancer in other studies22,24,28. First, we examined the positions of the three different missense mutations in the crystal structure of the PALB2 WD40 domain39. L939W, T1030I, and L1143P are present within blade 2, between blades 4 and 5, and between blades 5 and 6, respectively, and therefore potentially identify three independent binding surfaces on the PALB2 WD40 β-propeller structure (Figure 3A). This result is consistent with a potential role for the PALB2 WD40 domain in scaffolding a complex of HR and FA proteins, including RAD51C.
Figure 3. Analysis of the positions and potential effects of breast cancer mutations in the PALB2 WD40 domain.
(A) A ribbon diagram of the WD40 domain of PALB2 shown in grey (PDB ID 3EU739). The blades are numbered 1 to 7 and colored light grey to dark grey. The missense mutations/variants (L939, T1030 and L1143) found in breast cancer patients are shown in stick representation and in pink. (B–C) Surface representation of the region of PALB2 centered on Leu939 and Leu1143, respectively, colored in pink showing how both hydrophobic residues are solvent exposed. (D) Details of residues surrounding T1030 of blade 4. Residues E1011 and M1032, which make hydrogen bonds with the hydroxyl group of T1030, are highlighted. The figure was prepared with Pymol (http://www.pymol.org/).
We also modeled each mutation/variant based upon the structure of the PALB2 WD40 domain (Figure 3B–D). The L939 and L1143 residues are present on the surface of the WD40 domain structure and are therefore likely involved in protein-protein interactions. L939W, in particular, represents a change to a more bulky residue that may thereby perturb potential interactions. In contrast, the T1030I mutation likely disrupts hydrogen bonds between the hydroxyl group of threonine 1030 and the carboxylic and amide groups, respectively, of Glu1011 and Met1032. This mutation likely perturbs the pocket formed by amino acids Pro1009, Glu1011, Ile1013, Asn1034 and Lys1048 and is therefore predicted to be unstable.
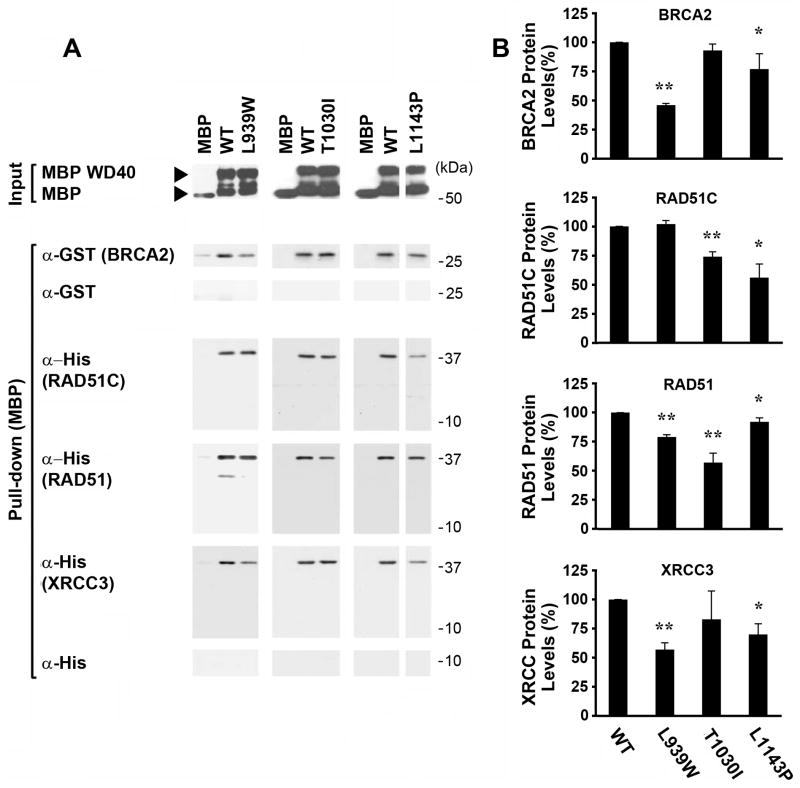
Next, we tested the effects of the L939W, T1030I and L1143P PALB2 alterations on in vitro interactions with various HR-associated proteins (Figure 4). Examples of a representative experiment, performed with proteins isolated following expression in bacteria, are shown in Figure 4A. Although none of the PALB2 mutations completely abrogated the interaction of its WD40 element with the HR proteins, the L939W variant showed a stronger disruption of direct binding to BRCA2 and the L1143P mutant protein had a weaker effect on the interaction with BRCA2. Both differences were reproducible and statistically significant in three independent experiments (Figure 4B). The T1030I mutant did not show any decrease in the interaction with BRCA2.
Figure 4. Different patterns of in vitro binding of RAD51C, BRCA2, RAD51, and XRCC3 to disease-associated missense mutants of PALB2 suggest that the PALB2 WD40 domain may scaffold these proteins into a complex.
(A) A representative in vitro binding experiment. MBP alone, or MBP-fused to the wild-type PALB2 WD40 domain or to different breast cancer-associated mutants/variants of this domain, were expressed in bacteria, purified, and immobilized on maltose beads. Isolated MBP-WD40 proteins or MBP alone are shown as “Inputs”. A GST-tagged BRCA2 fragment (amino acids 1–75), or His-tagged RAD51C, RAD51, or XRCC3, were purified from bacteria and incubated with the purified MBP fusion proteins. Proteins present in the MBP fusion protein pull-down were detected with anti-GST or Anti-His antibodies, as appropriate. GST alone served as a negative control for GST-BRCA2, while His alone served as a negative control for His-fusion to RAD51C, RAD51, or XRCC3. (B) A graph showing quantification of binding from three independent experiments. Values for each HR protein were determined by densitometry and adjusted relative to the levels of each form of PALB2 in the Input lanes and are shown normalized to the amounts that immunoprecipitated with the wild-type PALB2 WD40 domain. The mean and standard deviation are shown for each value (* = P<0.05; ** = P<0.01).
In contrast, RAD51C and RAD51 displayed patterns of interaction with each PALB2 mutant that were distinct from BRCA2 and which were distinct from each other (Figure 4). The L939W mutant had no effect on direct binding of the PALB2 WD40 domain by RAD51C, but the T1030I variant, and especially the L1143P PALB2 variant, were associated with a statistically significant decrease in the interaction with RAD51C. In contrast, RAD51 showed significantly less binding to the L939W mutant and especially the T1030I mutant, in addition to a more minor decrease in binding to the L1143P mutant.
The RAD51 paralog XRCC3 also directly bound to the PALB2 WD40 domain and had a pattern of binding to the mutants that resembled that of BRCA2, but which was clearly distinct from that of RAD51C (Figure 4). This suggests that RAD51C and XRCC3 independently interact with PALB2.
Together, results with missense mutants of PALB2, shown in Figures 3–4, appear to identify three distinct surfaces on the PALB2 WD40 β-propeller structure that are involved in binding RAD51C, RAD51, and BRCA2 and XRCC3. Importantly, the distinct patterns of binding of RAD51C, RAD51, and BRCA2 to these PALB2 missense mutants also suggests that each HR protein may directly bind to the PALB2 WD40 domain in an independent manner. The above results support the possibility that the PALB2 WD40 domain scaffolds a complex of HR proteins.
Given that RAD51C and XRCC3 can form a complex together34, we tested whether the presence of PALB2 influences the association of these proteins (Supplemental Figure 2). Co-immunoprecipitation of RAD51C with XRCC3 was clearly increased in cells with a deficiency for PALB2. This suggests that formation of the PALB2 complex with RAD51C and/or XRCC3 may compete with the RAD51C-XRCC3 complex.
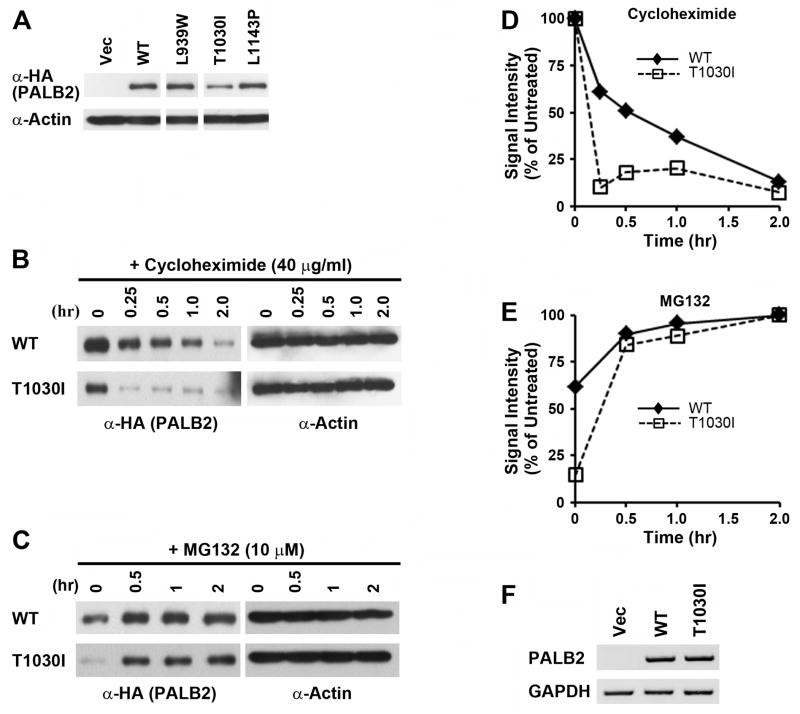
To test their function in DNA repair, we then expressed the three breast cancer-associated mutants along with N-terminal Flag-HA epitope tags in PALB2-deficient EUFA1341 cells derived from a FA patient13. While the L939W and L1143P mutants expressed at similar levels as the wild-type protein, the T1030I mutant was present at clearly decreased levels in multiple experiments (Figure 5A). We hypothesized that the T1030I mutant was unstable, as predicted in Figure 3D. To further test this possibility, we treated cells containing wild-type PALB2 or the T1030I mutant with cycloheximide to inhibit new protein synthesis (Figure 5B). The turnover of the mutant was more rapid than that of the wild-type protein. Further, treatment with MG132 more modestly increased the levels of wild-type PALB2 but restored the T1030I mutant to near the levels of wild-type protein (Figure 5C). This suggests that the T1030I mutant of PALB2 was degraded in a proteasome-dependent manner. Quantification of these results is shown in Figures 5D and 5E. The levels of mRNA for each form of PALB2 were similar, however (Figure 5F). Together, our results demonstrate that one way in which missense mutations of the PALB2 WD40 domain may act in breast cancer is by decreasing the stability of the encoded protein.
Figure 5. The breast cancer-associated T1030I mutant of PALB2 is unstable.
(A) Wild-type PALB2 or the L939W, T1030I, and L1143P mutants/variants were retrovirally expressed in EUFA1341 cells along with a N-terminal Flag-HA epitope tag. The level of each protein was determined by immunoblotting with α-HA antibodies. Actin was used as a loading control. (B) To examine relative rates of turnover, new protein synthesis was inhibited in EUFA1341 cells reconstituted with Flag-HA tagged wild-type PALB2 or with the T1030I mutant by treating with 40 μg/ml cycloheximide. Cell lysates were prepared at the indicated time points. (C) EUFA1341 cells reconstituted with wild-type PALB2 or the T1030I mutant were treated with 10 μM MG132 and harvested at the indicated time points. (D–E) Quantification of the levels of wild-type PALB2 or the T1030I mutant following treatment with cycloheximide (D) or MG132 (E) at various time points was determined by densitometry of immunoblots. Results for each protein were normalized to the values at 0 hr (D) or 2 hr (E) of treatment. (F) Levels of mRNA of wild-type or mutant PALB2 in reconstituted EUFA1341 cells were analyzed by RT-PCR. Primers that detected Flag-HA-tagged PALB2 but not endogenous PALB2 were utilized. GAPDH was used as a control for the levels of an unrelated endogenous protein.
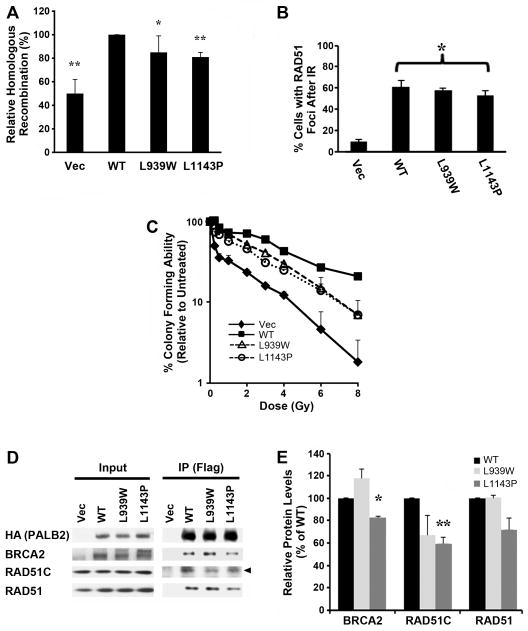
Unlike PALB2-T1030I protein, the L939W and L1143P mutants appeared to be stably expressed in EUFA1341 cells (Figure 5A). To measure the effects of the L939W and L1143P mutants/variants of PALB2 on DNA double-strand break (DSB)-initiated HR, we expressed each form in U2OS-DR cells that contained a GFP reporter for HR40 and depleted endogenous PALB2 with a siRNA directed against its 3′-UTR. Each mutant displayed modest but statistically significant decreases in DSB-initiated HR that were intermediate to those of cells which contained the empty vector alone and were depleted of endogenous PALB2 (Figure 6A).
Figure 6. The L939W and L1143P mutants of the PALB2 WD40 domain are associated with defective DSB-initiated HR and with increased sensitivity to ionizing radiation.
(A) Gene conversion assays were performed in U2OS-DR cells stably expressing wild-type PALB2, or the L939W or L1143P mutants, and depleted of endogenous PALB2 using a siRNA directed against its 3′-UTR. GFP-positive cells were quantified by flow cytometry. Each value represents the average of three independent experiments performed in duplicate. The data shown are the mean and standard deviation of three independent replicates (* = P < 0.05; ** = P < 01). (B) The percentage of cells with RAD51 foci at 16 hr after exposure to 10 Gy IR. U2OS-DR cells were stably reconstituted with the different forms of PALB2, as indicated, and endogenous PALB2 was depleted as described above. Each value represents the mean and standard deviation of 3 counts of 150 or more cells each with 3 or more RAD51 foci (* = P < 0.05). (C) IR colony assay performed in EUFA1341 cells reconstituted with wild-type PALB2, the L939W or L1143P mutants, or with vector alone. Cells were treated with the indicated doses of IR. Differences for EUFA1341 cells reconstituted with the vector or with either mutant, as compared to cells corrected with WT PALB2, were statistically significant (P < 0.05) at doses of IR ranging from 1–8 Gy. (D–E) Wild-type PALB2, the L939W or L1143P mutants, or the vector alone, were stably expressed in EUFA1341 cells along with a N-terminal Flag-HA epitope tag. Levels of associated HR proteins in extracts (Input), or in immunoprecipitates (IP) prepared using α-Flag M2 agarose beads, are shown in (D). These results are representative of two independent experiments. The position of RAD51C is indicated by an arrowhead. Immunoprecipitates represent 200-fold the levels loaded for Input lanes. Quantification of the levels of BRCA2, RAD51C or RAD51 which immunoprecipitated with the WT, L939W or L1143P forms of PALB2 in two independent experiments (E). Levels of immunoprecipitating protein were normalized to the input for each form of PALB2 and each interacting protein. The average +/− S.D. are shown relative to the values for PALB2-WT (which was set to 100%). Statistical significance: * = P < 0.05; ** = P < 0.01.
We also tested whether the L939W and L1143P mutants of PALB2 had any effect on RAD51 foci. We depleted endogenous PALB2 in U2OS-DR cells that expressed PALB2-WT or either mutant, as described above. The L1143P mutant displayed a modest but statistically significant decrease in the assembly of RAD51 foci (Figure 6B), consistent with the modest but significant decrease in DSB-HR that is associated with this mutant.
As an additional measure of the importance of altered interactions between the PALB2, BRCA2, and RAD51C proteins, we tested resistance of EUFA1341 cells reconstituted with the stable L939W and L1143P mutants/variants of PALB2 to ionizing radiation (IR) (Figure 6C). A role for PALB2 in promoting resistance to IR has not been reported previously. Interestingly, the L939W and L1143P PALB2 mutants showed a moderate, but significant increase in sensitivity to IR when compared to those which were isogenically-corrected with wild-type PALB2.
Consistent with our findings of compromised DNA repair, the L939W and L1143P mutants of PALB2 showed altered binding to HR proteins in EUFA1341 cells (Figure 6D). Quantification is shown in Figure 6E. The interactions of BRCA2 and RAD51C with PALB2-L1143P were statistically lower than those with PALB2-WT. Although not statistically different from binding to PALB2-WT, PALB2-L939W showed trends of increased binding to BRCA2 and decreased binding to RAD51C.
Finally, we also tested RAD51C mutants for perturbation of the complex with PALB2 and BRCA2. The L138F and D159N mutants were identified in breast/ovarian cancer patients, while the R258H mutant was identified in a FA patient. Each mutant was previously demonstrated to be functionally impaired in DNA repair-related assays8,38,41. As shown for a representative experiment, L138F, D159N and R258H all displayed decreased binding to BRCA2, and to a lesser degree to PALB2 and RAD51, when expressed transiently in 293T cells (Fig. 7). Thus, taken together with Figures 4 and 6, mutations of either PALB2 or RAD51C, found in breast/ovarian cancer or FA, perturb the function of a complex of HR proteins that includes PALB2, RAD51C, and BRCA2.
Figure 7. Disease-associated mutants of RAD51C have decreased interactions with BRCA2 and PALB2.
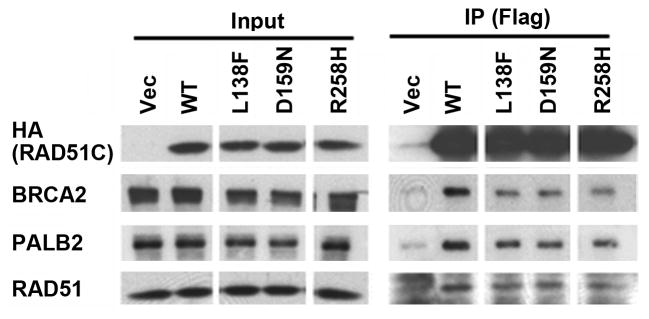
(A) Wild-type RAD51C, the L138F and D159N mutants found in breast cancer patients, or the R258H mutant found in a Fanconi anemia patient, were transiently expressed in 293T cells along with a N-terminal Flag-HA epitope tag. The levels of protein in extracts (Input), or in immunoprecipitates prepared using α-Flag M2 agarose beads (IP), were determined by immunoblotting with the indicated antibodies. Immunoprecipitates represent 500-fold the levels loaded for Input lanes.
DISCUSSION
Here, we demonstrate that the RAD51C protein interacts with, and can directly bind, PALB2. We term this complex, which also includes RAD51, BRCA2, and XRCC3, the “HR complex”. Our results suggest that these proteins interact and function in a common pathway of DNA repair by HR. This is particularly interesting given that PALB2, BRCA2 and RAD51C are all breast/ovarian cancer susceptibility (BRCA) genes8–12,42, as well as FA genes13,38,43,44. Notably, FA is associated with a predisposition to cancer36. Thus, together, our finding that patient-derived mutations of either RAD51C or the PALB2 WD40 domain disrupt the HR complex suggests the importance of this complex in preventing cancer.
The PALB2 WD40 domain coordinates a protein complex that contains both RAD51C and BRCA2
While the role of PALB2 in recruiting BRCA2 to sites of DNA damage has already been established13,15,17, RAD51C was only known to associate with other RAD51 paralogs, in two different complexes34, and with RAD1835. Thus, we demonstrate a novel protein complex that yields insight into the function of PALB2, RAD51C, and BRCA2 as tumor suppressors and FA proteins.
By showing that RAD51C and BRCA2, as well as RAD51, all directly bind to the WD40 domain of PALB2, we propose that a key function of PALB2 is to individually bind multiple proteins which have an essential role in DSB-initiated HR4,33,45. The role of the PALB2 WD40 domain in binding these HR proteins is supported by the finding that three different breast cancer-associated missense mutants/variants of PALB2, L939W, T1030I and L1143P, have distinct patterns of altered binding to BRCA2, RAD51C, and RAD51 in vitro. In fact, the WD40 domain could scaffold a complex that contains these HR proteins. Consistent with this possibility, previous work has shown that WD40 domains can coordinate simultaneous interactions with multiple proteins46.
We find that the PALB2-L1143P mutant is associated with a moderate but significant decrease in RAD51 foci after exposure to IR. In contrast, this mutant results in a modest but significant increase in the formation of BRCA2 foci (Supplemental Figure 3). It is possible that by binding to PALB2, RAD51C normally leads to a concomitant release of BRCA2 and recruitment of RAD51. This might explain the increased levels of BRCA2 foci and decreased RAD51 foci observed in cells reconstituted with the PALB2-L1143P mutant. This process could involve the DNA binding activity of RAD51C47 and/or a change in the composition of the complex with PALB2. The PALB2-L1143P mutant could have indirect effects on RAD51, since it had altered interactions with RAD51C and BRCA2 but not RAD51 both in vitro (Figure 4) and in cells (Figure 6). We expect that decreased recruitment of RAD51, despite increased levels of BRCA2 foci, underlies the significant decreases in HR and resistance to IR that are associated with the PALB2-L1143P mutant.
Contrasting reports have suggested that RAD51C and XRCC3 function either before or after RAD51 foci formation33,38,48,49 in various cell types, including human cells, following exposure to DNA damage. While the PALB2-L1143P mutant would appear to have some defect before RAD51 foci formation, this does not preclude an additional function afterwards. The interaction of RAD51C with PALB2 could have a role in promoting RAD51 foci formation, since the PALB2-L1143P mutant includes a defect in this interaction.
As added support for the effects of missense mutations of the PALB2 WD40 domain on DNA repair, the L939W variant but not L1143P is associated with an intermediate sensitivity to a PARP inhibitor (Supplemental Figure 4). Since both mutants are associated with increased sensitivity to IR, this coud reflect different patterns of interactions of each mutant with HR proteins (Figure 4).
RAD51 is unlikely to be required for the interactions of BRCA2 or RAD51C with the WD40 domain of PALB2. First, both BRCA2 and RAD51C directly, and thus potentially independently, bind the PALB2 WD40 domain (Figure 4). Second, siRNA-mediated depletion of RAD51 decreased the levels of RAD51, but not those of PALB2 or BRCA2, which immunoprecipitated with RAD51C (Supplemental Figure 5).
While the PALB2-BRCA2-RAD51C-RAD51 complex appears to be independent of the previously described RAD51B-C-D-XRCC2 complex34, another RAD51 paralog, XRCC3, appears to interact with this PALB2 complex. This is the third reported complex of RAD51 paralogs, but the first that also includes non-paralog proteins. Given that the L939W and L1143P mutants of PALB2 have altered interactions with RAD51C, XRCC3, BRCA2 and RAD51, and are defective for DNA repair, these RAD51 paralogs would appear to have a function in this PALB2 complex. While RAD51C-XRCC3 has been reported to have a Holliday junction resolvase activity in vitro50, it is currently unclear what function, if any, this complex by itself has in intact cells.
A protein complex has been reported that includes FANCG, FANCD2, BRCA2 and XRCC351. Given that we describe a PALB2 complex that contains BRCA2 and XRCC3, these complexes may be related. Future work will be required to examine this potential relationship.
Comparison of the effects of truncation mutants of the PALB2 WD40 domain to those of missense mutants
Mutants of PALB2 found in breast cancer and FA patients that result in a nonfunctional truncated protein, due to partial removal or absence of the WD40 domain, are completely deficient for DSB-initiated HR10,12,13. These truncations abrogate binding to BRCA2 and we report here that such large-scale truncations also eliminate binding to RAD51C and RAD51 (Fig. 2C), and thereby fully disrupt the HR complex.
More recently, missense mutations of PALB2 have been identified in breast/ovarian cancer patients and are also present in FA patients10,11,13,21–31. Because these mutants are not clearly pathogenic, it is therefore more difficult to predict any clinical outcome, especially in heterozygous carriers. Here, we have performed the first functional characterization of such PALB2 missense mutants/variants. In contrast to truncating mutants, selected missense mutants of the WD40 domain are found associated with partial but incomplete disruption of the HR complex. Correspondingly, DSB-initiated HR is only partially compromised.
The breast cancer-associated L939W and L1143P mutants/variants of PALB2 are linked to modest but significantly decreased DSB-initiated HR efficiencies and increased IR sensitivities (Figure 6). We therefore predict that missense mutants of PALB2 may confer lower levels of genomic instability, and thus a lower risk of cancer, than truncation mutants. Notably, our repair-related assays indicate that the L939W variant identified in various studies21,22,24,28, generally more frequently in breast cancer patients than healthy individuals, may be pathogenic. Importantly, our study establishes assays for the functional characterization of other missense mutants/variants of PALB2 in the future.
Results obtained with the L939W, T1030I and L1143P mutants/variants of PALB2 suggest that there are at least two different mechanisms by which missense mutants of the PALB2 WD40 domain may lead to increased genomic instability in breast cancer. First, we find that the T1030I mutant is unstable when expressed in human cells. Decreased levels of PALB2-T1030I due to instability are therefore expected to diminish its function in HR. A second means by which mutants of the PALB2 WD40 domain may lead to increased genomic instability is by altered binding to HR proteins, as illustrated by the L939W and L1143P mutants/variants. The L1143P mutant significantly decreased binding to RAD51C and BRCA2, both in vitro and in human cells.
PALB2, BRCA2, and RAD51C are linked to breast/ovarian cancer and to Fanconi anemia
FA is an inherited chromosome instability syndrome clinically characterized by bone marrow failure, congenital anomalies, and a predisposition to leukemia and various solid tumors36. Eight of the sixteen identified FA proteins (FANCA/B/C/E/F/G/L/M) form a core complex required for the monoubiquitination of FANCD2 and FANCI36,37. Other FA proteins, including PALB2, BRCA2, and RAD51C, are not required for the monoubiquitination of FANCD2 or FANCI, and are therefore believed to function downstream of FANCD213,38,43,44. Interestingly, BRCA252, RAD51C33, and as we show here, PALB2, are required for resistance to IR, unlike upstream FA proteins that lead to the monoubiquitination of FANCD2 and FANCI. Further, PALB2, BRCA2, and RAD51C are associated with inherited breast cancer, while upstream FA pathway proteins are not53. Thus, a more prominent role in DSB repair and in cellular resistance to IR may underlie the shared association of the PALB2, RAD51C, and BRCA2 subset of FA genes with breast cancer. Consistent with this possibility, the products of other breast cancer susceptibility genes, including BRCA1, BRIP1, ATM, CHEK2, NBS1 and RAD502, are also involved in mediating the repair of DSBs induced by IR.
In conclusion, we have identified a novel complex of HR and BRCA proteins. Importantly, our results yield insight into the function of PALB2 as a tumor suppressor by demonstrating that its WD40 domain may act to coordinate this complex, which contains two other tumor suppressor proteins: BRCA2 and RAD51C. Finally, the function of this complex in DNA repair may be the basis for the common association of the products of the PALB2, RAD51C and BRCA2 genes with three distinct diseases: breast cancer, ovarian cancer, and FA.
MATERIALS AND METHODS
Cell culture
HeLa S3, 293T and U2OS-DR cell lines, and EUFA1341 cells reconstituted with different forms of PALB2 were cultured and irradiated as described previously15. Stock solutions of cycloheximide (10 mg/ml in H2O; Sigma), MG132 (10 mM in DMSO; Calbiochem), and Olaparib (5 mg/ml in DMSO; Selleck Chemicals) were kept at −20°C.
Antibodies
Mouse anti-RAD51C (2H11) and rabbit anti-XRCC3 antibodies were purchased from Novus. Rabbit anti-RAD51 (H92) and mouse anti-His (H8) antibodies were purchased from Santa Cruz. Mouse anti-RAD51D (5B3) antibody was obtained from Chemicon. Anti-HA, anti-MBP, anti-GST, anti-β actin, and anti-PALB2 antibodies15, and anti-BRCA2 antibodies54, were described previously.
Immunofluorescence microscopy
Cells were fixed and processed with antibodies as described previously15.
Cloning and mutagenesis
Human RAD51C cDNA was purchased from Open Biosystems. RAD51C with N-terminal Flag-HA or His-Flag epitope tags were generated and subcloned into pcDNA3.1. Alternatively, RAD51C was cloned into the pMIEG3 retroviral vector with a N-terminal His6-Flag epitope tag55. PALB2 mutants (L939W, T1030I, and L1143P) were generated as described previously15. To generate fusion proteins for in vitro studies, the PALB2 WD40 domain (amino acids 859–1186), RAD51 or RAD51C were cloned into pMAL-c2X (NEB) with N-terminal maltose-binding protein (MBP). Alternatively, RAD51C was cloned into pET28b (Novagen) with C-terminal Histidine (His). An N-terminal fragment of BRCA2 (1–75 a.a) was cloned into pGEX4T-1, which contained N-terminal Glutatione S-transferase (GST).
Expression and purification of recombinant proteins
Fusion proteins were expressed in bacteria (4 h at 30°C with 0.3 mM isopropyl β-thiogalactopyranoside). Recombinant proteins were purified using amylose resins (NEB) for MBP, glutathione sepharose 4B beads (GE healthcare) for GST, and Talon metal affinity resins (Clontech) for His, according to the manufacturers’ instructions.
In vitro GST and MBP pull-downs
GST and MBP-tagged proteins (5 μg) were bound to glutathione beads and amylose resins, respectively, and mixed with 5 μg of each purified target protein in binding buffer (10 mM Tris pH 7.4, 75 mM NaCl, 0.5 mM EDTA, 0.025% Tween-20, 1 mM DTT, and 0.5 mg/ml BSA) including 0.5 mM PMSF and 1X protease inhibitor cocktail, and incubated overnight at 4°C. Beads were then washed and bound proteins eluted.
Transfection and viral transduction
Retroviral transduction with pMIEG3-RAD51C, or pMMP-PALB2 wild-type and its mutants with a N-terminal Flag-HA tag, was performed as described previously15,55. Transduced cells were selected with 0.5 μg/ml puromycin for 6 days after the last infection.
Identification of RAD51C-interacting proteins by mass spectrometry
RAD51C complexes were purified from HeLaS3 cells retrovirally transduced with His6-FLAG tagged RAD51C as described previously56. The immunopurified complex was analyzed by mass spectrometry.
Immunoprecipitation
Cell pellets were lysed in 420 NETN buffer as described previously15. Immunoprecipitations were performed with anti-Flag M2 Affinity Gel (Sigma) or specified antibodies as described previously15.
RT-PCR analysis
Total RNA was extracted using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. Synthesis of cDNAs from RNA with the Advantage RT for PCR Kit (Clontech) was by PCR-amplification using the following primers (Forward: 5′-gtgatgctgtactgttgtcttcct-3′; Reverse: 5′-cgacttgtcatcgtcgtccttg-3′).
Assay of DNA DSB-initiated HR
U2OS-DR cells with an integrated GFP reporter and containing PALB2 or its mutants were assayed as described previously15.
IR colony and PARP inhibitor sensitivity assays
EUFA1341 cells reconstituted with wild-type PALB2 or its mutants were seeded onto 6 cm dishes (2×103 cells/dish) in triplicate, and after allowing attachment cells were irradiated. Cells were incubated for 10–12 days to form colonies and washed with PBS. Colonies were fixed with Wright-Giemsa reagent (EMD), stained with Buffer solution Giordano (Fisher), dried, and counted manually.
To measure Olaparib sensitivity, cells were seeded into 96 well plates at 1 × 106 cells/well and were treated with a range of concentrations of olaparib beginning 4 hr later. Cells were incubated 4 days and relative survival was measured using a colorimetric assay15.
Supplementary Material
Acknowledgments
We are grateful to Dr. Hans Joenje (Vrije Universiteit Medical Center), Dr. Youngho Kim (Wonkwang University), Dr. James Lessard (Cincinnati Children’s Research Foundation), Dr. Yoshihiro Nakatani (Dana-Farber Cancer Institute), and Drs. Maria Jasin and Koji Nakanishi (Memorial Sloan-Kettering Cancer Center), for EUFA1341 fibroblasts, the human RAD51 cDNA, anti-actin antibodies, pOZ vectors, and U2OS-DR cells and pCBASce, respectively. This work was supported by the BMBF networks of “Inherited bone marrow failure syndromes” and “Foamyvirus-mediated genetic therapy for FNACA (FoneFA) and NIH R01 CA138237 and CA155294 (H.H.), NIH R01 HL084082 (A.R.M.), and NIH R01 HL085587 (P.R.A.).
Footnotes
CONFLICT OF INTEREST
MF and HH may receive royalties based on a licensing agreement with Myriad Genetics, Inc. for the use of RAD51C as a cancer susceptibility gene. All other authors declare that they have no potential conflicts of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://nature.com/onc).
References
- 1.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2012;12(1):68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walsh T, King MC. Ten genes for inherited breast cancer. Cancer Cell. 2007;11(2):103–105. doi: 10.1016/j.ccr.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 3.Turnbull C, Rahman N. Genetic predisposition to breast cancer: past, present, and future. Annu Rev Genomics Hum Genet. 2008;9:321–345. doi: 10.1146/annurev.genom.9.081307.164339. [DOI] [PubMed] [Google Scholar]
- 4.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7(2):263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 5.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4(4):511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 6.Davies AA, Masson JY, McIlwraith MJ, et al. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell. 2001;7(2):273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 7.Carreira A, Hilario J, Amitani I, et al. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell. 2009;136(6):1032–1043. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meindl A, Hellebrand H, Wiek C, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet. 2010;42(5):410–414. doi: 10.1038/ng.569. [DOI] [PubMed] [Google Scholar]
- 9.Vuorela M, Pylkas K, Hartikainen JM, et al. Further evidence for the contribution of the RAD51C gene in hereditary breast and ovarian cancer susceptibility. Breast Cancer Res Treat. 2011;130(3):1003–1010. doi: 10.1007/s10549-011-1677-x. [DOI] [PubMed] [Google Scholar]
- 10.Erkko H, Xia B, Nikkila J, et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007;446(7133):316–319. doi: 10.1038/nature05609. [DOI] [PubMed] [Google Scholar]
- 11.Rahman N, Seal S, Thompson D, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39(2):165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tischkowitz M, Xia B, Sabbaghian N, et al. Analysis of PALB2/FANCN-associated breast cancer families. Proc Natl Acad Sci U S A. 2007;104(16):6788–6793. doi: 10.1073/pnas.0701724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia B, Dorsman JC, Ameziane N, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39(2):159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 14.Sy SM, Huen MS, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci U S A. 2009;106(17):7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang F, Fan Q, Ren K, Andreassen PR. PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol Cancer Res. 2009;7(7):1110–1118. doi: 10.1158/1541-7786.MCR-09-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang F, Ma J, Wu J, et al. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol. 2009;19(6):524–529. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia B, Sheng Q, Nakanishi K, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22(6):719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 18.Stirnimann CU, Petsalaki E, Russell RB, Muller CW. WD40 proteins propel cellular networks. Trends Biochem Sci. 2010;35(10):565–574. doi: 10.1016/j.tibs.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Buisson R, Dion-Cote AM, Coulombe Y, et al. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat Struct Mol Biol. 2010;17(10):1247–1254. doi: 10.1038/nsmb.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dray E, Etchin J, Wiese C, et al. Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat Struct Mol Biol. 2010;17(10):1255–1259. doi: 10.1038/nsmb.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hellebrand H, Sutter C, Honisch E, et al. Germline mutations in the PALB2 gene are population specific and occur with low frequencies in familial breast cancer. Hum Mutat. 2011;32(6):E2176–2188. doi: 10.1002/humu.21478. [DOI] [PubMed] [Google Scholar]
- 22.Adank MA, van Mil SE, Gille JJ, Waisfisz Q, Meijers-Heijboer H. PALB2 analysis in BRCA2-like families. Breast Cancer Res Treat. 2011;127(2):357–362. doi: 10.1007/s10549-010-1001-1. [DOI] [PubMed] [Google Scholar]
- 23.Balia C, Sensi E, Lombardi G, Roncella M, Bevilacqua G, Caligo MA. PALB2: a novel inactivating mutation in a Italian breast cancer family. Fam Cancer. 2010;9(4):531–536. doi: 10.1007/s10689-010-9382-1. [DOI] [PubMed] [Google Scholar]
- 24.Bogdanova N, Sokolenko AP, Iyevleva AG, et al. PALB2 mutations in German and Russian patients with bilateral breast cancer. Breast Cancer Res Treat. 2011;126(2):545–550. doi: 10.1007/s10549-010-1290-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casadei S, Norquist BM, Walsh T, et al. Contribution of inherited mutations in the BRCA2-interacting protein PALB2 to familial breast cancer. Cancer Res. 2011;71(6):2222–2229. doi: 10.1158/0008-5472.CAN-10-3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Catucci I, Milgrom R, Kushnir A, et al. Germline mutations in BRIP1 and PALB2 in Jewish high cancer risk families. Fam Cancer. 2012 doi: 10.1007/s10689-012-9540-8. [DOI] [PubMed] [Google Scholar]
- 27.Ding YC, Steele L, Chu LH, et al. Germline mutations in PALB2 in African-American breast cancer cases. Breast Cancer Res Treat. 2011;126(1):227–230. doi: 10.1007/s10549-010-1271-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hofstatter EW, Domchek SM, Miron A, et al. PALB2 mutations in familial breast and pancreatic cancer. Fam Cancer. 2011;10(2):225–231. doi: 10.1007/s10689-011-9426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tischkowitz M, Capanu M, Sabbaghian N, et al. Rare germline mutations in PALB2 and breast cancer risk: a population-based study. Hum Mutat. 2012;33(4):674–680. doi: 10.1002/humu.22022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng Y, Zhang J, Niu Q, Huo D, Olopade OI. Novel germline PALB2 truncating mutations in African American breast cancer patients. Cancer. 2012;118(5):1362–1370. doi: 10.1002/cncr.26388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong MW, Nordfors C, Mossman D, et al. BRIP1, PALB2, and RAD51C mutation analysis reveals their relative importance as genetic susceptibility factors for breast cancer. Breast Cancer Res Treat. 2011;127(3):853–859. doi: 10.1007/s10549-011-1443-0. [DOI] [PubMed] [Google Scholar]
- 32.Miller KA, Sawicka D, Barsky D, Albala JS. Domain mapping of the Rad51 paralog protein complexes. Nucleic Acids Res. 2004;32(1):169–178. doi: 10.1093/nar/gkg925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takata M, Sasaki MS, Tachiiri S, et al. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol Cell Biol. 2001;21(8):2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masson JY, Tarsounas MC, Stasiak AZ, et al. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 2001;15(24):3296–3307. doi: 10.1101/gad.947001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang J, Huen MS, Kim H, et al. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol. 2009;11(5):592–603. doi: 10.1038/ncb1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kee Y, D’Andrea AD. Molecular pathogenesis and clinical management of Fanconi anemia. J Clin Invest. 2012;122(11):3799–3806. doi: 10.1172/JCI58321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bogliolo M, Schuster B, Stoepker C, et al. Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am J Hum Genet. 2013;92(5):800–806. doi: 10.1016/j.ajhg.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vaz F, Hanenberg H, Schuster B, et al. Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet. 2010;42(5):406–409. doi: 10.1038/ng.570. [DOI] [PubMed] [Google Scholar]
- 39.Oliver AW, Swift S, Lord CJ, Ashworth A, Pearl LH. Structural basis for recruitment of BRCA2 by PALB2. EMBO Rep. 2009;10(9):990–996. doi: 10.1038/embor.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakanishi K, Yang YG, Pierce AJ, et al. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci U S A. 2005;102(4):1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Somyajit K, Subramanya S, Nagaraju G. Distinct roles of FANCO/RAD51C protein in DNA damage signaling and repair: implications for Fanconi anemia and breast cancer susceptibility. J Biol Chem. 2012;287(5):3366–3380. doi: 10.1074/jbc.M111.311241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wooster R, Bignell G, Lancaster J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378(6559):789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 43.Reid S, Schindler D, Hanenberg H, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39(2):162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 44.Howlett NG, Taniguchi T, Olson S, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297(5581):606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 45.Stark JM, Hu P, Pierce AJ, Moynahan ME, Ellis N, Jasin M. ATP hydrolysis by mammalian RAD51 has a key role during homology-directed DNA repair. J Biol Chem. 2002;277(23):20185–20194. doi: 10.1074/jbc.M112132200. [DOI] [PubMed] [Google Scholar]
- 46.Makde RD, England JR, Yennawar HP, Tan S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature. 2010;467(7315):562–566. doi: 10.1038/nature09321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lio YC, Mazin AV, Kowalczykowski SC, Chen DJ. Complex formation by the human Rad51B and Rad51C DNA repair proteins and their activities in vitro. J Biol Chem. 2003;278(4):2469–2478. doi: 10.1074/jbc.M211038200. [DOI] [PubMed] [Google Scholar]
- 48.Chun J, Buechelmaier ES, Powell SN. Rad51 paralog complexes BCDX2 and CX3 act at different stages in the BRCA1-BRCA2-dependent homologous recombination pathway. Mol Cell Biol. 2013;33(2):387–395. doi: 10.1128/MCB.00465-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshihara T, Ishida M, Kinomura A, et al. XRCC3 deficiency results in a defect in recombination and increased endoreduplication in human cells. EMBO J. 2004;23(3):670–680. doi: 10.1038/sj.emboj.7600087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y, Tarsounas M, O’Regan P, West SC. Role of RAD51C and XRCC3 in genetic recombination and DNA repair. J Biol Chem. 2007;282(3):1973–1979. doi: 10.1074/jbc.M609066200. [DOI] [PubMed] [Google Scholar]
- 51.Wilson JB, Yamamoto K, Marriott AS, et al. FANCG promotes formation of a newly identified protein complex containing BRCA2, FANCD2 and XRCC3. Oncogene. 2008;27(26):3641–3652. doi: 10.1038/sj.onc.1211034. [DOI] [PubMed] [Google Scholar]
- 52.Abbott DW, Freeman ML, Holt JT. Double-strand break repair deficiency and radiation sensitivity in BRCA2 mutant cancer cells. J Natl Cancer Inst. 1998;90(13):978–985. doi: 10.1093/jnci/90.13.978. [DOI] [PubMed] [Google Scholar]
- 53.Andreassen PR, Ren K. Fanconi anemia proteins, DNA interstrand crosslink repair pathways, and cancer therapy. Curr Cancer Drug Targets. 2009;9(1):101–117. doi: 10.2174/156800909787314011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hayakawa T, Zhang F, Hayakawa N, et al. MRG15 binds directly to PALB2 and stimulates homology-directed repair of chromosomal breaks. J Cell Sci. 2010;123(Pt 7):1124–1130. doi: 10.1242/jcs.060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ali AM, Pradhan A, Singh TR, et al. FAAP20: a novel ubiquitin-binding FA nuclear core-complex protein required for functional integrity of the FA-BRCA DNA repair pathway. Blood. 2012;119(14):3285–3294. doi: 10.1182/blood-2011-10-385963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh TR, Saro D, Ali AM, et al. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol Cell. 2010;37(6):879–886. doi: 10.1016/j.molcel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.