Abstract
T and B cells share a common somatic gene rearrangement mechanism for assembling the genes that code for their antigen receptors and developmental pathways with many parallels. Shared usage of basic helix-loop-helix E proteins as transcriptional drivers underlies these common features. However, the transcription factor networks in which these E proteins are embedded are different both in membership and in architecture for T and B cell gene regulatory programs. These differences permit lineage commitment decisions to be made in different hierarchical orders. Furthermore, in a contrast to B-cell gene networks, the T-cell gene network architecture for effector differentiation is sufficiently modular so that E protein inputs can be removed. Complete “T-cell-like” effector differentiation can proceed without T-cell receptor rearrangement or selection when E proteins are neutralized, yielding natural killer and other innate lymphoid cells.
Introduction
From the first recognition of how somatic mutation, clonal receptor gene expression, and clonal selection operate in the adaptive immune system, T and B lymphocytes have appeared to be cell types with a self-evidently close relationship. Current evidence still indicates that they are the only cell types in the mammalian organism that use RAG1/RAG2-mediated programmed gene rearrangement in their development. The receptors that they use to recognize antigen are highly similar immunoglobulin superfamily structures which form the recognition surfaces for antigen when assembled into disulfide-bonded heterodimers. The development of the two lymphoid cell types presents even more striking parallels, as both pass through an ordered series of alternating proliferative phases, cell cycle arrest phases for gene rearrangement, and quality control checkpoints that operate to ensure a properly expanded population with a properly selected antigen recognition receptor repertoire. However, in development T- and B-cell precursors adopt strictly divergent paths at a surprisingly early stage of differentiation. Furthermore, recent evidence on the evolution of immune cell types indicates that the separation between T-cell-like and B-cell-like programs dates back more than 500 million years, before the use of immunoglobulin superfamily genes in antigen recognition (1). How can we understand the relationship between the shared and divergent features of these cell types? The answers lie in the use of distinct combinations of transcriptional regulatory network modules within the programs that generate these cell types, some of them mutually inhibitory, which this review will try to bring into focus.
Parallel, distinct, and more broadly shared developmental program elements
Parallel pathways for T and B cell precursor differentiation
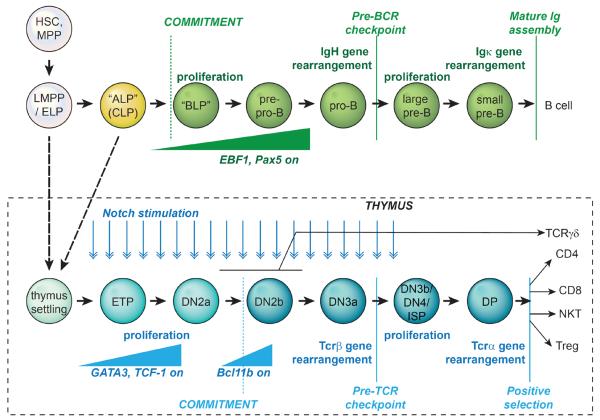
Major outlines of T and B cell development are well established and have been extensively reviewed as separate subjects (2–13). Figure 1 reviews the main pathways and stages for development of B cell and T cell precursors in mice, the system in which they have been most thoroughly dissected. Table 1 lists the markers by which successive stages are distinguished. Uncommitted hematopoietic precursors can develop into B cells in the bone marrow, primarily in the endosteal niche (14, 15), or in the fetal liver before birth. In contrast, uncommitted precursors must migrate first to the thymus in order to receive the signals that trigger T cell development, most importantly via ligands that activate the Notch signaling pathway. However, the two programs once under way are strikingly parallel, as shown in Figure 1, in which the program for B cells is compared with that for the major fraction of T cells that use TCRαβ-class receptors. From the earliest stages, the T and B cell programs display both shared and mutually exclusive characteristics.
FIGURE 1.
Schematic of major stages of B and T cell development. Consult Table 1 for definition of stage phenotypes. The figure introduces key stages and emphasizes the parallelism between B cell development stages and αβ T lineage cell stages in terms of immunoreceptor gene rearrangement timing, proliferative bursts and major developmental checkpoints. Specific regulatory genes important for lineage specification are turned on during the intervals shown. Stages are defined by ability to discriminate phenotypes and do not represent uniform lengths of time or numbers of cell cycles. Note that the T cell program unlike the B cell program generates at least five distinct types of T cells within the thymus (in fact the TCRγδ cells are further subdivided, not shown). “Thymus settling” = thymus settling precursors, which are thought to be derived from “ALP” type common lymphoid precursors and/or from certain LMPP or other ELP type cells in vivo. All of these, and even myeloid specified cells, can respond when introduced into the thymus by developing into T cells.
TABLE 1.
Phenotypic definitions of lymphoid precursor subsets
| Cell type | Name (if different) | Major markers | Comments |
|---|---|---|---|
| HSC | Hematopoietic stem cell | Lin− Kithigh Sca-1high Flt3− CD34low CD150+ CD48− | |
| MPP | Multipotent precursor | Lin− Kithigh Sca-1high Flt3−/+ CD34+ and/or CD150− CD48+ | These cells still have erythroid and megakaryocyte potential |
| LMPP | Lymphoid-primed multipotent precursor | Lin− Kithigh Sca-1high Flt3very high CD27+ Vcam-1− | For lymphoid precursor activity, Vcam− Flt3very high are important |
| ELP | Early lymphoid precursor | Like LMPP–ALP, Terminal transferase+, defined by Ragl reporter transgene | Ragl expression in LMPP/ALP type cells is low or intermittent |
| CLP or “ALP” | Common lymphoid precursor “A” type | Lin− Kitlow Sca-1low Flt3very high CD27+ IL7R+ Ly6d− | These are the true “common” lymphoid precursors |
| BLP | B-cell biased common lymphoid precursor | Lin− Kitlow Sca-1low Flt3int CD27+ IL7R+ Ly6d+ | Within this stage, Rag1 is upregulated, a λ5 transgene begins to be activated, and EBF1 is turned on |
| Pre-Pro B | B220+ CD43+ CD19− Kitlow Flt3low IL7R+ Ly6d+ | Continuum from late BLP marked by B220 upregulation and EBF1 and Pax5 up; caution that without Ly6d marker, NK cells have similar phenotype | |
| Pro-B | B220+ CD43+ CD19+ Kitlow CD27− Flt3low/− IL7R+ Ly6d+, IgH− | Pax5 signature; rearranging Ig heavy chain; Flt3 and CD27 off | |
| Large pre-B | B220+ CD19+ CD43− internal IgH+, surface IgM− | Kit turns off; cells with good IgH rearrangement cycle | |
| Small pre-B | B220+ CD19+ CD43−, internal IgH+, Surface IgM− | Cells stop proliferating, rearrange Igκ and λ | |
| B cell | B220+ CD19+ CD43− Surface IgM+ | ||
| Thymus settling | Thymus settling cell (not well defined) | CD4+/− CD8− CD24int CD25− CD44high, cKitvery high, Ccr9+, CD27high, Flt3+ | Inferential phenotype |
| ETP | Early T-cell precursor | CD4+/− CD8− CD24int CD25− CD44high, cKitvery high, CD27high | |
| DN2a | CD4− CD8− CD24+ CD25+ CD44high cKitvery high, CD27high | Not yet committed; good TCRγδ and TCRαβ potential | |
| DN2b | CD4− CD8− CD24+ CD25+ CD44+ cKitint, CD27high, internal TCRβ− | T lineage committed; good TCRγδ and TCRαβ potential | |
| DN3a | CD4− CD8− CD24+ CD25+ CD44− cKitlow, CD27low, internal TCRβ− | TCRβ rearranging; cells biased against TCRγδ | |
| DN3b | CD4− CD8− CD24+ CD25+ CD44− cKitlow, CD27high, CD28+, surface TCR−, internal TCRβ+ | Large proliferating transitional intermediate following success in TCRβ rearrangement | |
| DN4 | CD4− CD8− CD24+ CD25− CD44− cKitlow, CD28+, surface TCR−, internal TCRβ+ | Large proliferating transitional intermediate | |
| ISP | Immature single positive | CD8+ or CD4+, CD24+, CD28+, surface TCR−, internal TCRβ+ | Large proliferating transitional intermediate |
| DP | Double positive | CD4+ CD8+ TCRβ+, CD24+ | These cells acquire TCRα chains and cell-surface mature TCR complexes during the DP stage |
| CD4 SP | CD4 single-positive | CD4+ CD8− TCRαβ+ CD24+ to negative | |
| CD8 SP | CD8 single positive | CD4− CD8+ TCRαβ+ CD24+ to negative | |
| NKT | Invariant NK-like T cell | CD4+/−, CD8− TCRαβlow, CD24 negative, CD44+, NK1.1+ | Invariant TCR used; NK marker expression depends on allele |
| Treg | Natural (thymus-derived) regulatory T cell | Effectively identified with Foxp3 reporter gene |
Both T and B cell precursors begin their development with a period of transcriptional ground-breaking that turns on genes that contribute to antigen-receptor assembly and antigen receptor-dependent signaling, during the BLP, pre-pro-B and ProB stages of B-cell differentiation and the DN2a/2b and DN3a stages of T-cell differentiation. This early activation is important because antigen receptor gene rearrangement status and signaling competence must be repeatedly used at distinct checkpoints during development to determine the fate of the cells.
Immune receptor genes are assembled through tightly limited phases of somatic gene rearrangement, mostly at two specific stages that are equivalent in B and TCRαβ T cells (Fig. 1). For rearrangement of the immunoglobulin (Ig) and T-cell receptor (TCR) genes themselves, both T and B cells use the same gene products: RAG1 and RAG2 to recognize and cleave the recombination substrates, Terminal deoxynucleotidyl transferase (TdT=Dntt) for mutagenesis of the cleaved ends, and DNA-PK (Prkdc), Artemis, Ku70 and Ku80, and DNA ligase IV for resealing. Among these genes, expression of the Rag1, Rag2, and Dntt genes is specific to developing lymphocytes, and common to both T and B lineages.
In order to generate the signals that will enable cells to pass developmental checkpoints, the nascent antigen receptor gene products need to be assembled into signaling complexes. Thus, components that are not antigen receptors themselves must also be provided in early development. Such gene products include the CD3γ, δ, and ε and TCRζ chains (Cd3g, Cd3d, Cd3e, Cd247 respectively) for T cells and the Igα and Igβ chains (mb-1=Cd79a and B29=Cd79b, respectively) for B cells, and the transcriptional activation onsets for these genes represent useful landmarks for entry into each pathway. Also expressed transiently are immunoreceptor “surrogate chains”, pre-TCRα (Ptcra) for T cells and the combination of VpreB and λ5 (Igll1) for B cells. In each lineage, these supply a lineage-specific dimerization partner or partner complex to stabilize the first immune receptor chain to be expressed in each lineage, enabling a provisional immunoreceptor complex to be assembled at the cell surface even before the second chain of the immunoreceptor has been generated. Such antigen receptor assembly and signaling genes are turned on at parallel, corresponding stages of T and B cell development, i.e. the DN2a, 2b, 3a stages for T cells and the BLP, pre-proB, and proB stages for B cells. All of these events are respectively B or T lymphocyte specific.
Both T and B cell precursors begin their transcriptional priming for differentiation in coordination with a phase of stromal-cell dependent cell division. In mice, the activation of lineage-specific genes occurs in B and T cell precursors as they proliferate, initially under the influence of cytokines that include IL-7 (or TSLP) and Kit ligand or Flt3 ligand (16, 17). The involvement of the related receptor tyrosine kinases Kit and Flt3 in the progenitor cells and of IL-7R as the main growth factor receptor for early T cell and B cell precursors is another common link between the two programs. A role for CXCL12 (SDF) chemokine binding to the CXCR4 receptor is also likely to be crucial to support lymphoid precursors at early stages in the bone marrow (14, 15). Thus, similar growth factor receptor dependent signaling pathways begin to participate in both programs, long before antigen receptor-dependent signaling is possible. However, RAG1/2-mediated gene rearrangement is most efficient in the G1 stage of the cell cycle, and so proliferation needs to slow after initial expansion in order to enable antigen receptor gene rearrangement to proceed. Rearrangement in both B and T cell precursors focuses initially on genes that code for one of the heterodimer chains of the immune receptors: the TCRβ chain genes in TCRαβ T cell precursors or the Ig heavy chain (IgH) genes in B cell precursors, joining first one of the D to one of the J gene segments, then some V to the joined DJ segments. (A somewhat different path or set of paths is followed by T-cell precursors that rearrange TCRγ and TCRδ to form TCRγδ receptors. For reviews, see (18–22).) Many of the developing B cells or precursors of αβ T cells stop proliferating entirely to complete this round of gene rearrangement.
The ability of the cells to progress beyond this point now becomes tightly dependent on the success of the rearrangement event to generate a transcription unit that codes for a functionally competent TCRβ or IgH chain. The cells detect success by the assembly of the newly generated immune receptor chain with the pre-existing surrogate chain and signaling complex components. A new burst of proliferation can only be triggered by the ligand-independent signal generated when the new immune receptor gene product assembles successfully with the signaling complex components. This primary developmental checkpoint is the pro-B to pre-B transition in B cell development (pre-BCR checkpoint, in Fig. 1) and the β-selection checkpoint in T cell development (pre-TCR checkpoint).
Once IgH+ pre-B cells or T-cell precursors that have successfully passed through β-selection finish their proliferative burst, they are allowed to rearrange the gene segments coding for the second of the dimeric immune receptor chains, the Igκ or Igλ light chain genes for B cells and the TCRα chain for T cells. This process will create the cells' clonally distinct, mature antigen recognition structures, and only cells that generate successful mature immune receptors will be allowed to emerge as functional B or T cells. Success criteria for B cells include expression of a light chain competent to dimerize with the IgH heavy chain, to form a receptor that is not grossly reactive with autologous structures within the bone marrow environment. Success criteria for T cells include not only correct dimerization and avoidance of gross autoreactivity, but also the ability to form low-affinity but significant interactions with major histocompatibility complex (MHC) cell-surface structures in the thymic microenvironment.
Limitation on parallels between T and B cell differentiation pathways
Although the outlines of the pathways of differentiation are similar, the signaling molecules that complex with T and B cell immunoreceptors are highly lineage specific and are expressed in mutually exclusive developmental patterns. They provide some of the clearest case studies for the transcriptional regulatory mechanisms that define T or B cell identity. Furthermore, the pathways of differentiation themselves have very different character because the T-cell developmental program branches to offer many more effector specialization options (CD4, CD8, NKT, Treg, and multiple classes of TCRγδ cells) than there are B-cell options.
Once a full TCRαβ heterodimer is expressed, at the CD4+ CD8+ “Double Positive” stage, the interaction with MHC triggers the “positive selection” phase of T-cell development (Fig. 1). In a departure from the pattern of B cell development, this selection event not only rescues cells from programmed cell death or developmental arrest, but also results in functional program divergence, as cells become programmed to a CD4, CD8, NKT (innate-like T cell), or Treg fate. The details of these branching intrathymic selection pathways are deeply studied and well understood (23–33), and although they continue beyond the scope of this review, we note here that they involve new roles for some of the same transcriptional regulatory factors that establish T-cell identity in the first place. Different sublineages of T cells, including different types of T cells that use TCRγδ receptors instead of TCRαβ receptors, may vary according to the degree to which they are polarized to separate functional pathways before they leave the thymus, by interaction with self-ligands, or in the periphery, by interaction with non-self ligands. However, T cells need to choose among various alternative sets of effector genes to poise for future TCR-dependent activation, and this choice is as much a feature of T-cell development as is the expression of a particular TCR heterodimer.
Further diversification of T-cell effector roles can continue, after the cells leave the thymus, as individual T cells continue to shape their transcriptional profiles in response to the conditions in which they encounter antigen. These stimulation-triggered changes involve choice of one of several distinct, coherent transcriptional response programs, each based on a different, highly coordinated pattern of cytokine gene expression, and each at least partly exclusive of the others. In addition to the cytotoxic T lymphocyte (CTL) killer program, which is mostly determined intrathymically, these programs include: a mucosal surface-defensive inflammatory program (Th17); a program that triggers antiviral responses and destruction of intracellular bacteria (Th1); a program that enhances expulsion of parasites (Th2); a partially overlapping program that enhances B-cell stimulation and Ig secretion (Tfh); and a program for damping the responses of other immune cells (Treg) which can be induced either within the thymus or in the periphery. The existence of these distinct functional branches within the T-cell program, and the need to coordinate expression of genes at numerous non-TCR loci within and between these branches [e.g. (34–40)], together give the T-cell program its distinctive transcriptional regulatory complexity.
Developmental paths and choices
Shared cellular and regulatory histories for T and B cell precursors
There is good evidence for strong common program elements underlying the competence to make T and B cells as well as other types of lymphocytes. Three large subdivisions of hematopoietic differentiation are recognized: erythromegakaryocytic development, myeloid development, and lymphoid development. Stem cell differentiation spawns partially-restricted precursors as intermediates that have been channeled toward (or away from) one of these three general directions through cell-intrinsic mechanisms. From stem cells through most multipotent precursor cells, B and T cell developmental potentials initially cosegregate (Figure 2; HSC, MPP, LMPP, ELP, “ALP” stages). They also cosegregate with potential to give rise to non-B, non-T innate lymphocytes (ILC), including Natural Killer (NK) cells. Interestingly, lymphoid precursors generally reveal an ability to give rise to dendritic cells as well, until they become fully committed to a particular B, T, or Innate Lymphocyte fate. Cells that have lost all access to erythroid and megakaryocytic development (LMPP, ELP) remain capable of giving rise to all these cell types. In contrast, many if not all pan-lymphoid precursors still possess conditional myeloid potential (41–48), although there are clear differences between the physiological conditions that favor generating all types of lymphocytes and those that favor generation of myeloid cells from the same precursors (49, 50). This suggests that lymphocytes emerge through a jointly lymphoid-myeloid biased progenitor pathway before specializing to different lymphocyte fates.
FIGURE 2.
Major alternative fate branchpoints for B and T cell precursors. The figure explains the timing of commitment, by showing the demonstrated alternative fates that can still be adopted by B and T-cell precursors until the latest stages shown. Heavy line arrows: major pathways. Light solid line arrows: strong pathways, high precursor frequencies for the indicated fate alternative. Light dashed line arrows: variant pathways, not a default in vivo but readily demonstrable at high frequency under experimental conditions. Dotted line arrows: measurable experimentally but distinctly reduced precursor frequency for the indicated path at this stage as compared to immediate precursor. Mac, DC: macrophage and dendritic cells. In addition, granulocyte fates are also accessible to pre-commitment early T cells. For simplicity, within the T-cell lineage, fates that are still robustly accessible at the DN2a stage are not also shown for the antecedent thymus settling and ETP stages. BLP and DN2b cells are largely if not completely committed.
The most restricted precursors that can still generate both T and B cells are Lymphoid-Primed Multipotent Precursors (LMPP) and Common Lymphoid Precursors (CLP; those rigorously defined to be pan-lymphoid precursors are also termed “ALP”)(Figure 2). Both of these are distinguished from stem cells by their expression of transcription factors Ikaros (Ikzf1) and PU.1 (Sfpi1 in mouse, SPI1 in human), and the growth factor receptor protein tyrosine kinase Flt3 (Flk2). Whereas both have lost the ability to generate erythroid and megakaryocytic cells, LMPP are much better as myeloid precursors than CLP in vivo, whereas LMPP are slower to generate lymphoid progeny than CLP (45, 47, 51–54). Despite these consistent findings, several questions are unresolved. It is uncertain whether transition from an LMPP-like state through a CLP-like intermediate is an absolute prerequisite for lymphoid development (55). For example, Ikaros mutants that cannot generate CLP or early B cells at all can still enter the T-cell developmental pathway (56). It is also not completely clear yet what the regulatory difference is that explains these differences in myeloid potential, as lymphomyeloid bifunctional regulators like PU.1 and Flt3 (see below) seem to be expressed similarly in both. To some extent, the ability of cells to manifest myeloid potential is a function of the signaling environment, as Toll-like receptor signaling or alterations in G-protein coupled receptor signals can drive even the most restricted CLP to a nonlymphoid fate (57, 58). An obvious difference between LMPP and CLP has to do with the relative levels of growth factor receptors IL-7R and Kit they express on their surfaces. Both are defined by high expression of the growth factor receptor Flt3, but LMPP express more Kit and less IL-7R, whereas CLP express less Kit and more IL-7R. IL-7R signaling can defer or antagonize myeloid development in several contexts as well as supporting lymphoid development (59, 60). However, expression levels of these growth factor receptors substantially overlap, and the fractions of both kinds of progenitors that have T-cell potential appear to share expression of the chemokine receptor Ccr9 as well (46, 61–64). In accord with their overlapping lymphoid potentials, then, LMPP and CLP type cells thus probably share responsiveness to common signals.
Intrinsically, B and T cell potential depends on a common, indispensable set of transcription factors. Precursors for both lineages are intensely dependent on the Ets-family transcription factor PU.1 for their generation and survival, and they also require the zinc finger transcription factors Ikaros (Ikzf1) and Bcl11a and the basic helix-loop-helix (bHLH) “E protein” family member E2A (encoded by a gene known as Tcfe2a, bHLHb21, or Tcf3). PU.1 has other major roles in myeloid and dendritic cell development, but it is targeted in lymphoid precursors to distinctive genomic sites (65, 66). Ikaros, Bcl11a, and E2A are also expressed in stem or multipotent progenitors as well as lymphoid precursors (2, 44, 67–71), but they are most important for the lymphocyte developmental programs in general and absolutely required for B-cell development. In T cell development, where paralogs of Ikaros, Bcl11a, and E2A are also programmed for expression together with these three factors themselves, some compensation is possible for mutations in any one of these genes. Nevertheless, in Ikaros, Bcl11a, or E2A mutants the breakthrough T-cell development is suboptimal, abnormal, and prone to leukemic transformation (72–74).
Phenotypically recognizable CLPs only emerge in cells with intact function of Ikaros, Bcl11a, and E2A, and both B and T cell precursor activity is acutely dependent on E2A even under manipulations by which the precursors can be restored. Generation of both T and B cells also depends on a second group of factors which are important not only for producing lymphoid precursors but also for a broad range of other hematopoietic programs, including stem-cell, erythoid, neutrophil, and megakaryocyte development. In addition to the lymphomyeloid factor PU.1, already mentioned, these include the essential definitive hematopoiesis factor Myb (75–79), the zinc-finger repressive factor Gfi1 (80–85), the cytokine receptor signal transducing factor pair STAT5a/STAT5b (86, 87), and heterodimers of Runx family transcription factors with their common partner, CBFβ (88–92). Recent evidence suggests that a role for Hoxa9 may be important as well (93), while another broadly expressed zinc finger factor, Miz-1 (Zbtb17), appears important to potentiate responsiveness to IL-7 receptor signaling in early lymphoid precursors (94, 95). The requirements for programming of the CCR9+ LMPP and CLP precursors establish the ground state upon which both T and B cell gene regulatory networks are built.
Timing of lineage choices for T and B-cell precursors
In the bone marrow there is a continuum of B cell development from LMPP through CLP to EBF+, Pax5+ pro-B cells, and several combinations of surface markers and transgenic markers have helped to establish prospectively the developmental stages involved (Fig. 2, Table 1). Activation of several genes in otherwise LMPP-like cells act as harbingers of lymphoid bias. These include the first expression of Rag1 as well as IL-7R (96). Cells with clear IL-7R expression are defined as CLP (52, 97) (Figs. 1, 2). At the CLP stage, the cells do not yet express any B-lineage restricted surface markers, but they are primed to begin lymphocyte development. Assay systems based on transplantation in vivo or clonal growth in culture without stroma indicate that they are greatly diminished in ability to generate myeloid cells, although under other conditions with supportive stroma they do reveal significant latent myeloid potential (49, 50). Many CLPs still retain considerable T, NK, and/or DC potential; these are also Flt3high and Ly-6d-negative (“ALP”), whereas CLPs that have turned on Ly-6d expression are strongly biased to the B-cell fate (“BLP”) (98–100),(101). Progress towards B-cell commitment within these BLPs also correlates with downregulation of Kit. Complete commitment to the B-cell fate is accompanied by increased Rag1 expression and ability to express an Igλ transgene, even before expression of classic ProB cell surface markers (102, 103). The cells are then termed pre-pro-B cells when they turn on B220, and pro-B cells when they activate CD19, when CD27 expression is also extinguished.
Myeloid potential of all IL7R+ CLP is reduced in comparison with that of IL7R− LMPP. It is ambiguous whether the B-cell biased CLP have lost the last vestiges of myeloid potential any more absolutely than the Ly6d− CLPs (“ALP”), but they differ from ALP in showing a greatly reduced ability to generate T cells or NK cells (99). Thus, from a B-cell perspective, T- and NK-lineage fate exclusion is the most prominent aspect of lineage commitment, and closely linked with the initiation of the B-cell differentiation program.
For T lineage cells, the impact of Notch signaling causes lineage choices to be made in a different order. The events that cause loss of access to the B-cell fate are not the rate limiting events of T-cell commitment, but rather are the earliest responses that the cells make under the influence of thymic signals (Fig. 2, “thymus settling” precursors vs. ETP). In contrast, the final stages of T-cell lineage commitment clearly involve mechanisms that exclude fates other than the B cell fate. Myeloid and dendritic-cell potential is readily demonstrated in early T cell precursors from fetal or adult thymus, at stages many cell divisions after B-cell potential is lost. The T-cell precursors most related to bone marrow or fetal liver “ALPs” are the Early T-cell Precursors (ETP; Kit-high DN1 cells)(41, 56). In postnatal animals, evidence from lineage tracing reporters, intrathymic Notch signaling mutations, and actual cell purification and culture assays has converged to imply that many cells can enter the thymus with B-cell potential intact (61, 64, 104–108), but they lose this B-cell potential early during the expansion of the ETP population. In fetal thymus during the first wave of T-cell development, ETPs appear to enter the thymus devoid of B-cell potential already (109, 110). Thus, even in the postnatal thymus, the great majority of ETPs at any given time will already have become intrinsically unable to adopt the B cell fate, even when switched to a favorable B-cell promoting environment (61, 107, 108). Under the same conditions, in contrast, they retain readily demonstrable ability to generate macrophages, dendritic cells, granulocytes, and NK cells among other cell types (111–120). This natural plasticity coincides with the sustained relatively high expression of the myeloid and progenitor-cell factor PU.1 in the T-cell precursors throughout these cell cycles, while B-cell specific regulatory gene expression is strictly prohibited. Access to the dendritic cell pathway then decreases after the ETP stage, but the ability to generate innate-type lymphocytes and nonlymphoid cells alike persists for multiple additional generations, into the DN2 stage, and is only lost as the T-cell precursors transition from DN2a to DN2b (see Fig. 1). Loss of B-lineage access thus not only precedes loss of myeloid and dendritic potential; it also precedes the upregulation of recombinase factors Rag1, Rag2, and Dntt in T-lineage cells, which occurs in the DN3 and later stages of T cell development, as discussed below (for review, see (121)). Thus, even the features that are most shared by B and T cells are controlled through developmental programs that in fact differ sharply between B- and T-cell precursors.
Creating a B-cell specific regulatory state
Essential regulators of the B cell program
B cell development depends on a vital combination of transcription factors which are activated or upregulated early in B cell development and sustained in activity long into mature B cell immunological responses. The threshold to begin B cell development is crossed when the transcription factors EBF1 and Pax5 are turned on de novo, creating a self-stabilized and unique regulatory state (2, 4, 122). EBF1 expression, together with E2A, is needed to establish a recognizable B-lineage gene expression profile. Pax5 expression is needed to complete and stabilize B-cell lineage commitment, to open the full range of Ig heavy-chain V regions for rearrangement, and to complete the signaling complexes that enable the cells to receive B-cell receptor signals (5).
Both EBF1 and Pax5 are expressed only in B lineage cells among all hematopoietic cell types, even though they are representatives of transcription factor families with ancestral roles in completely different tissues including adipose tissue, stroma, kidney, liver, and the central nervous system. Because there are no paralogs for EBF1 and Pax5 used in other hematopoietic cells, the advent of these factors with their unique DNA binding specificities establishes a regulatory state that is clearly disjunct from those in any other hematopoietic cells. In combination with the pan-hematopoietic legacy of Myb, Runx, Ikaros and other factors, the addition of EBF1 and Pax5 gives early B-cell precursors a repertoire of DNA binding factors that is probably unique in the whole organism. Many if not all B-lineage specific genes appear to get direct regulatory contributions from EBF1 and/or Pax5. Only in the very terminal stages of B-cell immune responses, when the cell switches off most B-cell genes and transdifferentiates into a constitutively antibody-secreting plasmacell, do Ebf1 and Pax5 become silenced.
Starting the B-cell program
Activation of Ebf1 and Pax5 is triggered by a combination of E2A activity, FoxO1 activity, and Runx/CBFβ activity, under permissive conditions that depend on IL-7R signaling in the earliest CLP1. The cells must surpass a certain minimum net level of E2A activity before this can occur. The rise in activity from LMPP to CLP is not fully explained, but many uncommitted early lymphoid precursors already detectably express E2A target genes such as Rag1, Rag2 and Dntt at low levels (124–126), and expression of the diagnostic receptor genes Flt3 and Il7r itself appears to depend on E2A as well (98, 127). E2A then appears to upregulate one of the additional factors with which it will collaborate for EBF1 activation, namely FoxO1, creating a feed-forward network circuit (128, 129). Runx/CBFβ activity at this point is required independently (89), and as a prerequisite for direct positive regulation of EBF1 as well (88). Once EBF1 is present, it collaborates potently with E2A to turn on multiple early B-cell genes directly (124, 130, 131) and to activate Pax5 expression, with assistance from PU.1 working at another Pax5 enhancer element (132). This switch to turn on EBF1 and Pax5 also draws upon the background contributions of Ikaros (44, 133–136), Bcl11a (67, 137), Myb (76, 77, 138), and Gfi1 (81, 82) expression, for additional regulatory and supportive inputs.
Activation of EBF1 and Pax5 is not easy. Even if E2A, Runx1/CBFβ and FoxO1 are present, these B-cell specific regulatory genes can be kept silent if the myeloid transcription factor C/EBPα is induced (139–141). Excessive signals from a variety of cytokine receptors are sufficient to accomplish this in uncommitted lymphoid precursors (139, 142, 143). Forced ectopic imposition of C/EBPα rapidly turns EBF1 and Pax5 off, together with their target genes, not only in ALPs but even in later-stage cells that had already committed to a B-cell fate (140, 141), as the cells are reprogrammed to a myeloid fate. This may be facilitated by the presence of the myeloid-supportive factor PU.1 which is also required by these B-cell precursors. As already noted, Toll-like receptor signaling in the CLP may also abort B-cell development (57), and this may occur by redirecting the effects of PU.1 to activation of myeloid or dendritic-cell genes instead. In normal multilineage precursors, even if present, E2A activity can also be antagonized by signals that induce the E protein antagonists Id1, 2, or 3. Id1 and Id2 especially are well expressed in a variety of other hematopoietic progenitors and cell types (98, 144), and initially E2A itself can induce Id2 expression in a negative autoregulatory feedback loop (145). Thus, while the outlines of B-cell development can be understood once a critical threshold of E2A activity is crossed, there is competition for the fate of the cells. The SNAG-domain repressor, Gfi1, appears to be important not only to moderate levels of PU.1 in early precursors of B cells (82), but also to keep Id1 and Id2 expression in check (81, 85). Ikaros can directly limit PU.1 expression once cells have entered the B lineage pathway (146), and may also contribute to crossing the E protein:Id protein threshold, by helping to activate Gfi1 (82, 133).
Managing IL-7R signals may also require special regulation, to accommodate the cells' peculiar use of the global cell cycle arrest factor FoxO1 as a part of their cell-type specification machinery. IL-7R signals are crucial for early murine B cell development whether or not they directly activate Ebf1 (94, 123, 147, 148). Both Myb and another required transcription factor, Miz-1 (Zbtb17), act to promote IL-7R expression and sensitivity (76, 77, 94). However, signaling through a growth factor receptor like IL-7R would generally activate not only STAT5, which either activates or supports expression of EBF1, but also the phosphatidylinositol (PI) 3-kinase pathway. PI 3-kinase activation triggers the protein kinase Akt, which stimulates proliferation through a pathway that powerfully inhibits FoxO1. Yet FoxO1 is important not only to activate EBF1 but also for direct regulation of Rag genes, Dntt, and Il7r itself, in collaboration with E2A (149–151). Thus, in principle, if IL-7R strongly activated PI 3-kinase at a pre-specification stage it could retard activation of Ebf1 and a key group of B-cell genes. B lineage cells appear to be able to avert this problem by regulating the coupling of IL-7R signaling to PI 3-kinase as opposed to STAT5 activating pathways (149, 152). Later in pro-B cells, when cell cycle arrest is needed to permit efficient Ig gene rearrangement, IL-7R-mediated activation of STAT5 itself is curtailed (153), but shifts between FoxO1 protection and de-protection punctuate the later phases of B cell development as well (149, 154).
Mechanistic insights into transcription factor activity to define cell type in early B cells
As soon as Ebf1 and Pax5 are activated, these B-cell regulatory factors become entrained in a direct mutual positive feedback circuit which amplifies and locks down the B-lineage identity of the cells. Analyses of transcription factor mutants, epistasis tests, and direct analysis of transcription factor binding across the genome in pro-B and pre-B cells have provided a very strong picture of this circuit (75, 124, 128, 129, 132, 155–158). These results show that E2A has direct positive inputs into Ebf1 and Pax5 loci, EBF1 and PU.1 also have direct positive roles in regulating Pax5, EBF1 and E2A directly sustain FoxO1 expression, while Pax5 and FoxO1 positively regulate Ebf1 as well. Furthermore, although E2A is already broadly expressed in hematopoietic precursors, it is highly upregulated in B-cell precursors. Besides a high level of transcription of the gene coding for E2A, the effectiveness of the E2A protein is sharply increased once EBF1 is present, by the fact that EBF1 powerfully keeps Id2 repressed (157, 159). This inhibition of an inhibitor makes B-lineage specification irreversible, while the mutual positive feedbacks between EBF1 and Pax5 help to lock in a stably specified state.
Not only E2A and FoxO1, but also Runx1 and PU.1, can be drawn into the enterprise of activating B-cell specific differentiation genes together with EBF1 and Pax5, through corecruitment to sites of joint binding with lineage-specific factors (66, 89, 124, 132, 160). PU.1 binding in particular shows a very different distribution in pre-pro B cells and B cells than in myeloid cells, shifting in the B-lineage cells to a pattern dominated by linkage to E2A or EBF1 sites (66). In addition to their activating roles, moreover, EBF1, Pax5, and even E2A and FoxO1 acquire specific repressive roles in the B-cell context (75, 124, 128, 155, 157, 159, 161–163). The ability of the same factors to silence non-B cell genes as to activate B-cell genes gives the B-cell program its tight coherence.
B-cell gene networks begin to operate in the critical stages through which a CLP becomes a pro-B cell, using the upregulation of the marker Ly6d and the downregulation of Flt3 (Flk2) to dissect these early events as described above (99–101, 122, 164). Before initiation of the B-cell program, many E2A binding sites are associated with likely Runx target sites (124). The nature of the dimers that contain E2A in the earliest progenitors may in fact be different from the E-protein type homodimers in slightly later B-lineage precursor cells, since the total E2A pool in the progenitors likely includes a substantial fraction of E2A in heterodimers with SCL (Tal1) or especially Lyl1, class B bHLH factors which contribute their own fine-scale binding preferences. Lyl1 continues to be expressed throughout B cell development. (165, 166). In whatever dimer form it is found, however, E2A positively regulates FoxO1 in these early precursors, and then works with FoxO1 to turn on expression of EBF1 as already noted (128, 129). Individually, many cells activate EBF1 before they express Pax5, and this initial activation corresponds with the “ALP to BLP” transition (102). The presence of EBF1 enables E2A to redistribute to new sites: about half of the EBF1 sites are sites of co-binding with E2A, many of them sites where E2A was not bound before (124). FoxO1 collaborates intimately with both E2A and EBF1 in regulating genes that define the B-cell identity (128). As Pax5 is induced, it in turn creates new sites of recruitment for the other factors.
Like PU.1 and E2A (66, 124, 167), both Pax5 and EBF1 can play rate-limiting roles to recruit chromatin-modifying enzymes in the context of gene regulation (155, 157). Both EBF1 and Pax5 are clearly bifunctional. As is often seen by comparison of ChIP-seq with gene expression analysis of mutant cells, both transcription factors are found engaged at many-fold more high-quality binding sites than the number of genes that they functionally affect. However, their binding is implicated directly in both positive and negative gene regulatory responses.
Pax5 has a larger number of known binding sites across the genomes of developing B cells than EBF1, and these shift significantly as B-cell development proceeds, even between successive immature B-cell states. It binds to a large fraction of all active cis-regulatory elements at given stages of B-cell development, even though only a small percentage of the bound genes show Pax5-dependent activity patterns. Pax5 binding has been shown to result in the appearance of new DNase-hypersensitive sites in the genes that it activates, whereas it causes loss of DNase-hypersensitivity at sites in genes that it represses (164). In the case of EBF1, a comparison of binding, local histone modification, and gene expression across successive B-cell developmental stages shows that EBF1 binding can also poise a B-lineage gene's cis-regulatory site for future activation, long before the full requirements have been met for transcriptional activation. This activity is correlated with local histone modification to a histone H3K4me2+ state, which can persist through immature developmental stages until the EBF1-bound regulatory element is ready for full activation in the mature B cell (157). However, EBF1 cannot freely open chromatin at all of its own cognate sites across the genome. Its binding in early B cells is shaped by the hematopoietic context, and ectopic expression of EBF1 only turns on B-lineage target genes in a hematopoietic context. In fact, it binds a completely different spectrum of genomic sites in another context where it plays an equally important developmental role, in adipose cells, where it also interacts with a different set of regulatory partners. Even in mature B cells, EBF1 shifts to occupy a different spectrum of sites (75), where the rules for its collaborations with partners such as FoxO1 also appear to change. Thus, despite the continuing importance of this factor in mature B cells, its precise deployments vary.
One highly B-cell specific locus that has been studied in depth, the Cd79a locus (mb-1, Igα), has provided a detailed example of the mechanism through which these B-cell specific and broadly utilized factors can collaborate (2, 160). EBF1 supplies the critical rate-limiting element needed to complete a DNA-binding complex with E2A and Runx1/CBFβ, which can then initiate local DNA demethylation of the Cd79a promoter. As demethylation propagates from the site of EBF1, E2A, and Runx1 binding, it unmasks a key Pax/Ets composite site which can then cooperatively bind Pax5 with a pan-lymphoid factor, Ets1, to lock the promoter into an active state. Versions of this intricate transcription factor collaboration are likely to be occurring at many other sharply activated B-cell genes.
Another country: the leap into T-cell development
Three ambiguous regulators and an “outside agitator”
T-cell precursors begin with the same intrinsic regulatory factor complex as B-cell precursors, inherited from multipotent progenitor ancestors, and they initially express very similar cytokine and chemokine receptors to guide and mediate their interactions with environmental cues. However, the T-cell developmental program depends on the migration of the cells into a foreign environment, namely the thymus. The special feature of the thymus is its dense presentation of potent Delta-class Notch ligands, primarily Delta-like 4 (DLL4), which bombard the cells with signals that ultimately render most non-T-cell fates inaccessible (7, 13, 168). Notch signaling prohibits activation of the B-lineage specific factors EBF1 and Pax5 and sets in train a regulatory cascade that ultimately makes their silence irreversible. It also antagonizes myeloid development and blocks access to several other fates through mechanisms described below. But in order to forge a T-cell identity for the progenitors, it also needs to turn on T-cell specific regulatory factors. The most T-lineage specific of these factors in fact comprise a very short list: they are TCF-1 (encoded by the Tcf7 gene) (169–172), GATA-3 (173–177), and Bcl11b (178–182).
There are a number of central differences between the T-cell program governed by Notch, TCF-1, GATA-3, and Bcl11b and the B-cell program governed by EBF1 and Pax5.
First is that the Notch signaling is extrinsically controlled and non-cell-autonomous. Even so, it is not simply a transient trigger to activate the T-cell specific factors. Instead, it remains a sustained regulatory requirement throughout multiple cell cycles of the T-cell specification program, before and even for some time after commitment. Thus even after GATA-3, TCF-1, and Bcl11b are turned on, many T-cell differentiation genes continue to receive direct regulatory input from an environmental signal-dependent transcription factor and cannot continue to be expressed if the contact with the environment is interrupted (183–185).
Second, the essential T-cell specific transcription factors themselves are mostly confined to developing T cells, with instructive exceptions discussed below; but their paralogs are abundantly expressed and prominent in other hematopoietic lineages including hematopoietic stem and progenitor cells. GATA-3 is a close relative of the stem-cell and mast-cell factor GATA-2, and of the erythromegakaryocytic factor GATA-1. Bcl11b is a close relative of the pan-lymphoid required factor Bcl11a. TCF-1 itself is related to TCF-4 (Tcf7l2 gene product), which is also expressed in stem and progenitor cells, although the level to which TCF-1 rises in T-cell precursors is much higher. The progenitor factors are turned off in different patterns, but mostly at about the same time that their T-cell relatives are turned on (11). The close biochemical similarities between T-cell specific factors and their relatives expressed in multipotent progenitors should enable them to bind to similar or identical target sites, all other things being equal. This makes it more challenging than in B cells to understand how new cis-regulatory elements are activated in early T-cell development.
Third, many of the other transcription factors expressed by T-cell precursors are coexpressed along with their paralogs, providing considerable regulatory redundancy. Examples include Ikaros, which is coexpressed with 1–3 other family members during T-cell specification; E2A, which is coexpressed with two isoforms of its close relative HEB (Tcf12); all three Runx family members; more than 10 Ets family members, and even TCF-1, which after the initial stages is coexpressed with its close relative LEF-1 (186–188). The patterns of expression of these family members are not the same, and there are some examples of different family members that antagonistically cross-regulate each other in early T cells: e.g. Gfi1 and Gfi1b (189), PU.1 and Ets1 (190, 191), and particular isoforms of TCF-1 against LEF-1 (192, 193). Nevertheless, the cases of Ikaros/Helios and E2A/HEB make it clear that considerable redundancy is real. This systematic lack of parsimony in the cells' regulatory behavior may indicate that evolutionary selection has buttressed these regulatory functions so that they are not vulnerable to mutation in a single gene. Ironically, though, the weak single-gene knockout phenotypes that result may be interpreted to suggest the reverse.
Fourth, unlike the B-cell specification network, the T-cell specification gene regulatory network does not appear to be dominated by positive feedback relationships. Instead, at least three different network modules within the program can be discerned, and the interactions between them are rich with cross-inhibition, autoinhibition, and incoherent feed-forward circuit relationships.
Stages in progression toward T-lineage commitment
Because T-cell development occurs in a separate anatomical compartment from the rest of hematopoiesis, it is easier to define phenotypic stages in the process even before a distinctive set of T-lineage marker genes is turned on. Progressively finer stages have been distinguished by phenotype and placed in sequence by the development of increasingly powerful developmental assay systems. Whereas precursor-product relationships in the early pioneering studies had to be assayed by injection of precursor cells into the thymus of a recipient mouse, the cells could later be manipulated by reconstitution into fetal thymus organ cultures. Elegant modifications of these systems revealed a wealth of information not only about early development but also about later TCR-dependent selection events (194–196). The field of early T-cell development has taken a great step forward with the development of completely open monolayer coculture systems in which the thymus is replaced by OP9 (or Tst4) stromal cells transfected to express DLL1 or DLL4 (197–199). The ability to track, modify, interrupt and restart the T-cell development process at will has made the contingencies of progression and the single-cell developmental potentials at each stage in this system extremely well defined.
The cells that enter the thymus initially (thymus-settling precursors) are not committed to the T-cell lineage. Both in terms of regulatory genes and in terms of their targets, these cells appear very similar to lymphoid-primed multipotent precursors. As they expand in the thymus under the influence of Notch-DLL4 interaction and local Kit ligand expression, they initially express growth factor receptor genes, for example, which are most associated with myeloid and dendritic cell fate (Flt3, Kit, Csf2rb, Csf1r), and signal transduction kinases (Lyn, Btk) that are more commonly associated with B-cell function. However, interestingly, even when they are derived from IL-7R+ CLP (50) and/or Rag1+ Early Lymphocyte precursors (96), these ETP cells have no detectable IL-7R and virtually no Rag1 expression. Conceivably, therefore, their intrathymic development begins with an actual reversal of certain lymphoid-priming gene expression steps that had already occurred in their prethymic antecedents (see below).
More in parallel with B cell precursors, ETP then go on to downregulate Flt3 midway through this first phase. Initial repression of Il-7R and Flt3 downregulation in thymus-settling precursors may be important for the process leading to T-lineage restriction (108, 200) as described in a later section. The survival of these precursors thus becomes dependent on Kit/Kit ligand interaction (201–203), and the high Kit level maintained on ETPs and DN2a cells is one of their defining characteristics. As Flt3 is downregulated, some progenitor-associated transcription factors begin to be turned off (Meis1, Mef2c, Lmo2, and early steps in the downregulation of Bcl11a and Erg) (rev. in (11)), and the pro-myeloid Cebpa regulatory gene begins to be silenced (65, 204). As Notch-DLL4 signaling turns on Hes1, it also causes the cells to activate Gata3 and Tcf7 expression (205, 206). Tcf7, at least, is clearly a direct target of the Notch-RBPJ (CBF1/Suppressor of Hairless/Lag1) transcription factor complex (169, 170). Thus, by the time the first definitive cell-surface markers of entry into the T-cell pathway are expressed during the transition from ETP/DN1 to DN2a, the cells have already modified their progenitor inheritance in important ways.
The transition into DN2a is indicated by upregulation of several other genes that are at least partially driven by Notch signaling. Notch directly induces sharp de novo activation of cell-surface marker CD25 (IL2 receptor α, Il2ra) and the HEBalt promoter isoform of Tcf12 (187). This is accompanied by increases in levels of already-expressed Runx1, Gfi1, Ets1, and the canonical isoform of Tcf12, as well as further increases in Gata3 and Tcf7 expression. These factors promote the first increases in expression of numerous T-cell marker and functional genes including Thy1, Cd3g, Cd3d, and Cd3e, that will increase their expression more dramatically in the next stage. Now Rag1 is (re)activated and Il7r is also strongly upregulated, and the expansion of the cells becomes highly sensitive to IL-7 levels. Even at this early point, the T-cell program begins to offer cells various possible branches of specialization within the T-cell fate. It is at the DN2a/2b stages that the precursors may have their highest propensity to rearrange TCRγ and TCRδ genes instead of TCRβ (207, 208)(Fig. 1), and this too is known to be dependent on IL-7 (209, 210).
Despite their broad expression of T-lineage genes, however, DN2a stage cells remain uncommitted to the T-cell fate (Fig. 2). They have already lost the ability to make B cells, and this ability cannot be restored by removing them from the DLL-rich environment. However, upon transfer to recipient animals or to DLL-free cultures with appropriate cytokines, they can still develop into NK cells, dendritic cells, macrophages, or granulocytes (rev. in (121)). The DN2 population also includes rare cells with mast-cell potential(114). A fraction of them can even respond to the addition of cytokines IL-33 + IL-7 in the presence of Notch signaling, to become an innate lymphocyte type called ILC2 (“nuocyte”)(211). Note that these are not “reprogramming” experiments in which the regulatory state of the cell needs to be altered by transfection to force new transcription factors to be expressed. Instead, these assays explore the flexibility of the cell's intrinsic developmental gene network at that stage, simply providing the intact cell with supportive environmental conditions for a variety of options. The cells remain uncommitted through multiple rounds of cell division under the influence of Notch signaling (212), until they cross into the “DN2b” stage. They then become committed, unable to develop into anything but T cells, no matter what the environment.
Commitment was first detected as linked to activation of a particular Lck-GFP transgene (213), and is functionally dependent on onset of expression of the transcription factor Bcl11b (178, 179). Bcl11b is sharply upregulated in late DN2a stage and then strongly expressed in virtually all T cells from the DN2b stage onward (182, 214). Its advent causes the cytokine receptor Kit to be downregulated, while the CD3 cluster, Thy1, and Rag1 genes are now strongly upregulated. The surrogate TCR chain gene Ptcra, the surrogate partner for a newly rearranged Tcrb gene product to make the pre-TCR, also begins its steep rise in expression which will peak just before β-selection. As additional T-cell signaling genes such as Cd247, Prkcb1 and Itk are turned on, and Zap70 begins to be expressed, the prerequisites for TCR signaling approach completion (65, 215).
The drivers of these climax events in T-cell specification may come from a variety of regulatory inputs, working independently or jointly. From the DN2b stage to the DN3a stage, the cells appear to experience stronger Notch signaling, since the Notch signal-dependent target genes Dtx1 and Notch3 as well as Ptcra all shoot to their highest levels during this transition. Many of the genes activated at this stage are also thought to be E2A/HEB targets (216, 217). In fact, the DN3a phenotype depends on E2A and/or HEB maintenance (218), implying that the net activity of these bHLH factors reaches a maximum at this point as well. The E2A/HEB-dependent genes include the recombinase genes (Rag1, Dntt) that now reach a peak of expression. Meanwhile, GATA-3 reaches its highest permitted level, and TCF-1 is joined by its paralog, LEF-1, from DN2b stage onward. There is a sharp increase in expression of the pan-lymphoid factor Ets1, which among diverse other functions collaborates with Runx1 to activate the enhancer of the TCRβ gene (219). With these major actors on the stage, the regulatory requirements to undergo β-selection are complete.
Molecular switches in the T-cell lineage commitment transition
The exclusion of T-cell fate alternatives is caused at least in part by extinguishing expression of the “non-T” regulatory factors with which the cells began their intrathymic development. This is easy to understand for the factors that would otherwise provide access to myeloid development. C/EBPα is strongly downregulated starting within the ETP stage, and its coding locus Cebpa becomes progressively more heavily modified by repressive histone marks throughout commitment (65). PU.1 itself, although required for the generation of T-cell precursors, is an antagonist of the later T-cell program (190, 191, 220–222). It becomes silenced between the DN2a and DN3a stages (111, 222) through a mechanism dependent on Runx factors and possibly GATA-3 (114, 223–225). With the shutoff of PU.1, the cells lose a crucial prerequisite for both myeloid and dendritic-cell development2. Silenced in approximately the same interval are the stem- and progenitor-cell factor genes Tal1 (coding for SCL), Gfi1b, and Hhex; Lyl1, the paralog of Tal1, is repressed just a stage later, between DN2b and DN3a. Bcl11a, Tsc22d1, and the progenitor-cell-specific Ets-family gene Erg are also downregulated throughout this process, although their final extinction is not until DN3 (65, 215)(rev. in (11)).
The exclusion of the NK cell alternative is a little different, since well-known NK-cell regulatory genes by default begin T-cell development silent in a repressive chromatin context: i.e. Tbx21, Eomes, Zfp105 (65). However, they can be activated readily when the cells are exposed to supportive cytokines in reduced Notch signaling conditions (228). The pathway may involve STAT5 and its potential to induce Id2 (229), assisted by Ets1 activity (230), which is well established in the earliest T-cell precursors. Id2 itself is not a transcriptional activator, but Hhex, one of the progenitor-associated transcription factors expressed normally through the DN2a stage, has recently been found to participate in the NK cell program (231). Another NK program activator, Nfil3 (E4BP4) (232, 233), is also weakly expressed but still available for activation in early T cells, even beyond the ETP and DN2a stages. Conceivably these factors provide the thin end of a wedge for activating the other NK-cell regulators.
The window of opportunity for switching to NK lineage fate is most likely limited by the advent of Bcl11b expression (compare Fig. 1, Fig. 2). Once it is turned on, Bcl11b is a powerful, nonredundant repressor of NK differentiation and NK cell potential. During and even after commitment (179, 180), it is required for the ongoing repression of three regulatory genes involved in NK cell development (231), i.e. Hhex, Id2, and Nfil3. Loss of Bcl11b enables T-lineage cells rapidly to upregulate these genes, followed by Tbx21 and Eomes and their target differentiation genes, as well as other regulators stringently restricted to the NK pathway (180, 181, 234). Thus, whereas other fates are excluded by silencing their main activators, the NK cell fate may be excluded by the activation of a constitutive repressor.
In contrast to factors supporting the myeloid, DC and NK fates, the crucial B-cell factors which distinguish the B-cell pathway from the T-cell pathway, EBF1 and Pax5, are by default both silent and inaccessible in repressive chromatin in ETP populations (65). The mechanism that renders and keeps them silent therefore needs to be explained in terms of the earliest events occurring when cells experience the thymic microenvironment. One factor that is needed for B lineage exclusion is GATA-3 (235), as discussed below.
T-cell transcription factors and T-lineage precursor survival
Molecular mechanisms operating in the T-cell developmental program are not as well understood as those of the B-cell program, but it is clear that specification depends on positive regulatory inputs from GATA-3, TCF-1, and E proteins, under the sustained influence of Notch signals. The roles of these factors are all required, to the best of current knowledge, from the ETP stage through β-selection. During T-lineage specification, these requirements are enforced by linkage of GATA-3, TCF-1, E proteins, together with Notch signals not only to particular differentiation genes as targets, but also to proliferation and survival pathways.
Notch signaling is important for proliferation, and becomes increasingly important for viability as the cells progress through commitment and lose their non-T developmental options. Perhaps the maximum dependence on Notch signals for viability is seen in the committed DN3a cells as they await a successful gene rearrangement for β-selection (236). These cells are no longer proliferating and can only be saved from death by a successful TCRβ gene rearrangement. Yet during this interval they draw support from Notch signals for basic survival until they can qualify for β-selection.
After β-selection, Notch signaling becomes disabled and T-cell identity becomes Notch-independent, presumably sustained because the other factors and Bcl11b continue to play crucial roles.
For GATA-3 and TCF-1, as early as the ETP stage there are catastrophic declines in viability if these factors are removed (170, 237). This is striking because the immediate precursors of ETP, before thymic entry, do not express either factor yet. This newly acquired “addiction” emphasizes how much impact intrathymic signaling has on the programming of cells even before they progress beyond the ETP stage. However, it also means that some gene regulatory effects of these factors are hard to measure, since the cells in which loss of function has altered gene expression may not be viable.
E protein function is important also from the prethymic stages onward (71), and many of the T cell differentiation genes, e.g. Ptcra, the HEBalt promoter of Tcf12, and Rag1, are apparently directly activated by E proteins or E proteins in collaboration with Notch (187, 216, 217, 238–240). Despite partial redundancy with HEB (241), loss of E2A alone causes sharp population decreases in the ETP and DN2a compartments, as well as a severe impact on the regulation of the β-selection checkpoint for cells that manage to get that far (74, 242, 243). HEB (Tcf12) is also crucial for T-cell development, with a special positive role during the DN2-DN3 stages for the “HEBalt” variant isoform of HEB that is induced in response to Notch signaling (187, 244).
Balancing act: restrictive cross-regulation of T-cell transcription factors
These equally essential T-cell factors might be predicted to work in a positive regulatory feedback circuit similar to the E2A, FoxO1, EBF1, and Pax5 circuit in early B cells. Indeed, there is evidence for a positive feedback between E proteins and Notch within the T-cell context (227, 245). An “exception that proves the rule” is the case of TCRγδ cells, which are weaned from their Notch-dependence precociously (207, 246) when, or because, their assembly of TCRγδ complexes triggers many of them to upregulate the E protein antagonist, Id3 (247).
E2A itself helps to turn on Notch1 expression, as one key contribution to T cell development (71, 216). This may seem strange in view of E2A's crucial ruole in B cell development, which is aborted by Notch signals. However, the Notch1 transmembrane molecule is not itself a transcriptional regulator until it is cleaved after contact with a ligand (e.g. DLL4 or DLL1), and cells can express Notch1 very strongly without activating Notch target genes if the environment does not provide these ligands. Therefore, as long as cells are in a DLL-poor environment, there is no contradiction between E2A turning on Notch1 itself and E2A triggering the B cell pathway. Interestingly, in the context of T cell development, Notch1 signaling also may reciprocate to protect E protein activity. E protein antagonists Id2 and Id3 are inducible in T-cell precursors by cytokine or TCR signaling (248). However, Notch signaling in some T-lineage contexts can restrain the activation of these E protein antagonists (227). Thus, as T-cell specification is under way, an environment-dependent positive feedback can be established, and Notch and E proteins are able to work together.
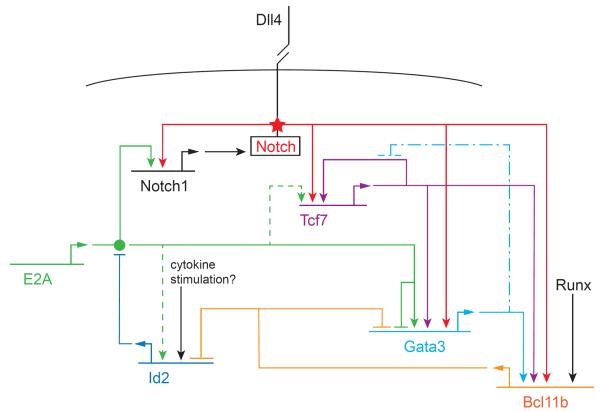
However, the roles of TCF-1 and GATA-3 appear to fall into a separate regulatory circuit module or modules from both Notch and E proteins. TCF-1 and GATA-3 are normally activated in parallel by Notch signaling (Figure 3), and they generally operate together. Tcf7 can also be positively regulated by E proteins, as shown in mature T cells (249). Both TCF-1 and GATA-3 play repeated roles in modulation of various T-cell functions during development and later function, usually coexpressed and often required in parallel. However, they are not obligate partners at the target gene level, but rather operate on distinct sets of target genes and provide inputs to the T cell pathway different from those provided by Notch signaling or E protein action. TCF-1 overexpression in precursors can bypass the need for Notch signals to activate a surprisingly large fraction of the T-cell developmental program(169). In multipotent hematopoietic precursor cells forced to express TCF-1 in the absence of Notch ligands, there is substantial activation of Gata3, endogenous Tcf7 and Lef1, and even Bcl11b, along with Il2ra and some Cd3g expression (Fig. 3; note that inputs from Notch and TCF-1 follow “OR” logic). Some of the target gene effects can be due to collaboration of the exogenous TCF-1 with endogenously activated GATA-3 and E proteins. However, the directly Notch-dependent targets Ptcra and Dtx1 remain completely silent, implying that these TCF-1 effects can be divorced from Notch activation pathways.
FIGURE 3.
Partial gene regulatory network for early T-cell specification. Horizontal lines: genes coding for regulatory factors. Bent arrows: transcription and translation of gene leading to product (itself a regulatory factor). Genes and their products have the same color code. Arrows: positive regulatory effects of factors on indicated genes. Blocked-end lines: negative regulatory effects of factors on indicated genes. Filled circle: ability of E2A to form a functionally active dimer: this assembly is blocked by Id2. Star: activation of Notch transmembrane protein by binding to its ligand Delta-like 4 (DLL4) on a neighboring cell; the effect is to cleave the Notch intracellular domain and allow it to be transported to the nucleus where it functions as a transcriptional coactivator on the indicated target genes. Note that none of these regulators acts in a strictly all-or-nothing way. Although many genes require Notch input for activation, those shown here do not require its continuation for maintenance of of expression. The repressors indicated here modulate rather than silence the indicated target genes. Solid lines: relationships with strong molecular and functional support within the context of T-lineage specification. Dashed lines: relationships seen in related cells but not directly demonstrated in DN T-cell precursors. Long dash-dot lines: inferred effect of GATA-3 on Tcf7 when GATA-3 is overexpressed. Details and sources are given in the text.
GATA-3 expression above a certain threshold level is important not only for ETP viability, but also to furnish DN2 cells with the capacity to advance to DN3 stage (225, 243). In complete knockouts, although a small number of cells still manage to reach a DN2-like stage, GATA-3 loss severely reduces their ability to turn on Bcl11b or Cd3e (235). However, GATA-3 loss does not inhibit activation of Notch target genes such as Ptcra or Il2ra (i.e. the CD25 marker that defines cells as “DN2”), yielding an abnormal phenotype with some genes at DN3-like levels while others are ETP-like (235). Thus, GATA-3 is not needed to sustain the T-cell genes activated in direct responses to Notch signals, nor to transduce Notch signaling itself. Furthermore, although GATA-3 binds to a conserved, active enhancer of the Tcf7 gene in early T cells (65), it is not needed for Tcf7 activation. In this case, although many TCF-1 targets may also require GATA-3, the Tcf7 gene itself may be activated by Notch (169, 170) without GATA-3 (Fig. 3).
GATA-3 can, however, act as an antagonist of T-cell differentiation, rather than as a driver. Evidence actually emerged several years ago, when GATA-3 was tested in gain-of-function scenarios similar to one used successfully to show the positive effects of TCF-1 (114, 250). When higher levels of GATA-3 were introduced to prethymic cells or early ETP/DN2 stage cells, T-cell development was catastrophically inhibited, not promoted. Overexpression did not simply kill the cells, but actually changed their developmental status to drive them out of the T-cell pathway. Survival became possible only if the cells were removed from Notch signaling; then, high-level GATA-3 synergized with the loss of Notch signals to repress Il7r and extinguish the T-cell gene expression profile. It repressed the Tcf7 expression that is normally sustained when Notch signals are withdrawn, suggesting that TCF-1 positive autoregulation might be disabled by high GATA-3 (Fig. 3, dash-dot line). If the cells were kept alive, high-level GATA-3 could convert them into mast cells (114). Thus, T cell identity depends on GATA-3 but also on the preservation of strict limits to GATA-3 expression.
If Gata3 is being activated by Notch signals initially, and then by positive inputs from TCF-1 as well, how can its expression be so carefully restrained during normal T cell development? The answer seems to be by antagonistic feedback from two other essential T-cell regulators: Bcl11b, and E2A (Fig. 3). Deletion of either of these factors causes Gata3 expression to rise (L. Li, J. A. Zhang, and E. V. R., unpublished). Although Bcl11b is activated in a GATA-3 dependent process (235), it also feeds back to limit Gata3 expression (L. Li, J. A. Zhang, and E. V. R., unpublished). In E2A knockout cells, the GATA-3 increase can be enough to disrupt T-cell development: surprisingly, a large part of the E2A-deletion defect can be corrected by shRNA against GATA-3 (243). Thus, E proteins have at least two qualitatively distinct roles in regulating GATA-3 activity. First, they help to turn it on: they may support its expression directly (216, 240), and they also make possible the Notch signaling that turns Gata3 on in the first place and initially protects it from being inhibited by PU.1 (227). Second, they enforce a damping repressive limit on Gata3 expression to prevent it from blocking the T cell pathway (243). Both the positive and the negative roles are critical for T-cell specification.
Special status of E proteins and Notch in the T and B lymphocyte developmental programs and their alternatives
E protein activity is the most important common strand that relates the T-cell and B-cell programs at the transcriptional level. Directly attributable to E protein positive regulation are expression of Rag1/Rag2 genes for immunoreceptor gene rearrangement in both lineages. However, in important ways the linkages of E proteins to other regulatory factors in gene regulatory networks are profoundly different, and this is crucial to explain how the two programs can diverge.
In the B cell program, E2A activity participates directly in the feed-forward regulatory network that confers and sustains B-cell identity (124), including the direct regulation of FoxO, EBF1, and Pax5. It is crucial throughout the bone marrow developmental stages of B-cell development with continuing roles in the periphery (251). Although the strict requirement for E2A in B-cell specification can be bypassed in part by forced expression of EBF1, this is only due to the presence of lower, but potentially functional, levels of the related bHLH factor HEB in E2A-knockout cells (129, 252). The case is different for T cells. In the T-cell program, E protein activity can be dissected from other parts of the specification network circuitry, working against as well as along with the other crucial T-cell factor, GATA-3. Even certain genes like Il7r that receive positive input from E protein activity at early stages can switch to be negatively regulated by E proteins at later stages (239). Working across the decisions mediated by lineage-selection factors like GATA-3 and TCF-1 in T-cell precursors and mature T cells alike, E protein inhibition and release will be used repeatedly as the switch between activated and resting states (239, 248, 253, 254). In particular, while E protein levels will be high during resting and memory states, Id activation to neutralize E proteins will be a broad feature of antigen-triggered T-cell effector activation (255). Thus, E protein dominance need not be a constant in T cell programming. As discussed below, this reversibility of E protein usage gives T cells a particularly close relationship to NK cells and other innate lymphocytes.
T vs. B lineage decision and its alternatives
Double locking mechanism for B cell exclusion of the T cell option
To block access to the T cell program in B-cell precursors, B cell regulatory factors must target the functions that distinguish the T-cell network from the B cell program. They cannot attack E proteins, since these are needed by both developmental programs. Other points of vulnerability in the T-cell program must be targeted for silencing by EBF1 and Pax5, during the transition to Ly-6d+ “BLP” and pro-B cell stage, as cells make the decision to exclude the T-cell fate (Fig. 4).
FIGURE 4.
Key regulatory targets for program maintenance and alternative program exclusion in development of T, B, and innate lymphocytes. The framework is a simplified version of Fig. 2. Green: stages of B-cell development in which EBF1 and Pax5 expression is dominant; i.e. the EBF1 and Pax5 “territory”. Light blue: stages of T-cell development in which GATA-3 and TCF-1 are dominant, i.e. the GATA3 and TCF-1 “territory”. Violet: Id2 “territory” of programs that generate innate cells. Note that the influence of Notch signaling (dark blue frame) does not extend throughout T-cell development but does extend to some ILCs. GATA-3 and TCF-1 also help generating certain ILCs, including a major type of NK cells. The figure depicts the selective cross-repression between the components of B and T-cell programs that uniquely distinguish them from each other. In contrast, E proteins including E2A are components of both.
Pax5 has long been of interest for its negative roles. The concept of commitment as an important stage in hematopoietic development was galvanized by the demonstration that Pax5-deficient pro-B cells could proliferate in vitro for months or years, yet never fully commit to the B cell fate (256, 257). The cells maintain a clearly B-lineage gene expression profile and even undergo significant Ig gene rearrangement as long as IL-7 is supplied. However, upon IL-7 removal, the cells reveal that they still have access to a variety of myeloid and dendritic cell fates (macrophages, granulocytes, dendritic cells, osteoclasts), and if exposed to Notch ligands, they efficiently switch to the T-cell fate. Thus, although it is turned on too late to explain the initial decrease in myeloid potential from LMPP to CLP, Pax5 is involved in the gene network that confirms loss of access to both the T and myeloid developmental pathways.
Although Pax5-knockout pro-B cells have a ready, spontaneous ability to develop into myeloid cells when IL-7 is removed, the fate which is most severely limited by Pax5 is the T cell fate. Ectopic Pax5 expression in hematopoietic precursors has a particularly strong repressive influence on the ability of multipotent precursors to give rise to T cells, and as B-cell development is amplified it is at the expense of T cell development (258, 259). This can be explained by the finding that Pax5 acts as a potent repressor of Notch1 expression itself.
In this respect, from the moment a B-cell precursor achieves an adequate level of Pax5, it should become the equivalent of a Notch1-deficient precursor, selectively blocked in T-cell potential. However, biasing against T-cell potential in Ly6d+ BLPs begins even before the cells fully express Pax5 (102, 103). Furthermore, even Pax5-deficient B-cell precursors can be completely blocked from T-cell development, provided that their EBF1 is sufficiently activated (159). If Ebf1 is deleted, pro-B cells with the Pax5 gene intact regain the ability to enter the T-cell pathway (260, 261). Thus EBF1 as well as Pax5 plays a role in blocking access to the T-cell program.
Recent evidence shows that EBF1 can downregulate both Gata3 and Tcf7 (260, 261). It binds directly to sites in the Tcf7 gene and at least two sites bracketing the transcriptional start site of Gata3, one or both of which are important for repression function. When EBF1 binding is blocked at these sites, Gata3 expression surges again (260). Notably, EBF1 appears to be repressing here by acting as an “anti-insulator”. The position of the upstream EBF1 site lies at the boundary between the open chromatin of the Gata3 gene and a large block of repressed chromatin (strongly marked with H3K27me3 modification), which constitutively covers the Gata3 “1a” promoter in precursors (65). In T-lineage cells, the main Gata3 “1b” promoter is clear to drive expression. EBF1 binding, however, allows the Gata3 1b promoter and first intron to become buried in repressive marks as well (260), i.e., presumably by allowing the repressive chromatin to spread. Gata3 and Tcf7 repression condemns to death any cell that may be embarking on the T cell program in response to Notch signals. Thus, EBF1 and Pax5 complementarily block access to the T cell fate, first by preventing Notch signaling from turning on the T-cell regulatory program even in precursors that still express substantial Notch1, and then by removing Notch1 itself (Fig. 4).
Divergence of T-cell precursors from CLP: undoing a B-cell promoting state
To avoid these repressive events, T-cell precursors themselves must apparently forego using the early B-lineage events broadly as a “head start”, even in cases where later they will turn on the same genes. It is likely that the initial expression of Ets1, Ikaros, Runx1, Myb, and even factors like PU.1 in LMPPs and CLPs may be equally efficient as a start for both. However, in specific ways the T-cell precursors must start by weakening the B-lineage biases that may already be imposed by expression of IL-7R and E proteins, and revealed by expression of Rag1.
Even strong IL-7R expression from the earliest stage can be a problem for T-cell precursors. Indeed, forced expression of either IL-7R or constitutively activated STAT5 can redirect ETP differentiation toward the B-cell fate even within the thymus (262). Interestingly, as noted above, both Il7r and Rag1/Rag2 are activated in early B-lineage cells in a FoxO1-dependent way. Thus, one way to downregulate both genes might be simply to switch growth factor receptor dependence so as to allow Akt activation to inhibit FoxO1. PI 3-kinase activation by signals from the growth factor receptor Kit, which is vital for early T-cell development (263), could play a role in establishing this condition (264), until the B-cell pathway is fully inaccessible.
The relationship of E proteins to Notch signals must be altered in order to allow the T lineage to develop. In the B cell program, Notch signaling itself has the power to antagonize E2A itself, by inducing degradation of the E2A protein (265). This mechanism has been invoked as one of the barriers to B cell development in the thymus. Indeed, Notch signaling applied to CLP may initially prime the cells for development into innate type lymphocytes, such as NK cells and lymphoid tissue inducer cells (discussed further below), i.e., differentiation fates which depend on antagonism or loss of E protein function (266, 267). Why does this not occur in developing T-cell precursors, then, or how do they escape from the late ETP stage on? One part of the answer is the regulation of the components that work downstream of Notch to target E2A for degradation. The critical step in Notch-triggered E2A degradation is the phosphorylation of E2A in a conserved domain by a MAP kinase (265). B cell precursors are richly endowed with MAP kinase activity. Peculiarly, though, T cell precursors maintain unusually low levels of MAP kinase throughout the specification stages (268). Thus E2A protein becomes much less sensitive to degradation by Notch at an early stage of the T-cell program, enabling it to establish its positive collaborative circuit with Notch described above (Fig. 3).
T cell exclusion of the B cell option
The crucial E2A target gene for activation of the B cell program is EBF1, as we have seen. Both FoxO1 and STAT5a/b have been invoked as collaborators in this process, yet neither one is excluded from the T-cell program. STAT5a and b are well expressed and activated by IL-7R signaling in the DN2a/2b stages particularly; and although FoxO1 expression is higher in B-lineage precursors, T-lineage cells compensate with high expression of its close relative, FoxO3a. Thus, all the expected requirements for E2A activation of EBF1 are there. However, despite co-activation of these potential collaborators with E2A in the early T-cell program, EBF1 and Pax5 are never activated from the closed state in chromatin in which they reside in multipotent precursors and stem cells (65, 269). One possibility is that the E2A levels simply fail to climb high enough in the early T-cell program to activate EBF1; it is true that Tcf3 (or Tcfe2a, codes for E2A) transcript levels are notably stable throughout T-cell development, ~3× fold lower than their peaks in pro-B and pre-B cells. This might suffice if EBF1 activation by E2A were strictly dose dependent, more so than other target genes. There are, however, variant CLP with particularly low E2A:Id2 ratios that are reported to be able to generate B cells and NK cells but not T cells (98). Extrapolating from the behavior of these cells, the E protein-dependent functions in T cell development would require even higher doses of E protein activity than those in B-cell development, not lower ones. A more satisfying explanation may therefore be that one of the early-activated genes in the T cell program actively interferes with the ability of E2A to activate EBF1, or directly singles EBF1 out for silencing.
The timing of the loss of B-cell potential in developing T cells implies that one of the earliest Notch-induced regulatory changes in these cells creates an intrinsic barrier to the B-cell program, long before the cells make the transition from ETP to DN2a (Fig. 4). Three T-lineage regulatory candidates that are induced strongly in this period are Hes1, TCF-1, and GATA-3. Hes1 is required for T-cell specification and population survival, but Hes1 conditional knockout cells do not expand the population of B cells in the thymus, showing that the inhibition of B-cell development by Notch signaling remains intact (270); and Hes1 is actually expressed in early B lineage precursors. Tcf7 (encoding TCF-1) knockout cells are also severely blocked in T cell development, but they do not display increased B or myeloid potential (169, 170). The later lineage commitment factor Bcl11b is expressed too late to account for B-cell exclusion (111, 182), and does not contribute to this aspect of commitment (225). It is GATA-3, rather, that appears to have an indispensable activity as an antagonist of B-cell development (235). The extreme impacts of Gata3 deletion on early T-lineage survival and proliferation have made it necessary to exclude alternative interpretations assiduously (237), but the evidence seems strong. The cells that can give rise to B cells may proceed as far as turning on Tcf7 and Notch target genes, showing that they are not deficient at entering the T-cell pathway(235). They become arrested in a DN2a-like state, as their expression of Bcl11b and the Cd3 genes remains low. However, it is the loss of Gata3 and not the loss of Bcl11b that restores access to the B-lineage program, as shown by analysis of T-cell precursors from single and double conditional deletion mutants (225). The exact mechanism through which GATA-3 is working to prohibit B-lineage specification is still under investigation, but its timing of expression and functional impact imply that it is the crucial mediator.
Common vs. exclusive transcription factor modules: linkages to “innate” lymphoid alternatives
The parallel aspects of the T and B cell programs reflect their shared use of gene rearrangement and selection in a particular order to enable the cells to acquire recognition specificities, and as already described in both lineages these shared features are regulated at least in part by E protein activity. Some of the most discordant aspects have to do with function. Programming for B-cell function is mostly concerned with making the cells responsive to Ig-based signals that trigger cellular decisions to proliferate, survive or die, and to express different variants of the cell's immunoglobulin genes (some of which require additional rounds of somatic mutagenesis). The same genes that confer B cells' main recognition specificity are the genes that confer most of their effector function. In contrast, the programming for T-cell function requires transcriptional poising and regulation of a considerably broader set of effector-related gene loci in addition to the genes that regulate recognition, proliferation, survival, and apoptosis decisions.
Importantly, these effector functional elements are not unique to T cells, but they are not shared with B cells; instead, they are shared with the newly appreciated group of non-T, non-B cells called innate lymphocytes (ILC), which do not use RAG-based gene rearrangement in their developmental programs. This creates a disjunction between recognition-based and effector-based criteria for defining the closest relatives of T cells, and this poses a challenge to any uniquely intimate relationship between T and B cell programs.
The ILC lineages of lymphocytes depend not on E protein activity but on the neutralization of E proteins by Id2 in order to develop (Fig. 4). ILCs include not only natural killer (NK) cells, similar to cytotoxic T lymphocytes, but also a range of other cytokine-producing innate lymphocyte subsets corresponding roughly to Th2 and Th17 T cells (271, 272). None of these correspond to “innate B cells” – it appears that the B cell program has too direct and intimate an involvement of E proteins in every aspect of cellular function to possess an “innate” equivalent. However, the T cell effector programs are not E protein dependent in the same way. They depend on GATA3, TCF-1/LEF-1, Runx factors, T-bet (Tbx21) or Eomesodermin (Eomes), Tox, the nuclear receptor Rorγt (Rorc), Maf, Batf, Ahr, and other factors in different combinations. To a large extent, these are the same factors which the innate-type lymphocytes deploy for their own corresponding functional programs. These effector transcriptional programs are thus separable from the developmental use of E proteins, which play their main T-cell developmental roles in setting up TCR gene rearrangement and selection checkpoints.
The common thread linking all the ILCs is the fact that they all depend on Id2 (Fig. 4). Because of their neutralization of E protein activity, ILCs do not express Rag genes. However, this does not give them a profoundly different heritage from T and B precursors. In fact, lineage tracing has shown that most NK cells descend from Rag1+ precursors (96). Thus, it is important to note that Id2 is not properly a lineage-specific identifier. It is readily inducible in common lymphoid precursors and early T-cell precursors until the activation of definitive commitment factors EBF1 and Bcl11b, respectively. Even after T-lineage commitment, it is regularly, albeit transiently, upregulated in mature T cell function (as Bcl11b levels temporarily drop) in the innate-like “NKT pathway and when an effector response is induced. Here, the modularity of the T-cell program is a great help to understand its relationship to ILC developmental lineages. As late as the DN2a stage, signals from IL-33 can redirect the effects of factors like GATA-3 and TCF-1 to work in the ILC2 program (Fig. 2), via induction of Id2 and the orphan nuclear receptor RORα (211, 273–275). Thus, while repression of PU.1 helps to explain the loss of dendritic cell or myeloid access during commitment, it is easiest to explain the loss of access to the innate lymphocyte fates by forcible imposition and protection of E protein activity, at a minimum in two phases to last just long enough for TCR gene rearrangement and TCR-based selection to be complete.
The connection between T and innate cell developmental programs is thus arguably much closer than that between the T cell program and the B cell program. Three different innate cell developmental programs share major aspects of their regulatory programs with three different branches of the T-cell development pathway. Nevertheless, the very bridge between the T-cell and innate cell programs is based on eliminating exactly those regulatory functions, the E protein dependent expression of Rag genes, that T and B cells most uniquely share. Thus, the regulatory factors and regulatory circuit modules that determine lymphocyte identity do not act in a simple hierarchy of relatedness, but rather in a matrix of regulatory interactions, which crosses valuable cellular features to generate a richly diversified immune functional repertoire.
Concluding Remarks
The T-cell and B-cell programs are linked by their shared use of E protein-dependent genes which include the functions they share for immunoreceptor gene rearrangement as well as lineage-specific functions. Their common progenitors share the need for IL-7 receptor signaling, which provides unique support functions, and the need for E protein activation and protection from inhibitors. Both the T-cell program and the B cell program use repressors of the E-protein antagonist Id2, Bcl11b and EBF1 respectively, at pivotal stages in their lineage commitment processes. However, the architectures of the gene networks in which these lineage commitment factors work are distinct. In the case of the B-cell program, E protein use is also tightly interlocked with lineage-specific factors in positive feedback and feed-forward circuits that control direct specification functions. In the case of the T-cell program, E protein use is modular and capable of being unlinked from the circuitry that maintains T-cell identity factors such as GATA-3 and TCF-1. Transcription factor deployment generates cell type identity, but transcription factors can be used in different combinations and may be expressed in cycles of repression and reactivation. There is much still to be learned to clarify the mechanisms involved, particularly to connect factor expression to profiles of genome-wide binding to transcriptional function, and to reveal the ways that environmental signals impinge on these relationships. The results already emphasize that single factors must be viewed not as “master regulators” but as participants in a richly endowed and interlinked regulatory context. To understand the relationship between lymphocyte developmental programs, it is important to understand the operating rules for the transcriptional regulatory networks at their hearts as well as the components from which they are built.
ACKNOWLEDGMENTS
I am grateful to many colleagues for inspiring this paper, and I especially wish to thank the attendees of the FASEB Conferences on Molecular Mechanisms of Lymphocyte Development and Function and the Aegean Conferences on Gene Regulation in Lymphocyte Development for discussions, clarifications, and insights. I also thank members of my group for letting me refer to their own work before publication. Development of these ideas was supported by NIH grants CA148278, CA90233, HL89123, AI083514 and AI95943 and by the Albert Billings Ruddock Professorship.
Footnotes
Although binding sites for STAT5 have been identified in the Ebf1 cis-regulatory regions, the role of IL-7 receptor signaling may be more to support viability of the cells or to enhance Ebf1 expression indirectly than to participate directly in Ebf1 activation (123). However, this dependence has been evaluated in cells that have already turned on substantial Rag1 expression, which may reflect primarily “BLP” and later cells.
This helps explain the loss of access to the “myeloid dendritic” or “conventional dendritic” cell pathways, but does not fully explain the cells' inability to generate plasmacytoid dendritic cells. Unlike other dendritic cells, plasmacytoid dendritic cells use the close PU.1 relative SpiB instead of PU.1 (226). SpiB expression actually seems to be antagonized by PU.1 in thymocytes (190) and shoots up in a transient spike in DN3a cells as they await β-selection, after the cells are already committed. In principle this might enable DN3 cells to become plasmacytoid dendritic cells (226), but it does not under normal conditions. The high levels of GATA-3 also expressed in the DN3 cells may provide the restraint on dendritic cell development (226, 227)
REFERENCES
- 1.Bajoghli B, Aghaallaei N, Hess I, Rode I, Netuschil N, Tay BH, Venkatesh B, Yu JK, Kaltenbach SL, Holland ND, Diekhoff D, Happe C, Schorpp M, Boehm T. Evolution of genetic networks underlying the emergence of thymopoiesis in vertebrates. Cell. 2009;138:186–97. doi: 10.1016/j.cell.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 2.Ramirez J, Lukin K, Hagman J. From hematopoietic progenitors to B cells: mechanisms of lineage restriction and commitment. Curr.Opin.Immunol. 2010;22:177–84. doi: 10.1016/j.coi.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zandi S, Bryder D, Sigvardsson M. Load and lock: the molecular mechanisms of B-lymphocyte commitment. Immunol Rev. 2010;238:47–62. doi: 10.1111/j.1600-065X.2010.00950.x. [DOI] [PubMed] [Google Scholar]
- 4.Mandel EM, Grosschedl R. Transcription control of early B cell differentiation. Curr.Opin.Immunol. 2010;22:161–7. doi: 10.1016/j.coi.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 5.Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat.Immunol. 2007;8:463–70. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- 6.Matthias P, Rolink AG. Transcriptional networks in developing and mature B cells. Nat Rev.Immunol. 2005;5:497–508. doi: 10.1038/nri1633. [DOI] [PubMed] [Google Scholar]
- 7.Thompson PK, Zúñiga-Pflücker JC. On becoming a T cell, a convergence of factors kick it up a Notch along the way. Semin Immunol. 2011;23:350–9. doi: 10.1016/j.smim.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Naito T, Tanaka H, Naoe Y, Taniuchi I. Transcriptional control of T-cell development. Int Immunol. 2011;23:661–8. doi: 10.1093/intimm/dxr078. [DOI] [PubMed] [Google Scholar]
- 9.Love PE, Bhandoola A. Signal integration and crosstalk during thymocyte migration and emigration. Nat Rev Immunol. 2011;11:469–77. doi: 10.1038/nri2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Q, Bell JJ, Bhandoola A. T-cell lineage determination. Immunol.Rev. 2010;238:12–22. doi: 10.1111/j.1600-065X.2010.00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rothenberg EV, Zhang J, Li L. Multilayered specification of the T-cell lineage fate. Immunol Rev. 2010;238:150–68. doi: 10.1111/j.1600-065X.2010.00964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrie HT, Zuniga-Pflucker JC. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu Rev Immunol. 2007;25:649–79. doi: 10.1146/annurev.immunol.23.021704.115715. [DOI] [PubMed] [Google Scholar]
- 14.Ding L, Morrison SJ. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 2013;495:231–5. doi: 10.1038/nature11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenbaum A, Hsu YM, Day RB, Schuettpelz LG, Christopher MJ, Borgerding JN, Nagasawa T, Link DC. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495:227–30. doi: 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sitnicka E, Buza-Vidas N, Ahlenius H, Cilio CM, Gekas C, Nygren JM, Mansson R, Cheng M, Jensen CT, Svensson M, Leandersson K, Agace WW, Sigvardsson M, Jacobsen SE. Critical role of FLT3 ligand in IL-7 receptor independent T lymphopoiesis and regulation of lymphoid-primed multipotent progenitors. Blood. 2007;110:2955–64. doi: 10.1182/blood-2006-10-054726. [DOI] [PubMed] [Google Scholar]
- 17.Dolence JJ, Gwin K, Frank E, Medina KL. Threshold levels of Flt3-ligand are required for the generation and survival of lymphoid progenitors and B-cell precursors. Eur J Immunol. 2011;41:324–34. doi: 10.1002/eji.201040710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narayan K, Sylvia KE, Malhotra N, Yin CC, Martens G, Vallerskog T, Kornfeld H, Xiong N, Cohen NR, Brenner MB, Berg LJ, Kang J, Immunological Genome Project C Intrathymic programming of effector fates in three molecularly distinct γδ T cell subtypes. Nat Immunol. 2012;13:511–8. doi: 10.1038/ni.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kreslavsky T, Gleimer M, Garbe AI, von Boehmer H. αβ versus γδ fate choice: counting the T-cell lineages at the branch point. Immunol Rev. 2010;238:169–81. doi: 10.1111/j.1600-065X.2010.00947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonneville M, O'Brien RL, Born WK. γδ T cell effector functions: a blend of innate programming and acquired plasticity. Nat Rev Immunol. 2010;10:467–78. doi: 10.1038/nri2781. [DOI] [PubMed] [Google Scholar]
- 21.Taghon T, Rothenberg EV. Molecular mechanisms that control mouse and human TCR-αβ and TCR-γδ T cell development. Semin Immunopathol. 2008;30:383–98. doi: 10.1007/s00281-008-0134-3. [DOI] [PubMed] [Google Scholar]
- 22.Xiong N, Raulet DH. Development and selection of γδ T cells. Immunol Rev. 2007;215:15–31. doi: 10.1111/j.1600-065X.2006.00478.x. [DOI] [PubMed] [Google Scholar]
- 23.Hsieh CS, Lee HM, Lio CW. Selection of regulatory T cells in the thymus. Nat Rev Immunol. 2012;12:157–67. doi: 10.1038/nri3155. [DOI] [PubMed] [Google Scholar]
- 24.Wirnsberger G, Hinterberger M, Klein L. Regulatory T-cell differentiation versus clonal deletion of autoreactive thymocytes. Immunol Cell Biol. 2011;89:45–53. doi: 10.1038/icb.2010.123. [DOI] [PubMed] [Google Scholar]
- 25.Alonzo ES, Sant'Angelo DB. Development of PLZF-expressing innate T cells. Curr Opin Immunol. 2011;23:220–7. doi: 10.1016/j.coi.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carpenter AC, Bosselut R. Decision checkpoints in the thymus. Nat Immunol. 2010;11:666–73. doi: 10.1038/ni.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naito T, Taniuchi I. The network of transcription factors that underlie the CD4 versus CD8 lineage decision. Int Immunol. 2010;22:791–6. doi: 10.1093/intimm/dxq436. [DOI] [PubMed] [Google Scholar]
- 28.He X, Park K, Kappes DJ. The role of ThPOK in control of CD4/CD8 lineage commitment. Annu.Rev.Immunol. 2010;28:295–320. doi: 10.1146/annurev.immunol.25.022106.141715. [DOI] [PubMed] [Google Scholar]
- 29.Das R, Sant'Angelo DB, Nichols KE. Transcriptional control of invariant NKT cell development. Immunol Rev. 2010;238:195–215. doi: 10.1111/j.1600-065X.2010.00962.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collins A, Littman DR, Taniuchi I. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat.Rev.Immunol. 2009;9:106–15. doi: 10.1038/nri2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwartzberg PL, Mueller KL, Qi H, Cannons JL. SLAM receptors and SAP influence lymphocyte interactions, development and function. Nat Rev Immunol. 2009;9:39–46. doi: 10.1038/nri2456. [DOI] [PubMed] [Google Scholar]
- 32.Wang L, Bosselut R. CD4−CD8 lineage differentiation: Thpok-ing into the nucleus. J.Immunol. 2009;183:2903–10. doi: 10.4049/jimmunol.0901041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singer A, Adoro S, Park JH. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat.Rev.Immunol. 2008;8:788–801. doi: 10.1038/nri2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yosef N, Shalek AK, Gaublomme JT, Jin H, Lee Y, Awasthi A, Wu C, Karwacz K, Xiao S, Jorgolli M, Gennert D, Satija R, Shakya A, Lu DY, Trombetta JJ, Pillai MR, Ratcliffe PJ, Coleman ML, Bix M, Tantin D, Park H, Kuchroo VK, Regev A. Dynamic regulatory network controlling TH17 cell differentiation. Nature. 2013;496:461–8. doi: 10.1038/nature11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkurst CN, Muratet M, Newberry KM, Meadows S, Greenfield A, Yang Y, Jain P, Kirigin FK, Birchmeier C, Wagner EF, Murphy KM, Myers RM, Bonneau R, Littman DR. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, Yan X, Zhong B, Nurieva RI, Wang A, Wang X, Martin-Orozco N, Wang Y, Chang SH, Esplugues E, Flavell RA, Tian Q, Dong C. Bcl6 expression specifies the T follicular helper cell program in vivo. J Exp Med. 2012;209:1841–52. S1–24. doi: 10.1084/jem.20120219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vahedi G, Takahashi H, Nakayamada S, Sun HW, Sartorelli V, Kanno Y, O'Shea JJ. STATs shape the active enhancer landscape of T cell populations. Cell. 2012;151:981–93. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samstein RM, Arvey A, Josefowicz SZ, Peng X, Reynolds A, Sandstrom R, Neph S, Sabo P, Kim JM, Liao W, Li MO, Leslie C, Stamatoyannopoulos JA, Rudensky AY. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151:153–66. doi: 10.1016/j.cell.2012.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei G, Abraham BJ, Yagi R, Jothi R, Cui K, Sharma S, Narlikar L, Northrup DL, Tang Q, Paul WE, Zhu J, Zhao K. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity. 2011;35:299–311. doi: 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, Schones DE, Peng W, Sun HW, Paul WE, O'Shea JJ, Zhao K. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–67. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luc S, Luis TC, Boukarabila H, Macaulay IC, Buza-Vidas N, Bouriez-Jones T, Lutteropp M, Woll PS, Loughran SJ, Mead AJ, Hultquist A, Brown J, Mizukami T, Matsuoka S, Ferry H, Anderson K, Duarte S, Atkinson D, Soneji S, Domanski A, Farley A, Sanjuan-Pla A, Carella C, Patient R, de Bruijn M, Enver T, Nerlov C, Blackburn C, Godin I, Jacobsen SE. The earliest thymic T cell progenitors sustain B cell and myeloid lineage potential. Nat Immunol. 2012;13:412–9. doi: 10.1038/ni.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doulatov S, Notta F, Eppert K, Nguyen LT, Ohashi PS, Dick JE. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat.Immunol. 2010;11:585–93. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- 43.Kawamoto H, Ikawa T, Masuda K, Wada H, Katsura Y. A map for lineage restriction of progenitors during hematopoiesis: the essence of the myeloid-based model. Immunol.Rev. 2010;238:23–36. doi: 10.1111/j.1600-065X.2010.00959.x. [DOI] [PubMed] [Google Scholar]
- 44.Ng SY, Yoshida T, Zhang J, Georgopoulos K. Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity. 2009;30:493–507. doi: 10.1016/j.immuni.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Månsson R, Hultquist A, Luc S, Yang L, Anderson K, Kharazi S, Al Hashmi S, Liuba K, Thorén L, Adolfsson J, Buza-Vidas N, Qian H, Soneji S, Enver T, Sigvardsson M, Jacobsen SEW. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity. 2007;26:407–19. doi: 10.1016/j.immuni.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 46.Lai AY, Kondo M. Identification of a bone marrow precursor of the earliest thymocytes in adult mouse. Proc.Natl.Acad.Sci.U.S.A. 2007;104:6311–6. doi: 10.1073/pnas.0609608104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lai AY, Kondo M. Asymmetrical lymphoid and myeloid lineage commitment in multipotent hematopoietic progenitors. J. Exp. Med. 2006;203:1867–73. doi: 10.1084/jem.20060697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perry SS, Welner RS, Kouro T, Kincade PW, Sun XH. Primitive lymphoid progenitors in bone marrow with T lineage reconstituting potential. J.Immunol. 2006;177:2880–7. doi: 10.4049/jimmunol.177.5.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richie Ehrlich LI, Serwold T, Weissman IL. In vitro assays misrepresent in vivo lineage potentials of murine lymphoid progenitors. Blood. 2011;117:2618–24. doi: 10.1182/blood-2010-05-287102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schlenner SM, Madan V, Busch K, Tietz A, Laufle C, Costa C, Blum C, Fehling HJ, Rodewald HR. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity. 2010;32:426–36. doi: 10.1016/j.immuni.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 51.Saran N, Lyszkiewicz M, Pommerencke J, Witzlau K, Vakilzadeh R, Ballmaier M, von Boehmer H, Krueger A. Multiple extrathymic precursors contribute to T-cell development with different kinetics. Blood. 2010;115:1137–44. doi: 10.1182/blood-2009-07-230821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Serwold T, Ehrlich LI, Weissman IL. Reductive isolation from bone marrow and blood implicates common lymphoid progenitors as the major source of thymopoiesis. Blood. 2009;113:807–15. doi: 10.1182/blood-2008-08-173682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luc S, Anderson K, Kharazi S, Buza-Vidas N, Boiers C, Jensen CT, Ma Z, Wittmann L, Jacobsen SE. Down-regulation of Mpl marks the transition to lymphoid-primed multipotent progenitors with gradual loss of granulocytemonocyte potential. Blood. 2008;111:3424–34. doi: 10.1182/blood-2007-08-108324. [DOI] [PubMed] [Google Scholar]
- 54.Adolfsson J, Månsson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA, Anderson K, Sitnicka E, Sasaki Y, Sigvardsson M, Jacobsen SE. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential: a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 55.Ichii M, Shimazu T, Welner RS, Garrett KP, Zhang Q, Esplin BL, Kincade PW. Functional diversity of stem and progenitor cells with B-lymphopoietic potential. Immunol.Rev. 2010;237:10–21. doi: 10.1111/j.1600-065X.2010.00933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Allman D, Sambandam A, Kim S, Miller JP, Pagan A, Well D, Meraz A, Bhandoola A. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol. 2003;4:168–74. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- 57.Welner RS, Pelayo R, Nagai Y, Garrett KP, Wuest TR, Carr DJ, Borghesi LA, Farrar MA, Kincade PW. Lymphoid precursors are directed to produce dendritic cells as a result of TLR9 ligation during herpes infection. Blood. 2008;112:3753–61. doi: 10.1182/blood-2008-04-151506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lai AY, Watanabe A, O'Brien T, Kondo M. Pertussis toxin-sensitive G proteins regulate lymphoid lineage specification in multipotent hematopoietic progenitors. Blood. 2009;113:5757–64. doi: 10.1182/blood-2009-01-201939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kondo M, Scherer DC, Miyamoto T, King AG, Akashi K, Sugamura K, Weissman IL. Cell-fate conversion of lymphoid-committed progenitors by instructive actions of cytokines. Nature. 2000;407:383–6. doi: 10.1038/35030112. [DOI] [PubMed] [Google Scholar]
- 60.Hsu CL, Kikuchi K, Kondo M. Activation of MEK/ERK signaling pathway is involved in the myeloid lineage commitment. Blood. 2007;110:1420–8. doi: 10.1182/blood-2007-02-071761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heinzel K, Benz C, Martins VC, Haidl ID, Bleul CC. Bone marrow-derived hemopoietic precursors commit to the T cell lineage only after arrival in the thymic microenvironment. J Immunol. 2007;178:858–68. doi: 10.4049/jimmunol.178.2.858. [DOI] [PubMed] [Google Scholar]
- 62.Krueger A, von Boehmer H. Identification of a T lineage-committed progenitor in adult blood. Immunity. 2007;26:105–16. doi: 10.1016/j.immuni.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwarz BA, Sambandam A, Maillard I, Harman BC, Love PE, Bhandoola A. Selective thymus settling regulated by cytokine and chemokine receptors. J.Immunol. 2007;178:2008–17. doi: 10.4049/jimmunol.178.4.2008. [DOI] [PubMed] [Google Scholar]
- 64.Benz C, Bleul CC. A multipotent precursor in the thymus maps to the branching point of the T versus B lineage decision. J. Exp. Med. 2005;202:21–31. doi: 10.1084/jem.20050146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 2012;149:467–82. doi: 10.1016/j.cell.2012.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu Y, Wang J, Khaled W, Burke S, Li P, Chen X, Yang W, Jenkins NA, Copeland NG, Zhang S, Liu P. Bcl11a is essential for lymphoid development and negatively regulates p53. J Exp Med. 2012;209:2467–83. doi: 10.1084/jem.20121846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mercer EM, Lin YC, Benner C, Jhunjhunwala S, Dutkowski J, Flores M, Sigvardsson M, Ideker T, Glass CK, Murre C. Multilineage priming of enhancer repertoires precedes commitment to the B and myeloid cell lineages in hematopoietic progenitors. Immunity. 2011;35:413–25. doi: 10.1016/j.immuni.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Pooter RF, Kee BL. E proteins and the regulation of early lymphocyte development. Immunol.Rev. 2010;238:93–109. doi: 10.1111/j.1600-065X.2010.00957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Semerad CL, Mercer EM, Inlay MA, Weissman IL, Murre C. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proc.Natl.Acad.Sci.U.S.A. 2009;106:1930–5. doi: 10.1073/pnas.0808866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dias S, Mansson R, Gurbuxani S, Sigvardsson M, Kee BL. E2A proteins promote development of lymphoid-primed multipotent progenitors. Immunity. 2008;29:217–27. doi: 10.1016/j.immuni.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Winandy S, Wu P, Georgopoulos K. A dominant mutation in the Ikaros gene leads to rapid development of leukemia and lymphoma. Cell. 1995;83:289–99. doi: 10.1016/0092-8674(95)90170-1. [DOI] [PubMed] [Google Scholar]
- 73.Liu P, Keller JR, Ortiz M, Tessarollo L, Rachel RA, Nakamura T, Jenkins NA, Copeland NG. Bcl11a is essential for normal lymphoid development. Nat.Immunol. 2003;4:525–32. doi: 10.1038/ni925. [DOI] [PubMed] [Google Scholar]
- 74.Bain B, Engel I, Robanus Maandag EC, te Riele HP, Voland JR, Sharp LL, Chun J, Huey B, Pinkel D, Murre C. E2A deficiency leads to abnormalities in αβ T-cell development and to rapid development of T-cell lymphomas. Mol. Cell. Biol. 1997;17:4782–91. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Györy I, Boller S, Nechanitzky R, Mandel E, Pott S, Liu E, Grosschedl R. Transcription factor Ebf1 regulates differentiation stage-specific signaling, proliferation, and survival of B cells. Genes Dev. 2012;26:668–82. doi: 10.1101/gad.187328.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fahl SP, Crittenden RB, Allman D, Bender TP. c-Myb is required for pro-B cell differentiation. J.Immunol. 2009;183:5582–92. doi: 10.4049/jimmunol.0901187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Greig KT, Carotta S, Nutt SL. Critical roles for c-Myb in hematopoietic progenitor cells. Semin.Immunol. 2008;20:247–56. doi: 10.1016/j.smim.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 78.Lieu YK, Kumar A, Pajerowski AG, Rogers TJ, Reddy EP. Requirement of cmyb in T cell development and in mature T cell function. Proc Natl Acad Sci U S A. 2004;101:14853–8. doi: 10.1073/pnas.0405338101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bender TP, Kremer CS, Kraus M, Buch T, Rajewsky K. Critical functions for c-Myb at three checkpoints during thymocyte development. Nat.Immunol. 2004;5:721–9. doi: 10.1038/ni1085. [DOI] [PubMed] [Google Scholar]
- 80.Zohren F, Souroullas GP, Luo M, Gerdemann U, Imperato MR, Wilson NK, Göttgens B, Lukov GL, Goodell MA. The transcription factor Lyl-1 regulates lymphoid specification and the maintenance of early T lineage progenitors. Nat Immunol. 2012;13:761–9. doi: 10.1038/ni.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li H, Ji M, Klarmann KD, Keller JR. Repression of Id2 expression by Gfi-1 is required for B-cell and myeloid development. Blood. 2010;116:1060–9. doi: 10.1182/blood-2009-11-255075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Spooner CJ, Cheng JX, Pujadas E, Laslo P, Singh H. A recurrent network involving the transcription factors PU.1 and Gfi1 orchestrates innate and adaptive immune cell fates. Immunity. 2009;31:576–86. doi: 10.1016/j.immuni.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hock H, Orkin SH. Zinc-finger transcription factor Gfi-1: versatile regulator of lymphocytes, neutrophils and hematopoietic stem cells. Curr.Opin.Hematol. 2006;13:1–6. doi: 10.1097/01.moh.0000190111.85284.8f. [DOI] [PubMed] [Google Scholar]
- 84.Möröy T. The zinc finger transcription factor Growth factor independence 1 (Gfi1) Int.J Biochem.Cell Biol. 2005;37:541–6. doi: 10.1016/j.biocel.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 85.Yücel R, Karsunky H, Klein-Hitpass L, Möröy T. The transcriptional repressor Gfi1 affects development of early, uncommitted c-Kit + T cell progenitors and CD4/CD8 lineage decision in the thymus. J. Exp. Med. 2003;197:831–44. doi: 10.1084/jem.20021417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Malin S, McManus S, Busslinger M. STAT5 in B cell development and leukemia. Curr Opin Immunol. 2010;22:168–76. doi: 10.1016/j.coi.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 87.Yao Z, Cui Y, Watford WT, Bream JH, Yamaoka K, Hissong BD, Li D, Durum SK, Jiang Q, Bhandoola A, Hennighausen L, O'Shea JJ. Stat5a/b are essential for normal lymphoid development and differentiation. Proc Natl Acad Sci U S A. 2006;103:1000–5. doi: 10.1073/pnas.0507350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Seo W, Ikawa T, Kawamoto H, Taniuchi I. Runx1-Cbfβ facilitates early B lymphocyte development by regulating expression of Ebf1. J Exp Med. 2012;209:1255–62. doi: 10.1084/jem.20112745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lukin K, Fields S, Lopez D, Cherrier M, Ternyak K, Ramirez J, Feeney AJ, Hagman J. Compound haploinsufficiencies of Ebf1 and Runx1 genes impede B cell lineage progression. Proc.Natl.Acad.Sci.U.S.A. 2010;107:7869–74. doi: 10.1073/pnas.1003525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guo Y, Maillard I, Chakraborti S, Rothenberg EV, Speck NA. Core binding factors are necessary for natural killer cell development, and cooperate with Notch signaling during T cell specification. Blood. 2008;112:480–92. doi: 10.1182/blood-2007-10-120261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Talebian L, Li Z, Guo Y, Gaudet J, Speck ME, Sugiyama D, Kaur P, Pear WS, Maillard I, Speck NA. T-lymphoid, megakaryocyte, and granulocyte development are sensitive to decreases in CBFβ dosage. Blood. 2007;109:11–21. doi: 10.1182/blood-2006-05-021188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Growney JD, Shigematsu H, Li Z, Lee BH, Adelsperger J, Rowan R, Curley DP, Kutok JL, Akashi K, Williams IR, Speck NA, Gilliland DG. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106:494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gwin KA, Shapiro MB, Dolence JJ, Huang ZL, Medina KL. Hoxa9 and flt3 signaling synergistically regulate an early checkpoint in lymphopoiesis. J Immunol. 2013;191:745–54. doi: 10.4049/jimmunol.1203294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kosan C, Saba I, Godmann M, Herold S, Herkert B, Eilers M, Möröy T. Transcription factor Miz-1 is required to regulate interleukin-7 receptor signaling at early commitment stages of B cell differentiation. Immunity. 2010;33:917–28. doi: 10.1016/j.immuni.2010.11.028. [DOI] [PubMed] [Google Scholar]
- 95.Saba I, Kosan C, Vassen L, Möröy T. IL-7R-dependent survival and differentiation of early T-lineage progenitors is regulated by the BTB/POZ domain transcription factor Miz-1. Blood. 2011;117:3370–81. doi: 10.1182/blood-2010-09-310680. [DOI] [PubMed] [Google Scholar]
- 96.Igarashi H, Gregory SC, Yokota T, Sakaguchi N, Kincade PW. Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity. 2002;17:117–30. doi: 10.1016/s1074-7613(02)00366-7. [DOI] [PubMed] [Google Scholar]
- 97.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–72. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 98.Pereira de Sousa A, Berthault C, Granato A, Dias S, Ramond C, Kee BL, Cumano A, Vieira P. Inhibitors of DNA binding proteins restrict T cell potential by repressing Notch1 expression in Flt3-negative common lymphoid progenitors. J Immunol. 2012;189:3822–30. doi: 10.4049/jimmunol.1103723. [DOI] [PubMed] [Google Scholar]
- 99.Inlay MA, Bhattacharya D, Sahoo D, Serwold T, Seita J, Karsunky H, Plevritis SK, Dill DL, Weissman IL. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev. 2009;23:2376–81. doi: 10.1101/gad.1836009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Karsunky H, Inlay MA, Serwold T, Bhattacharya D, Weissman IL. Flk2+ common lymphoid progenitors possess equivalent differentiation potential for the B and T lineages. Blood. 2008;111:5562–70. doi: 10.1182/blood-2007-11-126219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Holmes ML, Carotta S, Corcoran LM, Nutt SL. Repression of Flt3 by Pax5 is crucial for B-cell lineage commitment. Genes Dev. 2006;20:933–8. doi: 10.1101/gad.1396206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zandi S, Åhsberg J, Tsapogas P, Stjernberg J, Qian H, Sigvardsson M. Single-cell analysis of early B-lymphocyte development suggests independent regulation of lineage specification and commitment in vivo. Proc Natl Acad Sci U S A. 2012;109:15871–6. doi: 10.1073/pnas.1210144109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mansson R, Zandi S, Anderson K, Martensson IL, Jacobsen SE, Bryder D, Sigvardsson M. B-lineage commitment prior to surface expression of B220 and CD19 on hematopoietic progenitor cells. Blood. 2008;112:1048–55. doi: 10.1182/blood-2007-11-125385. [DOI] [PubMed] [Google Scholar]
- 104.Feyerabend TB, Terszowski G, Tietz A, Blum C, Luche H, Gossler A, Gale NW, Radtke F, Fehling HJ, Rodewald HR. Deletion of Notch1 converts pro-T cells to dendritic cells and promotes thymic B cells by cell-extrinsic and cell-intrinsic mechanisms. Immunity. 2009;30:67–79. doi: 10.1016/j.immuni.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 105.Weerkamp F, Baert MR, Brugman MH, Dik WA, de Haas EF, Visser TP, de Groot CJ, Wagemaker G, van Dongen JJ, Staal FJ. Human thymus contains multipotent progenitors with T/B lymphoid, myeloid, and erythroid lineage potential. Blood. 2006;107:3131–7. doi: 10.1182/blood-2005-08-3412. [DOI] [PubMed] [Google Scholar]
- 106.De Smedt M, Hoebeke I, Reynvoet K, Leclercq G, Plum J. Different thresholds of Notch signaling bias human precursor cells toward B-, NK-, monocytic/dendritic-, or T-cell lineage in thymus microenvironment. Blood. 2005;106:3498–506. doi: 10.1182/blood-2005-02-0496. [DOI] [PubMed] [Google Scholar]
- 107.Sambandam A, Maillard I, Zediak VP, Xu L, Gerstein RM, Aster JC, Pear WS, Bhandoola A. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat.Immunol. 2005;6:663–70. doi: 10.1038/ni1216. [DOI] [PubMed] [Google Scholar]
- 108.Tan JB, Visan I, Yuan JS, Guidos CJ. Requirement for Notch1 signals at sequential early stages of intrathymic T cell development. Nat.Immunol. 2005;6:671–9. doi: 10.1038/ni1217. [DOI] [PubMed] [Google Scholar]
- 109.Harman BC, Jenkinson WE, Parnell SM, Rossi SW, Jenkinson EJ, Anderson G. T/B lineage choice occurs prior to intrathymic Notch signalling. Blood. 2005;106:886–92. doi: 10.1182/blood-2004-12-4881. [DOI] [PubMed] [Google Scholar]
- 110.Masuda K, Itoi M, Amagai T, Minato N, Katsura Y, Kawamoto H. Thymic anlage is colonized by progenitors restricted to T, NK, and dendritic cell lineages. J.Immunol. 2005;174:2525–32. doi: 10.4049/jimmunol.174.5.2525. [DOI] [PubMed] [Google Scholar]
- 111.Yui MA, Feng N, Rothenberg EV. Fine-scale staging of T cell lineage commitment in adult mouse thymus. J Immunol. 2010;185:284–93. doi: 10.4049/jimmunol.1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–7. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- 113.Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, Kawamoto H. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–72. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- 114.Taghon T, Yui MA, Rothenberg EV. Mast cell lineage diversion of T lineage precursors by the essential T cell transcription factor GATA-3. Nat Immunol. 2007;8:845–55. doi: 10.1038/ni1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Balciunaite G, Ceredig R, Rolink AG. The earliest subpopulation of mouse thymocytes contains potent T, significant macrophage, and natural killer cell but no B-lymphocyte potential. Blood. 2005;105:1930–6. doi: 10.1182/blood-2004-08-3087. [DOI] [PubMed] [Google Scholar]
- 116.Lu M, Tayu R, Ikawa T, Masuda K, Matsumoto I, Mugishima H, Kawamoto H, Katsura Y. The earliest thymic progenitors in adults are restricted to T, NK, and dendritic cell lineage and have a potential to form more diverse TCRβ chains than fetal progenitors. J Immunol. 2005;175:5848–56. doi: 10.4049/jimmunol.175.9.5848. [DOI] [PubMed] [Google Scholar]
- 117.Shen HQ, Lu M, Ikawa T, Masuda K, Ohmura K, Minato N, Katsura Y, Kawamoto H. T/NK bipotent progenitors in the thymus retain the potential to generate dendritic cells. J Immunol. 2003;171:3401–6. doi: 10.4049/jimmunol.171.7.3401. [DOI] [PubMed] [Google Scholar]
- 118.Ikawa T, Fujimoto S, Kawamoto H, Katsura Y, Yokota Y. Commitment to natural killer cells requires the helix-loop-helix inhibitor Id2. Proc.Natl.Acad.Sci.U.S.A. 2001;98:5164–9. doi: 10.1073/pnas.091537598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Michie AM, Carlyle JR, Schmitt TM, Ljutic B, Cho SK, Fong Q, Zúñiga-Pflücker JC. Clonal characterization of a bipotent T cell and NK cell progenitor in the mouse fetal thymus. J.Immunol. 2000;164:1730–3. doi: 10.4049/jimmunol.164.4.1730. [DOI] [PubMed] [Google Scholar]
- 120.Wu L, Li CL, Shortman K. Thymic dendritic cell precursors: relationship to the T lymphocyte lineage and phenotype of the dendritic cell progeny. J. Exp. Med. 1996;184:903–11. doi: 10.1084/jem.184.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rothenberg EV. T cell lineage commitment: identity and renunciation. J Immunol. 2011;186:6649–55. doi: 10.4049/jimmunol.1003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Welinder E, Åhsberg J, Sigvardsson M. B-lymphocyte commitment: identifying the point of no return. Semin Immunol. 2011;23:335–40. doi: 10.1016/j.smim.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 123.Malin S, McManus S, Cobaleda C, Novatchkova M, Delogu A, Bouillet P, Strasser A, Busslinger M. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat Immunol. 2010;11:171–9. doi: 10.1038/ni.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, Ideker T, Glass CK, Murre C. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat.Immunol. 2010;11:635–43. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jones ME, Zhuang Y. Regulation of V(D)J recombination by E-protein transcription factors. Adv Exp Med Biol. 2009;650:148–56. doi: 10.1007/978-1-4419-0296-2_12. [DOI] [PubMed] [Google Scholar]
- 126.Hsu LY, Lauring J, Liang HE, Greenbaum S, Cado D, Zhuang Y, Schlissel MS. A conserved transcriptional enhancer regulates RAG gene expression in developing B cells. Immunity. 2003;19:105–17. doi: 10.1016/s1074-7613(03)00181-x. [DOI] [PubMed] [Google Scholar]
- 127.Kee BL. E and ID proteins branch out. Nat.Rev.Immunol. 2009;9:175–84. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- 128.Mansson R, Welinder E, Åhsberg J, Lin YC, Benner C, Glass CK, Lucas JS, Sigvardsson M, Murre C. Positive intergenic feedback circuitry, involving EBF1 and FOXO1, orchestrates B-cell fate. Proc Natl Acad Sci U S A. 2012;109:21028–33. doi: 10.1073/pnas.1211427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Welinder E, Mansson R, Mercer EM, Bryder D, Sigvardsson M, Murre C. The transcription factors E2A and HEB act in concert to induce the expression of FOXO1 in the common lymphoid progenitor. Proc Natl Acad Sci U S A. 2011;108:17402–7. doi: 10.1073/pnas.1111766108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.O'Riordan M, Grosschedl R. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 1999;11:21–31. doi: 10.1016/s1074-7613(00)80078-3. [DOI] [PubMed] [Google Scholar]
- 131.Sigvardsson M, O'Riordan M, Grosschedl R. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity. 1997;7:25–36. doi: 10.1016/s1074-7613(00)80507-5. [DOI] [PubMed] [Google Scholar]
- 132.Decker T, Pasca dM, McManus S, Sun Q, Bonifer C, Tagoh H, Busslinger M. Stepwise activation of enhancer and promoter regions of the B cell commitment gene Pax5 in early lymphopoiesis. Immunity. 2009;30:508–20. doi: 10.1016/j.immuni.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 133.Ferreiros-Vidal I, Carroll T, Taylor B, Terry A, Liang Z, Bruno L, Dharmalingam G, Khadayate S, Cobb BS, Smale ST, Spivakov M, Srivastava P, Petretto E, Fisher AG, Merkenschlager M. Genome-wide identification of Ikaros targets elucidates its contribution to mouse B-cell lineage specification and pre-B-cell differentiation. Blood. 2013;121:1769–82. doi: 10.1182/blood-2012-08-450114. [DOI] [PubMed] [Google Scholar]
- 134.Reynaud D, Demarco IA, Reddy KL, Schjerven H, Bertolino E, Chen Z, Smale ST, Winandy S, Singh H. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat.Immunol. 2008;9:927–36. doi: 10.1038/ni.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang JH, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, Bigby M, Georgopoulos K. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–49. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- 136.Kirstetter P, Thomas M, Dierich A, Kastner P, Chan S. Ikaros is critical for B cell differentiation and function. Eur J Immunol. 2002;32:720–30. doi: 10.1002/1521-4141(200203)32:3<720::AID-IMMU720>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 137.Lee BS, Dekker JD, Lee BK, Iyer VR, Sleckman BP, Shaffer AL, 3rd, Ippolito GC, Tucker PW. The BCL11A transcription factor directly activates RAG gene expression and V(D)J recombination. Mol Cell Biol. 2013;33:1768–81. doi: 10.1128/MCB.00987-12. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 138.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–59. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 139.Hsu CL, King-Fleischman AG, Lai AY, Matsumoto Y, Weissman IL, Kondo M. Antagonistic effect of CCAAT enhancer-binding protein-α and Pax5 in myeloid or lymphoid lineage choice in common lymphoid progenitors. Proc.Natl.Acad.Sci.U.S.A. 2006;103:672–7. doi: 10.1073/pnas.0510304103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bussmann LH, Schubert A, Vu Manh TP, De Andres L, Desbordes SC, Parra M, Zimmermann T, Rapino F, Rodriguez-Ubreva J, Ballestar E, Graf T. A robust and highly efficient immune cell reprogramming system. Cell Stem Cell. 2009;5:554–66. doi: 10.1016/j.stem.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 141.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–76. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 142.Iwasaki-Arai J, Iwasaki H, Miyamoto T, Watanabe S, Akashi K. Enforced Granulocyte/Macrophage Colony-Stimulating Factor signals do not support lymphopoiesis, but instruct lymphoid to myelomonocytic lineage conversion. J. Exp. Med. 2003;197:1311–22. doi: 10.1084/jem.20021843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kondo M, Scherer DC, Miyamoto T, King AG, Akashi K, Sugamura K, Weissman IL. Cell-fate conversion of lymphoid-committed progenitors by instructive actions of cytokines. Nature. 2000;407:383–6. doi: 10.1038/35030112. [DOI] [PubMed] [Google Scholar]
- 144.Leeanansaksiri W, Wang H, Gooya JM, Renn K, Abshari M, Tsai S, Keller JR. IL-3 induces inhibitor of DNA-binding protein-1 in hemopoietic progenitor cells and promotes myeloid cell development. J.Immunol. 2005;174:7014–21. doi: 10.4049/jimmunol.174.11.7014. [DOI] [PubMed] [Google Scholar]
- 145.Ikawa T, Kawamoto H, Goldrath AW, Murre C. E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. J Exp. Med. 2006;203:1329–42. doi: 10.1084/jem.20060268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zarnegar MA, Rothenberg EV. Ikaros represses and activates PU.1 cell-type-specifically through the multifunctional Sfpi1 URE and a myeloid specific enhancer. Oncogene. 2012;31:4647–54. doi: 10.1038/onc.2011.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tsapogas P, Zandi S, Ahsberg J, Zetterblad J, Welinder E, Jonsson JI, Mansson R, Qian H, Sigvardsson M. IL-7 mediates Ebf-1-dependent lineage restriction in early lymphoid progenitors. Blood. 2011;118:1283–90. doi: 10.1182/blood-2011-01-332189. [DOI] [PubMed] [Google Scholar]
- 148.Kikuchi K, Kasai H, Watanabe A, Lai AY, Kondo M. IL-7 specifies B cell fate at the common lymphoid progenitor to pre-proB transition stage by maintaining early B cell factor expression. J.Immunol. 2008;181:383–92. doi: 10.4049/jimmunol.181.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ochiai K, Maienschein-Cline M, Mandal M, Triggs JR, Bertolino E, Sciammas R, Dinner AR, Clark MR, Singh H. A self-reinforcing regulatory network triggered by limiting IL-7 activates pre-BCR signaling and differentiation. Nat Immunol. 2012;13:300–7. doi: 10.1038/ni.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Dengler HS, Baracho GV, Omori SA, Bruckner S, Arden KC, Castrillon DH, DePinho RA, Rickert RC. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat.Immunol. 2008;9:1388–98. doi: 10.1038/ni.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Amin RH, Schlissel MS. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat.Immunol. 2008;9:613–22. doi: 10.1038/ni.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Johnson SE, Shah N, Bajer AA, LeBien TW. IL-7 activates the phosphatidylinositol 3-kinase/AKT pathway in normal human thymocytes but not normal human B cell precursors. J Immunol. 2008;180:8109–17. doi: 10.4049/jimmunol.180.12.8109. [DOI] [PubMed] [Google Scholar]
- 153.Johnson K, Chaumeil J, Micsinai M, Wang JM, Ramsey LB, Baracho GV, Rickert RC, Strino F, Kluger Y, Farrar MA, Skok JA. IL-7 functionally segregates the pro-B cell stage by regulating transcription of recombination mediators across cell cycle. J Immunol. 2012;188:6084–92. doi: 10.4049/jimmunol.1200368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Herzog S, Hug E, Meixlsperger S, Paik JH, DePinho RA, Reth M, Jumaa H. SLP-65 regulates immunoglobulin light chain gene recombination through the PI(3)K-PKB-Foxo pathway. Nat.Immunol. 2008;9:623–31. doi: 10.1038/ni.1616. [DOI] [PubMed] [Google Scholar]
- 155.McManus S, Ebert A, Salvagiotto G, Medvedovic J, Sun Q, Tamir I, Jaritz M, Tagoh H, Busslinger M. The transcription factor Pax5 regulates its target genes by recruiting chromatin-modifying proteins in committed B cells. EMBO J. 2011;30:2388–404. doi: 10.1038/emboj.2011.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mansson R, Zandi S, Welinder E, Tsapogas P, Sakaguchi N, Bryder D, Sigvardsson M. Single-cell analysis of the common lymphoid progenitor compartment reveals functional and molecular heterogeneity. Blood. 2010;115:2601–9. doi: 10.1182/blood-2009-08-236398. [DOI] [PubMed] [Google Scholar]
- 157.Treiber T, Mandel EM, Pott S, Gyory I, Firner S, Liu ET, Grosschedl R. Early B cell Factor 1 regulates B cell gene networks by activation, repression, and transcription- independent poising of chromatin. Immunity. 2010;32:714–25. doi: 10.1016/j.immuni.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 158.Roessler S, Gyory I, Imhof S, Spivakov M, Williams RR, Busslinger M, Fisher AG, Grosschedl R. Distinct promoters mediate the regulation of Ebf1 gene expression by Interleukin-7 and Pax5. Mol.Cell Biol. 2007;27:579–94. doi: 10.1128/MCB.01192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pongubala JM, Northrup DL, Lancki DW, Medina KL, Treiber T, Bertolino E, Thomas M, Grosschedl R, Allman D, Singh H. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat.Immunol. 2008;9:203–15. doi: 10.1038/ni1555. [DOI] [PubMed] [Google Scholar]
- 160.Maier H, Ostraat R, Gao H, Fields S, Shinton SA, Medina KL, Ikawa T, Murre C, Singh H, Hardy RR, Hagman J. Early B cell factor cooperates with Runx1 and mediates epigenetic changes associated with mb-1 transcription. Nat.Immunol. 2004;5:1069–77. doi: 10.1038/ni1119. [DOI] [PubMed] [Google Scholar]
- 161.Schebesta A, McManus S, Salvagiotto G, Delogu A, Busslinger GA, Busslinger M. Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration, and immune function. Immunity. 2007;27:49–63. doi: 10.1016/j.immuni.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 162.Thal MA, Carvalho TL, He T, Kim HG, Gao H, Hagman J, Klug CA. Ebf1-mediated down-regulation of Id2 and Id3 is essential for specification of the B cell lineage. Proc.Natl.Acad.Sci.U.S.A. 2009;106:552–7. doi: 10.1073/pnas.0802550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bhalla S, Spaulding C, Brumbaugh RL, Zagort DE, Massari ME, Murre C, Kee BL. Differential roles for the E2A activation domains in B lymphocytes and macrophages. J.Immunol. 2008;180:1694–703. doi: 10.4049/jimmunol.180.3.1694. [DOI] [PubMed] [Google Scholar]
- 164.Revilla-i-Domingo R, Bilic I, Vilagos B, Tagoh H, Ebert A, Tamir IM, Smeenk L, Trupke J, Sommer A, Jaritz M, Busslinger M. The B-cell identity factor Pax5 regulates distinct transcriptional programmes in early and late B lymphopoiesis. EMBO J. 2012;31:3130–46. doi: 10.1038/emboj.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Capron C, Lecluse Y, Kaushik AL, Foudi A, Lacout C, Sekkai D, Godin I, Albagli O, Poullion I, Svinartchouk F, Schanze E, Vainchenker W, Sablitzky F, Bennaceur-Griscelli A, Dumenil D. The SCL relative LYL-1 is required for fetal and adult hematopoietic stem cell function and B-cell differentiation. Blood. 2006;107:4678–86. doi: 10.1182/blood-2005-08-3145. [DOI] [PubMed] [Google Scholar]
- 166.Painter MW, Davis S, Hardy RR, Mathis D, Benoist C. Transcriptomes of the B and T lineages compared by multiplatform microarray profiling. J.Immunol. 2011;186:3047–57. doi: 10.4049/jimmunol.1002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, Ragoussis J, Natoli G. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–28. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 168.Yashiro-Ohtani Y, Ohtani T, Pear WS. Notch regulation of early thymocyte development. Semin Immunol. 2010;22:261–9. doi: 10.1016/j.smim.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 169.Weber BN, Chi AW, Chavez A, Yashiro-Ohtani Y, Yang Q, Shestova O, Bhandoola A. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–8. doi: 10.1038/nature10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Germar K, Dose M, Konstantinou T, Zhang J, Wang H, Lobry C, Arnett KL, Blacklow SC, Aifantis I, Aster JC, Gounari F. T-cell factor 1 is a gatekeeper for T-cell specification in response to Notch signaling. Proc Natl Acad Sci U S A. 2011;108:20060–5. doi: 10.1073/pnas.1110230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Okamura RM, Sigvardsson M, Galceran J, Verbeek S, Clevers H, Grosschedl R. Redundant regulation of T cell differentiation and TCRβ gene expression by the transcription factors LEF-1 and TCF-1. Immunity. 1998;8:11–20. doi: 10.1016/s1074-7613(00)80454-9. [DOI] [PubMed] [Google Scholar]
- 172.Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, Van de Wetering M, Oosterwegel M, Wilson A, MacDonald HR, Clevers H. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374:70–4. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- 173.Hosoya T, Maillard I, Engel JD. From the cradle to the grave: activities of GATA-3 throughout T-cell development and differentiation. Immunol.Rev. 2010;238:110–25. doi: 10.1111/j.1600-065X.2010.00954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ho IC, Tai TS, Pai SY. GATA3 and the T-cell lineage: essential functions before and after T-helper-2-cell differentiation. Nat.Rev.Immunol. 2009;9:125–35. doi: 10.1038/nri2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Hozumi K, Negishi N, Tsuchiya I, Abe N, Hirano K, Suzuki D, Yamamoto M, Engel JD, Habu S. Notch signaling is necessary for GATA3 function in the initiation of T cell development. Eur J Immunol. 2008;38:977–85. doi: 10.1002/eji.200737688. [DOI] [PubMed] [Google Scholar]
- 176.Hattori N, Kawamoto H, Fujimoto S, Kuno K, Katsura Y. Involvement of transcription factors TCF-1 and GATA-3 in the initiation of the earliest step of T cell development in the thymus. J Exp Med. 1996;184:1137–47. doi: 10.1084/jem.184.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ting CN, Olson MC, Barton KP, Leiden JM. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature. 1996;384:474–8. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- 178.Ikawa T, Hirose S, Masuda K, Kakugawa K, Satoh R, Shibano-Satoh A, Kominami R, Katsura Y, Kawamoto H. An essential developmental checkpoint for production of the T cell lineage. Science. 2010;329:93–6. doi: 10.1126/science.1188995. [DOI] [PubMed] [Google Scholar]
- 179.Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 2010;329:89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Li P, Burke S, Wang J, Chen X, Ortiz M, Lee SC, Lu D, Campos L, Goulding D, Ng BL, Dougan G, Huntly B, Gottgens B, Jenkins NA, Copeland NG, Colucci F, Liu P. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 2010;329:85–9. doi: 10.1126/science.1188063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Albu DI, Feng D, Bhattacharya D, Jenkins NA, Copeland NG, Liu P, Avram D. BCL11B is required for positive selection and survival of double-positive thymocytes. J Exp Med. 2007;204:3003–15. doi: 10.1084/jem.20070863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tydell CC, David-Fung ES, Moore JE, Rowen L, Taghon T, Rothenberg EV. Molecular dissection of prethymic progenitor entry into the T lymphocyte developmental pathway. J Immunol. 2007;179:421–38. doi: 10.4049/jimmunol.179.1.421. [DOI] [PubMed] [Google Scholar]
- 183.Georgescu C, Longabaugh WJ, Scripture-Adams DD, David-Fung ES, Yui MA, Zarnegar MA, Bolouri H, Rothenberg EV. A gene regulatory network armature for T lymphocyte specification. Proc.Natl.Acad.Sci.U.S.A. 2008;105:20100–5. doi: 10.1073/pnas.0806501105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Maillard I, Tu L, Sambandam A, Yashiro-Ohtani Y, Millholland J, Keeshan K, Shestova O, Xu L, Bhandoola A, Pear WS. The requirement for Notch signaling at the β-selection checkpoint in vivo is absolute and independent of the pre-T cell receptor. J Exp Med. 2006;203:2239–45. doi: 10.1084/jem.20061020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Reizis B, Leder P. Direct induction of T lymphocyte-specific gene expression by the mammalian Notch signaling pathway. Genes Dev. 2002;16:295–300. doi: 10.1101/gad.960702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.David-Fung ES, Butler R, Buzi G, Yui MA, Diamond RA, Anderson MK, Rowen L, Rothenberg EV. Transcription factor expression dynamics of early T-lymphocyte specification and commitment. Dev Biol. 2009;325:444–67. doi: 10.1016/j.ydbio.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wang D, Claus CL, Vaccarelli G, Braunstein M, Schmitt TM, Zuniga-Pflucker JC, Rothenberg EV, Anderson MK. The basic helix-loop-helix transcription factor HEBAlt is expressed in pro-T cells and enhances the generation of T cell precursors. J Immunol. 2006;177:109–19. doi: 10.4049/jimmunol.177.1.109. [DOI] [PubMed] [Google Scholar]
- 188.Anderson MK, Hernandez-Hoyos G, Diamond RA, Rothenberg EV. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development. 1999;126:3131–48. doi: 10.1242/dev.126.14.3131. [DOI] [PubMed] [Google Scholar]
- 189.Doan LL, Porter SD, Duan Z, Flubacher MM, Montoya D, Tsichlis PN, Horwitz M, Gilks CB, Grimes HL. Targeted transcriptional repression of Gfi1 by GFI1 and GFI1B in lymphoid cells. Nucleic Acids Res. 2004;32:2508–19. doi: 10.1093/nar/gkh570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Franco CB, Scripture-Adams DD, Proekt I, Taghon T, Weiss AH, Yui MA, Adams SL, Diamond RA, Rothenberg EV. Notch/Delta signaling constrains reengineering of pro-T cells by PU.1. Proc Natl Acad Sci U S A. 2006;103:11993–8. doi: 10.1073/pnas.0601188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Dionne CJ, Tse KY, Weiss AH, Franco CB, Wiest DL, Anderson MK, Rothenberg EV. Subversion of T lineage commitment by PU.1 in a clonal cell line system. Dev Biol. 2005;280:448–66. doi: 10.1016/j.ydbio.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 192.Tiemessen MM, Baert MR, Schonewille T, Brugman MH, Famili F, Salvatori DC, Meijerink JP, Ozbek U, Clevers H, van Dongen JJ, Staal FJ. The nuclear effector of Wnt-signaling, Tcf1, functions as a T-cell-specific tumor suppressor for development of lymphomas. PLoS Biol. 2012;10:e1001430. doi: 10.1371/journal.pbio.1001430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Yu S, Zhou X, Steinke FC, Liu C, Chen SC, Zagorodna O, Jing X, Yokota Y, Meyerholz DK, Mullighan CG, Knudson CM, Zhao DM, Xue HH. The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity. 2012;37:813–26. doi: 10.1016/j.immuni.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Anderson G, Jenkinson EJ. Investigating central tolerance with reaggregate thymus organ cultures. Methods Mol Biol. 2007;380:185–96. doi: 10.1007/978-1-59745-395-0_11. [DOI] [PubMed] [Google Scholar]
- 195.Katsura Y, Kawamoto H. Stepwise lineage restriction of progenitors in lympho-myelopoiesis. Int Rev Immunol. 2001;20:1–20. doi: 10.3109/08830180109056720. [DOI] [PubMed] [Google Scholar]
- 196.Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–76. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- 197.Schmitt TM, Zúñiga-Pflücker JC. Induction of T cell development from hematopoietic progenitor cells by Delta-like-1 in vitro. Immunity. 2002;17:749–56. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 198.Besseyrias V, Fiorini E, Strobl LJ, Zimber-Strobl U, Dumortier A, Koch U, Arcangeli ML, Ezine S, MacDonald HR, Radtke F. Hierarchy of Notch-Delta interactions promoting T cell lineage commitment and maturation. J. Exp. Med. 2007;204:331–43. doi: 10.1084/jem.20061442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Mohtashami M, Shah DK, Nakase H, Kianizad K, Petrie HT, Zúñiga-Pflücker JC. Direct comparison of Dll1- and Dll4-mediated Notch activation levels shows differential lymphomyeloid lineage commitment outcomes. J Immunol. 2010;185:867–76. doi: 10.4049/jimmunol.1000782. [DOI] [PubMed] [Google Scholar]
- 200.Sambandam A, Maillard I, Zediak VP, Xu L, Gerstein RM, Aster JC, Pear WS, Bhandoola A. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat Immunol. 2005;6:663–70. doi: 10.1038/ni1216. [DOI] [PubMed] [Google Scholar]
- 201.Waskow C, Paul S, Haller C, Gassmann M, Rodewald HR. Viable c-KitW/W mutants reveal pivotal role for c-kit in the maintenance of lymphopoiesis. Immunity. 2002;17:277–88. doi: 10.1016/s1074-7613(02)00386-2. [DOI] [PubMed] [Google Scholar]
- 202.Massa S, Balciunaite G, Ceredig R, Rolink AG. Critical role for c-kit (CD117) in T cell lineage commitment and early thymocyte development in vitro. Eur J Immunol. 2006;36:526–32. doi: 10.1002/eji.200535760. [DOI] [PubMed] [Google Scholar]
- 203.Wang H, Pierce LJ, Spangrude GJ. Distinct roles of IL-7 and stem cell factor in the OP9-DL1 T-cell differentiation culture system. Exp.Hematol. 2006;34:1730–40. doi: 10.1016/j.exphem.2006.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Laiosa CV, Stadtfeld M, Xie H, de Andres-Aguayo L, Graf T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBPα and PU.1 transcription factors. Immunity. 2006;25:731–44. doi: 10.1016/j.immuni.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 205.Schmitt TM, De Pooter RF, Gronski MA, Cho SK, Ohashi PS, Zúñiga-Pflücker JC. Induction of T cell development and establishment of T cell competence from embryonic stem cells differentiated in vitro. Nat.Immunol. 2004;5:410–7. doi: 10.1038/ni1055. [DOI] [PubMed] [Google Scholar]
- 206.Taghon TN, David ES, Zuniga-Pflucker JC, Rothenberg EV. Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling. Genes Dev. 2005;19:965–78. doi: 10.1101/gad.1298305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ciofani M, Knowles GC, Wiest DL, von Boehmer H, Zuniga-Pflucker JC. Stage-specific and differential Notch dependency at the αβ and γδ T lineage bifurcation. Immunity. 2006;25:105–16. doi: 10.1016/j.immuni.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 208.Feng N, Vegh P, Rothenberg EV, Yui MA. Lineage divergence at the first TCR-dependent checkpoint: preferential γδ and impaired αβ T cell development in nonobese diabetic mice. J Immunol. 2011;186:826–37. doi: 10.4049/jimmunol.1002630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Huang J, Durum SK, Muegge K. Cutting edge: histone acetylation and recombination at the TCRγ locus follows IL-7 induction. J.Immunol. 2001;167:6073–7. doi: 10.4049/jimmunol.167.11.6073. [DOI] [PubMed] [Google Scholar]
- 210.Ye SK, Agata Y, Lee HC, Kurooka H, Kitamura T, Shimizu A, Honjo T, Ikuta K. The IL-7 Receptor controls the accessibility of the TCRγ Locus by Stat5 and histone acetylation. Immunity. 2001;15:813–23. doi: 10.1016/s1074-7613(01)00230-8. [DOI] [PubMed] [Google Scholar]
- 211.Wong SH, Walker JA, Jolin HE, Drynan LF, Hams E, Camelo A, Barlow JL, Neill DR, Panova V, Koch U, Radtke F, Hardman CS, Hwang YY, Fallon PG, McKenzie ANJ. Transcription factor RORα is critical for nuocyte development. Nat Immunol. 2012;13:229–36. doi: 10.1038/ni.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Manesso E, Chickarmane V, Kueh HY, Rothenberg EV, Peterson C. Computational modelling of T-cell formation kinetics: output regulated by initial proliferation-linked deferral of developmental competence. J R Soc Interface. 2013;10:20120774. doi: 10.1098/rsif.2012.0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Masuda K, Kakugawa K, Nakayama T, Minato M, Katsura Y, Kawamoto H. T cell lineage determination precedes the initiation of TCRβ rearrangement. J Immunol. 2007;179:3699–706. doi: 10.4049/jimmunol.179.6.3699. [DOI] [PubMed] [Google Scholar]
- 214.Wu X, Satpathy AT, Kc W, Liu P, Murphy TL, Murphy KM. Bcl11a controls Flt3 expression in early hematopoietic progenitors and is required for pDC development in vivo. PLoS One. 2013;8:e64800. doi: 10.1371/journal.pone.0064800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Mingueneau M, Kreslavsky T, Gray D, Heng T, Cruse R, Ericson J, Bendall S, Spitzer MH, Nolan GP, Kobayashi K, von Boehmer H, Mathis D, Benoist C, Immunological Genome C, Best AJ, Knell J, Goldrath A, Jojic V, Koller D, Shay T, Regev A, Cohen N, Brennan P, Brenner M, Kim F, Rao TN, Wagers A, Heng T, Ericson J, Rothamel K, Ortiz-Lopez A, Mathis D, Benoist C, Bezman NA, Sun JC, Min-Oo G, Kim CC, Lanier LL, Miller J, Brown B, Merad M, Gautier EL, Jakubzick C, Randolph GJ, Monach P, Blair DA, Dustin ML, Shinton SA, Hardy RR, Laidlaw D, Collins J, Gazit R, Rossi DJ, Malhotra N, Sylvia K, Kang J, Kreslavsky T, Fletcher A, Elpek K, Bellemare-Pelletier A, Malhotra D, Turley S. The transcriptional landscape of αβ T cell differentiation. Nat Immunol. 2013;14:619–32. doi: 10.1038/ni.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ikawa T, Kawamoto H, Goldrath AW, Murre C. E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. J Exp Med. 2006;203:1329–42. doi: 10.1084/jem.20060268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Miyazaki M, Rivera RR, Miyazaki K, Lin YC, Agata Y, Murre C. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nat Immunol. 2011;12:992–1001. doi: 10.1038/ni.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wojciechowski J, Lai A, Kondo M, Zhuang Y. E2A and HEB are required to block thymocyte proliferation prior to pre-TCR expression. J Immunol. 2007;178:5717–26. doi: 10.4049/jimmunol.178.9.5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007;21:1882–94. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Laiosa CV, Stadtfeld M, Graf T. Determinants of lymphoid-myeloid lineage diversification. Annu Rev Immunol. 2006;24:705–38. doi: 10.1146/annurev.immunol.24.021605.090742. [DOI] [PubMed] [Google Scholar]
- 221.Lefebvre JM, Haks MC, Carleton MO, Rhodes M, Sinnathamby G, Simon MC, Eisenlohr LC, Garrett-Sinha LA, Wiest DL. Enforced expression of Spi-B reverses T lineage commitment and blocks β-selection. J Immunol. 2005;174:6184–94. doi: 10.4049/jimmunol.174.10.6184. [DOI] [PubMed] [Google Scholar]
- 222.Anderson MK, Weiss AH, Hernandez-Hoyos G, Dionne CJ, Rothenberg EV. Constitutive expression of PU.1 in fetal hematopoietic progenitors blocks T cell development at the pro-T cell stage. Immunity. 2002;16:285–96. doi: 10.1016/s1074-7613(02)00277-7. [DOI] [PubMed] [Google Scholar]
- 223.Zarnegar MA, Chen J, Rothenberg EV. Cell-type-specific activation and repression of PU.1 by a complex of discrete, functionally specialized cis-regulatory elements. Mol Cell Biol. 2010;30:4922–39. doi: 10.1128/MCB.00354-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Huang G, Zhang P, Hirai H, Elf S, Yan X, Chen Z, Koschmieder S, Okuno Y, Dayaram T, Growney JD, Shivdasani RA, Gilliland DG, Speck NA, Nimer SD, Tenen DG. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat Genet. 2008;40:51–60. doi: 10.1038/ng.2007.7. [DOI] [PubMed] [Google Scholar]
- 225.Scripture-Adams DD, Li L, Arias AM, Elihu KJ, Butler I, R. R., Champhekar A, Zhang JA, Rothenberg EV. GATA-3 dose-dependent checkpoints in early T cell commitment. J Immunol. 2013 doi: 10.4049/jimmunol.1301663. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Dontje W, Schotte R, Cupedo T, Nagasawa M, Scheeren F, Gimeno R, Spits H, Blom B. Delta-Like1-induced Notch1 signalling regulates the human plasmacytoid dendritic cell versus T cell lineage decision through control of GATA-3 and Spi-B. Blood. 2006;107:2446–52. doi: 10.1182/blood-2005-05-2090. [DOI] [PubMed] [Google Scholar]
- 227.Del Real MM, Rothenberg EV. Architecture of a lymphomyeloid developmental switch controlled by PU.1, Notch and Gata3. Development. 2013;140:1207–19. doi: 10.1242/dev.088559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Schmitt TM, Ciofani M, Petrie HT, Zúñiga-Pflücker JC. Maintenance of T cell specification and differentiation requires recurrent Notch receptor-ligand interactions. J Exp Med. 2004;200:469–79. doi: 10.1084/jem.20040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Li HS, Yang CY, Nallaparaju KC, Zhang H, Liu YJ, Goldrath AW, Watowich SS. The signal transducers STAT5 and STAT3 control expression of Id2 and E2-2 during dendritic cell development. Blood. 2012;120:4363–73. doi: 10.1182/blood-2012-07-441311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Ramirez K, Chandler KJ, Spaulding C, Zandi S, Sigvardsson M, Graves BJ, Kee BL. Gene deregulation and chronic activation in natural killer cells deficient in the transcription factor ETS1. Immunity. 2012;36:921–32. doi: 10.1016/j.immuni.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Cohen NR, Brennan PJ, Shay T, Watts GF, Brigl M, Kang J, Brenner MB, ImmGen Project C, Monach P, Shinton SA, Hardy RR, Jianu R, Koller D, Collins J, Gazit R, Garrison BS, Rossi DJ, Narayan K, Sylvia K, Kang J, Fletcher A, Elpek K, Bellemare-Pelletier A, Malhotra D, Turley S, Best AJ, Knell J, Goldrath A, Jojic V, Koller D, Shay T, Regev A, Cohen N, Brennan P, Brenner M, Kreslavsky T, Bezman NA, Sun JC, Kim CC, Lanier LL, Miller J, Brown B, Merad M, Gautier EL, Jakubzick C, Randolph GJ, Kim F, Rao TN, Wagers A, Heng T, Painter M, Ericson J, Davis S, Ergun A, Mingueneau M, Mathis D, Benoist C. Shared and distinct transcriptional programs underlie the hybrid nature of iNKT cells. Nat Immunol. 2013;14:90–9. doi: 10.1038/ni.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, Seddon B, Coles M, Kioussis D, Brady HJM. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat.Immunol. 2009;10:1118–24. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- 233.Kamizono S, Duncan GS, Seidel MG, Morimoto A, Hamada K, Grosveld G, Akashi K, Lind EF, Haight JP, Ohashi PS, Look AT, Mak TW. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J. Exp. Med. 2009;206:2977–86. doi: 10.1084/jem.20092176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Kastner P, Chan S, Vogel WK, Zhang LJ, Topark-Ngarm A, Golonzhka O, Jost B, Le Gras S, Gross MK, Leid M. Bcl11b represses a mature T-cell gene expression program in immature CD4+CD8+ thymocytes. Eur J Immunol. 2010;40:2143–54. doi: 10.1002/eji.200940258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.García-Ojeda ME, Klein Wolterink RG, Lemaître F, Richard-Le Goff O, Hasan M, Hendriks RW, Cumano A, Di Santo JP. GATA-3 promotes T cell specification by repressing B cell potential in pro-T cells. Blood. 2013;121:1749–59. doi: 10.1182/blood-2012-06-440065. [DOI] [PubMed] [Google Scholar]
- 236.Ciofani M, Zúñiga-Pflücker JC. Notch promotes survival of pre-T cells at the β-selection checkpoint by regulating cellular metabolism. Nat.Immunol. 2005;6:881–8. doi: 10.1038/ni1234. [DOI] [PubMed] [Google Scholar]
- 237.Hosoya T, Kuroha T, Moriguchi T, Cummings D, Maillard I, Lim KC, Engel JD. GATA-3 is required for early T lineage progenitor development. J Exp Med. 2009;206:2987–3000. doi: 10.1084/jem.20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Jones ME, Zhuang Y. Stage-specific functions of E-proteins at the β-selection and T-cell receptor checkpoints during thymocyte development. Immunol Res. 2011;49:202–15. doi: 10.1007/s12026-010-8182-x. [DOI] [PubMed] [Google Scholar]
- 239.Jones ME, Zhuang Y. Acquisition of a functional T cell receptor during T lymphocyte development is enforced by HEB and E2A transcription factors. Immunity. 2007;27:860–70. doi: 10.1016/j.immuni.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Schwartz R, Engel I, Fallahi-Sichani M, Petrie HT, Murre C. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediated signaling, and T lineage development. Proc Natl Acad Sci U S A. 2006;103:9976–81. doi: 10.1073/pnas.0603728103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Barndt RJ, Dai M, Zhuang Y. Functions of E2A-HEB heterodimers in T-cell development revealed by a dominant negative mutation of HEB. Mol Cell Biol. 2000;20:6677–85. doi: 10.1128/mcb.20.18.6677-6685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Engel I, Johns C, Bain G, Rivera RR, Murre C. Early thymocyte development is regulated by modulation of E2A protein activity. J. Exp. Med. 2001;194:733–46. doi: 10.1084/jem.194.6.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Xu W, Carr T, Ramirez K, McGregor S, Sigvardsson M, Kee BL. E2A transcription factors limit expression of Gata3 to facilitate T lymphocyte lineage commitment. Blood. 2013;121:1534–42. doi: 10.1182/blood-2012-08-449447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Braunstein M, Anderson MK. HEB in the spotlight: Transcriptional regulation of T-cell specification, commitment, and developmental plasticity. Clin Dev Immunol. 2012;2012:678705. doi: 10.1155/2012/678705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Yashiro-Ohtani Y, He Y, Ohtani T, Jones ME, Shestova O, Xu L, Fang TC, Chiang MY, Intlekofer AM, Blacklow SC, Zhuang Y, Pear WS. Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes Dev. 2009;23:1665–76. doi: 10.1101/gad.1793709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Developmental and molecular characterization of emerging β- and γδ-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24:53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 247.Lauritsen JP, Wong GW, Lee SY, Lefebvre JM, Ciofani M, Rhodes M, Kappes DJ, Zuniga-Pflucker JC, Wiest DL. Marked induction of the helix-loop-helix protein Id3 promotes the γδ T cell fate and renders their functional maturation Notch independent. Immunity. 2009;31:565–75. doi: 10.1016/j.immuni.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Bain G, Cravatt CB, Loomans C, Alberola-Ila J, Hedrick SM, Murre C. Regulation of the helix-loop-helix proteins, E2A and Id3, by the Ras-ERK MAPK cascade. Nat Immunol. 2001;2:165–71. doi: 10.1038/84273. [DOI] [PubMed] [Google Scholar]
- 249.Masson F, Minnich M, Olshansky M, Bilic I, Mount AM, Kallies A, Speed TP, Busslinger M, Nutt SL, Belz GT. Id2-mediated inhibition of E2A represses memory CD8+ T cell differentiation. J Immunol. 2013;190:4585–94. doi: 10.4049/jimmunol.1300099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Anderson MK, Hernandez-Hoyos G, Dionne CJ, Arias AM, Chen D, Rothenberg EV. Definition of regulatory network elements for T cell development by perturbation analysis with PU.1 and GATA-3. Dev Biol. 2002;246:103–21. doi: 10.1006/dbio.2002.0674. [DOI] [PubMed] [Google Scholar]
- 251.Kwon K, Hutter C, Sun Q, Bilic I, Cobaleda C, Malin S, Busslinger M. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity. 2008;28:751–62. doi: 10.1016/j.immuni.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 252.Seet CS, Brumbaugh RL, Kee BL. Early B Cell Factor promotes B lymphopoiesis with reduced Interleukin 7 responsiveness in the absence of E2A. J. Exp. Med. 2004;199:1689–700. doi: 10.1084/jem.20032202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Yashiro-Ohtani Y, He Y, Ohtani T, Jones ME, Shestova O, Xu L, Fang TC, Chiang MY, Intlekofer AM, Blacklow SC, Zhuang Y, Pear WS. Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes Dev. 2009;23:1665–76. doi: 10.1101/gad.1793709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Koltsova EK, Ciofani M, Benezra R, Miyazaki T, Clipstone N, Zuniga-Pflucker JC, Wiest DL. Early growth response 1 and NF-ATc1 act in concert to promote thymocyte development beyond the β-selection checkpoint. J Immunol. 2007;179:4694–703. doi: 10.4049/jimmunol.179.7.4694. [DOI] [PubMed] [Google Scholar]
- 255.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. 2012;12:749–61. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–62. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 257.Rolink AG, Nutt SL, Melchers F, Busslinger M. Long-term in vivo reconstitution of T-cell development by Pax5-deficient B-cell progenitors. Nature. 1999;401:603–6. doi: 10.1038/44164. [DOI] [PubMed] [Google Scholar]
- 258.Souabni A, Cobaleda C, Schebesta M, Busslinger M. Pax5 promotes B lymphopoiesis and blocks T cell development by repressing Notch1. Immunity. 2002;17:781–93. doi: 10.1016/s1074-7613(02)00472-7. [DOI] [PubMed] [Google Scholar]
- 259.Cotta CV, Zhang Z, Kim HG, Klug CA. Pax5 determines B- versus T-cell fate and does not block early myeloid-lineage development. Blood. 2003;101:4342–6. doi: 10.1182/blood-2002-10-3139. [DOI] [PubMed] [Google Scholar]
- 260.Banerjee A, Northrup D, Boukarabila H, Jacobsen SE, Allman D. Transcriptional repression of Gata3 is essential for early B cell commitment. Immunity. 2013;38:930–42. doi: 10.1016/j.immuni.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Nechanitzky R, Akbas D, Scherer S, Györy I, Hoyler T, Ramamoorthy S, Diefenbach A, Grosschedl R. Transcription factor EBF1 is essential for the maintenance of B cell identity and prevention of alternative fates in committed cells. Nat Immunol. 2013 doi: 10.1038/ni.2641. [DOI] [PubMed] [Google Scholar]
- 262.Goetz CA, Harmon IR, O'Neil JJ, Burchill MA, Johanns TM, Farrar MA. Restricted STAT5 activation dictates appropriate thymic B versus T cell lineage commitment. J.Immunol. 2005;174:7753–63. doi: 10.4049/jimmunol.174.12.7753. [DOI] [PubMed] [Google Scholar]
- 263.Massa S, Balciunaite G, Ceredig R, Rolink AG. Critical role for c-kit (CD117) in T cell lineage commitment and early thymocyte development in vitro. Eur.J.Immunol. 2006;36:526–32. doi: 10.1002/eji.200535760. [DOI] [PubMed] [Google Scholar]
- 264.Moller C, Alfredsson J, Engstrom M, Wootz H, Xiang Z, Lennartsson J, Jonsson JI, Nilsson G. Stem cell factor promotes mast cell survival via inactivation of FOXO3a-mediated transcriptional induction and MEK-regulated phosphorylation of the proapoptotic protein Bim. Blood. 2005;106:1330–6. doi: 10.1182/blood-2004-12-4792. [DOI] [PubMed] [Google Scholar]
- 265.Nie L, Xu M, Vladimirova A, Sun XH. Notch-induced E2A ubiquitination and degradation are controlled by MAP kinase activities. EMBO J. 2003;22:5780–92. doi: 10.1093/emboj/cdg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Cherrier M, Sawa S, Eberl G. Notch, Id2, and RORγt sequentially orchestrate the fetal development of lymphoid tissue inducer cells. J Exp Med. 2012;209:729–40. doi: 10.1084/jem.20111594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Boos MD, Yokota Y, Eberl G, Kee BL. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J. Exp. Med. 2007;204:1119–30. doi: 10.1084/jem.20061959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Nie L, Perry SS, Zhao Y, Huang J, Kincade PW, Farrar MA, Sun XH. Regulation of lymphocyte development by cell-type-specific interpretation of Notch signals. Mol Cell Biol. 2008;28:2078–90. doi: 10.1128/MCB.00844-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Weishaupt H, Sigvardsson M, Attema JL. Epigenetic chromatin states uniquely define the developmental plasticity of murine hematopoietic stem cells. Blood. 2010;115:247–56. doi: 10.1182/blood-2009-07-235176. [DOI] [PubMed] [Google Scholar]
- 270.Wendorff AA, Koch U, Wunderlich FT, Wirth S, Dubey C, Bruning JC, MacDonald HR, Radtke F. Hes1 is a critical but context-dependent mediator of canonical Notch signaling in lymphocyte development and transformation. Immunity. 2010;33:671–84. doi: 10.1016/j.immuni.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 271.Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie AN, Mebius RE, Powrie F, Vivier E. Innate lymphoid cells - a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–9. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 272.Rankin L, Groom J, Mielke LA, Seillet C, Belz GT. Diversity, function, and transcriptional regulation of gut innate lymphocytes. Front Immunol. 2013;4:22. doi: 10.3389/fimmu.2013.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Yang Q, Monticelli LA, Saenz SA, Chi AW, Sonnenberg GF, Tang J, De Obaldia ME, Bailis W, Bryson JL, Toscano K, Huang J, Haczku A, Pear WS, Artis D, Bhandoola A. T cell factor 1 is required for group 2 innate lymphoid cell generation. Immunity. 2013;38:694–704. doi: 10.1016/j.immuni.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Hoyler T, Klose CS, Souabni A, Turqueti-Neves A, Pfeifer D, Rawlins EL, Voehringer D, Busslinger M, Diefenbach A. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 2012;37:634–48. doi: 10.1016/j.immuni.2012.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Mjösberg J, Bernink J, Peters C, Spits H. Transcriptional control of innate lymphoid cells. Eur J Immunol. 2012;42:1916–23. doi: 10.1002/eji.201242639. [DOI] [PubMed] [Google Scholar]