Abstract
During cell stress, the transcription and translation of immediate early genes are prioritized, while most other messenger RNAs (mRNAs) are stored away in stress granules or degraded in processing bodies (P-bodies). TIA-1 is an mRNA-binding protein that needs to translocate from the nucleus to seed the formation of stress granules in the cytoplasm. Because other stress granule components such as TDP-43, FUS, ATXN2, SMN, MAPT, HNRNPA2B1, and HNRNPA1 are crucial for the motor neuron diseases amyotrophic lateral sclerosis (ALS)/spinal muscular atrophy (SMA) and for the frontotemporal dementia (FTD), here we studied mouse nervous tissue to identify mRNAs with selective dependence on Tia1 deletion. Transcriptome profiling with oligonucleotide microarrays in comparison of spinal cord and cerebellum, together with independent validation in quantitative reverse transcriptase PCR and immunoblots demonstrated several strong and consistent dysregulations. In agreement with previously reported TIA1 knock down effects, cell cycle and apoptosis regulators were affected markedly with expression changes up to +2-fold, exhibiting increased levels for Cdkn1a, Ccnf, and Tprkb vs. decreased levels for Bid and Inca1 transcripts. Novel and surprisingly strong expression alterations were detected for fat storage and membrane trafficking factors, with prominent +3-fold upregulations of Plin4, Wdfy1, Tbc1d24, and Pnpla2 vs. a −2.4-fold downregulation of Cntn4 transcript, encoding an axonal membrane adhesion factor with established haploinsufficiency. In comparison, subtle effects on the RNA processing machinery included up to 1.2-fold upregulations of Dcp1b and Tial1. The effect on lipid dynamics factors is noteworthy, since also the gene deletion of Tardbp (encoding TDP-43) and Atxn2 led to fat metabolism phenotypes in mouse. In conclusion, genetic ablation of the stress granule nucleator TIA-1 has a novel major effect on mRNAs encoding lipid homeostasis factors in the brain, similar to the fasting effect.
Electronic supplementary material
The online version of this article (doi:10.1007/s10048-014-0397-x) contains supplementary material, which is available to authorized users.
Keywords: TIA-1, Transcriptome, Cell cycle, Lipid trafficking, RNA processing machinery, Motor neuron disease, Frontotemporal dementia, Cerebellar ataxia
Introduction
Cells have evolved various mechanisms to compensate different types of environmental stress like UV irradiation, oxidative stress, or heat. Cytoplasmic stress responses include the formation of stress granules (SGs) and processing bodies (P-bodies) [1–4]. During stress, most messenger RNAs (mRNAs) are removed from ribosomal translation, thus conserving energy and allowing stress-induced damage repair or degradation [5]. While SGs are thought to be a place where the bulk of mRNAs, as well as some proteins, undergoes storage and triage, P-bodies contain mRNAs dedicated for decay [6]. This is compatible with the observation that SGs contain mRNAs within stalled translation initiation complexes including 40S ribosomal subunits but are devoid of eIF2, whereas P-bodies contain multiple mRNA decapping enzymes [6]. Both SGs and P-bodies are dynamic structures that assemble and disassemble rapidly [7]. They share a common pool of components and can fuse to exchange mRNAs [2, 6, 8]. In contrast to P-bodies, SGs only exist transiently during stress conditions [6].
This formation of cytoplasmic SGs depends on the shuttling of the 43 kDa protein TIA-1 from the nucleus and on the aggregation of a C-terminal proteolytic TIA-1 fragment of 15 kDa that includes a glutamine-rich prion-related domain (PRD) [1, 9–11]. TIA-1 was initially identified as T-cell-restricted intracellular antigen 1 and was subsequently investigated particularly in immunological cell types [12]. It contains also three RNA-recognition motifs (RRM) and binds to adenine/uridine-rich elements (AREs) in the 3’-untranslated region of mRNAs. TIA-1 (gene symbol TIA1) and its homolog TIAR (gene symbol TIAL1) have roles not only in the nucleus for gene transcription and pre-mRNA splicing [13, 14], but also in the cytoplasm for mRNA stability and translation regulation [5, 15, 16]. TIA-1 is associated with diverse cell processes including inflammation [16], apoptosis [17], autophagy [18], and cell proliferation [18, 19].
The role of SGs in human pathology have become increasingly clear, since mutations in several SG components are responsible for hereditary degeneration syndromes of peripheral and central motor neurons, namely amyotrophic lateral sclerosis (ALS), spinal muscular atrophy (SMA), and frontotemporal dementia (FTD). SG component proteins with a causal role for motor neuron diseases include TDP-43 (gene symbol TARDBP) [20–23], FUS [24–26], ATXN2 [27–29], SMN [30, 31], Tau (gene symbol MAPT) [32, 33], HNRNPA2B1, and HNRNPA1 [34]. In the SG component ATXN2, the presence of a polyglutamine domain mutation may lead to pathogenic unstable expansions. Intermediate size ATXN2 expansions comprise a risk factor for ALS through mRNA-mediated TDP-43 interaction [27, 35–37], while larger polyglutamine expansions in ATXN2 lead to Levodopa-responsive Parkinsonism [38] or to prominent cerebellar involvement with later progression to a multisystem atrophy of the nervous system, known as spinocerebellar ataxia type 2 (SCA2) [39]. Like TIA-1, several of these RNA-binding proteins shuttle from the nucleus to the cytoplasm during cell stress, and for TDP-43, it is known that its cytoplasmic accumulation depends on cyclin-dependent kinases [40]. While the protein composition of SGs is under intense investigation [41], much work remains to be done for the identification of mRNAs regulated by SGs, particularly in the vulnerable nervous tissue.
While all of these disease-associated proteins and their target RNAs shuttle to preformed SGs, the initial stress-induced nucleation of SGs appears dependent on TIA1, TIAL1, TTP, G3BP1/2, and FMRP [10, 32]. G3BP1 deletion results in massive neuronal death during embryogenesis, suggesting that it has a developmental role independent from its role(s) in the stress response [42]. TIA-1 is well characterized as a SG-nucleating protein, and Tia1 knockout (KO) mice not only exhibit grossly normal brain development, but also exhibit high embryonic lethality, consistent with dysregulation of a stress response [16]. We now used these mice for a transcriptome screen of nervous tissue at adult age, aiming to define the consequences of defective SG formation on RNA processing. The results confirm previous results obtained from human TIA1 knock down experiments in HeLa cells about cell cycle regulator modulation [19]. Importantly, our data documented novel strong effects on lipid storage and membrane dynamics factors. These insights may help to understand the disordered mRNA regulation, which makes a major contribution to the pathology underlying motor neuron diseases [43, 44].
Material and methods
Animals
Tia1 KO mice (bred into C57BL6/J background for more than 10 generations) were obtained from Harvard University, Dana Farber Cancer Institute. In these mice, homologous recombination of exon 4 results in a shortened Tia1 mRNA and absence of the 43 kDa TIA-1 protein [16]. C57BL6/J wild-type (WT) mice from The Jackson Laboratory were used as control. The animals were housed and kept in individually ventilated cages under routine health monitoring until the appropriate adult ages at the FELASA-certified mfd Diagnostics GmbH in Wendelsheim, Germany. They were fed ad libitum, were bred in homozygous matings, and were sacrificed by cervical dislocation. Nervous tissues and liver were removed in minimal time, frozen in liquid nitrogen, and stored at −80 °C. Genotypes were controlled by tail biopsy and DNA analysis. DNA was isolated from tail biopsies of Tia1 KO mice by Proteinase K (Ambion) treatment. PCR was performed using 50 ng DNA, 17 μl Pink Juice [125 μM Cresol Red sodium salt (Sigma Aldrich), 12.5 % 10× PCR buffer with 15 mM MgCl2 (Applied Biosystems), 250 μM dNTPs (Thermo Scientific), 25 % sucrose], 0.25 μl Taq Polymerase (AmpliTaq® DNA Polymerase, Applied Biosystems) and 1 μl of the primers KO1 5′-GTCGTGACAAGCCACACTTG-3′ and KO2 5′-AATTCCATCAGAAGCTTATCGAT-3′. The following conditions were applied: initial denaturation at 94 °C for 2 min, 33 cycles of 94 °C for 15 s denaturation, 58 °C for 30 s annealing, 72 °C for 1 min elongation, and a final elongation step at 72 °C for 10 min. The predicted length of the KO allele is 400 bp. Genotypes were further confirmed by quantitative real-time reverse transcriptase polymerase chain reaction (qPCR) measurement of Tia1 mRNA in the tissues under study. All procedures were in accordance with the German Animal Welfare Act, the Council Directive of 24 November 1986 (86/609/EWG) with Annex II and the ETS123 (European Convention for the Protection of Vertebrate Animals).
Transcriptome profiling
The dissected tissues cerebellum, spinal cord, midbrain, and liver from Tia1 KO mice and WT C57BL6/J mice at the age of 12 and 24 weeks (n = 3 vs. 3 mice/age) were sent to MFT Services (Tübingen, Germany). After RNA extraction, linear amplification and biotinylation of 100 ng of total RNA was performed with the GeneChip HT 3′IVT Express Kit (Affymetrix, Santa Clara, CA, USA) according to the manufacturer’s instructions. GeneChip HT Mouse Genome 430 2.0 Array Plates (Affymetrix) were used to hybridize fifteen micrograms of labeled and fragmented cRNA, to wash, stain, and scan automatically in a GeneTitan instrument (Affymetrix). Each of these oligonucleotide microarray chips is able to detect more than 39,000 transcripts with multiple probes for each mRNA. Visual inspection of scanned images was used to control for hybridization artifacts and proper grid alignment. AGCC 3.0 (Affymetrix) processed results were stored in CEL files. Further data analysis steps were carried out with the software platform R 2.14.0 and Bioconductor 2.14.0 [45]. First, the complete expression information from every chip was background corrected, quantile normalized, and summarized with Robust Multichip Average [46]. Empirical Bayes shrinkage of the standard errors was employed to derive the moderated F-statistic [47]. The resulting p values underwent multiple testing corrections according to “Benjamini-Hochberg” [48]. A decision matrix was produced through the function “decide tests” within the limma package, to attribute significant changes to individual contrasts. Thus, significant up- or downregulations were encoded by values of 1 or −1, respectively, to compare the consistency of significant expression changes across tissues and ages. All original transcriptome data were deposited at the public database Gene Expression Omnibus (GEO series accession # GSE54418, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE54418).
RNA isolation and expression analysis
RNA for qPCR expression analysis was extracted from cerebellar tissue (25 mg) of 12-week-old mice with Trizol® reagent (Invitrogen). Before cDNA synthesis, 1 μg of RNA was digested with DNase I Amplification Grade (Invitrogen). Reverse transcription was performed with SuperScript III Reverse Transcriptase (Invitrogen). Subsequently, expression levels were measured with the StepOnePlus Real-Time PCR System (Applied Biosystems) using 25 ng cDNA, 10 μl of FastStart Universal Probe Master (Rox) Mix (04914058001, Roche), and 1 μl of one of the following TaqMan Assays (Applied Biosystems): Atxn2 (Mm01199894_m1), Bid (Mm00432073_m1), Ccnf (Mm00432385_m1), Cdkn1a (Mm00432448_m1), Cntn4 (Mm00476065_m1), Dcp1b (Mm01183995_m1), Inca1 (Mm01243670_m1), Pabpc1 (Mm00849569_s1), Plin4 (Mm01272159_m1), Pnpla2 (Mm00503046_g1), Tbc1d24 (Mm00557451_m1), Tardbp (Mm00523870_g1), Tia1 Exon 3–4 (Mm01183616_m1), Tial1 (Mm00437049_m1), Tprkb (Mm00616325_m1), Tsen2 (Mm01184390_m1), Wdfy1 (Mm00840455_m1), and Tbp (Mm00446973_m1) as endogenous control. The PCR conditions were 50 °C for 2 min, 95 °C for 10 min, and 40 cycles of 95 °C for 15 s and 60 °C for 60 s. Analysis of the gene expression data was conducted using the 2−ΔΔCt method [49].
Protein extraction and quantitative immunoblots
For SDS-PAGE followed by immunoblotting, protein was extracted from 25 mg cerebellar tissue of 12-week-old mice. The tissue was homogenized with a motor pestle in 10 vol. RIPA buffer [50 mM Tris–HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 % Igepal CA-630 (Sigma), 0.5 % sodium deoxycholate, 0.1 % SDS, 1 mM PMSF, Complete Protease Inhibitor Cocktail (Roche)] and incubated on ice for 15 min. After centrifugation at 4 °C and 16,000×g for 20 min, the supernatant was stored (RIPA-soluble fraction), and the remaining pellet was dissolved in ½ vol. 2 × SDS buffer [137 mM Tris–HCl (pH 6.8), 4 % SDS, 20 % glycerol, Complete Protease Inhibitor Cocktail (Roche)] by sonification followed by 10 min of centrifugation at 16,000×g. The resulting supernatant was stored as RIPA-insoluble fraction. Protein concentration was determined with the BCA protein assay kit (Interchim, France), and 20 μg of each sample were loaded onto a 7.5 % polyacrylamide gel. After gel electrophoresis, the proteins were transferred to a PVDF membrane by wet blotting. The membranes were blocked with 5 % slim milk powder in PBST and incubated with antibodies against PLIN4 (1:500, Novus Biologicals), WDFY1 (1:500, Life Span BioSciences), CNTN4 (1:1,000, Abcam), or β-ACTIN (1:10,000, Sigma). ECL (Pierce) was used for visualizing the bands, which were subsequently quantified via densitometric analysis with ImageJ.
Statistical analysis
Data were analyzed with GraphPad Prism software version 5.04 (2010) using Student’s t test. Error bars indicate SEM. Significant p values (<0.05) were marked as follows: p < 0.05 *, p < 0.01 **, p < 0.001 ***.
Results
Transcriptome survey identifies strong changes of specific mRNAs in spinal cord
Microarray chip profiling of the transcriptome detected the loss of Tia1 correctly by one oligonucleotide (1431708_PM_a_at) corresponding to sequences at exon 4, whereas Tia1 oligonucleotides covering exons 9–11 (1416813_PM_at, 1416812_PM_at, 1416814_PM_at, 1437934_PM_at) detected significant upregulation of expression. These observations are in good agreement with a previous report [16] stating that the homologous recombination event within the Tia1 gene deletes sequences at exon 4, resulting in a shortened stable mRNA and in absence of TIA-1 protein. In the spinal cord, the expression profiling documented 115 oligonucleotides with significant upregulation both at 12 and 24 weeks of age vs. 70 oligonucleotides with significant downregulation at both ages, upon comparison of 3 KO and 3 WT tissues. The strongest three upregulations in spinal cord concerned Plin4 (3.3-fold), Wdfy1 (average 2.3-fold, detected consistently by three oligonucleotide spots), and Cdkn1a (average 2.2-fold, detected consistently by two oligonucleotide spots), while the strongest three downregulations concerned Gkn3 (in human only a pseudogene is conserved [50]), Bid (−1.9-fold), and Tsen2 (−1.8-fold) (Table 1). To further eliminate false positive candidates and to focus the investigation on mRNAs with relevance also for other tissues, the consistency of significant expression changes was compared from spinal cord to cerebellum, midbrain, and liver at both ages. Transcripts with significant expression change in the same direction in at least six out of the eight conditions under study were selected. They constituted 32 upregulations and 20 downregulations. All these Tia1 KO transcriptome data were made publically available via the GEO database. We concentrated further research on 17 transcripts with known function in shared pathways (Table 1).
Table 1.
Transcriptome profiling in four Tia1 KO mice tissues at two ages identifies consistent expression dysregulations. Tia1 KO and WT mice (3 vs. 3 at age 12 and 24 weeks) were compared, the significance of expression changes was determined, and consistently dysregulated transcript levels were shown with average fold changes. Negative values represent reduced expression (with green color highlighting its significance), while positive values represent induced expression (with red color highlighting its significance). Bold values illustrates transcripts with established induction by fasting conditions. The transcripts were grouped to reflect the convergent functions of the corresponding gene products in three pathways and were shown in alphabetical order
| Gene symbol | Gene name | Oligo spot ID | Fold change | |||
|---|---|---|---|---|---|---|
| Spinal cord 12 weeks | Spinal cord 24 weeks | Cerebellum 12 weeks | Cerebellum 24 weeks | |||
| Tia1 | Cytotoxic granule-associated RNA-binding protein 1 (TIA-1) | 1431708_PM_a_at | −4.78 | −4.72 | −3.60 | −3.45 |
| Cell cycle control | ||||||
| Bid | BH3 interacting domain death agonist | 1417045_PM_at | −1.98 | −1.82 | −1.67 | −1.70 |
| Ccnf | Cyclin F | 1422513_PM_at | 1.44 | 1.35 | 1.65 | 1.40 |
| Cdkn1a | Cyclin-dependent kinase inhibitor 1A (P21/Cip1) | 1421679_PM_a_at | 1.83 | 2.48 | 3.07 | 1.98 |
| 1424638_PM_at | 1.88 | 2.57 | 2.74 | 1.88 | ||
| Fgfrl1 | Fibroblast growth factor receptor-like 1 | 1447878_PM_s_at | −1.36 | −1.35 | −1.21 | −1.59 |
| Inca1 | Inhibitor of CDK, cyclin A1 interacting protein 1 | 1448034_PM_at | −1.28 | −1.15 | −1.26 | −1.33 |
| Nde1 | Nuclear distribution gene E homolog 1 (A. nidulans) | 1435737_PM_a_at | 1.32 | 1.28 | 1.53 | 1.31 |
| Tprkb | Tp53rk binding protein | 1425410_PM_at | 1.32 | 1.28 | 1.55 | 1.54 |
| Lipid storage and membrane trafficking | ||||||
| Angptl4 | Angiopoietin-like 4 | 1417130_PM_s_at | 2.30 | 2.50 | 2.05 | 1.79 |
| Cntn4 | Contactin-4 | 1438782_PM_at | −1.31 | −1.50 | −2.32 | −2.61 |
| Mfsd2a | Major facilitator superfamily domain containing 2A | 1428223_PM_at | 1.51 | 1.37 | 1.58 | 1.49 |
| Plin4 | Perilipin-4 | 1418595_PM_at | 3.51 | 3.05 | 2.62 | 2.11 |
| Pnpla2 | Patatin-like phospholipase domain containing 2 | 1428143_PM_a_at | 1.42 | 1.41 | 1.40 | 1.20 |
| Pnpla7 | Patatin-like phospholipase domain containing 7 | 1451361_PM_a_at | 1.24 | 1.42 | 1.28 | 1.37 |
| Tbc1d24 | TBC1 domain family, member 24 | 1448028_PM_at | 1.68 | 1.42 | 1.95 | 1.54 |
| 1442325_PM_at | 1.97 | 1.46 | 1.73 | 1.55 | ||
| Wdfy1 | WD repeat and FYVE domain-containing 1 | 1424749_PM_at | 1.36 | 3.15 | 1.38 | 2.94 |
| 1437358_PM_at | 1.37 | 3.32 | 1.31 | 2.91 | ||
| 1435588_PM_at | 1.34 | 3.17 | 1.39 | 2.75 | ||
| RNA processing machinery | ||||||
| Dcp1b | DCP1 decapping enzyme homolog b (S. cerevisiae) | 1444030_PM_at | 1.93 | 1.94 | 2.73 | 1.71 |
| Tsen2 | tRNA splicing endonuclease 2 homolog (S. cerevisiae) | 1459346_PM_at | −1.66 | −1.87 | −1.83 | −1.67 |
qPCR validates dysregulated levels of several transcripts in three pathways
Convergent effects were evident for the pathways of lipid storage and membrane trafficking, of cell cycle control, and additionally of the RNA processing machinery. The changes in expression levels of such genes were reassessed by the independent technique qPCR in cerebellum (Suppl. Figure 1). The results on the lipid pathway confirmed upregulations for Plin4 which encodes a lipid droplet storage factor (3.2-fold), for Wdfy1 encoding a modulator of PI3K control over endosome membrane trafficking (3.2-fold), for Tbc1d24 as Rab-GTPase activating vesicle dynamics factor (2.1-fold), and for Pnpla2 as component of the lipolytic cascade and as regulator of adiposome size (1.5-fold). A membrane pathway relevant downregulation was observed for Cntn4 as a glycosylphosphatidylinositol-anchored membrane adhesion factor implicated in axon network connections and synaptogenesis (−2.4-fold). Regarding the cell cycle pathway, upregulations were confirmed for Cdkn1a as cycle progression inhibitor (1.5-fold), Ccnf as a centrosome reduplication inhibitor during G2 phase (1.6-fold), and Tprkb as an ADP-ribose activated and p53-related protein kinase that transduces the PI3K/TOR pathway (1.1-fold). Cell cycle pathway relevant downregulations were confirmed for Bid as an ATM-effector that also activates the S-phase checkpoint (−1.7-fold), and Inca1 as an interactor of cyclin A1 that inhibits cyclin-dependent kinase and proliferation (−1.3-fold). Regarding the RNA processing pathway, the upregulation was confirmed for Dcp1b (1.2-fold) as a component of the RNA decapping and degradation machinery in P-bodies. In contrast, for Tsen2, the qPCR results suggested a significant upregulation (1.1-fold) instead of the downregulation previously observed by oligonucleotide microarray chips, a puzzling result since alternative splicing isoforms for this transcript are not documented. Since microarray chip data depend on the oligonucleotide choice and quality, additional hypothesis-driven qPCR were performed for important SG components with relevance for neurodegeneration and general mRNA translation. These experiments revealed a significant increase in the levels of Tial1 (1.2-fold), but did not detect major changes for the Pabpc1, Tardbp, or Atxn2 transcript levels. Altogether, most strong candidates from the transcriptome screening could be validated upon individual reassessment.
Quantitative immunoblots demonstrate altered levels for PLIN4, WDFY1, and CNTN4
To test whether these alterations of mRNA levels are compensated, for example by increased translation rates, or possess downstream consequences for the respective protein levels, quantitative immunoblots of cerebellar tissue were performed for three factors in the membrane dynamics pathway. Corresponding to the upregulation of the Plin4 transcript, the perilipin-4 protein levels were significantly upregulated (2.2-fold) in the RIPA-soluble tissue fraction that contains the freely soluble proteins (Fig. 1a), while they were undetectable in the SDS-soluble tissue fraction that contains membranes and more insoluble proteins. This observation is consistent with previous reports that perilipin-4 is recruited onto ER-membranes and lipid droplets only when factors such as diacylglycerol become abundant [51]. Again, in parallel to the upregulation of the Wdfy1 transcript, the WD repeat and FYVE domain-containing 1 protein levels were significantly upregulated (1.5-fold) in the SDS-soluble tissue fraction, while its presence in the RIPA-soluble tissue fraction was not significantly altered (Fig. 1b). The localization of WDFY1 to the SDS fraction is consistent with the FYVE domain association with the phosphatidylinositol 3-phosphates of endosomal membranes [52]. In agreement with the downregulation of the Cntn4 transcript, the contactin-4 protein levels were significantly decreased (−1.9-fold) in the SDS-soluble tissue fraction (Fig. 1c). Thus, the Tia1 knockout has selective effects on mRNA levels, resulting in abnormal levels of at least three proteins in the pathway of membrane dynamics and lipid storage.
Fig. 1.
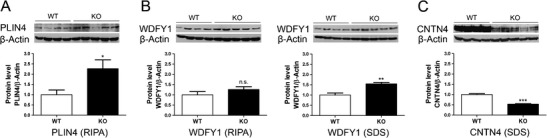
Quantitative immunoblots demonstrate significantly increased levels of perilipin-4 and WDFY1, but decreased levels of CNTN4 in Tia1 KO tissue. In cerebellum of 12-week-old mice (a), the PLIN4 levels were elevated in the RIPA-soluble protein fraction, whereas (b) the WDFY1 levels were elevated in the SDS fraction and (c) the CNTN4 levels were decreased in the SDS fraction (n = 4 WT vs. 5 KO mice)
Discussion
In the past, transcriptome profiling has been helpful to document changes in overall transcription and RNA processing, leading to the discovery of altered pathways and signaling networks in human disease [53]. While it is usually cumbersome in an organism to unravel how stressors impact neuronal function in molecular detail, this study of knockout tissues identifies novel selective RNA effects of TIA-1, which cause altered levels of the corresponding proteins that modulate membrane dynamics and lipid storage.
TIA-1 is a key stress granule component, capable of nucleating SGs when overexpressed and inhibiting SG formation when absent [10]. As a consequence, one might have expected an alteration in the levels of other stress granule components when TIA-1 is depleted. However, this assumption was not corroborated in the Tia1 KO mouse tissues for most of the SG-associated genes tested. In the transcriptome data, there was no obvious dysregulation for any other known SG components. There are several possible explanations for this: (1) the loss of Tia1 might be compensated by expression changes in other genes that were not present on the chip, by alternative splicing changes that are not represented on the microarray chip or by expression changes with bare significance (e.g. Tial1); (2) a Tia1 KO could have severe effects on the localization of stress granule components without influencing their expression; or (3) Tia1 deletion might only have an effect on their expression levels under acute stress, which was absent from the tissue of young mice that were kept in a pathogen-free environment and were allowed to eat ad libitum. The slight upregulation of Tial1 mRNA levels is probably a compensatory effort, since TIA-1 overexpression was observed to substitute for Tial1 deletion and to correlate inversely with Tial1 expression levels [54]. Interestingly, a relatively stronger upregulation of Dcp1b, encoding a core component of the mRNA decapping complex in P-bodies, may indicate increased mRNA decay in the absence of TIA-1.
The more substantial effects of the Tia1 KO on cell cycle and apoptosis-related factors are in agreement with previous reports [19]. A team investigating the effects of TIA1 knock down in human HeLa cells observed proliferative effects with increased cell numbers in S- or G2/M-phases and an induction of anchorage-independent growth, in parallel to upregulation of interleukin/chemokine transcripts and downregulation of transcript levels for the tumor necrosis factor superfamily member 10 and the P21protein/CDKN1A-activated kinase PAK3 [19]. In partial accord, a recent study of Tia1 KO effects in murine embryonic fibroblasts observed again a prominent cell cycle effect, but documented reduced rates of cell proliferation, cell cycle progression delay, increased cell size, and apoptosis enhancement [18]. Our data documented downregulated transcript levels for apoptosis-promoting factors such as Bid and Fgfrl1. The downregulation of Bid was previously described to occur after serum starvation and to induce autophagy [55, 56]. The downregulated transcript levels of cell cycle inhibitors such as Fgfrl1 and Inca1 on the one hand, together with the upregulated transcript levels of cell cycle enhancers like Ccnf and Nde1 transcripts, seem difficult to integrate with the upregulation of the cell cycle inhibitor Cdkn1a on the other hand. Beyond possible consequences for neurogenesis, there is a clear role of CDKN1A/p21 for glia proliferation [57]. The upregulation of CDKN1A expression is a known response to starvation, which arrests the cell cycle and thus protects from cell death [58, 59]. Beyond glia cells, an additional role of CDKN1A/p21 in adult neurons regarding DNA damage response, neuroprotection, neuronal senescence, motor neuron regeneration, and tauopathy is established [60–66]. In this context, also the upregulation of Nde1 is interesting, since it encodes a modulator of mitotic spindle function and progenitor migration, which is responsible for neuron number in cortical layers II-IV [67]. Altogether, the role of TIA-1 for regulating cell cycle, cell death, and stress responses in adult nervous tissue is credible.
Our transcriptome profiling highlighted an unknown function for TIA-1 in membrane dynamics and lipid storage. One-fifth of the altered transcripts detected are involved in lipid storage, transport, or membrane trafficking, a number far exceeding stochastic expectations even in view of the high lipid content of brain tissue. Several dysregulated factors are involved in the formation of lipid droplets. These structures store neutral lipids in their core and are important for lipid transportation [68], vesicle trafficking, and cell signaling [69]. Perilipin 4 (encoded by Plin4) was shown in adipocytes to coat the nascent lipid droplets [70]. Accordingly, an upregulation of Plin4 in the Tia-1 KO mice might correlate with an enhanced formation or turnover of lipid droplets. This notion is strengthened by the fact that two other lipid droplet components, Pnpla2 and Pnpla7 (encoding patatin-like phospholipase domain containing 2 and 7, respectively) also show increased transcript levels. Pnpla2 hydrolyzes triglycerides, thus providing the organism with energy through the supply of free fatty acids and altering membrane composition [68, 71]. This mechanism becomes important during starvation stress. Furthermore, it has been shown that Pnpla7 levels are increased by fasting and that PNPLA7 may be involved in organophosphorus compound-induced motor neuron degeneration [72, 73]. Although our animals were not fasting, two other transcripts that are normally increased under this condition were also upregulated, namely Mfsd2a and Angptl4 [74–76]. These data suggest that there are fasting-like stress conditions in the Tia1 KO mouse model, which are independent of food availability, but balance the organism towards gaining energy from fatty acids. Thus, deletion of Tia1 increases the levels of transcripts that are normally induced by fasting conditions and are involved in lipid transport and membrane trafficking.
The Tia1 KO-induced upregulation of Wdfy1 and downregulation of Cntn4 levels modulate two factors implicated in phosphoinositide-dependent membrane binding. The WD repeat and FYVE domain-containing 1 protein interacts with phosphoinositide-3-phosphate enriched endosomal membranes, in particular under stress-induced acidic conditions, helping to recruit other proteins involved in membrane trafficking [52, 77]. Upregulation of Wdfy1 can be induced by pharmacological inhibition of autophagy during starvation stress [78]. Interestingly, Wdfy1 level upregulation and Tia1 dysregulation were among the 16 most promising biomarkers that characterized the brain of mouse model of Alzheimer’s disease, with Wdfy1 showing the changes earlier than Tia1 [79]. Similarly, the upregulation detected consistently by two oligonucleotide spots for Tbc1d24 encodes an activator of small GTPases involved in the regulation of membrane trafficking, which was shown to act as potent modulator of primary axonal arborization [80, 81]. Its homolog Tbc1d1 was linked to human obesity and a Tbc1d1 mutation underlies the absence of diet-induced obesity in the lean mouse strain [82–84]. A perhaps even more intriguing finding regarding medical relevance is the downregulation of contactin-4, since this glycosylphosphatidylinositol-anchored neuronal adhesion protein is involved in axon guidance and synaptic plasticity [85–88] and interacts with the Alzheimer’s disease mediator amyloid precursor protein [89]. Genetic haploinsufficiency of contactin-4 was demonstrated to cause developmental delay [90]. Other members of the contactin protein family have been implicated in selective motor neuron pathology, namely contactin-1 in human [91] and the contactin-2 ortholog in zebrafish [92, 93]. It is noteworthy that contactin-2/TAG1 is a strong regulator of diet-induced obesity [94]. Thus, these data emphasize the role of TIA-1 for the stress-dependent composition and trafficking of membranes as well as their protein interactions.
It is important to note that the effect of TIA-1 on lipid and membrane dynamics is paralleled by similar effect of two other SG components. A genetic ablation of the RNA-binding protein ATXN2 in mice leads to obesity, appearance of lipid droplets in the liver, increased blood cholesterol, cerebellar gangliosides, and sulfatides [95]. Conversely, gain-of-function mutations of ATXN2 lead to a multisystem atrophy of the nervous system [39]. This scenario with ATXN2 loss-of-function affecting lipid homeostasis, while its excess causes neurodegenerative diseases, shows a striking similarity to the effects of TDP-43. Postnatal deletion of the TDP-43-encoding Tardbp gene was shown to cause dramatic loss of body fat and weight together with a downregulation of the leanness factor Tbc1d1 [96]. Conversely again, the overexpression of Tardbp leads to increased fat deposition and adipocyte hypertrophy together with an upregulation of Tbc1d1 [97]. A representative TDP-43 mutation that causes neurodegenerative diseases was shown to enhance normal TDP-43 splicing function for some RNA targets but loss-of-function for others, in the absence of aggregation or nuclear depletion of TDP-43 [98]. Jointly, these data underscore a prominent role of three SG components for mRNAs that regulate lipid metabolism and membrane composition under stress.
In conclusion, our data show that ablation of Tia1 in mouse tissues leads to changed expression levels of few constituents of the mRNA processing machinery, of specific cell cycle and apoptosis pathways components, and of various lipid storage and membrane dynamics factors. We propose that TIA-1 depletion induces starvation-like conditions as a trigger for the upregulation of these transcripts. These findings may be relevant to elucidate the role of stress granules and aberrant RNA processing for the prominent axon transport pathology in motor neuron diseases such as ALS, SMA, FTD, and SCA2.
Electronic supplementary material
Below is the link to the electronic supplementary material.
qPCR analysis of cerebellar expression in 12-week-old WT and Tia1 KO mice validates most microarray findings. Several transcripts involved in (A) cell cycle regulation, (B) lipid storage and membrane dynamics, and (C) RNA processing were demonstrated to show significant expression changes (n = 6 WT vs. 6 KO mice, the order within each pathway is alphabetical). The >3-fold upregulation of Plin4 and Wdfy1 and >−2-fold downregulation of Cntn4 transcript levels were prominent. (JPEG 1581 kb)
Acknowledgment
The project was financed by the DFG project AU 96/13-1. We are grateful to the staff at ZFE University Hospital Frankfurt and at mfd Wendelsheim for technical assistance and to Dr. M. Jendrach for manuscript proofreading.
Abbreviations
- Angptl4
Angiopoietin-like 4
- Atxn2
Ataxin-2
- Bid
BH3 interacting-domain death agonist
- Ccnf
Cyclin F
- Cdkn1a
Cyclin-dependent kinase inhibitor 1A (P21/Cip1)
- Cntn4
Contactin-4
- Dcp1b
DCP1 decapping enzyme homolog B (S. cerevisiae)
- Fgfrl1
Fibroblast growth factor receptor-like 1
- Inca1
Inhibitor of CDK cyclin A1 interacting protein 1
- Mfsd2a
Major facilitator superfamily domain containing 2A and angiopoietin-like 4
- Nde1
Nuclear distribution gene E homolog 1 (NudE neurodevelopment protein 1)
- Pabpc1
Poly(A)-binding protein cytoplasmic 1
- Plin4
Perilipin-4
- Pnpla2
Patatin-like phospholipase domain containing 2
- Pnpla7
Patatin-like phospholipase domain containing 7
- Tbc1d24
TBC1 domain family member 24
- Tbp
TATA-box-binding protein
- Tardbp
TAR DNA-binding protein-43 (TDP-43)
- Tia1
T-cell-restricted intracellular antigen-1 cytotoxic granule-associated RNA-binding protein (TIA-1)
- Tial1
TIA1 cytotoxic granule-associated RNA binding protein-like 1
- Tprkb
Tp53rk binding protein
- Tsen2
tRNA splicing endonuclease 2 homolog (S. cerevisiae)
- Wdfy1
WD repeat and FYVE domain-containing 1 (FENS-1)
References
- 1.Kedersha NL, et al. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147(7):1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kedersha N, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169(6):871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teixeira D, et al. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11(4):371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nature reviews. Mol Cell Biol. 2009;10(6):430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 5.Anderson P, Kedersha N. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones. 2002;7(2):213–221. doi: 10.1379/1466-1268(2002)007<0213:vstroe>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172(6):803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kedersha N, Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans. 2002;30(Pt 6):963–969. doi: 10.1042/bst0300963. [DOI] [PubMed] [Google Scholar]
- 8.Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Biol. 2009;21(3):403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang T, et al. Identification of the sequence determinants mediating the nucleo-cytoplasmic shuttling of TIAR and TIA-1 RNA-binding proteins. J Cell Sci. 2005;118(Pt 23):5453–5463. doi: 10.1242/jcs.02669. [DOI] [PubMed] [Google Scholar]
- 10.Gilks N, et al. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15(12):5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson P, et al. A monoclonal antibody reactive with a 15-kDa cytoplasmic granule-associated protein defines a subpopulation of CD8+ T lymphocytes. J Immunol. 1990;144(2):574–582. [PubMed] [Google Scholar]
- 12.Anderson P, et al. Post-transcriptional regulation of proinflammatory proteins. J Leukoc Biol. 2004;76(1):42–47. doi: 10.1189/jlb.1103536. [DOI] [PubMed] [Google Scholar]
- 13.Del Gatto-Konczak F, et al. The RNA-binding protein TIA-1 is a novel mammalian splicing regulator acting through intron sequences adjacent to a 5’ splice site. Mol Cell Biol. 2000;20(17):6287–6299. doi: 10.1128/mcb.20.17.6287-6299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forch P, et al. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol Cell. 2000;6(5):1089–1098. doi: 10.1016/s1097-2765(00)00107-6. [DOI] [PubMed] [Google Scholar]
- 15.Dixon DA, et al. Regulation of cyclooxygenase-2 expression by the translational silencer TIA-1. J Exp Med. 2003;198(3):475–481. doi: 10.1084/jem.20030616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piecyk M, et al. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 2000;19(15):4154–4163. doi: 10.1093/emboj/19.15.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawakami A, et al. Identification and functional characterization of a TIA-1-related nucleolysin. Proc Natl Acad Sci U S A. 1992;89(18):8681–8685. doi: 10.1073/pnas.89.18.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez-Jimenez C, Izquierdo JM. T-cell intracellular antigen (TIA)-proteins deficiency in murine embryonic fibroblasts alters cell cycle progression and induces autophagy. PLoS One. 2013;8(9):e75127. doi: 10.1371/journal.pone.0075127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reyes R, Alcalde J, Izquierdo JM. Depletion of T-cell intracellular antigen proteins promotes cell proliferation. Genome Biol. 2009;10(8):R87. doi: 10.1186/gb-2009-10-8-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li YR, et al. Stress granules as crucibles of ALS pathogenesis. J Cell Biol. 2013;201(3):361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu-Yesucevitz L, et al. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS One. 2010;5(10):e13250. doi: 10.1371/journal.pone.0013250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDonald KK, et al. TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum Mol Genet. 2011;20(7):1400–1410. doi: 10.1093/hmg/ddr021. [DOI] [PubMed] [Google Scholar]
- 23.Bentmann E, Haass C, Dormann D. Stress granules in neurodegeneration—lessons learnt from TAR DNA binding protein of 43 kDa and fused in sarcoma. FEBS J. 2013;280(18):4348–4370. doi: 10.1111/febs.12287. [DOI] [PubMed] [Google Scholar]
- 24.Bosco DA, et al. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum Mol Genet. 2010;19(21):4160–4175. doi: 10.1093/hmg/ddq335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daigle JG, et al. RNA-binding ability of FUS regulates neurodegeneration, cytoplasmic mislocalization and incorporation into stress granules associated with FUS carrying ALS-linked mutations. Hum Mol Genet. 2013;22(6):1193–1205. doi: 10.1093/hmg/dds526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vance C, et al. ALS mutant FUS disrupts nuclear localization and sequesters wild-type FUS within cytoplasmic stress granules. Hum Mol Genet. 2013;22(13):2676–2688. doi: 10.1093/hmg/ddt117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elden AC, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466(7310):1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nonhoff U, et al. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol Biol Cell. 2007;18(4):1385–1396. doi: 10.1091/mbc.E06-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farg MA, et al. Ataxin-2 interacts with FUS and intermediate-length polyglutamine expansions enhance FUS-related pathology in amyotrophic lateral sclerosis. Hum Mol Genet. 2013;22(4):717–728. doi: 10.1093/hmg/dds479. [DOI] [PubMed] [Google Scholar]
- 30.Dewey CM, et al. TDP-43 aggregation in neurodegeneration: are stress granules the key? Brain Res. 2012;1462:16–25. doi: 10.1016/j.brainres.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hua Y, Zhou J. Survival motor neuron protein facilitates assembly of stress granules. FEBS Lett. 2004;572(1–3):69–74. doi: 10.1016/j.febslet.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 32.Wolozin B. Regulated protein aggregation: stress granules and neurodegeneration. Mol Neurodegener. 2012;7:56. doi: 10.1186/1750-1326-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vanderweyde T, et al. Contrasting pathology of the stress granule proteins TIA-1 and G3BP in tauopathies. J Neurosci Off J Soc Neurosci. 2012;32(24):8270–8283. doi: 10.1523/JNEUROSCI.1592-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim HJ, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495(7442):467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gispert S, et al. The modulation of Amyotrophic Lateral Sclerosis risk by ataxin-2 intermediate polyglutamine expansions is a specific effect. Neurobiol Dis. 2012;45(1):356–361. doi: 10.1016/j.nbd.2011.08.021. [DOI] [PubMed] [Google Scholar]
- 36.Lee T, et al. Ataxin-2 intermediate-length polyglutamine expansions in European ALS patients. Hum Mol Genet. 2011;20(9):1697–1700. doi: 10.1093/hmg/ddr045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lahut S, et al. ATXN2 and its neighbouring gene SH2B3 are associated with increased ALS risk in the Turkish population. PLoS One. 2012;7(8):e42956. doi: 10.1371/journal.pone.0042956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Charles P, et al. Are interrupted SCA2 CAG repeat expansions responsible for Parkinsonism? Neurology. 2007;69(21):1970–1975. doi: 10.1212/01.wnl.0000269323.21969.db. [DOI] [PubMed] [Google Scholar]
- 39.Auburger GW. Spinocerebellar ataxia type 2. Handb Clin Neurol. 2012;103:423–436. doi: 10.1016/B978-0-444-51892-7.00026-7. [DOI] [PubMed] [Google Scholar]
- 40.Moujalled D, et al. Kinase inhibitor screening identifies cyclin-dependent kinases and glycogen synthase kinase 3 as potential modulators of TDP-43 cytosolic accumulation during cell stress. PLoS One. 2013;8(6):e67433. doi: 10.1371/journal.pone.0067433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohn T, et al. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol. 2008;10(10):1224–1231. doi: 10.1038/ncb1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zekri L, et al. Control of fetal growth and neonatal survival by the RasGAP-associated endoribonuclease G3BP. Mol Cell Biol. 2005;25(19):8703–8716. doi: 10.1128/MCB.25.19.8703-8716.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferraiuolo L, et al. Molecular pathways of motor neuron injury in amyotrophic lateral sclerosis. Nat Rev Neurol. 2011;7(11):616–630. doi: 10.1038/nrneurol.2011.152. [DOI] [PubMed] [Google Scholar]
- 44.Polymenidou M, et al. Misregulated RNA processing in amyotrophic lateral sclerosis. Brain Res. 2012;1462:3–15. doi: 10.1016/j.brainres.2012.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Irizarry RA, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smyth GK (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: p. Article 3 [DOI] [PubMed]
- 48.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 49.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 50.Geahlen JH, et al. Evolution of the human gastrokine locus and confounding factors regarding the pseudogenicity of GKN3. Physiol Genomics. 2013;45(15):667–683. doi: 10.1152/physiolgenomics.00169.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skinner JR, et al. Diacylglycerol enrichment of endoplasmic reticulum or lipid droplets recruits perilipin 3/TIP47 during lipid storage and mobilization. J Biol Chem. 2009;284(45):30941–30948. doi: 10.1074/jbc.M109.013995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He J, et al. Membrane insertion of the FYVE domain is modulated by pH. Proteins. 2009;76(4):852–860. doi: 10.1002/prot.22392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Voineagu I, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474(7351):380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Le Guiner C, Gesnel MC, Breathnach R. TIA-1 or TIAR is required for DT40 cell viability. J Biol Chem. 2003;278(12):10465–10476. doi: 10.1074/jbc.M212378200. [DOI] [PubMed] [Google Scholar]
- 55.Goyeneche AA, Harmon JM, Telleria CM. Cell death induced by serum deprivation in luteal cells involves the intrinsic pathway of apoptosis. Reproduction. 2006;131(1):103–111. doi: 10.1530/rep.1.00751. [DOI] [PubMed] [Google Scholar]
- 56.Lamparska-Przybysz M, Gajkowska B, Motyl T. BID-deficient breast cancer MCF-7 cells as a model for the study of autophagy in cancer therapy. Autophagy. 2006;2(1):47–48. doi: 10.4161/auto.2149. [DOI] [PubMed] [Google Scholar]
- 57.Atanasoski S, et al. Cell cycle inhibitors p21 and p16 are required for the regulation of Schwann cell proliferation. Glia. 2006;53(2):147–157. doi: 10.1002/glia.20263. [DOI] [PubMed] [Google Scholar]
- 58.Kim MJ, et al. Mitochondrial ribosomal protein L41 mediates serum starvation-induced cell-cycle arrest through an increase of p21(WAF1/CIP1) Biochem Biophys Res Commun. 2005;338(2):1179–1184. doi: 10.1016/j.bbrc.2005.10.064. [DOI] [PubMed] [Google Scholar]
- 59.Braun F, et al. Serum-nutrient starvation induces cell death mediated by Bax and Puma that is counteracted by p21 and unmasked by Bcl-x(L) inhibition. PLoS One. 2011;6(8):e23577. doi: 10.1371/journal.pone.0023577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jurk D, et al. Postmitotic neurons develop a p21-dependent senescence-like phenotype driven by a DNA damage response. Aging Cell. 2012;11(6):996–1004. doi: 10.1111/j.1474-9726.2012.00870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langley B, et al. Pulse inhibition of histone deacetylases induces complete resistance to oxidative death in cortical neurons without toxicity and reveals a role for cytoplasmic p21(waf1/cip1) in cell cycle-independent neuroprotection. J Neurosci Off J Soc Neurosci. 2008;28(1):163–176. doi: 10.1523/JNEUROSCI.3200-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harms C, et al. Phosphatidylinositol 3-Akt-kinase-dependent phosphorylation of p21(Waf1/Cip1) as a novel mechanism of neuroprotection by glucocorticoids. J Neurosci Off J Soc Neurosci. 2007;27(17):4562–4571. doi: 10.1523/JNEUROSCI.5110-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Delobel P, et al. Cell-cycle markers in a transgenic mouse model of human tauopathy: increased levels of cyclin-dependent kinase inhibitors p21Cip1 and p27Kip1. Am J Pathol. 2006;168(3):878–887. doi: 10.2353/ajpath.2006.050540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tanaka H, et al. Cytoplasmic p21(Cip1/WAF1) enhances axonal regeneration and functional recovery after spinal cord injury in rats. Neuroscience. 2004;127(1):155–164. doi: 10.1016/j.neuroscience.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 65.Zaman K, et al. Protection from oxidative stress-induced apoptosis in cortical neuronal cultures by iron chelators is associated with enhanced DNA binding of hypoxia-inducible factor-1 and ATF-1/CREB and increased expression of glycolytic enzymes, p21(waf1/cip1), and erythropoietin. J Neurosci Off J Soc Neurosci. 1999;19(22):9821–9830. doi: 10.1523/JNEUROSCI.19-22-09821.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park DS, et al. Cyclin dependent kinase inhibitors and dominant negative cyclin dependent kinase 4 and 6 promote survival of NGF-deprived sympathetic neurons. J Neurosci Off J Soc Neurosci. 1997;17(23):8975–8983. doi: 10.1523/JNEUROSCI.17-23-08975.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Feng Y, Walsh CA. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 2004;44(2):279–293. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 68.Zehmer JK, et al. A role for lipid droplets in inter-membrane lipid traffic. Proteomics. 2009;9(4):914–921. doi: 10.1002/pmic.200800584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu P, et al. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J Biol Chem. 2004;279(5):3787–3792. doi: 10.1074/jbc.M311945200. [DOI] [PubMed] [Google Scholar]
- 70.Wolins NE, et al. Adipocyte protein S3-12 coats nascent lipid droplets. J Biol Chem. 2003;278(39):37713–37721. doi: 10.1074/jbc.M304025200. [DOI] [PubMed] [Google Scholar]
- 71.Jenkins CM, et al. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem. 2004;279(47):48968–48975. doi: 10.1074/jbc.M407841200. [DOI] [PubMed] [Google Scholar]
- 72.Richardson RJ, et al. Neuropathy target esterase (NTE): overview and future. Chem Biol Interact. 2013;203(1):238–244. doi: 10.1016/j.cbi.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 73.Kienesberger PC, et al. Identification of an insulin-regulated lysophospholipase with homology to neuropathy target esterase. J Biol Chem. 2008;283(9):5908–5917. doi: 10.1074/jbc.M709598200. [DOI] [PubMed] [Google Scholar]
- 74.Gonzalez-Muniesa P, et al. Fatty acids and hypoxia stimulate the expression and secretion of the adipokine ANGPTL4 (angiopoietin-like protein 4/ fasting-induced adipose factor) by human adipocytes. J Nutrigenet Nutrigenom. 2011;4(3):146–153. doi: 10.1159/000327774. [DOI] [PubMed] [Google Scholar]
- 75.Angers M, et al. Mfsd2a encodes a novel major facilitator superfamily domain-containing protein highly induced in brown adipose tissue during fasting and adaptive thermogenesis. Biochem J. 2008;416(3):347–355. doi: 10.1042/BJ20080165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koliwad SK, et al. Angiopoietin-like 4 (ANGPTL4, fasting-induced adipose factor) is a direct glucocorticoid receptor target and participates in glucocorticoid-regulated triglyceride metabolism. J Biol Chem. 2009;284(38):25593–25601. doi: 10.1074/jbc.M109.025452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ridley SH, et al. FENS-1 and DFCP1 are FYVE domain-containing proteins with distinct functions in the endosomal and Golgi compartments. J Cell Sci. 2001;114(Pt 22):3991–4000. doi: 10.1242/jcs.114.22.3991. [DOI] [PubMed] [Google Scholar]
- 78.Stanton MJ, et al. Angiogenic growth factor axis in autophagy regulation. Autophagy. 2013;9(5):789–790. doi: 10.4161/auto.23783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arisi I, et al. Gene expression biomarkers in the brain of a mouse model for Alzheimer’s disease: mining of microarray data by logic classification and feature selection. J Alzheimer Dis. 2011;24(4):721–738. doi: 10.3233/JAD-2011-101881. [DOI] [PubMed] [Google Scholar]
- 80.Falace A, et al. TBC1D24, an ARF6-interacting protein, is mutated in familial infantile myoclonic epilepsy. Am J Hum Genet. 2010;87(3):365–370. doi: 10.1016/j.ajhg.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Corbett MA, et al. A focal epilepsy and intellectual disability syndrome is due to a mutation in TBC1D24. Am J Hum Genet. 2010;87(3):371–375. doi: 10.1016/j.ajhg.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stone S, et al. TBC1D1 is a candidate for a severe obesity gene and evidence for a gene/gene interaction in obesity predisposition. Hum Mol Genet. 2006;15(18):2709–2720. doi: 10.1093/hmg/ddl204. [DOI] [PubMed] [Google Scholar]
- 83.Meyre D, et al. R125W coding variant in TBC1D1 confers risk for familial obesity and contributes to linkage on chromosome 4p14 in the French population. Hum Mol Genet. 2008;17(12):1798–1802. doi: 10.1093/hmg/ddn070. [DOI] [PubMed] [Google Scholar]
- 84.Chadt A, et al. Tbc1d1 mutation in lean mouse strain confers leanness and protects from diet-induced obesity. Nat Genet. 2008;40(11):1354–1359. doi: 10.1038/ng.244. [DOI] [PubMed] [Google Scholar]
- 85.Kaneko-Goto T, et al. BIG-2 mediates olfactory axon convergence to target glomeruli. Neuron. 2008;57(6):834–846. doi: 10.1016/j.neuron.2008.01.023. [DOI] [PubMed] [Google Scholar]
- 86.Murai KK, Misner D, Ranscht B. Contactin supports synaptic plasticity associated with hippocampal long-term depression but not potentiation. Curr Biol : CB. 2002;12(3):181–190. doi: 10.1016/s0960-9822(02)00680-2. [DOI] [PubMed] [Google Scholar]
- 87.Saito H, et al. Expression of olfactory receptors. G-proteins and AxCAMs during the development and maturation of olfactory sensory neurons in the mouse. Brain Res Dev Brain Res. 1998;110(1):69–81. doi: 10.1016/s0165-3806(98)00096-0. [DOI] [PubMed] [Google Scholar]
- 88.Yoshihara Y, et al. Overlapping and differential expression of BIG-2, BIG-1, TAG-1, and F3: four members of an axon-associated cell adhesion molecule subgroup of the immunoglobulin superfamily. J Neurobiol. 1995;28(1):51–69. doi: 10.1002/neu.480280106. [DOI] [PubMed] [Google Scholar]
- 89.Osterfield M, et al. Interaction of amyloid precursor protein with contactins and NgCAM in the retinotectal system. Development. 2008;135(6):1189–1199. doi: 10.1242/dev.007401. [DOI] [PubMed] [Google Scholar]
- 90.Fernandez T, et al. Disruption of contactin 4 (CNTN4) results in developmental delay and other features of 3p deletion syndrome. Am J Hum Genet. 2004;74(6):1286–1293. doi: 10.1086/421474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Querol L, et al. Antibodies to contactin-1 in chronic inflammatory demyelinating polyneuropathy. Ann Neurol. 2013;73(3):370–380. doi: 10.1002/ana.23794. [DOI] [PubMed] [Google Scholar]
- 92.Sittaramane V, et al. The cell adhesion molecule Tag1, transmembrane protein Stbm/Vangl2, and Lamininalpha1 exhibit genetic interactions during migration of facial branchiomotor neurons in zebrafish. Dev Biol. 2009;325(2):363–373. doi: 10.1016/j.ydbio.2008.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lin JF, et al. The cell neural adhesion molecule contactin-2 (TAG-1) is beneficial for functional recovery after spinal cord injury in adult zebrafish. PLoS One. 2012;7(12):e52376. doi: 10.1371/journal.pone.0052376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Buchner DA, et al. The juxtaparanodal proteins CNTNAP2 and TAG1 regulate diet-induced obesity. Mamm Genome Off J Int Mamm Genome Soc. 2012;23(7–8):431–442. doi: 10.1007/s00335-012-9400-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lastres-Becker I, et al. Insulin receptor and lipid metabolism pathology in ataxin-2 knock-out mice. Hum Mol Genet. 2008;17(10):1465–1481. doi: 10.1093/hmg/ddn035. [DOI] [PubMed] [Google Scholar]
- 96.Chiang PM, et al. Deletion of TDP-43 down-regulates Tbc1d1, a gene linked to obesity, and alters body fat metabolism. Proc Natl Acad Sci U S A. 2010;107(37):16320–16324. doi: 10.1073/pnas.1002176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stallings NR, et al. TDP-43, an ALS linked protein, regulates fat deposition and glucose homeostasis. PLoS One. 2013;8(8):e71793. doi: 10.1371/journal.pone.0071793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Arnold ES, et al. ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc Natl Acad Sci U S A. 2013;110(8):E736–E745. doi: 10.1073/pnas.1222809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
qPCR analysis of cerebellar expression in 12-week-old WT and Tia1 KO mice validates most microarray findings. Several transcripts involved in (A) cell cycle regulation, (B) lipid storage and membrane dynamics, and (C) RNA processing were demonstrated to show significant expression changes (n = 6 WT vs. 6 KO mice, the order within each pathway is alphabetical). The >3-fold upregulation of Plin4 and Wdfy1 and >−2-fold downregulation of Cntn4 transcript levels were prominent. (JPEG 1581 kb)


