Abstract
Plant shoots display indeterminate growth, while their evolutionary decedents, the leaves, are determinate. Determinate leaf growth is conditioned by the CIN-TCP transcription factors, which promote leaf maturation and which are negatively regulated by miR319 in leaf primordia. Here we show that CIN-TCPs reduce leaf sensitivity to cytokinin (CK), a phytohormone implicated in inhibition of differentiation in the shoot. We identify the SWI/SNF chromatin remodeling ATPase BRAHMA (BRM) as a genetic mediator of CIN-TCP activities and CK responses. An interactome screen further revealed that SWI/SNF complex components including BRM preferentially interacted with bHLH transcription factors and the bHLH-related CIN-TCPs. Indeed, TCP4 and BRM interacted in planta. Both TCP4 and BRM bound the promoter of an inhibitor of CK responses, ARR16, and induced its expression. Reconstituting ARR16 levels in leaves with reduced CIN-TCP activity restored normal growth. Thus, CIN-TCP and BRM together promote determinate leaf growth by stage-specific modification of CK responses.
Keywords: leaf maturation, CIN-TCP, chromatin remodeling, cytokinin, BRAHMA
Introduction
Plant shoots are characterized by indeterminate growth, while leaves undergo a gradual differentiation to reach a finite size and shape. Leaves are initiated at the flanks of the shoot apical meristem (SAM), and following the establishment of dorsiventrality they begin to expand laterally to generate a flat lamina (reviewed in Efroni et al., 2010). Lamina expansion is associated with progressive loss of morphogenetic potential in regions at leaf margins termed “marginal blastozones”, which generate the lamina and its lateral elaborations; e.g. serrations and leaflets (Hagemann and Gleissberg, 1996). These aspects of leaf maturation are correlated with protracted changes in gene expression (Efroni et al., 2008). The relationship between the gradual gene expression changes and the progressive loss of morphogenetic potential is not understood.
The earliest known markers of lamina formation are the CIN-TCP transcription factors whose induction requires the establishment of leaf dorsiventrality (Sarojam et al., 2010). The CIN-TCPs form a sub-clade of the class II TCPs, a family of non-canonical bHLH transcription factor. Five of the eight Arabidopsis CIN-TCPs are regulated by the miR319 microRNA (Palatnik et al., 2003). The CIN-TCPs jointly promote leaf maturation and the progression of the cell division arrest front (Nath et al., 2003; Ori et al., 2007; Efroni et al., 2008). In addition, CIN-TCPs promote leaf senescence by direct induction of LOX2, the product of which catalyses the first dedicated step in jasmonic acid (JA) biosynthesis (Schommer et al., 2008). In Arabidopsis, simultaneous downregulation of five or all eight CIN-TCPs results in large, crinkly leaves with extended leaf margin proliferation and a delay in the sequential progression of gene expression profiles that characterize normal leaf maturation schedule (Efroni et al., 2008).
The plant hormone cytokinin (CK) plays a role in leaf maturation that is opposite to that of the CIN-TCPs. CK promotes mitotic cell divisions, formation of marginal leaf serrations and marginal blastozone activity; moreover CK inhibits leaf senescence and delays leaf differentiation (Miller et al., 1955; Gan and Amasino 1995; Werner et al., 2003). CK sensing by the AHK receptor leads to the activation of B-class Arabidopsis response regulators (ARRs; Muller and Sheen, 2007). B-class ARRs promote transcription of CK downstream genes including A-class ARRs (D’Agostino et al., 2000). A-class ARRs in turn inhibit B-class ARRs, forming a negative feedback loop. Unraveling the developmental roles of A-class ARRs has been hindered by their extensive redundancy (To et al., 2004). For example, downregulation of multiple A-class ARRs is required for the indeterminate growth of the SAM (Leibfried et al., 2005).
In metazoans, as well as in plants, proper execution of many developmental programs depends on the accessible genome in the context of chromatin. Recent studies have highlighted the role of SWI/SNF complexes in this process (Ho and Crabtree 2010, Kwon and Wagner, 2007). These chromatin remodeling complexes use the energy derived from ATP hydrolysis to direct nucleosome disassembly, or to alter the position or conformation of the nucleosome (Clapier and Cairns, 2009). SWI/SNF complexes do not have DNA binding specificity on their own but are frequently recruited to their target loci by interaction with DNA-binding transcription factors. SWI/SNF ATPases alter nucleosome position or conformation to allow access of sequence specific binding proteins to the genomic DNA (Ho and Crabtree, 2010). In Arabidopsis, mutants in the SWI/SNF ATPase BRAHMA (BRM) fail to repress the embryonic/seed-specific programs and display other developmental defects in leaves (Tang et al., 2008, Hurtado et al., 2006). To date, few direct leaf targets of BRM are known, and little information is available about the sequence specific DNA binding proteins BRM acts in concert with. Here we show that BRM and the CIN-TCPs modulate leaf responses to CK and hence promote determinate leaf growth.
Results and Discussion
CIN-TCPs regulate the leaf sensitivity to CK
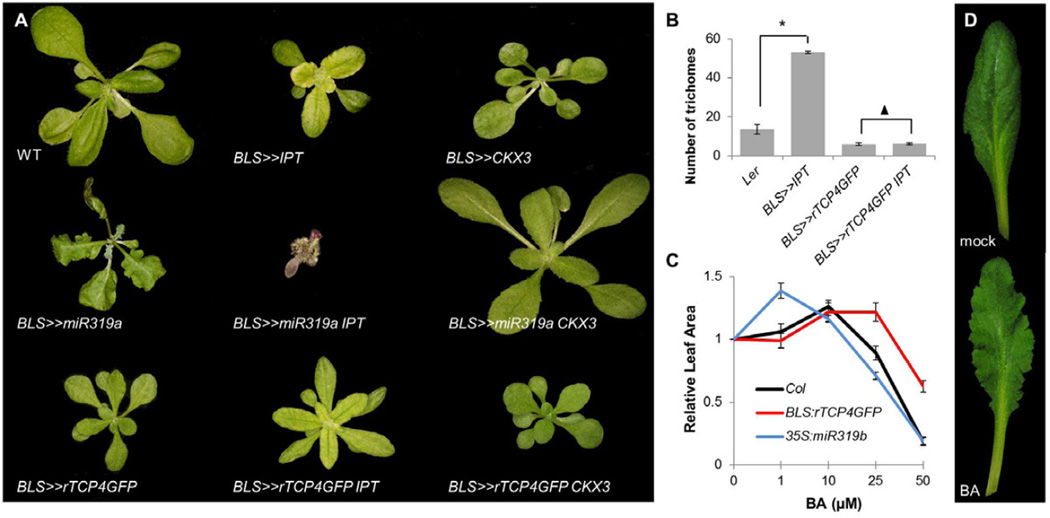
Leaves with reduced CIN-TCP activity display delayed maturation and, as a consequence, extended proliferation and maintenance of morphogenetic potential (Ori et al., 2007; Efroni et al., 2008), similarly to plants with elevated CK levels. To examine whether enhanced CK responses account for the delayed leaf maturation in cin-tcp mutants, we modulated CK levels in the developing leaf by expressing the enzyme IPT, which catalyzes CK production (Kakimoto, 2001), or CKX3, which catalyzes irreversible CK inactivation (Werner et al., 2003). To restrict manipulations to the relevant leaf tissue, the BLS promoter that drives expression in young leaves (Lifschitz et al., 2006) was used. Increased CK levels in leaves of BLS>>IPT plants resulted in small yellow leaves with excessive serrations, dense trichomes and anthocyanin accumulation typical of external CK application (Figure 1A-B; Figure S1A,C; Li et al., 1992). By contrast, reduction of CK levels by BLS>>CKX3 resulted in plants with smaller, rounder leaves, as previously reported (Figure 1A; Werner et al., 2003).
Figure 1. CIN-TCPs regulate Arabidopsis leaf response to CK.
A) 21-day-old plants expressing the CK biosynthesis enzyme IPT or CK deactivating enzyme CKX3 from a promoter active in young leaves (BLS). Plants with reduced or increased CIN-TCP levels overexpress miR319 or a miRNA insensitive form of TCP4, respectively. B) Prevalence of trichomes, a marker for CK activity, on leaf 3. *P<0.01, Student’s t-test. Triangle marks insignificant difference (P>0.3, Student’s t-test). Error bars are SE, n=15. C) Effects of bi-weekly exogenous application of BA on the relative growth of leaf 4 (mean leaf size at 0 μM BA is 137.9, 94.3 and 114.7 mm2 for Col, BLS:rTCP4GFP and 35S:miR319b, respectively). Error bars are SE, n=7 to 10. D) Expanded leaf 10 from short-day grown Col plants after twelve weeks of 40μM BA application. See also Figure S1.
Leaves of plants that overexpress miR319 from the BLS or from the 35S promoters are large and curly (Figure 1A; Efroni et al., 2008). IPT overexpression in BLS>>miR319a plants resulted in severely dwarfed purple plants that failed to reach maturity (Figure 1A), suggesting that these plants are hypersensitive to CK. Reducing CK levels in the BLS>>miR319a background by overexpression of CKX3 resulted in strong suppression of the leaf buckling phenotype and elimination of the excessive serrations (Figure 1A), suggesting that some phenotypes caused by a reduction of CIN-TCP function are due to elevated CK responses.
Constitutive overexpression of a miR319-insensitive version of the CIN-TCP gene TCP4 (rTCP4 hereafter) results in precocious arrest of shoot and leaf growth (Palatnik et al., 2003; Ori et al., 2007). By contrast, BLS>>rTCP4 plants are fertile and are characterized by small, smooth edged, dark green leaves with very few trichomes (Figure 1A; Figure S1B; Efroni et al., 2008). When IPT and rTCP4 were co-expressed, no additional serrations or trichomes were formed on the leaves of BLS>>IPT rTCP4 relative to BLS>>rTCP4 plants (Figure 1A-B; Figure S1B, D). Likewise, expressing CKX3 in BLS>>rTCP4 plants had little effect on plant morphology (Figure 1A).
We next assayed the effects of CIN-TCP levels on leaf growth in response to CK application by repeatedly spraying Col seedlings with varying concentrations of the CK 6-Benzylaminopurine (BA) followed by measuring the area of leaf 4. A bell-shaped response curve was obtained; low CK concentrations promoted development of larger leaves whereas higher CK concentrations inhibited leaf growth (Figure 1C). Moreover, the dose response curve of leaf 4 to exogenous CK application was dependent on CIN-TCP activity. 25μM BA increased leaf growth in BLS:rTCP4GFP, while that of wild type and 35S:miR319b was already inhibited by this dose of CK (Figure 1C; P<0.05; Student’s t-test). In contrast, leaf growth of 35S:miR319b was inhibited by 10 μM BA, a concentration that still promoted leaf growth in the wild type (Figure 1C; P<0.05; Student’s t-test). That the bell-shaped dose response curve to CK was maintained in all genotypes tested suggests that plants with altered CIN-TCP activity display altered leaf sensitivity to CK, rather than altered steady-state CK levels. As an independent test of leaf CK responses, we performed a callus induction assay. Here too, CK responses were enhanced in 35S:miR319b and reduced in BLS:rTCP4GFP plants (see Figure S1E and its legend for details). Finally, we sprayed leaves of short day-grown wild-type plants, a condition that delays leaf maturation, repeatedly with 40uM BA. This caused marginal elaborations, generating buckling leaves similar to those of 35S:miR319 plants (Figure 1D and Palatnik et al., 2003). Taken together, our studies suggest that the CIN-TCPs, including TCP4, dampen leaf responses to CK.
BRAHMA activity is required for promotion of leaf maturation by TCP4
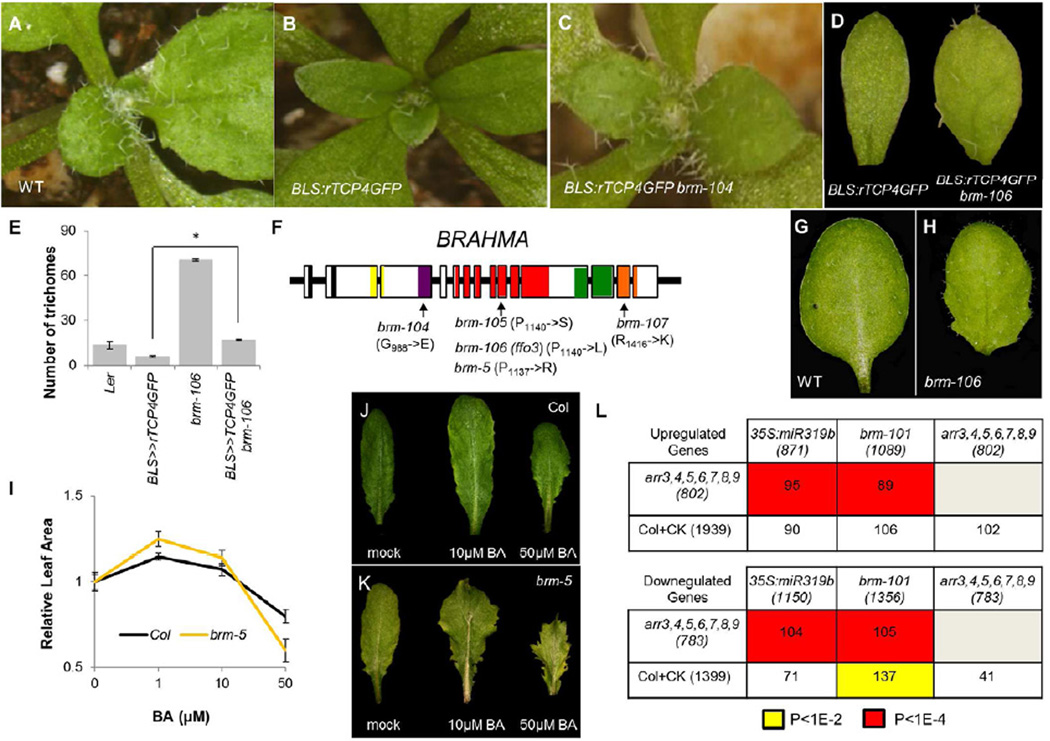
To understand how these CIN-TCPs modulate leaf CK responses, we identified factors required for the TCP4 activity. Towards this end, we mutagenized seeds of plants that displayed precocious leaf maturation due to rTCP4GFP overexpression from the BLS promoter. Leaves of BLS:rTCP4GFP formed few adaxial trichomes and had smooth margins (Figure 2A-B; Figure 1A; Figure S1A-B). Three extragenic suppressors were identified from progeny pools of 1000 M1 plants; their leaves were wider and larger, lighter green in color, and had many more trichomes and serrations than the parental BLS:rTCP4GFP plants (Figure 2C-E). The three mutants also had a short stature with compact inflorescences and short pedicels. Complementation tests revealed them to be allelic to a previously described mutant, ffo3 (Levin et al., 1998). Map based cloning (Supplemental methods) identified all suppressors as new brahma (brm) alleles. We named these brm-104 to brm-107, to match common terminology for BRM mutants (Figure 2F; Bezhani et al. 2008).
Figure 2. TCP4 and BRAHMA jointly promote leaf maturation.
A-D) The effect of TCP4 on leaf trichome production (A-C) or leaf growth (D) was suppressed by weak alleles of the SWI/SNF ATPase BRAHMA (B, D). E) brm-106 restores the number of trichomes of BLS:rTCP4GFP to wild type levels, while CK application does not (*P<1E-10, Student’s t-test). Error bars are SE, n=15. F) Hypomorph brm alleles identified. Domains: yellow (QLQ), purple (HSA), red (ATPase), green (AT-hook), orange (Bromodomain) G-H) Weak serrations and uneven lamina in short-day-grown brm-106 leaf 3. I-K) Effects of bi-weekly exogenous BA application on the relative growth of leaf 4 (I), and on serrations and marginal growth of leaf 6 (J-K) for long-day-grown Col and brm-5 plants. Mean leaf size at 0 μM BA:127.6 mm2 (Col), 127.4 mm2 (brm-5). Error bars are SE, n=7 to 10. L) The overlap in genes differentially expressed in plants with altered TCP or BRM activity, and in plants with genetic (arr mutants) or chemical (+CK) alteration of CK responses. P-values: hypergeometric test.
Null mutations in BRM cause severe phenotypes and are sterile (Hurtado et al., 2006; Kwon et al., 2006), suggesting that the new alleles are hypomorphs. In agreement with this, all four mutants carried missense mutations in important BRM domains (Figure 2F; Clapier and Cairns, 2009). The brm hypomorph alleles were small, fertile, early flowering and had curled leaves when grown in long days (this study; Farrona et al., 2004). However, when grown in short days, brm-106 leaves exhibited excessive intervein leaf growth resulting in an uneven lamina, as well as pronounced serrations of the leaf margins and light green color (Figure 2G-H). These phenotypes resemble partial loss of CIN-TCP activities (Schommer et al., 2008) and are consistent with a delay in leaf maturation.
The phenotypes of the brm hypomorphs suggested that CK sensitivity might be altered in these mutants. To test this, we treated wild type Ler, brm-106, BLS:rTCP4GFP and brm-106 BLS:rTCP4GFP seedlings with BA as described above (Figure S2A). Seedlings of the Ler cultivar plants proved more resistant than those of the Col cultivar to CK application and showed clear leaf growth inhibition only at 100μM BA (P<1E-4; Student’s t-test; compare Figure S2A with Figure 1C). By contrast, growth of brm-106 leaves was already significantly inhibited at a CK dose of 25μM BA (P<0.01; Student’s t-test). While leaves of BLS:rTCP4GFP did not show a significant response at any of the BA treatments used (P>0.15; ANOVA), CK responsiveness was restored to BLS:rTCP4GFP leaves with reduced BRM activity (BLS:rTCP4GFP brm-106; P<1E-7; ANOVA), with a characteristic bell shape dose response curve (Figure S2A).
Given the stronger responses seedlings of the Col cultivar to BA treatment, we next compared the BA dose response of brm-5, a hypomorph allele in Col background (Tang et al., 2008), to that of Col. Like 35S:miR319b leaves, brm-5 leaves showed increased growth relative to the wild type at low CK concentration (1μM BA; P<0.05; Student’s t-test; Figure 2I) and reduced growth compared to the wild type at high concentrations (50 μM BA; P<0.05; Student’s t-test). brm mutants also had more pronounced leaf serrations and increased leaf width compared to the wild type when similar CK concentrations were applied to both (Figure 2J-K). Additional support for altered CK response of brm mutants and of plants with altered TCP levels comes from the significant overlap of genes differentially expressed in brm and CK response mutants (Figure 2L).
TCP4 interacts with the BRAHMA and its complex member SWI3C
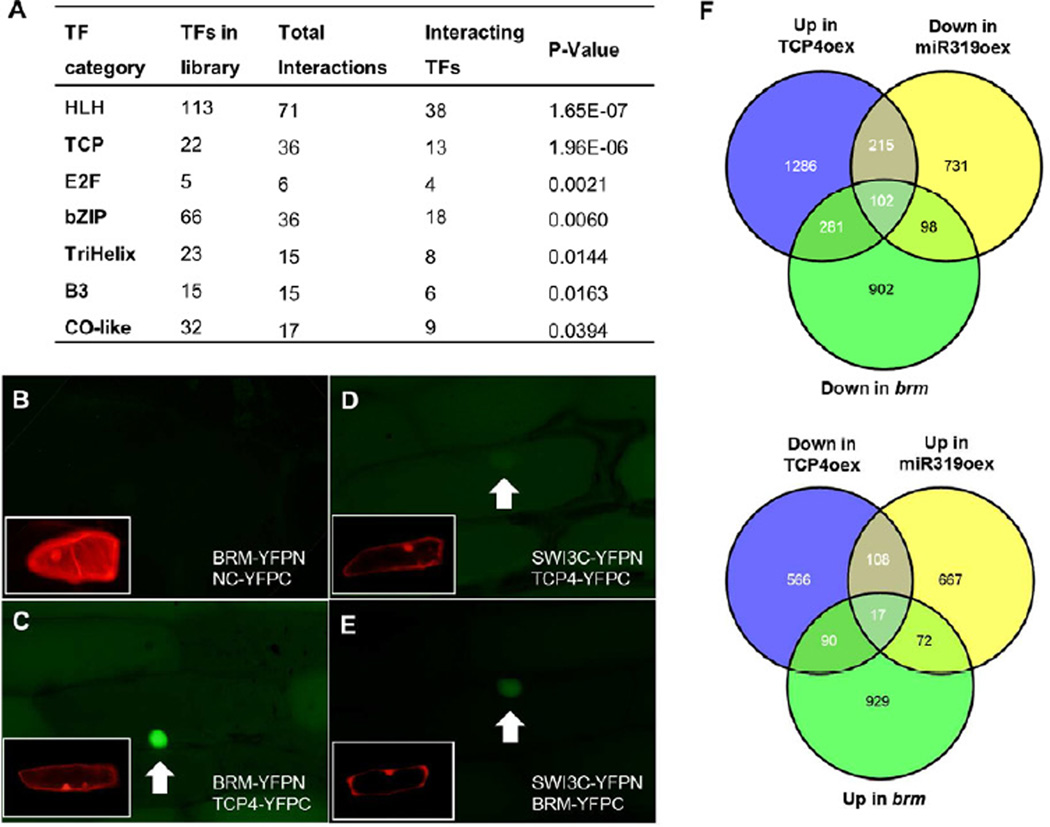
A reduction in BRM activity suppressed TCP4 overexpression and both brm and cin-tcp mutants were more sensitive to CK than the WT, suggesting that both factors may act together to coordinately regulate downstream targets. We conducted a yeast-two-hybrid interactome study aimed at identifying transcription factors that can recruit SWI/SNF complexes to the genomic loci they regulate using a library of 1,400 Arabidopsis transcription factors as prey (Song et al., 2008). This identified a total of 400 pair-wise interactions involving 210 unique transcription factors from 25 different families (Table S1). Transcription factors from seven families were significantly enriched as SWI2/SNF2SWI/SNF interacting (Figure 3A). The highest enrichment was observed for the bHLH and the TCP transcription factor families (P<1E-4, Fischer’s exact one-tailed). bHLH and bHLH-related DNA binding proteins such as TCPs may thus play an important role in SWI/SNF complex recruitment in Arabidopsis.
Figure 3. CIN-TCPs and BRM interact to regulate a common set of genes.
A) Transcription factor families with significantly enriched SWI/SNF interacting transcription factors (TFs). TFs from 32 different families were tested (Table S1). 7 families were significantly enriched (Fisher’s exact one-tailed P<0.05). B-E) Transfected epidermal onion cells with negative control (NC) (B), or interaction tests (C-E). RFP marks transformed cells and nuclei (insets). Arrows point to nuclei. F) Overlap of genes differentially expressed in apices with altered TCP4, miR319 and BRM levels. See also Figure S2 and Tables S1 and S2.
The SWI/SNF core complex in yeast and metazoans consists of four proteins: one catalytic subunit (a SWI/SNF ATPase), two SWI3 proteins and one SNF5 subunit (Clapier and Cairns, 2009; Kwon and Wagner, 2007, Jerzmanowski, 2007). The interactome screen included as baits the SWI/SNF ATPases BRM and SYD, three SWI3 proteins (A, B and C), and the SNF5 subunit BUSHY. Intriguingly, BRM and the proposed BRM complex subunit SWI3C (Archacki et al., 2009; Hurtado et al., 2006) interacted frequently with TCP transcription factors (Table S1). Interaction tests were repeated for seven of the eight CIN-TCPs; TCP3, TCP4 and TCP5 interacted with BRM and SWI3C (Figure S2B). The physical interaction between BRM or SWI3C and TCP4 was further verified in planta using bifluorescence molecular complementation. We observed strong binding of TCP4 to BRM and weaker binding of TCP4 to SWI3C (Figure 3B-E). Our combined data suggest that TCP4 and other CIN-TCPs may act together with the BRM chromatin remodeling complex to regulate downstream target genes. Genetic support for this hypothesis comes from the finding that brm-106 did not dramatically enhance the leaf phenotypes of 35S:miR319a plants; an enhancement would be expected if both were modifying CK responses independently (Figure S2C).
An extensive overlap in the genes altered in both CIN-TCP and BRM mutants
If the CIN-TCPs and BRM act in concert, we would expect a significant overlap between the genes with altered expression in each mutant. Indeed, there was a significant overlap in genes coordinately down- or upregulated in young 35S:miR319a and brm-101 seedlings (Efroni et al., 2008, Bezhani et al., 2007): 200 genes were downregulated in both conditions, while 89 genes were upregulated in both conditions (P<0.001; hypergeometric test; Figure 2F). In contrast, the overlap between genes upregulated in one genotype and downregulated in the other was as expected by chance alone (data not shown). When we probed for genes coordinately regulated in brm-101, 35S:miR319a plants and in BLS>>rTCP4 plants, 102 genes were identified as positively regulated (P<1E-39; Hypergeometric test) and 17 as negatively regulated (P<1E-5; Hypergeometric test; Figure 3A; Table S2). Interestingly, of these 119 putative TCP4 and BRM target genes, 52 (44%) were also differentially expressed in the expected manner in leaves of TCP5 overexpressors (Table S2), consistent with the observed physical interaction between TCP5 and BRM (Figure S2B).
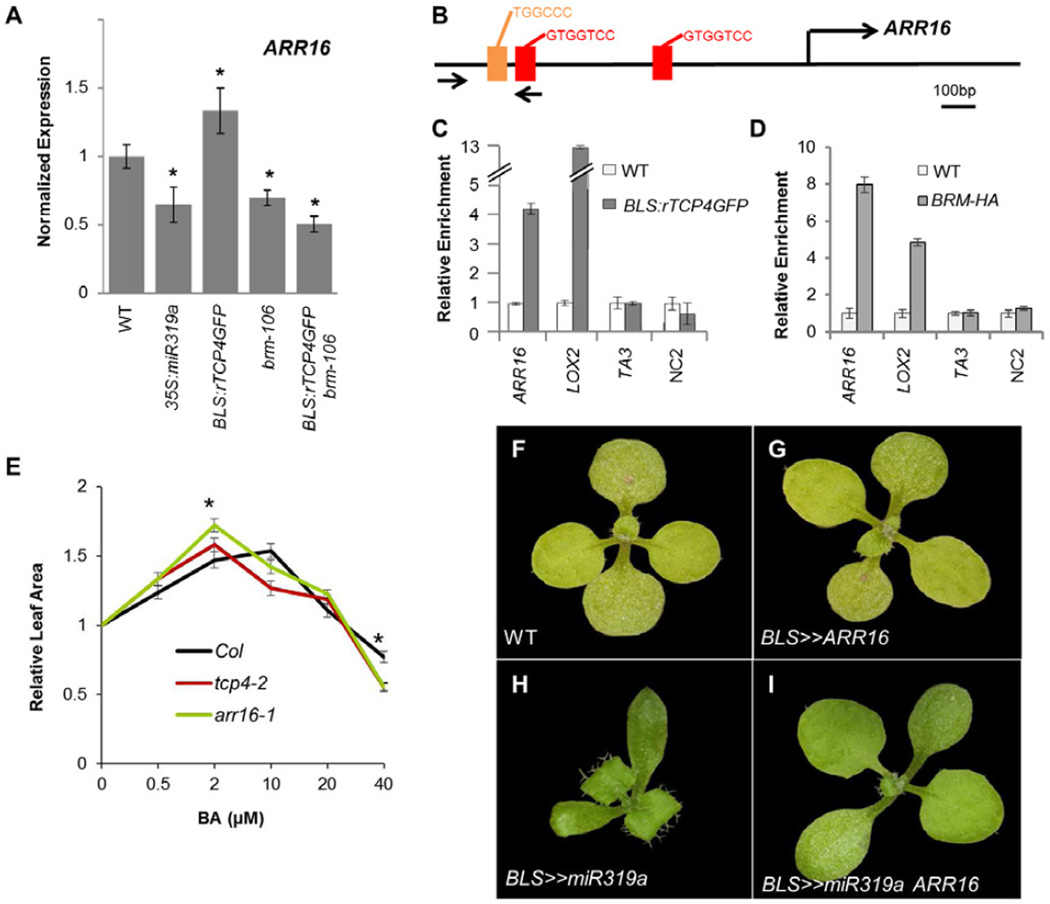
Given the physical interaction between TCP4 and BRM, and the similarity in the transcriptional response of the two mutants, we hypothesized that TCP4 and BRM might reside together on the promoter of common target genes. Among the common putative BRM and TCP4 targets (Table S2), we identified ARR16, an A-class response regulator and inhibitor of CK responses (To et al., 2004; Ren et al., 2009). An independent qRT-PCR experiment verified the microarray result (Figure 4A). In addition, we found that brm-106 abolished the increased ARR16 RNA accumulation in BLS:rTCP4GFP leaves (Figure 4A). Analysis of A-class ARR expression across leaf development based on a published dataset (Schmid et al., 2005; Efroni et al., 2008) showed that ARR16 and several CIN-TCPs were most highly expressed in young expanding leaves (Figure S3A-B). Moreover, the expression of five CIN-TCPs, including TCP4, was correlated with that of ARR16 across leaf development (R=0.61, P<0.05; Figure S3C).
Figure 4. CIN-TCPs and BRM modulate the expression of a CK response gene.
A) qRT-PCR measurement of ARR16 expression. *significant difference from WT (P<0.05, Student’s t-test). Error bars are SE, n=3. B) TCP4 binding motifs in the promoter of ARR16. Red: full motif; orange: core motif. C-D) ChIP from pBLS:TCP4GFP (C) or pBRM:BRM-HA (D) followed by qPCR of the ARR16 promoter (arrows in (B) mark the primers used), the LOX2 promoter, and two negative controls (TA3 and NC2). ChIP was repeated at least 3 times, and a representative result is shown. Error bars are SE for 3 technical repeats. E) Effect of bi-weekly exogenous BA application on the size of leaf 4 (mean leaf size at 0 μM BA is 98.2, 106.9 and 105.8 mm2 for Col, tcp4-2 and arr16-1, respectively). *P<0.01; Student’s t-test. Error bars are SE, n=10. F-I) Overexpression of ARR16 rescues the growth defects of leaves expressing miR319. See also Figure S3.
TCP4 and BRM bind the promoters of genes that direct hormonal responses
The 5' promoter sequence of ARR16 has two repeats of the TCP4 binding motif GTGGTCCA and a repeat of the core TCP motif TGGTCC (Figure 4B; Schommer et al., 2008), providing potential sites for TCP4 recruitment to the ARR16 promoter. To test whether ARR16 is a direct TCP4 target gene, we performed chromatin immunoprecipitation (ChIP) with GFP-tagged TCP4 (BLS:rTCP4GFP). Indeed, TCP4 associated with the 5' intergenic region of the ARR16 gene (Figure 4C). As a positive control, we confirmed association of TCP4 with the LOX2 promoter, previously demonstrated in-vitro (Schommer et al., 2008). Next, we employed a epitope tagged version of BRM, pBRM:BRM-HA, which fully rescues the morphological defects of brm-1 null mutants (Han et al., 2012) for ChIP. BRM-HA strongly bound to the 5’ intergenic region of both ARR16 and LOX2 (Figure 4D). Thus, TCP4 and BRM associated with common regulatory regions in Arabidopsis leaves, among which was the promoter of ARR16, a gene downregulated in plants overexpressing miR319 and in brm mutants.
Given the significant role of BRM and TCP4 in leaf CK responsiveness, they may coordinately regulate additional ARRs besides ARR16. Consistent with this hypothesis, we detected TCP4 and BRM association with the promoter of ARR6, a gene whose expression was high in young expanding leaves (Figure S3A) and responsive to the level of BRM or TCP4 activity (Figure S3D-F).
Leaf expression of ARR16 can partially substitute for CIN-TCPs
Since CK delays differentiation in leaf cells (Shani et al., 2010), the delayed maturation of 35S:miR319a leaves may be due to a compromised negative CK signaling feedback. We therefore wished to examine the effect of altered ARR16 activity on plants with different CIN-TCP levels. arr16-1 plants did not show morphological defects (Figure S3G-I). However, when tested for leaf CK responses, arr16-1 leaves were more sensitive to CK, displaying enhanced growth at 2μM BA, and reduced growth at 40μM BA relative to the wild type, respectively (P<1E-4 and P<0.01; Student’s t-test; Figure 4E). Notably, tcp4-2 single mutants exhibited similar hypersensitivity to CK (P<0.01 and P<0.01, respectively; Student’s t-test; Figure 4E).
We next tested whether ARR16 expression driven from a heterologous promoter could revert the leaf maturation defects of plants with compromised CIN-TCP activity. Expression of ARR16 from the BLS promoter resulted in plants with essentially unchanged leaves (Figure 4E-F). In contrast, expression of BLS>>ARR16 in BLS>>miR319a leaves resulted in a significant rescue of the excessive growth phenotype, flattening of the leaf lamina, and a reduction in the buckling typical of miR319 overexpressing plants (Figure 4G-H). The same result was obtained when both ARR16 and miR319 were expressed from the 35S promoter (Figure S3J). Moreover, expression of BLS>>ARR16 could largely rescue the growth inhibition of the CK overproducing BLS>>IPT plants (Figure S3K-L). To test whether ARR16-mediated rescue of CIN-TCP loss-of-function resulted from a general inhibition of the CK response, we crossed 35S:miR319 to previously described plants overexpressing another A-class ARR, ARR5 (Salome et al., 2006; Ren et al., 2009). In contrast to ARR16 overexpressors (Figure 4G-H, Figure S3J), ARR5 overexpressors did not alter the leaf phenotypes of plants with elevated miR319 levels (Figure S3M-P), supporting the previous finding (Ren et al., 2009) of functional specificity among A-class ARRs.
CK, chromatin, differentiation and organ size
CK responses are critical for the balance between indeterminate growth and differentiation in multiple plant tissues. In the Arabidopsis shoot, indeterminacy is maintained by the homeodomain transcription factor WUSCHEL, which represses the expression of several class A ARRs (Leibfried et al., 2005). Similarly, maintenance of the tomato leaf marginal blastozone is aided by CK activity (Shani et al., 2010), and partial loss of CK degradation results in larger Arabidopsis organs (Bartrina et al., 2011). We propose here that the TCP4 CIN-TCP, which is expressed at the onset of lamina formation, regulates leaf maturation through interaction with a BRM chromatin-remodeling complex and modification of the chromatin state of promoters of common targets such as ARR16 and ARR6. This provides a temporal cue to dampen CK responses, thus restricting morphogenetic programs that initially are active throughout leaf primordia and are later restricted to leaf margins (blastozones). Consistent with this hypothesis, minor changes in TCP4 levels, like those in CK levels, can have dramatic effects on overall leaf growth (Efroni et al., 2008).
The class I TCPs TCP14 and TCP15 have recently been shown to sensitize Arabidopsis leaf responses to CK (Steiner et al., 2012), a function opposite to that we describe for a Class II CIN-TCP in leaf development. Consistent with our findings, the BRANCHED Class II CIN-TCP gene, which is specifically expressed in axillary meristems, dampens apical dominance release - a classical CK response (Braun et al., 2012). These combined findings implicate the TCP family, as a whole, in regulating developmental responses to CK. It was previously suggested that a balance of the antagonistic activities of Class I and Class II TCPs may regulate the cell cycle and plant growth, possibly via opposite effects on common target genes (Li et al., 2005). We propose here that one of the interaction points of these opposing classes of TCP transcription factors is the CK response pathway.
Leaf maturation rate and hence the duration of leaf growth is highly variable even within the same plant, depending on the growth conditions. This plasticity requires that input signals, such as CK, could be modulated in response to the environment. Such modulation may rely, at least in part, on the chromatin status of target genes. In agreement with this idea, mutations in chromatin remodeling complexes, such as PICKLE (Furuta et al., 2011) or BRM (this study), can modulate CK responses, providing a potential for environmental regulation of leaf maturation schedule.
Experimental Procedures
Plant material
Plants were grown on soil under fluorescent light at 20°C in long day (16 hours light), unless short day is indicated (10 hours light). Op:CKX3 seeds were provided by Eilon Shani. Op:IPT seeds were previously described (Greenboim-Wainberg et al., 2005). Plants were of Ler ecotype, expect for brm-5, 35S:miR319b and BLS:rTCP4GFP that were used for CK response and callus induction experiments and the two 35S:ARR5 lines that were crossed with 35S:miR319b (Salome et al., 2006; Ren et al., 2009). Plasmid construction is detailed in Supplementary Experimental Procedures. Transgenic lines were generated as described by (Pekker et al., 2005). A representative single T-DNA insertion line was selected for further analysis. For the suppressor screen, BLS:rTCP4GFP seeds (0.2g) were incubated in 0.3% Ethyl methanesulfonate (EMS). M2 seeds were collected in pools of five M1 plants. To ensure absence of transgene silencing in plant carrying multiple transgenes, we monitored for presence of the morphological defects caused by each transgene in all genetic backgrounds.
Yeast two-hybrid screens
Six different yeast hosts, each carrying a SWI/SNF chromatin remodeling complex component as bait, were transformed with one of 1,400 transcription factors in the prey vector (Song et al., 2008). See Supplemental Experimental Procedures for further details on the interactome screen. Interactions between CIN-TCPs and BRM/SWI3C were confirmed by co-transforming bait and prey plasmids into yeast.
Tissue Collection, RNA Preparation, and RT-qPCR
To measure ARR16 or ARR6 levels, RNA was extracted using the RNA easy kit (Qiagen) either from 7 DAS long-day or 21 DAS short-day-grown plants with qualitatively similar results. qRT-PCR was performed on 1μg of total RNA according to (Steiner et al., 2012) on an Applied Biosystems 7300 RT-PCR system. For 21 DAS short-day-grown plants, 2 μg of purified RNA was used, and qRT-PCR was performed as in (Han et al., 2012). UBI21 (AT5G25760) or EIF4A1 (AT3G13920) served as internal controls. Primer sequences can be found in Supplemental Experimental Methods.
Hormonal Treatment and Callus Induction
For CK treatment, 7–8 plants of each genotype were sprayed twice a week with different concentrations of BA (Sigma) or with water from the appearance of the first two leaves. The area of the fully expanded leaf 4 was measured. For callus induction, plants were germinated of ½ MS, 1% sucrose agar plates. Leaf 3 was removed from 14-day-old seedlings and transferred to ½ MS sucrose plates containing varying amounts of 2–4D (Sigma) and kinetin (Sigma). Plates were sealed and kept at constant light at 23°C for four weeks.
ChIP
ChIP procedure and ChIP-qPCR were performed as previously described (Han et al., 2012) on 21-day-old short-day-grown plants. 500 mg of BLS:rTCP4GFP seedlings were used for GFP ChIP using 5 μl of anti-GFP antibody (Invitrogen, A6455). For Anti-HA ChIP, 1000 mg brm-1 BRM:BRM-HA plants and 20 μl of anti-HA antibody (Roche, 12CA5) were used. Negative controls were the retrotransposon TA3 (Han et al., 2012) and NC2 (genomic region 3’ of ARR16 (AT2G40660)). Primer sequences can be found in Supplemental Experimental Methods.
Bimolecular fluorescence complementation
Bimolecular fluorescent complementation (BiFC) plasmids were introduced into onion epidermal cells using particle bombardment with BioRad PDS-1000/He. 35S:2xmCherry was used as a transformation control and to mark the nuclei. Images were taken using an Olympus MVX100 epifluorescence microscope. The negative control construct pCL113 Tdy1-NLS was previously described (Ma et al., 2009).
Bioinformatics Analysis
Raw microarray data was analyzed in R (2.12.0), and bioconductor (2.5). MAS5 expression values were normalized to a median of 50 (Except for data from Buechel et al., 2010, which was processed with GC-RMA). Genes with normalized expression of less than 30 were considered absent. An arbitrarily log2 value cutoff of >|0.8| (1.74 fold change) was selected to identify genes that were significantly differentially expressed.
Supplementary Material
Highlights.
CIN-TCP transcription factors such as TCP4 modulate Arabidopsis leaf response to CK
A genetic screen identifies the chromatin remodeler BRM as required for TCP4 action
Interactome screen identifies TCPs as preferential BRM interactors.
TCP4 and BRM directly induce ARR16, a negative CK response regulator ARR16.
Acknowledgments
We thank D. Weiss, J. Bowman and N. Ori for comments, J. Reyes for pDBLeu-SYD, J. Pfluger for help with the interaction assays, C. Winter for statistical analyses, N. Ohad for technical advice, B. Ren and E. Shani for seeds and J. Lohman for sharing expression datasets. This work was supported in part by a grant from the Next-Generation BioGreen 21 Program (SSAC No. 2011), Rural Development Administration, Republic of Korea, and Basic Science Research Program through the National Research Foundation of Korea of MEST (2010-0012801) to JCH, by MINERVA, ISF 1294-10 to YE, and by NIH R01 GM64650-01 and NSF grant IOS 0849298 to DW. IE was supported by EMBO LTF185-2010.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Archacki R, Sarnowski TJ, Halibart-Puzio J, Brzeska K, Buszewicz D, Prymakowska-Bosak M, Koncz C, Jerzmanowski A. Genetic analysis of functional redundancy of BRM ATPase and ATSWI3C subunits of Arabidopsis SWI/SNF chromatin remodelling complexes. Planta. 2009;229:1281–1292. doi: 10.1007/s00425-009-0915-5. [DOI] [PubMed] [Google Scholar]
- Bezhani S, Winter C, Hershman S, Wagner JD, Kennedy JF, Kwon CS, Pfluger J, Su Y, Wagner D. Unique, shared, and redundant roles for the Arabidopsis SWI/SNF chromatin remodeling ATPases BRAHMA and SPLAYED. Plant Cell. 2007;19:403–416. doi: 10.1105/tpc.106.048272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartrina I, Otto E, Strnad M, Werner T, Schmülling T. Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell. 2011;23:69–80. doi: 10.1105/tpc.110.079079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun N, Saint de Germain A, Pillot J, Boutet-Mercey S, Dalmais M, Antoniadi I, Li X, Maia-Grondard A, Le Signor C, Bouteiller N, et al. The pea TCP transcription factor PsBRC1 acts downstream of Strigolactones to control shoot branching. Plant Physiol. 2012;158:225–238. doi: 10.1104/pp.111.182725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- D'Agostino IB, Deruère J, Kieber JJ. Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 2000;124:1706–1717. doi: 10.1104/pp.124.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Blum E, Goldshmidt A, Eshed Y. A protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell. 2008;20:2293–2306. doi: 10.1105/tpc.107.057521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Eshed Y, Lifschitz E. Morphogenesis of Simple and Compound Leaves: A Critical Review. Plant Cell. 2010;22:1019–1032. doi: 10.1105/tpc.109.073601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrona S, Hurtado L, Bowman JL, Reyes JC. The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development. 2004;130:4965–4975. doi: 10.1242/dev.01363. [DOI] [PubMed] [Google Scholar]
- Furuta K, Kubo M, Sano K, Demura T, Fukuda H, Liu Y, Shibata D, Kakimoto T. The CKH2/PKL chromatin remodeling factor negatively regulates cytokinin responses in Arabidopsis calli. Plant Cell Physiol. 2011;52:618–628. doi: 10.1093/pcp/pcr022. [DOI] [PubMed] [Google Scholar]
- Gan S, Amasino RM. Inhibition of leaf senescence by autoregulated production of cytokinin. Science. 1995;270:1986–1988. doi: 10.1126/science.270.5244.1986. [DOI] [PubMed] [Google Scholar]
- Greenboim-Wainberg Y, Maymon I, Borochov R, Alvarez J, Olszewski N, Ori N, Eshed Y, Weiss D. Cross talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell. 2005;17:92–102. doi: 10.1105/tpc.104.028472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann W, Gleissberg S. Organogenetic capacity of leaves: The significance of marginal blastozones in angiosperms. Plant Syst. Evol. 1996;199:121–152. [Google Scholar]
- Han SK, Wu M-F, Rodrigues A, BIOL425 Fall2010, Rodriguez PR, Wagner D. The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses Abscisic Acid Responses in the Absence of the Stress Stimulus in Arabidopsis. Plant Cell. doi: 10.1105/tpc.112.105114. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Crabtree GR. Chromatin remodeling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado L, Farrona S, Reyes JC. The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Mol Biol. 2006;62:291–304. doi: 10.1007/s11103-006-9021-2. [DOI] [PubMed] [Google Scholar]
- Jerzmanowski A. SWI/SNF chromatin remodeling and linker histones in plants. Biochim Biophys Acta. 2007;1769:330–345. doi: 10.1016/j.bbaexp.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Kakimoto T. Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate:ATP/ADP isopentenyltransferases. Plant Cell Physiol. 2001;42:677–685. doi: 10.1093/pcp/pce112. [DOI] [PubMed] [Google Scholar]
- Kwon CS, Hibara K, Pfluger J, Bezhani S, Metha H, Aida M, Tasaka M, Wagner D. A role for chromatin remodeling in regulation of CUC gene expression in the Arabidopsis cotyledon boundary. Development. 2006;133:3223–3230. doi: 10.1242/dev.02508. [DOI] [PubMed] [Google Scholar]
- Kwon CS, Wagner D. Unwinding chromatin for development and growth: a few genes at a time. Trends Genet. 2007;23:403–412. doi: 10.1016/j.tig.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Leibfried A, To JP, Busch W, Stehling S, Kehle A, Demar M, Kieber JJ, Lohmann JU. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature. 2005;438:1172–1175. doi: 10.1038/nature04270. [DOI] [PubMed] [Google Scholar]
- Levin JZ, Fletcher JC, Chen X, Meyerowitz EM. A genetic screen for modifiers of UFO meristem activity identifies three novel FUSED FLORAL ORGANS genes required for early flower development in Arabidopsis. Genetics. 1998;149:579–595. doi: 10.1093/genetics/149.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Potuschak T, Colón-Carmona A, Gutiérrez RA, Doerner P. Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc Natl Acad Sci U S A. 2005;102:12978–12983. doi: 10.1073/pnas.0504039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschitz E, Eviatar T, Rozman A, Shalit A, Goldshmidt A, Amsellem Z, Alvarez JP, Eshed Y. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc Natl Acad Sci U S A. 2006;103:6398–6403. doi: 10.1073/pnas.0601620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Slewinski TL, Baker RF, Braun DM. Tie-dyed1 encodes a novel, phloem-expressed transmembrane protein that functions in carbohydrate partitioning. Plant Physiol. 2009;149:181–194. doi: 10.1104/pp.108.130971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CO, Skoog F, Von Saltza MH, Strong F. Kinetin, a cell division factor from deoxyribonucleic acid. J Am Chem Soc. 1955;77:1392. [Google Scholar]
- Müller B, Sheen J. Advances in cytokinin signaling. Science. 2007;318:68–69. doi: 10.1126/science.1145461. [DOI] [PubMed] [Google Scholar]
- Nath U, Crawford BC, Carpenter R, Coen E. Genetic control of surface curvature. Science. 2003;299:1404–1407. doi: 10.1126/science.1079354. [DOI] [PubMed] [Google Scholar]
- Ori N, Refael-Cohen A, Etzioni A, Brand A, Yanai O, Shleizer S, Menda N, Amsellem Z, Efroni I, Pekker, et al. Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 2007;39:787–791. doi: 10.1038/ng2036. [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D. Control of leaf morphogenesis by microRNAs. Nature. 2003;425:257–263. doi: 10.1038/nature01958. [DOI] [PubMed] [Google Scholar]
- Pekker I, Alvarez JP, Eshed Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell. 2005;17:2899–2910. doi: 10.1105/tpc.105.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Liang Y, Deng Y, Chen Q, Zhang J, Yang X, Zuo J. Genome-wide comparative analysis of type-A Arabidopsis response regulator genes by overexpression studies reveals their diverse roles and regulatory mechanisms in cytokinin signaling. Cell Res. 2009;19:1178–1190. doi: 10.1038/cr.2009.88. [DOI] [PubMed] [Google Scholar]
- Salomé PA, To JP, Kieber JJ, McClung CR. Arabidopsis response regulators ARR3 and ARR4 play cytokinin-independent roles in the control of circadian period. Plant Cell. 2006;18:55–69. doi: 10.1105/tpc.105.037994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarojam R, Sappl PG, Goldshmidt A, Efroni I, Floyd SK, Eshed Y, Bowman JL. Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell. 2010;22:2113–2130. doi: 10.1105/tpc.110.075853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008;6:e230. doi: 10.1371/journal.pbio.0060230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shani E, Ben-Gera H, Shleizer-Burko S, Burko Y, Weiss D, Ori N. Cytokinin regulates compound leaf development in tomato. Plant Cell. 2010;22:3206–3217. doi: 10.1105/tpc.110.078253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Song NY, Shin SY, Kim HJ, Yun D, Lim CO, Lee SY, Kang KY, Hong JC. Isolation of CONSTANS as a TGA4/OBF4 interacting protein. Molecules and Cells. 2008;25:559–565. [PubMed] [Google Scholar]
- Steiner E, Efroni I, Gopalraj M, Saathoff K, Tseng T, Kieffer M, Eshed Y, Olszewski N, Weiss D. The Arabidopsis O-Linked N-Acetylglucosamine Transferase SPINDLY Interacts with Class I TCPs to Facilitate Cytokinin Responses in Leaves and Flowers. Plant Cell. 2012;24:96–108. doi: 10.1105/tpc.111.093518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To JP, Haberer G, Ferreira FJ, Deruère J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ. Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signaling. Plant Cell. 2004;16:1–17. doi: 10.1105/tpc.018978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, van Onckelen H, Schmülling T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell. 2003;15:2532–2550. doi: 10.1105/tpc.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.