Abstract
PURPOSE
Merkel cell carcinoma (MCC) is a highly aggressive skin cancer with strong evidence of viral carcinogenesis. The association of MCC with the Merkel cell polyomavirus (MCPyV) may explain the explicit immunogenicity of MCC. Indeed, MCPyV-encoded proteins are likely targets for cytotoxic immune responses to MCC as they are both foreign to the host and necessary to maintain the oncogenic phenotype. However, to date only a single MCPyV-derived CD8 T-cell epitope has been described, thus impeding specific monitoring of T-cell responses to MCC.
METHOD
To overcome this limitation, we scanned the MCPyV oncoprotein large T and small T antigens and the virus-capsid protein VP1 for potential T-cell epitopes, and tested for major histocompatibility complex (MHC) class I affinity. We confirmed the relevance of these epitopes using a high-throughput platform for T-cell enrichment and combinatorial encoding of MHC class I multimers.
RESULTS
In peripheral blood from 38 MCC patients and 30 healthy donors we identified 53 MCPyV-directed CD8 T-cell responses against 35 different peptide sequences. Strikingly, T-cell responses against oncoproteins were exclusively present in MCC patients, but not in healthy donors. We further demonstrate both the processing and presentation of the oncoprotein-derived epitopes, as well as the lytic activity of oncoprotein-specific T cells towards MHC-matched MCC cells. Demonstrating the presence of oncoprotein-specific T cells among tumor infiltrating lymphocytes further substantiated the relevance of the identified epitopes.
CONCLUSION
These T-cell epitopes represent ideal targets for antigen specific immune therapy of MCC, and enable tracking and characterization of MCPyV specific immune responses.
Keywords: Combinatorial encoding, Major histocompatibility complex class I multimers, Large T antigen, Small T antigen, Tumor infiltrating lymphocytes
Introduction
Merkel cell carcinoma (MCC) is a rare, aggressive skin cancer with an estimated 1600 cases/year in the US (1). MCC is more frequently observed in the elderly or immunocompromised populations, suggesting a role for immune surveillance in the development of MCC. Indeed, in iatrogenically immunocompromised patients, spontaneous regressions of MCC have been observed following cessation of immune suppression (2). While the hazard ratio for MCC is above 10 for immunosuppressed individuals, more than 90% of MCC patients are not obviously immune compromised (3). Moreover, intratumoral CD8 T-cell infiltration is strongly correlated to the survival of MCC patients (4). Nevertheless, once metastasized, MCC has a very poor prognosis; the 5-year relative survival for patients with stage IV metastatic disease is 18% (1). To date there is no established effective treatment to offer these patients. Due to the apparent function of the immune system to control this tumor, ongoing clinical trials are focusing on immune modulating approaches such as immune checkpoint blocking antibodies or targeted delivery of interleukin-2 (e.g. http://www.immomec.eu).
In 2008 Merkel cell polyomavirus (MCPyV) was discovered in association with 80% of MCCs. Subsequent work confirmed this association and further indicated that most, if not all, MCCs are associated with this polyomavirus (5). The expression of MCPyV-encoded proteins in MCC may explain the explicit immunogenicity of MCC. In MCC, the MCPyV genome is clonally integrated into the host cell chromosome with no preferred integration site (6–9). Like other polyomaviruses, MCPyV contains two early genes, i.e. the large T (LTA) and small T (STA) antigens, which are regarded as oncogenes. LTA and STA share 78 amino acids in the N terminus and their expression is necessary for the maintenance of this cancer (10–13). The LTA is truncated by a tumor specific mutation eliminating the helicase domain; this truncation is believed to be essential for the transforming activity of LTA (10). MCPyV also encodes viral capsid proteins (VP) that ensure encapsulation of viral DNA before lysis of the infected cells. Serologic studies have demonstrated the presence of anti-MCPyV antibodies in up to 88% of adults and more than 40% of children younger than 5 years (14, 15), indicating that infection with MCPyV is very common. However, antibodies against LTA and STA are largely restricted to MCC patients, and titers correlate with tumor burden (16). Cellular immunity to MCPyV is much less described. MCPyV-specific T cells have been detected in tumor-infiltrating lymphocytes and in peripheral blood of MCC patients, representing primarily CD4 T-cell responses. To date, only one single CD8 T-cell epitope restricted to HLA-A24 has been identified (17); thus, limiting the possibility to characterize and monitor cytotoxic immune responses in MCC patients.
To overcome this limitation, we took advantage of a high-throughput platform that we developed for T-cell epitope identification to characterize MCPyV-specific CD8 T-cell epitopes in LTA, STA and VP1. This platform is based on a multi-parallel enrichment of peptide-MHC reactive T cells and the detection of specific responses by combinatorial encoding with peptide-MHC multimers, enabling multi-epitope identification using limited patient material (18–21). Using this approach we have detected T-cell responses among 398 predicted T-cell epitopes restricted to HLA-A1, -A2, -A3, -A11 and -B7. In total, 53 MCPyV-specific T-cell responses were detected in 27 individuals, representing 35 different specificities. While T cells specific for VP1-derived epitopes were detected both in MCC patients and healthy donors, T cells recognizing LTA- and STA-derived epitopes were only present in MCC patients. Importantly, LTA- and STA-specific T cells killed target cells expressing LTA, including HLA-matched MCPyV-positive MCC cell lines.
Materials and Methods
Samples
PBL samples were collected from 30 healthy donors and 38 patients with the histologically diagnosis of MCC at various time points during their course of disease (see Supplementary Table 1). No information on age, gender or the presence of MCPyV DNA was available for the control subjects. From two patients we analyzed both PBL and TIL samples. Additionally we analyzed TIL cultures from another 17 MCC patients. PBLs were isolated by density centrifugation on Lymphoprep™ (Axis-Shield PoC) and cryopreserved at −150°C in FBS and 10% DMSO. Tumors were categorized as MCPyV positive based on immunohistochemical staining for LTA and/or PCR-based detection of MCPyV DNA in the tumor (16). Tumors were categorized as MCPyV negative if tumor material was available for testing but gave a negative result even though some of the patients were seropositive for antibodies recognizing LTA/STA, described by Paulson, et al. (16). Tumors were not available for staining in 9 of 38 patients analyzed. Patient material was collected based on informed written consent according to the Declaration of Helsinki and with an approved IRB (FHCRC IR#6585). For the establishment of TIL cultures tumors were removed, cut and incubated in T cell medium in 48 well plates with the addition of 3.4ug/ml PHA. On the second day IL-2 (50U/ml) and IL-15 (20ng/ml) were added. Media was refreshed every few days. TILs were cultured for 2–3 weeks and frozen prior to analysis. Healthy donor PBLs were obtained from buffy coats available from the central blood bank of the capital region of Copenhagen (RegionH), and isolated using the same procedure as for the patient PBLs.
The peptide library
For selection of LTA- and STA-derived peptides, the MCC 348 (Genebank id. FJ173809.1) sequence was used. For selection of VP1-derived peptides the MKL-1 (Genebank id. FJ173815) sequence was used. Peptides were purchased from Pepscan presto, Lelystad, NL, with >70% purity. Predicted peptides were tested for experimental binding affinity by an MHC ELISA (22). Briefly, rescue of the MHC complex after UV-mediated cleavage of the conditional ligand depends on the affinity of the rescue peptide. The level of MHC rescue was determined by an MHC ELISA. Plates were coated with streptavidin (2µg/mL) (Invitrogen, cat#S888) and biotinylated MHC complexes were added after the UV-mediated peptide exchange reaction (in 2.5nM, based on 100% rescue). Plates were incubated for 1 hour and washed, and correctly folded MHC molecules were detected by binding to HRP-conjugated β2-m antibodies (1.4µg/mL) (Acris GmbH, cat# 604HRP). After incubation and washing, a colorimetric reaction was initiated by addition of ABTS peroxidase substrate (KPL, cat# 506606). HLA-binding peptides were selected based on the ability to rescue the MHC molecule after UV-mediated cleavage of the conditional ligand, compared to that of a virus-derived control. The threshold for selection was set to 60% of the virus control peptides.
MHC monomer production and combinatorial encoding of MHC multimers
The production of monomers was performed as described in Hadrup et al. (23). In brief, HLA heavy chain is expressed in bacterial Bl21 (DE3) pLysS strain (Novagen, cat# 69451) and purified as inclusion-bodies. After solubilization, inclusion-bodies are refolded with β2m light chain and a UV-sensitive ligand (24, 25). Following refolding, monomers are biotinylated (Avidity cat#BirA) and purified using size-exclusion column (Waters, BioSuite125, 13µm SEC 21, 5×300mm column) HPLC (Waters 2489). Specific MHC monomers were generated by UV-induced peptide exchange (25), and thereafter multimerized with fluorochrome-conjugated streptavidin.
The generation of combinatorial encoded MHC multimers and the gating strategy is described in detail by Andersen et al. (19). Briefly, UV exchange was carried out for all selected HLA ligands and each multimer was assigned with a unique color combination, such that a T-cell response was identified by two colors enabling the simultaneous detection of 27 different specificities in a single sample. Data acquisition was performed on an LSR II flow cytometer (Becton Dickinson) with FacsDiva software. Positive responses were defined by a minimum of 10 events observed in two different two-color combinations. We used eight streptavidin conjugated fluorocromes: Qdot- 585(Q10111MD), 605 (Q10101MD), 625 (A10196), 655 (Q10121MD), 705 (Q10161MD) all from Invitrogen and PE-Cy7 (405206), APC (405207) and PE (405203) from Biolegend to encode the specific MHC multimers. For the screening of TILs, additional colors were used: BV421 (Biolegend cat# 405225) and PE-CF594 (BD cat# 562284). MHC multimers were prepared at 100ug/ml and cells were stained using 2ul of each double colored MHC multimer. First, we gated on lymphocytes followed by single cells, live cells, and CD8 cells. Gates were made for each single MHC multimer color and then combined to gate out events positive for only one MHC multimer color and three or more MHC multimer colors, while showing events positive for exactly two MHC multimer colors as exemplified in (19).
Enrichment of peptide-specific T cells
PBLs were thawed in media containing 25U/ml DNase (Stemcell cat# 07900) and left in X-vivo (Lonza, cat# Be04-418Q) supplemented with 5% HS (Sigma # H5422) over night in the incubator. Cells were washed in PBS supplemented with 2% FBS (FACS buffer) prior to staining with 0.1ug of each specific PE-coupled MHC multimer and incubated for 1h at 4°C. Cells were washed followed by 15 min of incubation with anti-PE microbeads (Miltenyi Biotec, cat# 130-048-801). After washing, cells were resuspended in 0.5ml RPMI with DNase and applied to the magnetic separation columns (25MS, Miltenyi Biotec, cat# 130-042-201) through a 30µm pre-separation filter. After washing, cells were flushed out in 2 ml of X-vivo, 5% human serum, 100 U/ml IL-2 (Proleukin, Novartis cat# 200-02), 15 ng/ml IL-15 (Peprotech, cat# 200-15)), centrifuged and resuspended in 200µl of the same X-vivo based media containing 5×104 feeder cells prepared from the negative fraction during the separation and 5×103 anti-CD3/CD28-coated Dynabeads (Invitrogen, cat# 111.31.D). Enriched cells were cultured in 96-well plates, and medium was refreshed at least twice a week. After 2–3 weeks of culturing antigen-specific T-cell responses were analyzed by flow cytometry using combinatorial encoding with MHC multimers.
Cell sorting and culturing
T cells were stained with relevant MHC multimers and sorted on a FACSAria (Becton Dickinson) into 96 well plates containing 105 irradiated feeder cells incubated with 2ug/ml PHA overnight in X-vivo supplemented with 5% human serum, 500U/ml IL-2 and 30ng/ml anti-CD3 (eBioscience, clone OKT3, cat# 14-0037-82). Cultures were left untouched for 5 days; media were refreshed to a high IL-2 containing media (3000 U/ml IL-2) which was maintained through the remaining of the culturing period. Established cultures were tested for antigen-specificity by MHC multimer staining. MCC cell lines, MKL-2, WaGa and MCC 13 and K562 cells carrying HLA-A1, -A2, -A3, -A11 and -B7 were cultured in RPMI 16 (Gibco#72400-021), 5% FBS (Gibco, cat#10270), 100ug/ml pen/strep (Gibco, cat# 15140-122).
Production of in-vitro transcribed mRNA
The cDNA encoding truncated LTA (Genebank nr: FJ 173809.1) was synthesized and cloned into pSP73-SphA64 (kindly provided by Professor E Gilboa, Duke University Medical Center, Durham, NC) using 5`PuvII/ 3`KpnI restriction sites (Geneart/Life Technologies, Regensburg, Germany). Prior to serving as DNA template for in-vitro transcription, pSP73-Sph/LTA/A64 was linearized with SpeI and purified using Wizard DNA Clean-Up System (Promega). The in-vitro transcription was performed with mMESSAGE mMACHINE T7 Ultra kit (Ambion, Austin TX, USA) and mRNA was purified with MEGAclear kit (Ambion) according to the manufacturer’s instructions. The mRNA length, concentration and purity was evaluated with the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto CA, USA), using RNA 6000 Nano LabChip Kit (Agilent Technologies), according to the manufacturer’s instructions. Data analysis was performed with 2100 Bioanalyzer software (Agilent Technologies).
Electroporation of K562 cells
K562 cells stably transduced with the relevant HLA (A2, A11 and B7, kindly provided by Miriam Heemskerk, University Hospital Leiden, The Netherlands), were washed twice, suspended in PBS (Invitrogen) and adjusted to a final cell density of 40×106 cells/ml. The cell suspensions (400 µl) were pre-incubated in a 2-mm gap electroporation cuvette for 5 min on ice. 8 µg of mRNA encoding LTA was transferred to the cuvette and K562 cells were pulsed using a BTX 830 square-wave electroporator (Harvard Apparatus, Holliston MA, USA). Electroporation settings were adjusted to 6 pulses, 560 V, 99 µs. After electroporation, K562 cells were rested in 37°C and subsequently transferred to pre-heated culture medium (RPMI 16 + 5% FBS). HLA-transduced K562 cells were analyzed for mycoplasma negativity, HLA expression and expression of the LTA mRNA.
Cytotoxicity assays
To investigate the cytotoxic capacity of T-cell cultures, a VITAL-FR cytotox assay was performed as previously described (26). In short, relevant K562 cells transfected with LTA were used as target cells, they were labeled with 10µM CSFE (Invitrogen, cat# C34554) for 5 min, washed and mixed 1:1 with mock-transfected control K562 cells that had been labeled with 5µM FR (Invitrogen, cat# C34553). The cell mixture (1:1) was loaded into a 96 well plate with 1000 target cells per well followed by addition of effector T cells. The background ratio between CFSE and FR stained cells was obtained from wells with no effector cells. The ratio between CFSE and FR in wells containing effector cells was normalized to the ratio in wells without effector cells. The experiment was performed once in duplicates, and cultures were incubated for 48 h before analysis. To investigate cytotoxicity against MCC cell lines (Waga, MKL-2 and MCC-13) a chromium release assay was performed. The MCPyV positive MCC cell line Waga was stimulated with INFγ (Imukin, cat# 001748A, 2000U/ml for 72h) for expression of HLA class I. No stimulation of the MCPyV positive MCC cell line, MKL-2 and the MCPyV negative MCC cell line MCC-13 was necessary (Supplementary Fig S4). 105 cells were stained with 40ul 5mCi 51Cr (Perkin Elmer cat#NEZ030005MC) for 2h and mixed every 15 minutes. T cells specific for A2-LTA/STAKLL (21% CD8 of total cells, 92% specific T cells of CD8) were added to the labeled cells in an actual ratio of 1:1 and incubated for 4h. 100ul of the supernatant was counted in a γ-counter (Perkin Elmer precisely, Wizard, 1470 Automatic gamma counter). The experiment was performed once in duplicate. MCC cell lines were generated as primary cell lines from patients with a confirmed MCC tumor after surgical removal. Cell lines were analyzed for LTA and STA expression, mycoplasma negativity, HLA expression and INFy induced expression of HLA.
Statistical analysis
Statistical analysis is performed as described in the result section. Asterisk in the figures indicate significant differences: *: p<0.05, **: p < 0.01, *** p < 0.001.
Results
Generating peptide libraries of HLA ligands derived from LTA, STA and VP1
To create a library of potential T-cell epitopes derived from LTA, STA and VP1, we first performed an in silico selection of MHC class I binding peptides based on prediction databases, SYFPEITHI (www.syfpeithi.de) (27) and netMHC (www.cbs.dtu.dk/services/NetMHC) (28). The selected LTA sequence corresponds to a truncated version of LTA lacking the helicase binding domain. Nine-, ten- and eleven-mer peptides potentially binding to HLA-A1, -A2, -A3, -A11 and -B7 with prediction values > 19 in SYFPEITHI and < 1000 in netMHC were selected for further analyses, i.e. a total of 398 peptides (Supplementary Table S2). All 398 candidate peptides were synthesized and functionally evaluated using an MHC-ELISA assay (22, 25). For each peptide the affinity to the cognate HLA molecule was calculated relative to established T-cell epitopes binding with high affinity to the respective HLA class I molecules. Based on an arbitrary chosen threshold of 60% stabilization efficiency relative to the high-affinity control peptide derived from cytomegalovirus (CMV), Influenza (FLU) or Epstein Barr virus (EBV), we identified medium to high-affinity MHC-binding peptides derived from the MCPyV-encoded proteins (Table 1 and Supplementary Fig. S1). Based on these results, 196 selected peptide ligands were subsequently used to screen peripheral blood lymphocytes (PBLs) obtained from MCC patients and healthy donors for reactive T cells. Some peptides could bind multiple HLA-types and thus we tested 237 different peptide-MHC complexes: 9 for HLA-A1, 59 for HLA-A2, 82 for HLA-A3, 53 for HLA-A11 and 34 for HLA-B7 (Supplementary Table S3).
Table 1.
Selection of MHC binding MCPyV peptides
| HLA control peptide |
Selection threshold |
No. of predicted peptides |
No. of selected peptides |
|
|---|---|---|---|---|
| HLA-A1 | CMV-pp50VTE | 60% | 40 | 9 |
| HLA-A2 | CMV-pp65 NLV | 60% | 102 | 59 |
| HLA-A3 | FLU-NPILR | 60% | 189 | 82 |
| HLA-A11 | EBV-EBNA4 AVF | 60% | 189 | 53 |
| HLA-B7 | CMV-pp65TRP | 60% | 84 | 34 |
Multiple MCPyV derived peptides are recognized by T cells
As an initial step, we prepared phycoerythrin (PE)-labeled peptide-MHC multimers for specific enrichment of reactive T cells. T-cell enrichments were performed on PBLs obtained from 38 MCC patients and 30 healthy donors by capturing PE-labeled MHC multimer binding T cells with anti-PE magnetic beads. Previous analyses have established that up to 400 different T-cell specificities can be enriched simultaneously by this procedure (20, 29). After enrichment for MCPyV-protein reactivity, T cells were cultured for 3 weeks in the presence of CD3/CD28-coated expander beads, IL-2 and IL-15 before further analysis by combinatorial encoding with peptide-MHC multimers and flow cytometry (21, 23). This method enables the simultaneous detection of 27 different T-cell populations per sample (20). Each specific response was verified by either a second experiment using differently labeled combinatorial encoded peptide-MHC multimers or a secondary enrichment (Fig. 1A).
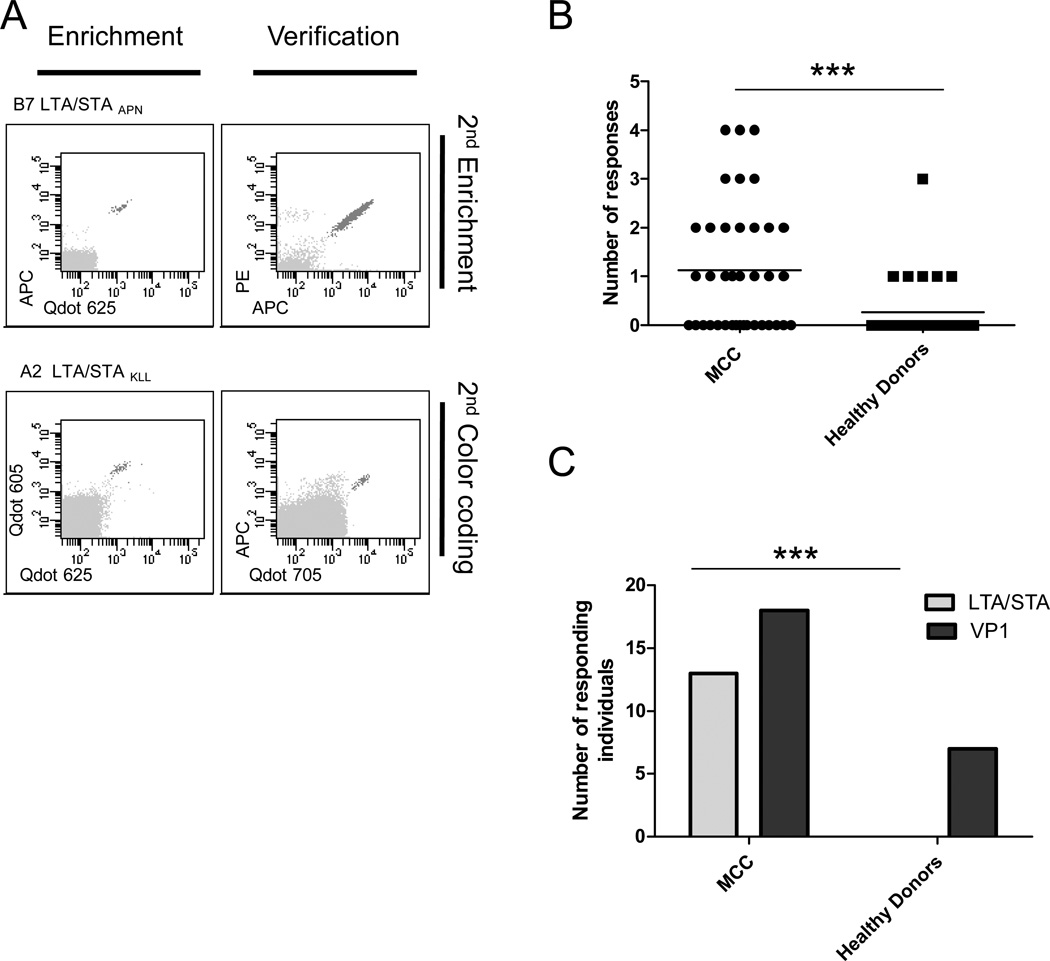
Fig. 1.
T-cell responses against MCPyV in MCC patients and healthy donors. (A) After T-cell enrichment, specific responses were detected using flow cytometry and combinatorial encoded MHC- multimers followed by verification with either a 2nd enrichment (top) or a 2nd MHC-multimer detection using a new color code (bottom). Representative examples from two MCC patients showing LTA/STA directed T-cell responses: MCC#27 B7-LTA/STAAPN (top row) and MCC#8 A2-LTA/STAKLL (bottom row). (B) The number of MCPyV specific T-cell responses per individual in MCC patients and healthy donors, every dot represents one individual. Asterisks indicate significant levels, two-tailed unpaired T-test. (C) The number of MCC patients or healthy donors where LTA/STA or VP1 specific T-cell responses were found. Asterisks indicate significant levels, Fisher`s exact test.
We detected T-cell responses against LTA-, STA- and VP1-derived epitopes for all MHC restriction elements included. In total, 53 MCPyV-directed T-cell responses were identified and confirmed in 27 individuals, recognizing 35 different peptide sequences (Table 2). For each specificity, representative examples of peptide-MHC reactive T-cell populations are depicted in Supplementary Fig S2. Notably, for several MCPyV-derived epitopes we detected peptide-MHC reactive T cells in multiple individuals, including: A3-VP1TVL (code shown: HLA restriction, viral protein with subscript of first three amino acids), A3/A11-VP1QMW, -VP1SLF, -VP1LQM, -VP1SLI, A2-LTASMF, A2-LTAKLL, and B7-LTAAPI, indicating that these could be immune dominant epitopes MCPyV-specific T-cell responses were present in both MCC patients and healthy donors, but were more frequently detected in MCC patients (21 of 38 MCC patients versus 6 of 30 healthy donors, p=0.006, Fisher`s Exact test) and patients further displayed a broader response in terms of number of recognized epitopes (p=0.0007, unpaired T-test) (Fig. 1B). Indeed, T-cell responses against epitopes of LTA and STA were exclusively detected in MCC patients (13/38) in contrast to none of the healthy donors (p<0.0001, Fisher`s Exact test) (Fig. 1C). VP1 responses were detected in both groups, but with a tendency of less frequent detection among healthy donors (p=0.08, Fisher`s Exact test) (Fig 1C). There were no correlation between LTA- and STA-specific T-cell responses in MCC patients with the viral status of the tumors (Supplementary Fig. S3). Since LTA- or STA-specific T-cell responses were not present in any of the healthy individuals and MCC samples may have been classified as MCPyV-negative due to the detection threshold of the method used (5) or may have lost the virus (30), our observation indicates that those MCC patients with detectable LTA- or STA-specific T-cell responses were exposed to relevant amounts of LTA and/or STA protein, albeit the presence of MCPyV in the tumor could not be confirmed. Data on MCPyV copy numbers were available from 20 patients, but no correlation was found with the number of LTA/STA responses (Pearson correlation p=0.83, R2= 0.0025). Also, the number of total MCPyV responses and LTA/STA responses were independent of stage of disease on the time of sampling.
Table 2.
Peptides eliciting CD8 T cell responses in MCC patients and healthy donors.
| Antigen | Peptide sequence |
AA position |
Length | MCC | Healthy Donors |
HLA restriction |
|---|---|---|---|---|---|---|
| Vp1 | SITQIELYL | 51 | 9 | 1/26 | A2 | |
| NLPAYSVARV | 97 | 10 | 1/26 | 1/16 | A2 | |
| NLINSLFSNL | 326 | 9 | 1/26 | A2 | ||
| FLDKFGQEKT | 372 | 9 | 1/26 | A2 | ||
| ALHGLPRYFNV | 301 | 11 | 1/26 | A2 | ||
| LLVKGGVEV | 34 | 9 | 1/26 | A2 | ||
| KPGCCPNVA | 21 | 9 | 1/NA | B7 | ||
| LPAYSVARVSL | 98 | 11 | 1/NA | B7 | ||
| LPRYFNVTLRK | 305 | 11 | 1/NA | B7 | ||
| LPRYFNVTL | 305 | 9 | 1/13 | B7 | ||
| ITIETVLGR | 195 | 9 | 1/16 | A3 | ||
| GLDPQAKAK | 211 | 9 | 1/16 | A3 | ||
| TLRKRWVKN | 312 | 9 | 1/16 | A3 | ||
| SLFSNLMPK | 330 | 9 | 2/16A3 | 2/7A11 | A3/A11 | |
| KMALHGLPR | 299 | 9 | 1/14 | A3 | ||
| TVLGRKMTPK | 199 | 10 | 4/16 | 1/14 | A3 | |
| NSLFSNLMPK | 329 | 10 | 1/16 | A3 | ||
| VTLRKRWVK | 311 | 9 | 1/7 | A11 | ||
| SLINVHYWDMK | 137 | 11 | 1/16A3, 1/7A11* | A3/A11 | ||
| QMWEAISVK | 121 | 9 | 3/16A3, 1/7A11* | A3/A11 | ||
| LQMWEAISVK | 120 | 10 | 1/16A3, 1/7A11* | A3/A11 | ||
| DLQGLVLDY | 174 | 9 | 1/7A11 | A11 | ||
| VLGRKMTPK | 200 | 9 | 1/7A11 | A11 | ||
| LTA | HSQSSSSGY | 137 | 9 | 1/14 | A1 | |
| VIMMELNTL | 50 | 9 | 1/26 | A2 | ||
| SMFDEVDEAPI | 74 | 11 | 2/26 | A2 | ||
| KLLEIAPNC | 15 | 9 | 2/26 | A2 | ||
| ASFTSTPPK | 294 | 9 | 1/16 | A3 | ||
| AAFKRSCLK | 32 | 9 | 1/7 | A11 | ||
| APIYGTTKF | 82 | 9 | 2/NA | B7 | ||
| APNCYGNIPL | 20 | 10 | 1/NA | B7 | ||
| APNCYGNIPLM | 20 | 11 | 1/NA | B7 | ||
| STA | KTLEETDYCLL | 171 | 11 | 1/26 | A2 | |
| TLEETDYCL | 172 | 9 | 1/26 | A2 | ||
| QLRDSKCACI | 113 | 10 | 1/26 | A2 |
Peptides in bold: T cell epitopes where processing and presentation is verified (see fig 3).
MCC and Healthy donor column: Number of times a response was detected/numbers of investigated HLA matched individuals.
Detected in the same individual.
NA: Not available. B7 tissue type was not available for the MCC patients; they were all tested for B7 responses.
MCPyV specific T cells have cytotoxic potential
The next series of experiments addressed the functional activity of LTA- or STA-specific T cells and the clinical relevance of such responses in MCC patients. Specific CD8 T-cell lines were established for three LTA-derived peptides: A2-LTAKLL, A11-LTAAAF and B7-LTAAPN.
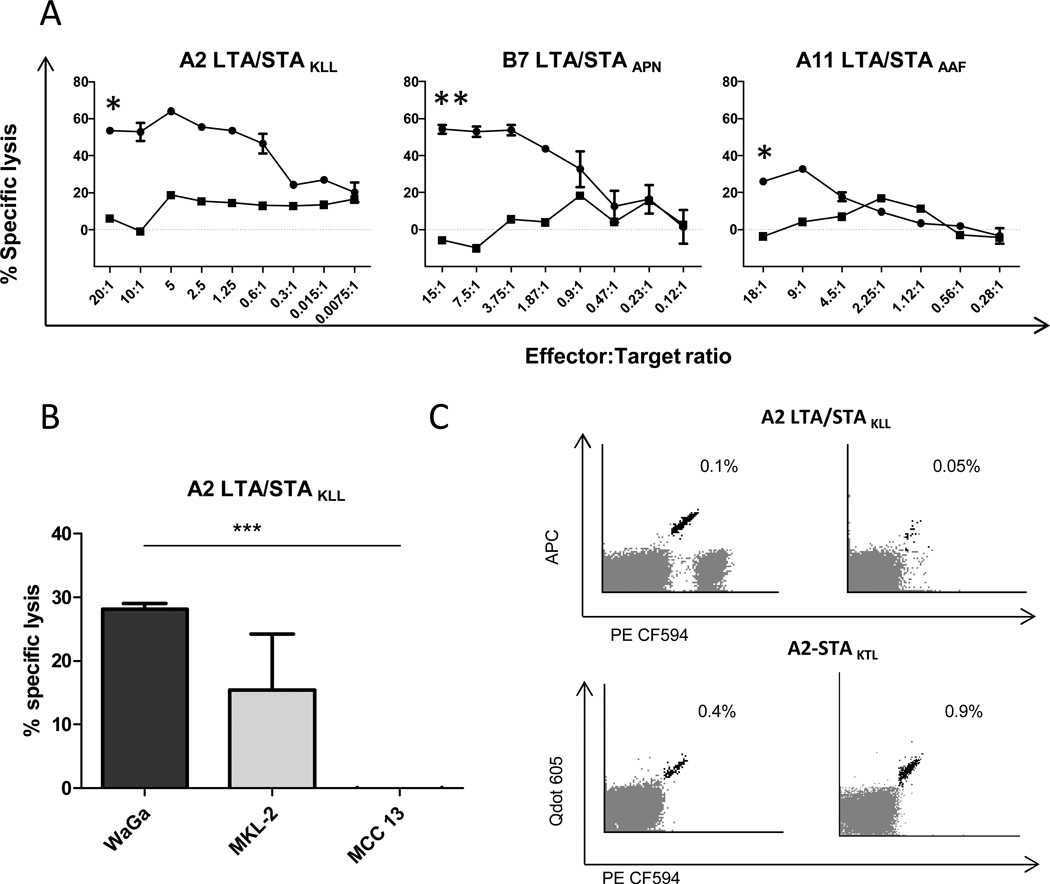
We scrutinized both the processing of LTA to the relevant peptide epitopes as well as the effector function of the respective T cells by means of genetically modified K562 cells expressing HLA-A2, -A11 or -B7 transfected with LTA mRNA (Supplementary Fig. S4A), which were used as target cells in cytotoxicity assays. Cytotoxicity was determined using a flow cytometry based assay comparing the ratio of differentially labeled target cells to non-target cells after co-culture with and without effector T cells. These assays clearly demonstrated, that every LTA-reactive T-cell line specifically killed the LTA-transfected but not the mock-transfected K562 cells (Fig. 2A) thus, demonstrating LTA processing and presentation in context of MHC class I. However, the clinically relevant question is whether the LTA-specific T cells exert cytotoxicity against MCC. Consequently, we next used HLA-A2-positive MCPyV-positive MCC tumor cell lines (i.e. WaGa and MKL-2) as target cells for the A2-LTAKLL-specific CD8 T-cell line; as control, we used a HLA-matched MCPyV-negative MCC cell line (MCC13) (Fig. 2B). MCC tumors and MCC cell lines are known to down regulate MHC class I expression, which can be re-induced by interferons. Consequently, re-induction or amplification of MHC class I expression was attained by culture in low doses of interferon prior to the assay (Supplementary Fig. S4B). These experiments revealed that A2-LTAKLL-specific CD8 T cells efficiently killed the virus-positive MCC cell lines (Waga vs. MCC13, p< 0.001. MKL-2 vs. MCC13, non significant. Two-tailed unpaired T-test. To date it is not entirely clear if T-cell responses to solid tumors detected in peripheral blood do indeed reflect ongoing responses in the tumor microenvironment, as there are several immune escape mechanisms operating in the latter (31). Hence, to further substantiate the clinical relevance of such LTA/STA-specific T cells we analyzed if these would be present among tumor infiltrating lymphocytes (TILs). Notably, when screening TILs from 19 MCC patients, by combinatorial encoded peptide-MHC multimers using the panel of 35 MCPyV-derived peptides previously found to enable T-cell recognition in peripheral blood (Table 2), we detected A2-LTAKLL-specific CD8 T cells in TILs from two patients and A2-STAKTL specific CD8 T cells from another two patients. We found no VP1 directed T-cell responses in TILs. PBL had also been analyzed from one of these patients where no specific T cells were identified (Fig. 2C). In conclusion, these observations underscore the clinical relevance of the identified T-cell epitopes for MCC.
Fig. 2.
Processing and presentation of MCPyV LTA allows T-cell mediated killing by MCPyV-specific T cells. (A) T-cell mediated killing of HLA-expressing LTA transfected K562 cells (Fig. S4A) detected by a flow cytometry-based cytotoxicity assay. The Y-axis shows percent specific lysis at different effector:target ratios with T cells recognizing A2-LTA/STAKLL (left), B7-LTA/STAAPN (middle), and A11-LTS/STAAAF (right). Reactive T-cell populations were enriched to purities ranging from 92–95% of CD8 T cells. Circles: K562 HLA+ LTA were stained with CSFE and K562 HLA were stained with FR. Squares: Unspecific lysis of HLA mis-matched K562 cells stained with CSFE and a second HLA mis-matched K562 was stained with FR for identifying the background lysis. Specific lysis was measured and calculated as described in material and methods. Each point represent the mean of two replicates and error bars the SEM, n=1. Asterisk indicates significant levels. one-tailed unpaired T-test. (B) Chromium release assay was performed using the 51Cr-labeled target cells: MCPyV+/HLA-A2+ MCC cell line WaGa preincubated with INFγ (Supplementary Fig. S4B), MCPyV+/HLA-A2+ MCC cell line MKL-2, and the MCPyV-negative A2+ MCC cell line MCC 13. Target cells were incubated at effector/target cell ratio of 1:1 in duplicates for 4 h before 51Cr release was measured. Bars represent the mean of two replicates and error bars the SEM, n=1. Asterisk indicates significant levels, two-tailed unpaired T-test. (C) Dot plot showing A2-LTAKLL or A2-STAKTL specific CD8 T cells as detected in TIL cultures from a total of 4 MCC patients with MHC mulitmers (APC/PE-CF594 for A2-LTA/STAKLL and Qdot605/ PE-CF594 for A2-STAKTL). The frequency of LTA/STA specific T cells in CD8 T cells is given.
T cell epitopes are shared among different MCPyV strains
To investigate viral strain specificity, we tested if the respective epitopes are encoded by the described MCPyV sequences. Alignment of all 88 MCPyV LTA sequences deposited at the NCBI nucleotide database revealed that the three LTA epitopes identified are encoded by most MCPyV strains. The variation within the T-cell epitope sequences are relatively low; only 17% carry a SNP at amino acid position 20 located in both the A2-LTAKLL and B7-LTAAPN epitopes, which results in a change from alanine to serine in 14%, to threonine in 2%, and to phenylalanine in 1%. Further, 1% carried a SNP at amino acid position 26 located only in the B7-LTAAPN epitope; this SNP causes a change from asparagine to aspartic acid. No SNPs were identified in the A11-LTAAFF epitope. (Fig 3A and Supplementary Fig. S5). Thus, the identified epitopes are highly conserved among the known MCPyV strains, and even in the cases where single amino acid variations are present, the recognition of these epitopes are likely maintained in most cases (32).
Fig. 3.
Sequence homology of MCPyV-derived CD8 T-cell epitopes within different strains of MCPyV and among other human polyomaviruses. (A) Circle diagram illustrating the variation within the T-cell epitope sequences of LTA from 88 different strains of MCPyV. 17% of the sequences carried a SNP at amino acid position 20, located in both the A2-LTAKLL and B7-LTAAPN epitope causing the indicated changes in the amino acid sequence. (B) Alignment of the MCPyV LTA amino-acid sequence (top) with the corresponding amino-acid sequences from the 10 other known human polyomaviruses, BKPyV, JCPyV, KIPyV, WUPyV, HPyV6, HPyV7, HPyV8, HPyV9, MWPyV and STLPyV. Grey boxes represent the position of the MCPyV-derived A2-, A11-, and B7-restricted epitopes. Amino acids from the other human polyomaviruses that differ from the MCPyV sequence are highlighted in bold. Below, in each epitope the anchor residues are in bold and the auxiliary anchor residues are underlined.
Next we tested for homology in the LTA/STA sequences of MCPyV compared to the additional 10 currently described human polyomaviridae (33, 34). In the region of the A2-, A11- and B7-restricted LTA/STA derived epitopes, we found a minimum of five differences in the amino-acid sequence of the T-cell receptor recognition site. Thus, although anchor residues are in some cases shared, this difference makes cross-recognition between the different polyomaviruses highly unlikely (Fig 3B)(32).
Discussion
Recent clinical achievements in immunotherapy for the treatment of solid tumors confirmed the relevance of the immune system in general and the role of cytotoxic T cells in particular for control of cancer (35–38). Although the impact of cytotoxic T-cell responses for disease control is variable among different cancers, multiple lines of evidence suggest that cellular immune function is unusually important for MCC: (i) intratumoral CD8 lymphocyte infiltration is significantly associated with improved survival (4), (ii) spontaneous regression, particularly after cessation of immune suppression, is observed even in metastatic MCC (2), and (iii) immune suppression is a strong risk factor for MCC and an independent predictor of diminished survival (3, 39). Indeed, when comparing the relevance of the immune system for disease control between cutaneous melanoma (i.e. the tumor entity for which the best evidence for the impact of therapeutic immune modulation has been reported) and MCC, it should be noted that the ratio of melanoma to MCC in the immune competent host is 60:1, whereas only 6:1 in the immune comprised. The association of MCC with MCPyV explains the explicit immunogenicity of this tumor. Indeed, MCPyV-encoded proteins are likely targets for cytotoxic immune responses as they are both foreign to the host and their expression is necessary to maintain the oncogenic phenotype (10–13). To date only a single MCPyV-derived MHC class I-restricted epitope has been described (17), thus impeding specific monitoring of T-cell responses to MCC. Consequently, the purpose of this study was to identify and validate a sufficient number of MCPyV-derived T-cell epitopes for future characterization of spontaneous, modulated or induced immune responses to MCC. This goal was achieved by means of a high-throughput flow cytometry based platform whereby CD8 T-cell responses against 35 MCPyV derived peptide epitopes with a broad range of MHC class I restriction elements were identified in peripheral blood of MCC patients. T-cell cultures specific for the three peptides derived from the region shared between MCPyV LTA and STA killed HLA-matched LTA-expressing K562 as well as MCC cell lines. This observation demonstrates that the respective peptides are indeed processed and presented. Furthermore, we could demonstrate the presence of A2-LTAKLL- and A2-STAKTL-specific CD8 T cells among TILs indicating the presence of such responses in the tumor microenvironment. T-cell responses in TILs were detected without any prior enrichment strategy of MCPyV reactive T cells, which may account for the difference in response frequency observed (4 in 19 TIL samples, versus, 21 in 38 PBL samples, p<0.05, Fisher`s Exact test). However, it may also reflect the lack of VP1 specific responses in the TILs, possibly due to lack of intra-tumoral expression of this protein (14), as VP1 responses constitute a large fraction of the T-cell responses observed in the PBL samples. When considering only LTA/STA directed responses, no significant difference is observed between the two sources of cells (4 in 19 TIL samples, versus 13 in 38 PBL samples, p>0.05, Fisher`s Exact test).
Interestingly, while T cells recognizing the MCPyV-encoded VP1 were present in both MCC patients and healthy donors, T-cell responses to the oncogenic proteins LTA and STA were exclusively detected in MCC patients. This notion is in accordance with recent observations concerning humoral immune responses, i.e. antibodies to MCPyV capsid antigens are frequently present in the general population, whereas anti-LTA and anti-STA responses are largely restricted to MCC patients (16). Notably, for patients who have no evidence of disease after therapy (n=9) LTA/STA T-cell reactivity was apparent in two patients on average three years after removal of the primary lesion, suggesting the generation of a memory T-cell response.
We observed a tendency towards a lower VP1 directed T-cell response frequency in the group of healthy donors compared to MCC, which may be explained by the fact that our healthy donor cohort was not selected of MCPyV positivity.
MCPyV is currently accepted to be a critical causative agent at least for MCPyV-positive MCC cases. Speculations about the origin of MCPyV-negative MCC cases have been proposed (30, 40). However, advances in detection methods for MCPyV implies that the number of ‘true’ MCPyV-negative MCC is smaller than initially assumed, e.g. using improved PCR-based methods the MCPyV genome was detected in 100% of the MCC tumors investigated (5). The observation that LTA- and STA-specific T-cell responses are detected exclusively in MCC patients, irrespective of their virus status, support the hypothesis that most, if not all MCC patients have indeed been subjected to LTA/STA protein expression during the course of their disease. A recent report describes CD4 T cell recognition of LTA from the JC polyomavirus in healthy donors, with mapping of epitopes in the region of LTA that is truncated in MCC, and thus not included in the present study (41). It is possible that this region of MCPyV LTA may likewise be immunogenic in MCPyV positive healthy donors.
For 16 MCC patients we investigated the presence of MCPyV-specific responses in PBLs directly ex-vivo, revealing no MCPyV-specific responses. Since MCPyV-specific responses were detected in most of the samples following T-cell enrichment, these responses are below the detection limit for MHC multimer analyses (0.005% of CD8 T cells). It should be noted that this observation also raises the concern that T cells from the naïve repertoire can also be selected, expanded and subsequently detected after the peptide-MHC based enrichment (29). However, the complete lack of responses against LTA/STA in healthy donors in conjunction to the readily detected VP1 response indicates that this is not the case in the current setting.
Evasion of immune responses is one of the hallmarks of cancer (42) and tumor cells exploit numerous of these mechanisms in order to create a tumor-promoting immune-suppressive microenvironment (4). Indeed, it was recently reported that MCC tumors utilize a spectrum of immune evasion strategies to locally suppress T-cell responses by inducing high concentrations of both CD4+ and CD8 Tregs and the expression of PD-L1 and PD-L2 within the tumor microenvironment (43). This immune suppression seems to not be confined to the tumor microenvironment, but extents to antigen-specific T cells in circulation, as these cells display an exhausted phenotypic profile (44). However, to date immunological studies in MCC were hindered by the limited number of known MCPyV-specific T-cell epitopes. Our present report provides the tools for further in depth analysis of such mechanisms covering a broad range of possible T-cell specificities. This is particularly important with the advent of efficient immune modulatory therapies such as immune checkpoint blocking antibodies or antibody-targeted cytokine therapy as it allows tracking and monitoring of T-cell responses (45–47). Moreover, the T-cell epitopes described here are of viral origin and – in the case of MCPyV LTA and STA - exclusively recognized in MCC; therefore they represent an ideal target for immunotherapy. Since these antigens are foreign to the patient’s immune system, it is highly feasible to generate high-affinity T-cell populations that recognize these antigens and subsequently use them in adoptive cell therapies along with immune modulating therapies.
Supplementary Material
Statement of translational relevance.
Merkel cell carcinoma is an often-lethal skin cancer of increasing interest due to the strong association with a polyomavirus. New data suggest that virtually all Merkel cell carcinoma cases are associated with expression of Merkel cell polyomavirus oncogenes. These oncogenes represent ideal targets for immunotherapy of Merkel cell carcinoma – being both specific and essential for the cancer cells, and foreign to the immune system.
We have identified multiple T-cell epitopes in Merkel cell polyomavirus encoded proteins, and show that T-cell responses to Merkel cell polyomavirus oncogenic- versus capsid-proteins clearly distinguish Merkel cell carcinoma patients from healthy donors. These epitopes can serve as important targets for immunotherapy, and will improve our ability to track immune responses in Merkel cell carcinoma patients. Furthermore, polyomavirus specific T-cell responses can be targeted by immune modulatory agents such as anti-CTLA-4 and anti-PD-1 for treatment of Merkel cell carcinoma.
Acknowledgments
We thank Miriam Heemskerk, University hospital Leiden, The Netherlands for providing the HLA transduced K562 cells and we appreciate the gift of the pSP73-SphA64 vector used in the in vitro transcription of LTA mRNA from Professor E Gilboa, Duke University Medical Center, Durham, NC. Furthermore we acknowledge the technical assistance provided by Tina Seremet, CCIT, Department of Hematology, University Hospital, DK and the statistical support from Tobias W. Klausen, CCIT, Department of Hematology, University Hospital Herlev, DK. We also thank Natalie Miller, University of Washington, Seattle, USA for careful proofreading our manuscript.
Grant support: FP7-Health 2011 – IMMOMEC; The Danish Cancer Society; The capital region of Denmark (regionH)
Footnotes
The authors declare no conflicts of interest
References
- 1.Lemos BD, Storer BE, Iyer JG, Phillips JL, Bichakjian CK, Fang LC, et al. Pathologic nodal evaluation improves prognostic accuracy in Merkel cell carcinoma: analysis of 5823 cases as the basis of the first consensus staging system. J Am Acad Dermatol. 2010;63:751–761. doi: 10.1016/j.jaad.2010.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedlaender MM, Rubinger D, Rosenbaum E, Amir G, Siguencia E. Temporary regression of Merkel cell carcinoma metastases after cessation of cyclosporine. Transplantation. 2002;73:1849–1850. doi: 10.1097/00007890-200206150-00028. [DOI] [PubMed] [Google Scholar]
- 3.Heath M, Jaimes N, Lemos B, Mostaghimi A, Wang LC, Peñas PF, et al. Clinical characteristics of Merkel cell carcinoma at diagnosis in 195 patients: the AEIOU features. J Am Acad Dermatol. 2008;58:375–381. doi: 10.1016/j.jaad.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paulson KG, Iyer JG, Tegeder AR, Thibodeau R, Schelter J, Koba S, et al. Transcriptome-wide studies of merkel cell carcinoma and validation of intratumoral CD8+ lymphocyte invasion as an independent predictor of survival. J Clin Oncol. 2011;29:1539–1546. doi: 10.1200/JCO.2010.30.6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodig SJ, Cheng J, Wardzala J, Dorosario A, Scanlon JJ, Laga AC, et al. Improved detection suggests all Merkel cell carcinomas harbor Merkel polyomavirus. J Clin Invest. 2012;122:4645–4653. doi: 10.1172/JCI64116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sastre-garau X, Peter M, Avril M-F, Laude H, Couturier J, Rozenberg F, et al. Merkel cell carcinoma of the skin: pathological and molecular evidence for a causative role of MCV in oncogenesis. J Pathol. 2009;218:48–56. doi: 10.1002/path.2532. [DOI] [PubMed] [Google Scholar]
- 8.Laude HC, Jonchère B, Maubec E, Carlotti A, Marinho E, Couturaud B, et al. Distinct merkel cell polyomavirus molecular features in tumour and non tumour specimens from patients with merkel cell carcinoma. PLoS Pathogen. 2010;6:e1001076. doi: 10.1371/journal.ppat.1001076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martel-Jantin C, Filippone C, Cassar O, Peter M, Tomasic G, Vielh P, et al. Genetic variability and integration of Merkel cell polyomavirus in Merkel cell carcinoma. J Virol. 2012;426:134–142. doi: 10.1016/j.virol.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 10.Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, et al. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc Natl Acad Sci USA. 2008;105:16272–16277. doi: 10.1073/pnas.0806526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shuda M, Kwun HJ, Feng H, Chang Y, Moore PS. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J Clin Invest. 2011;121:3623–3634. doi: 10.1172/JCI46323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Houben R, Shuda M, Weinkam R, Schrama D, Feng H, Chang Y, et al. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J Virol. 2010;84:7064–7072. doi: 10.1128/JVI.02400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houben R, Adam C, Baeurle A, Hesbacher S, Grimm J, Angermeyer S, et al. An intact retinoblastoma protein-binding site in Merkel cell polyomavirus large T antigen is required for promoting growth of Merkel cell carcinoma cells. Int J Cancer. 2012;130:847–856. doi: 10.1002/ijc.26076. [DOI] [PubMed] [Google Scholar]
- 14.Pastrana DV, Tolstov YL, Becker JC, Moore PS, Chang Y, Buck CB. Quantitation of human seroresponsiveness to Merkel cell polyomavirus. PLoS pathogens. 2009;5:e1000578. doi: 10.1371/journal.ppat.1000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen T, Hedman L, Mattila PS, Jartti T, Ruuskanen O, Söderlund-Venermo M, et al. Serological evidence of Merkel cell polyomavirus primary infections in childhood. J Clin Virol. 2011;50:125–129. doi: 10.1016/j.jcv.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 16.Paulson KG, Carter JJ, Johnson LG, Cahill KW, Iyer JG, Schrama D, et al. Antibodies to merkel cell polyomavirus T antigen oncoproteins reflect tumor burden in merkel cell carcinoma patients. Cancer Res. 2010;70:8388–8397. doi: 10.1158/0008-5472.CAN-10-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iyer JG, Afanasiev OK, McClurkan C, Paulson K, Nagase K, Jing L, et al. Merkel cell polyomavirus-specific CD8+ and CD4+ T-cell responses identified in Merkel cell carcinomas and blood. Clin Cancer Res. 2011;17:6671–6680. doi: 10.1158/1078-0432.CCR-11-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hadrup SR, Bakker AH, Shu CJ, Andersen RS, van Veluw J, Hombrink P, et al. Parallel detection of antigen-specific T-cell responses by multidimensional encoding of MHC multimers. Nat Method. 2009;6:520–526. doi: 10.1038/nmeth.1345. [DOI] [PubMed] [Google Scholar]
- 19.Andersen RS, Kvistborg P, Frøsig TM, Pedersen NW, Lyngaa R, Bakker AH, et al. Parallel detection of antigen-specific T cell responses by combinatorial encoding of MHC multimers. Nat Protoc. 2012;7:891–902. doi: 10.1038/nprot.2012.037. [DOI] [PubMed] [Google Scholar]
- 20.Hadrup SR, Schumacher TN. MHC-based detection of antigen-specific CD8+ T cell responses. Cancer immunol immunother. 2010;59:1425–1433. doi: 10.1007/s00262-010-0824-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersen RS, Thrue CA, Junker N, Lyngaa R, Donia M, Ellebæk E, et al. Dissection of T-cell antigen specificity in human melanoma. Cancer Res. 2012;72:1642–1650. doi: 10.1158/0008-5472.CAN-11-2614. [DOI] [PubMed] [Google Scholar]
- 22.Rodenko B, Toebes M, Hadrup SR, van Esch WJE, Molenaar AM, Schumacher TNM, et al. Generation of peptide-MHC class I complexes through UV-mediated ligand exchange. Nat Protoc. 2006;1:1120–1132. doi: 10.1038/nprot.2006.121. [DOI] [PubMed] [Google Scholar]
- 23.Hadrup SR, Toebes M, Rodenko B, Bakker AH, Egan DA, Ovaa H, et al. High-throughput T-cell epitope discovery through MHC peptide exchange. Schutkowski M, Reineke U, Methods in Mol Bio. 2009;524:383–405. doi: 10.1007/978-1-59745-450-6_28. [DOI] [PubMed] [Google Scholar]
- 24.Bakker AH, Hoppes R, Linnemann C, Toebes M, Rodenko B, Berkers CR, et al. Conditional MHC class I ligands and peptide exchange technology for the human MHC gene products HLA-A1, -A3, -A11, and -B7. Proc Natl Acad Sci USA. 2008;105:3825–3830. doi: 10.1073/pnas.0709717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toebes M, Coccoris M, Bins A, Rodenko B, Gomez R, Nieuwkoop NJ, et al. Design and use of conditional MHC class I ligands. Nat Med. 2006;12:246–251. doi: 10.1038/nm1360. [DOI] [PubMed] [Google Scholar]
- 26.Stanke J, Hoffmann C, Erben U, von Keyserling H, Stevanovic S, Cichon G, et al. A flow cytometry-based assay to assess minute frequencies of CD8+ T cells by their cytolytic function. J Immunol Methods. 2010;360:56–65. doi: 10.1016/j.jim.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 27.Rammensee H, Bachmann J, Emmerich NP, Bachor OA SS. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;Nov 50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 28.Lundegaard C, Lund O, Nielsen M. Accurate approximation method for prediction of class I MHC affinities for peptides of length 8, 10 and 11 using prediction tools trained on 9mers. Bioinformatics. 2008;24:1397–1398. doi: 10.1093/bioinformatics/btn128. [DOI] [PubMed] [Google Scholar]
- 29.Hombrink P, Hadrup SR, Bakker A, Kester MGD, Falkenburg JHF, von dem Borne Pa, et al. High-throughput identification of potential minor histocompatibility antigens by MHC tetramer-based screening: feasibility and limitations. PloS one. 2011;6:e22523. doi: 10.1371/journal.pone.0022523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Houben R, Grimm J, Willmes C, Weinkam R, Becker JC, Schrama D. Merkel cell carcinoma and Merkel cell polyomavirus: evidence for hit-and-run oncogenesis. J Dermatol Sci. 2012;132:254–256. doi: 10.1038/jid.2011.260. [DOI] [PubMed] [Google Scholar]
- 31.Becker JC, Andersen MH, Schrama D, Thor Straten P. Immune-suppressive properties of the tumor microenvironment. Cancer immunol immunother. 2013;62:1137–1148. doi: 10.1007/s00262-013-1434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gotch F, McMichael A, Rothbard J. Recognition of influenza A matrix protein by HLA-A2-restricted cytotoxic T lymphocytes. Use of analogues to orientate the matrix peptide in the HLA-A2 binding site. J Exp Med. 1988;168:2045–2057. doi: 10.1084/jem.168.6.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spurgeon ME, Lambert PF. Merkel cell polyomavirus: a newly discovered human virus with oncogenic potential. J Virol. 2013;435:118–130. doi: 10.1016/j.virol.2012.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeCaprio JA, Garcea RL. A cornucopia of human polyomaviruses. Nat Rev Microbiology. 2013;11:264–276. doi: 10.1038/nrmicro2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenberg Sa, Dudley ME. Adoptive cell therapy for the treatment of patients with metastatic melanoma. Current opinion in immunology. 2009;21:233–240. doi: 10.1016/j.coi.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ellebaek E, Iversen TZ, Junker N, Donia M, Engell-Noerregaard L, Met Ö, et al. Adoptive cell therapy with autologous tumor infiltrating lymphocytes and low-dose Interleukin-2 in metastatic melanoma patients. Journal of translational medicine. 2012;10:169. doi: 10.1186/1479-5876-10-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamid O, Robert C, Daud A, Hodi FS, Hwu W-J, Kefford R, et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med. 2013;369:134–144. doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radvanyi LG, Bernatchez C, Zhang M, Fox PS, Miller P, Chacon J, et al. Specific lymphocyte subsets predict response to adoptive cell therapy using expanded autologous tumor-infiltrating lymphocytes in metastatic melanoma patients. Clin Cancer Res. 2012;18:6758–6770. doi: 10.1158/1078-0432.CCR-12-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paulson KG, Iyer JG, Blom A, Warton EM, Sokil M, Yelistratova L, et al. Systemic immune suppression predicts diminished Merkel cell carcinoma-specific survival independent of stage. J invest dermatol. 2013;133:642–646. doi: 10.1038/jid.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Touzé A, Le Bidre E, Laude H, Fleury MJJ, Cazal R, Arnold F, et al. High levels of antibodies against merkel cell polyomavirus identify a subset of patients with merkel cell carcinoma with better clinical outcome. J Clin Invest. 2011;29:1612–1619. doi: 10.1200/JCO.2010.31.1704. [DOI] [PubMed] [Google Scholar]
- 41.Jelcic I, Aly L, Binder TMC, Jelcic I, Bofill-Mas S, Planas R, et al. T cell epitope mapping of JC polyoma virus-encoded proteome reveals reduced T cell responses in HLA-DRB1*04:01+ donors. J Virol. 2013;87:3393–3408. doi: 10.1128/JVI.02803-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanahan D, Weinberg Ra. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 43.Dowlatshahi M, Huang V, Gehad AE, Jiang Y, Calarese A, Teague JE, et al. Tumor-specific T cells in human Merkel cell carcinomas: a possible role for Tregs and T-cell exhaustion in reducing T-cell responses. J invest dermatol. 2013;133:1879–1889. doi: 10.1038/jid.2013.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Afanasiev OK, Yelistratova L, Miller N, Nagase K, Paulson K, Iyer J, et al. Merkel polyomavirus-specific T cells fluctuate with Merkel cell carcinoma burden and express therapeutically targetable PD-1 and Tim-3 exhaustion markers. Clin Cancer Res. 2013 doi: 10.1158/1078-0432.CCR-13-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.List T, Neri D. Immunocytokines: a review of molecules in clinical development for cancer therapy. Clinical pharmacology. 2013;5:29–45. doi: 10.2147/CPAA.S49231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.