ABSTRACT
Triosephosphate isomerase (TPI) catalyzes the interconversion of dihydroxyacetone phosphate (DHAP) and glyceraldehyde-3-phosphate (G3P). This reaction is required for glycolysis and gluconeogenesis, and tpi has been predicted to be essential for growth of Mycobacterium tuberculosis. However, when studying a conditionally regulated tpi knockdown mutant, we noticed that depletion of TPI reduced growth of M. tuberculosis in media containing a single carbon source but not in media that contained both a glycolytic and a gluconeogenic carbon source. We used such two-carbon-source media to isolate a tpi deletion (Δtpi) mutant. The Δtpi mutant did not survive with single carbon substrates but grew like wild-type (WT) M. tuberculosis in the presence of both a glycolytic and a gluconeogenic carbon source. 13C metabolite tracing revealed the accumulation of TPI substrates in Δtpi and the absence of alternative triosephosphate isomerases and metabolic bypass reactions, which confirmed the requirement of TPI for glycolysis and gluconeogenesis in M. tuberculosis. The Δtpi strain was furthermore severely attenuated in the mouse model of tuberculosis, suggesting that M. tuberculosis cannot simultaneously access sufficient quantities of glycolytic and gluconeogenic carbon substrates to establish infection in mice.
IMPORTANCE
The importance of central carbon metabolism for the pathogenesis of M. tuberculosis has recently been recognized, but the consequences of depleting specific metabolic enzymes remain to be identified for many enzymes. We investigated triosephosphate isomerase (TPI) because it is central to both glycolysis and gluconeogenesis and had been predicted to be essential for growth of M. tuberculosis. This work identified metabolic conditions that make TPI dispensable for M. tuberculosis growth in culture and proved that M. tuberculosis relies on a single TPI enzyme and has no metabolic bypass for the TPI-dependent interconversion of dihydroxyacetone phosphate and glyceraldehyde-3-phosphate in glycolysis and gluconeogenesis. Finally, we demonstrate that TPI is essential for growth of the pathogen in mouse lungs.
INTRODUCTION
Mycobacterium tuberculosis has evolved to replicate and survive within its only natural host and reservoir, the human body (1). This includes adaptation of its metabolism to sustain energy and biomass production during acute and chronic infections. As a consequence, M. tuberculosis is metabolically flexible and able to cocatabolize multiple carbon substrates in a compartmentalized manner (2). Thus, when given both a glycolytic carbon substrate such as glucose and a gluconeogenic substrate such as acetate, it metabolizes each carbon source simultaneously to its distinct metabolic fate through glycolysis and gluconeogenesis. In contrast, bacteria such as Escherichia coli and Bacillus subtilis exhibit catabolite repression and utilize multiple carbon substrates sequentially (3, 4). The ability of M. tuberculosis to efficiently cocatabolize multiple carbon substrates likely supports its pathogenic lifestyle, because the macrophage phagosome, where the bacilli largely reside within an infected host, is nutritionally restricted (5, 6).
Multiple lines of evidence point toward lipids and fatty acids as dominant carbon sources for growth and persistence of M. tuberculosis during infections (7–10). A role for carbohydrates was illustrated by the requirement of glucose phosphorylation during persistence for M. tuberculosis in chronic mouse infections (11). However, the mechanism by which phosphorylated glucose supports the pathogen’s survival remains unclear. M. tuberculosis lacking detectable phosphofructokinase activity replicated and persisted normally in mice, suggesting that energy metabolism through glycolysis may not be the major function of carbohydrates (12). Rather, glycolytic enzymes may contribute to metabolic homeostasis by rerouting metabolic intermediates to biosynthetic pathways such as the pentose phosphate pathway or/and alleviating the effect of toxic phosphorylated metabolites. For example, phosphofructokinase deficiency resulted in loss of viability of hypoxic, nonreplicating M. tuberculosis that was associated with accumulation of glucose-6-phosphate (12), while under aerobic conditions, accumulation of glucose-6-phosphate has been implicated as a source of reducing power protecting Mycobacterium smegmatis against oxidative stress (13).
Triosephosphate isomerase (TPI) catalyzes the interconversion of dihydroxyacetone phosphate (DHAP) and glyceraldehyde-3-phosphate (G3P), which is one of the seven reversible enzymatic reactions participating in both glycolysis and gluconeogenesis. The TPI-catalyzed reaction constitutes a convergence point in central carbon metabolism. It branches glycolysis, gluconeogenesis, the pentose phosphate pathway, and the entry point of glycerol. During glycolysis, TPI activity is required to channel triose phosphates produced by fructose bisphosphate aldolase to pyruvate. During gluconeogenesis, TPI provides the two substrates required for the aldolase to generate fructose-1,6-bisphosphate, which is further converted through phosphatase and isomerase activities to fructose-6-phosphate and glucose-6-phosphate, important biosynthetic precursors for cell wall components and nucleic acids.
TPIs are homodimeric enzymes, with each monomer consisting of a single 8-stranded α/β barrel domain (14) and represent some of the most efficient enzymes in nature (15, 16). In E. coli, TPI deficiency can be compensated for by activation of a normally inactive metabolic bypass reaction (17) or overexpression of a promiscuous isomerase (18), while Klebsiella pneumoniae and Sinorhizobium meliloti express two TPI enzymes (19, 20).
The M. tuberculosis genome contains a single tpi gene (rv1438), predicted to be required for optimal growth on agar plates (21–24). TPI from M. tuberculosis has been biochemically and structurally characterized (25, 26). It shares high structural similarity with other bacterial TPIs and exhibits a very high specific activity and a preference for the G3P-to-DHAP reaction over the reverse.
We investigated the role of TPI in central carbon metabolism of M. tuberculosis and confirmed that it is indeed required for glycolysis and gluconeogenesis. We furthermore demonstrated that essentiality of TPI for growth of M. tuberculosis is conditional; the enzyme can be inactivated completely without affecting growth as long as a mixture of glycolytic and gluconeogenic carbon substrates is provided. Metabolomic analyses confirmed that TPI is the only enzyme with triosephosphate isomerase activity in M. tuberculosis and ruled out a metabolic bypass reaction. Mouse infection experiments indicate that M. tuberculosis does not have access to a combination of glycolytic and gluconeogenic carbon substrates sufficient to establish infection.
RESULTS
tpi is required for growth of M. tuberculosis on standard agar plates.
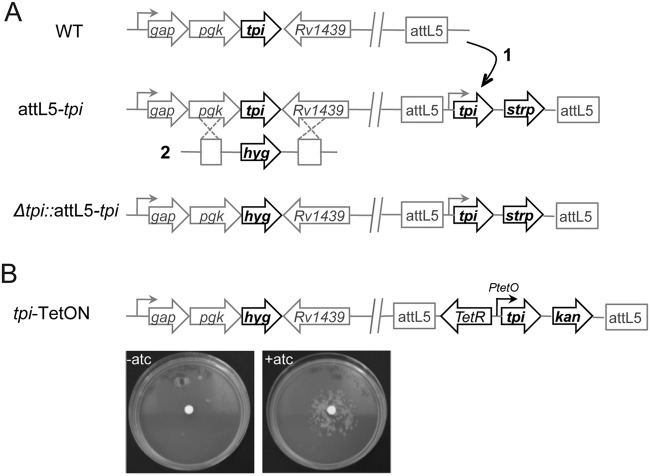
Genome-wide transposon mutagenesis studies predicted tpi to be required for optimal growth of M. tuberculosis on agar plates (22–24). We therefore first generated a conditional tpi knockdown mutant (tpi-TetON) to investigate the role of tpi in central carbon metabolism. We cloned tpi and generated an M. tuberculosis strain that is merodiploid for tpi (attL5-tpi). We then replaced the native copy of tpi by homologous recombination with a hygromycin resistance cassette, which resulted in a strain (the Δtpi::attL5-tpi mutant) that contains a single, constitutively expressed copy of tpi in the L5 mycobacteriophage attachment (attL5) site (Fig. 1A). Deletion of the wild-type copy of tpi in the Δtpi::attL5-tpi mutant and integration of plasmids into the attL5 site were confirmed by Southern blotting (see Fig. S1 in the supplemental material). Next, we utilized the Δtpi::attL5-tpi strain to generate a mutant, the tpi-TetON mutant, in which transcription of tpi could be repressed by the tetracycline repressor (TetR) (27, 28). As expected, the tpi-TetON mutant grew on agar plates only in the presence of the inducer anhydrotetracycline (ATC) (Fig. 1B). We also attempted to generate a tpi deletion mutant but failed to isolate such mutants, which confirmed the essentiality of tpi for growth on standard agar plates containing glucose, oleic acid, and glycerol.
FIG 1 .
Construction of M. tuberculosis tpi knockdown and conditional mutants. (A) Diagrams illustrate genomic regions of the indicated strains and required steps for the construction of tpi mutants: (1) integration of a plasmid containing a second copy of tpi driven by the predictive native promoter into the L5 mycobacteriophage attachment (attL5) site; (2) deletion of the native tpi copy by homologous recombination generating the Δtpi::attL5-tpi strain. (B) Replacement of the tpi attL5 site copy generated the tpi-TetON mutant, in which tpi transcription is induced by anhydrotetracycline (ATC) delivered on a filter in the center of the plate.
Generation of M. tuberculosis Δtpi.
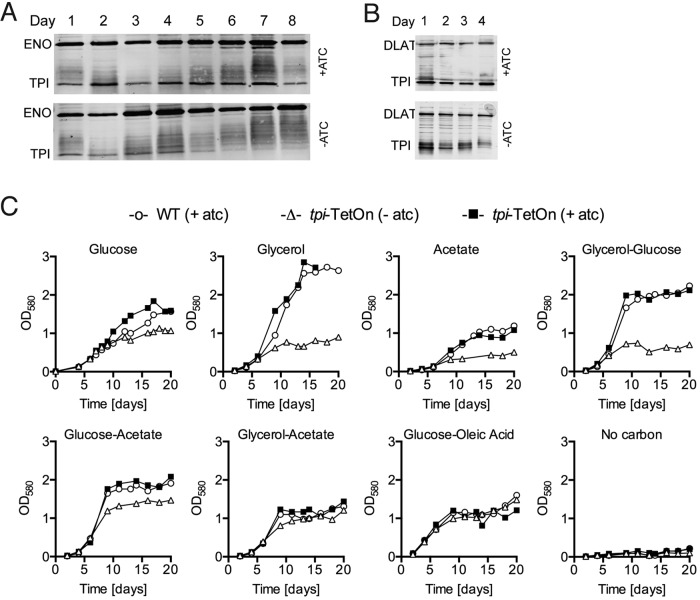
M. tuberculosis does not employ classical glucose-dependent catabolite repression to regulate carbon utilization and is capable of consuming more than one carbon source simultaneously. We therefore analyzed the impact of tpi silencing on TPI protein abundance and growth of M. tuberculosis in both single- and dual-carbon-source media. In media that contained glucose and oleic acid or glycerol and glucose but lacked ATC, levels of TPI protein decreased over time, and TPI was below the limit of detection after four to five days (Fig. 2A and B). TPI depletion impaired replication of M. tuberculosis with single carbon substrates (glucose, glycerol, and acetate) and the combination of glycerol and glucose (Fig. 2C). However, tpi silencing did not significantly affect growth in media containing combinations of glucose and acetate, glycerol and acetate, or glucose and oleic acid. To determine whether tpi silencing and TPI depletion indeed reduced the overall triosephosphate isomerase activity, we measured enzyme activity in cell lysates. Compared to wild-type (WT) M. tuberculosis, TPI enzymatic activity in lysates of a tpi-TetON strain grown in the presence of ATC was 3-fold reduced and 18-fold reduced in lysates from silenced tpi-TetON cultures (Table 1). Thus, tpi silencing significantly reduced but did not abolish TPI activity.
FIG 2 .
Impact of tpi silencing on growth of M. tuberculosis in culture. Immunoblot analysis of TPI (27 kDa) levels in the tpi-TetON mutant growing in 7H9 base medium with 0.1% glucose and 0.06% oleic acid (A) or 0.1% glucose and 0.1% glycerol (B) in the presence or absence of 200 ng/ml ATC. Samples were collected on the indicated days. Enolase (ENO; 45 kDa) and dihydrolipoamide acyltransferase (DLAT; 57 kDa) served as loading controls. (C) Growth of WT and the tpi-TetON mutant in carbon-defined 7H9 base medium in the presence or absence of ATC. Carbon sources were supplied at 0.2% (wt/vol) as single substrates or 0.1% (wt/vol) in media with two substrates, except oleic acid, which was added at 0.06%. ATC (200 ng/ml) was added at the beginning of the assay and replenished every week. Data are representative of at least two independent experiments.
TABLE 1 .
Triosephosphate isomerase activity in protein extracts of indicated M. tuberculosis strains
| Strain genotype | Activity (nmol min−1 mg−1)a |
|---|---|
| WT | 528.1 ± 101.8 |
| tpi-TetON + ATC | 179.1 ± 2.4 |
| tpi-TetOn − ATC | 29.3 ± 6.5 |
| Δtpi | ND |
| tpi-Comp | 149.2 ± 9.1 |
ND, not detected.
We sought to understand if the residual TPI activity was sufficient for normal growth of M. tuberculosis in the presence of a glycolytic and gluconeogenic carbon source or if TPI is truly dispensable for growth in these medium conditions. To address this, we again attempted to delete tpi but this time utilized agar plates containing carbon source combinations that were permissive for growth of the tpi-TetON mutant in liquid culture and isolated the Δtpi mutant from agar plates that contained glucose and oleic acid as sole carbon substrates. Deletion of tpi was confirmed by Southern blot analysis (see Fig. S1 in the supplemental material). We also confirmed loss of TPI by immunoblotting (see Fig. S2 in the supplemental material) and complete loss of triosephosphate isomerase activity in Δtpi cell lysates (Table 1). In the complemented mutant (tpi-Comp), which contains tpi transcribed from its putative native promoter, TPI expression and activity were partially restored.
TPI is required for growth and survival of M. tuberculosis with single carbon sources.
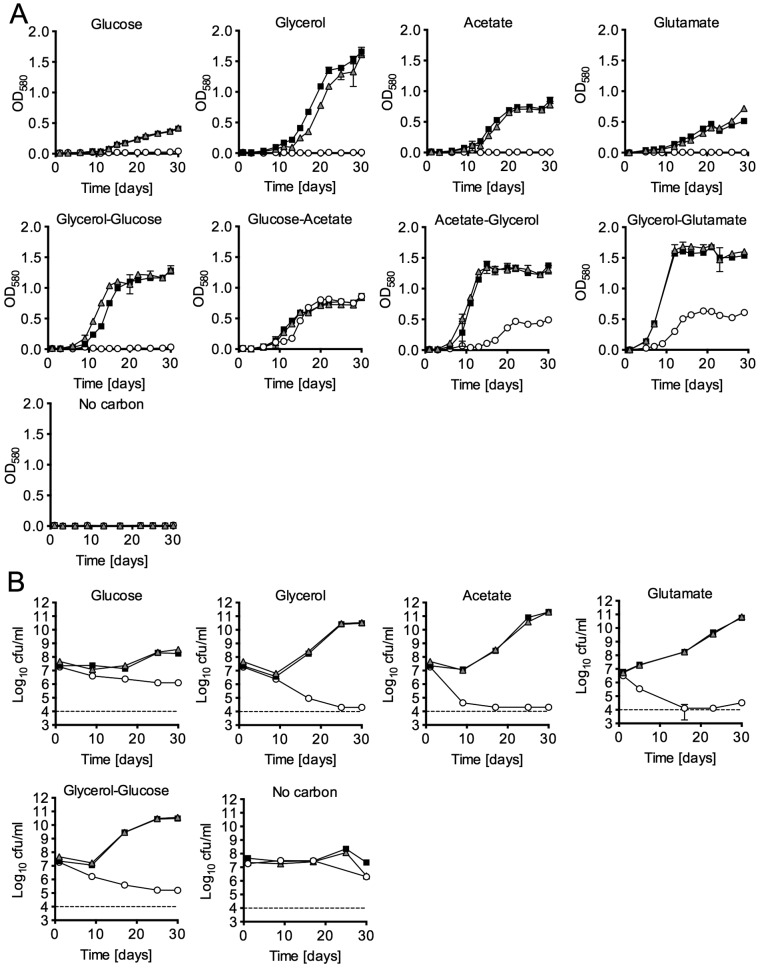
The growth curves of the tpi-TetON mutant suggested that TPI is dispensable for replication in the presence of a glycolytic and a gluconeogenic carbon source yet required if only either one is provided. To confirm this, we followed growth and survival of WT, Δtpi, and tpi-Comp strains in carbon-defined media. Deletion of tpi eliminated growth of M. tuberculosis with single carbon sources, including glucose, acetate, glycerol, and glutamate (Fig. 3A). Longer-chain fatty acids, such as propionate, butyrate, valerate, and hexanoic acid, also failed to support growth of the Δtpi strain (data not shown). Culture in single carbon sources decreased survival of the bacilli, albeit with different death rates (Fig. 3B). In media with acetate as the sole carbon source, the Δtpi strain died most rapidly, and CFU were reduced by almost 3 orders of magnitude in 9 days. In media with glycerol, the mutant’s CFU declined more slowly, and in glucose there was only about a 10-fold reduction over a period of 20 days. In the absence of a carbon source, the Δtpi mutant’s survival was similar to that of the WT, suggesting that killing in the presence of carbon sources did not result from starvation but might be due to a state of metabolic imbalance.
FIG 3 .
TPI is required for growth and survival of M. tuberculosis with single carbon sources. Growth and survival followed by absorbance measurement (A) and CFU assay (B) of WT (squares), Δtpi (circles), and tpi-Comp (triangles) in carbon-defined Sauton’s base medium containing the indicated carbon sources (0.4% glucose, 0.2% glycerol, 0.2% acetate, 0.2% glutamate, or a mix of 0.1% of each of the indicated two carbon sources). Dashed lines represent the limit of detection of the CFU determinations. Data are means ± standard deviations (SD) for triplicate cultures (error bars in panel B are smaller than the symbols). Data are representative of at least two independent experiments except for the growth curves with glutamate, which were performed once.
As predicted by the data generated with the tpi-TetON mutant, the Δtpi strain replicated in media containing glucose and acetate, with a rate indistinguishable from that of the WT. In contrast, when provided with glycerol and acetate, the Δtpi strain grew much more slowly than the WT and stopped replicating at a reduced final biomass. The Δtpi strain died in media containing glycerol and glucose, both carbon sources that enter central carbon metabolism above the TPI-catalyzed step. Although complementation with an integrative plasmid containing tpi expressed from its putative promoter did not result in WT TPI levels and activity (see Fig. S2 in the supplemental material and Table 1), it was sufficient to restore growth and survival similar to those of the WT under all conditions tested.
In summary, these data suggest that tpi is essential for metabolism of glucose through glycolysis and that of glycerol and acetate through gluconeogenesis. However, the metabolic block in the Δtpi strain can be overcome by supplying metabolites that feed central carbon metabolism simultaneously above and below the TPI-catalyzed step. The in vitro essentiality of TPI is thus defined by its metabolic products.
Absence of TPI blocks glycolytic and gluconeogenic carbon flow.
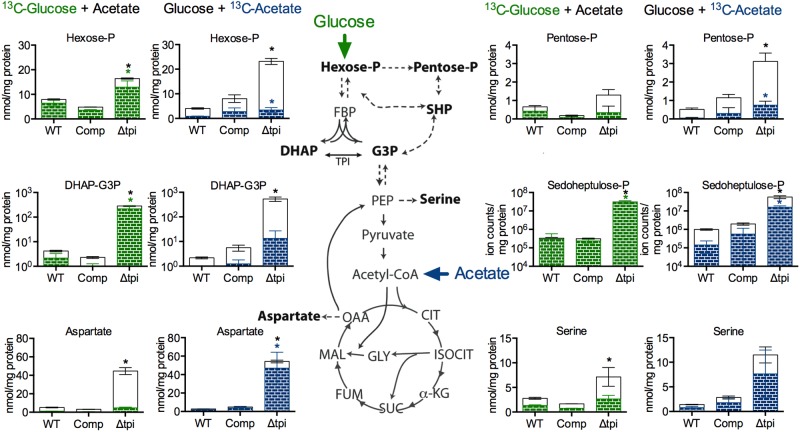
To better understand the metabolic consequences of loss of TPI, we traced the specific metabolic fates of uniformly (U) 13C-labeled glucose and acetate through glycolysis and gluconeogenesis, the pentose phosphate (PP) pathway, and the tricarboxylic acid (TCA) cycle. In the Δtpi strain grown on glucose and acetate, the triose phosphate pool consisting of DHAP and G3P increased 100-fold compared to those in the WT and the complemented mutant, which is consistent with the absence of triosephosphate isomerase activity (Fig. 4). The accumulating triose phosphates were almost fully labeled when the Δtpi strain was cultured in [U13-C]glucose, but label incorporation from [U-13C]acetate was drastically reduced, indicating that the accumulating DHAP/G3P was mostly derived from glucose.
FIG 4 .
Impact of tpi deletion on glycolytic and gluconeogenic carbon flow. Intrabacterial pool sizes and isotopic labeling of metabolites in the indicated M. tuberculosis strains were determined after a 24-h incubation on media containing [U-13C]glucose and acetate or glucose and [U-13C]acetate. Total bar heights indicate the intrabacterial concentration, whereas the colored area of each bar denotes the extent of 13C labeling achieved from [U-13C]glucose (green) or [U-13C]acetate (blue). All values are averages ± SD of measurements from independent triplicate cultures. *, P < 0.05 (Student’s t test, compared to the corresponding WT pool). Data are representative of two independent experiments. α-KG, α-ketoglutarate; CIT, citrate; DHAP, dihydroxyacetone phosphate; FBP, fructose bisphosphatase; FUM, fumarate; G3P, glyceraldehyde 3-phosphate; GLY, glyoxylate; MAL, malate; OAA, oxaloacetate, PEP, phosphoenolpyruvate; SHP, sedoheptulose phosphate; SUC, succinate.
The accumulation of metabolites was not limited to TPI substrates but was also observed for other metabolites upstream and downstream of the isomerase step, including the pools of hexose phosphate (hexose-P), pentose-P, and sedoheptulose-P, as well as serine and aspartate, which were used as indicators of the abundance of phosphoenolpyruvate (PEP) and oxaloacetate, respectively. Sedoheptulose-P pools increased 50- to 100-fold in the Δtpi strain consistent with increased flux through the pentose phosphate (PP) pathway. In Saccharomyces cerevisiae, inhibition of TPI has been associated with PP pathway activation and increased resistance to oxidants (29); however, the Δtpi strain was not more resistant to toxic concentrations of hydrogen peroxide than the WT and the complemented strain (see Fig. S3 in the supplemental material), suggesting that in M. tuberculosis, increased PP pathway metabolite pools are not associated with increased resistance to oxidative stress.
In contrast to the WT-like growth we observed for the Δtpi strain with glucose and acetate, the mutant grew poorly with glycerol and acetate. Upon uptake, glycerol is phosphorylated and directly converted to DHAP. DHAP can spontaneously and enzymatically produce methylglyoxal (MG) (30, 31), which as an electrophile can covalently react with the nucleophilic centers of both proteins and nucleic acids, some of which may be essential for cell growth and viability (32–35). We therefore examined levels of MG in the WT and Δtpi strains. MG concentrations in the Δtpi strain were 6-fold higher than those in the WT when the bacteria were grown in media containing both glycerol and acetate but not glucose and acetate (see Fig. S4 in the supplemental material). However, while this intracellular accumulation of MG was associated with a growth defect in glycerol- and acetate-containing medium (Fig. 3A), we observed no evidence of the accompanying formation of the predicted MG adducts, carboxyethyllysine, argpyrimidine, and carboxyethylcysteine, leaving the mechanistic significance of MG accumulation experimentally unresolved.
To confirm that tpi encodes the only triosephosphate isomerase and to identify which of the two metabolites, DHAP or G3P, is responsible for the increased triose-P pool, we determined the isotopomeric distribution of 13C-labeled triose-P and hexose-P in the Δtpi strain following growth with a combination of glycerol and acetate, with either carbon source being 13C labeled (Fig. 5). The triose-P pools were 100- to 150-fold increased in the Δtpi strain, as previously observed after growth on glucose and acetate, while there was no significant difference in hexose-P pool sizes between the strains (Fig. 5A). Deletion of TPI is predicted to change the distribution of 13C-labeled isotopologues of hexose-P when the cells are metabolizing [U-13C]glycerol (Fig. 5B). Consistent with this prediction, we observed a shift from the dominant 13C6-labeled hexose-P isotopologue in WT and complemented mutant to the 13C3 isotopologue in the Δtpi strain when the cells were given [U-13C]glycerol and unlabeled acetate (Fig. 5C). As expected, there were no changes in hexose-P isotopologue distribution between the strains when acetate was labeled and glycerol was unlabeled. While triose-P was fully labeled from [U-13C]glycerol, there was no significant label incorporation from [U-13C]acetate, indicating that the triose-P pool in the Δtpi strain consists predominantly of DHAP. Thus, formation of G3P from acetate was less efficient than production of DHAP from glycerol, resulting in a triose-P pool that is skewed toward DHAP when the bacteria are grown in the glycerol-and-acetate medium.
FIG 5 .

TPI is the only triosephosphate isomerase in M. tuberculosis. (A) Intrabacterial pool sizes and isotopic labeling of triose phosphates and hexose phosphate in the indicated M. tuberculosis strains after a 24-h incubation on media containing [U-13C]glycerol and acetate or glycerol and [U-13C]acetate. Total bar heights indicate the intrabacterial concentration, whereas the colored area of each bar denotes the extent of 13C labeling achieved from [U-13C]glycerol (purple) or [U-13C]acetate (blue). All values are averages ± SD from independent triplicate cultures. *, P < 0.05 Student’s t test. Data are representative of two independent experiments. (B) Schematic representation of the predicted isotopomeric distribution of 13C-labeled hexose phosphate and 13C-labeled triose phosphate in the WT and Δtpi strains after growth on [U-13C]glycerol- and acetate-containing media. 13C-labeled carbons are in pink. (C) Isotopomeric profile of triose phosphate and hexose phosphate of the samples used for panel A. m+0, unlabeled; m+1, singly 13C labeled; m+2, doubly 13C labeled, and so on. Differences in abundance between the m+6 isotopomer and all other isotopomers in WT were statistically significant (P < 0.05) except for the difference in abundance between the m+6 and m+0 isotopomers (P = 0.06). Differences in abundance of the m+3 isotopomer and all other detectable isotopomers in the Δtpi strain were statistically significant (P < 0.0001).
Together, these data establish TPI as the only enzyme with triosephosphate isomerase activity in M. tuberculosis and demonstrate that there is no metabolic bypass of the TPI-catalyzed step in glycolysis and gluconeogenesis.
TPI is required to establish infection in mice.
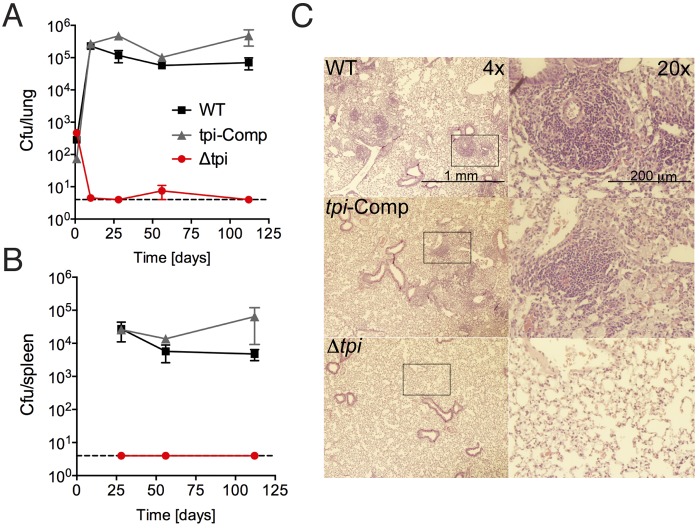
Carbohydrates and fatty acids have both been demonstrated to be important in M. tuberculosis mouse infections; however, growth in vivo seems to depend on fatty acid metabolism (8–11, 36). M. tuberculosis lacking TPI replicated normally in media with glucose and acetate (Fig. 3), and we reasoned that the mutant would replicate in macrophages and in mice if a glycolytic carbon source together with a gluconeogenic carbon source was available to the bacilli in sufficient quantities to support growth. In bone marrow-derived mouse macrophages, the Δtpi strain failed to replicate (Fig. 6), suggesting that the bacilli do not have access to a growth-permissive combination of glycolytic and gluconeogenic carbon substrates. In mice infected by aerosol, the Δtpi strain was rapidly eliminated from the lungs and unable to colonize the spleen (Fig. 7A and B). In all except one animal, both organs remained culture negative (limit of detection, 4 CFU/organ) throughout the experiment (112 days). Mice infected with the Δtpi strain did not show any lung pathology (Fig. 7C). In contrast, infection with the complemented mutant resulted in growth in lungs and spleens as well as lung pathology similar to that observed in WT-infected mice despite the reduced TPI levels detected in this strain (see Fig. S2 in the supplemental material). These results revealed that M. tuberculosis requires TPI to replicate in vivo and successfully establish an infection.
FIG 6 .
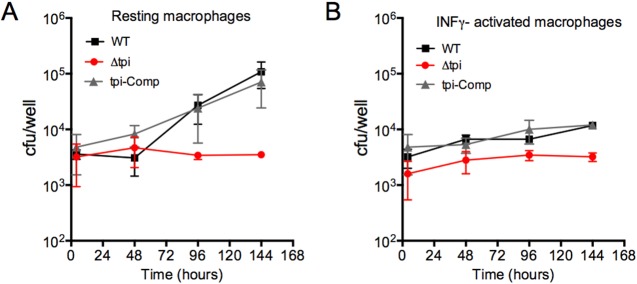
TPI is required for replication in macrophages. Bacterial loads in resting (A) and gamma interferon (IFN-γ)-activated (B) bone marrow-derived macrophages infected with the WT, Δtpi, and tpi-Comp strains were determined. Data are means ± SD of triplicate cultures and are representative of 2 independent experiments.
FIG 7 .
TPI is required to establish infection in mice. Bacterial titers in lungs (A) and spleens (B) of C57BL/6 mice infected with the indicated strains were determined. The dashed line represents the limit of detection (4 CFU/organ). After day 1, no CFU were detected in the lungs or spleens from mice infected with the Δtpi strain except in the lung of one animal at day 56, which contained 18 CFU. Data are means ± SD from 4 mice per group and time point and are representative of two independent experiments. (C) Lung sections stained with hematoxylin and eosin from mice infected with the indicated strains at day 56 postinfection. The boxed areas in the left panels (magnification, ×4) are shown in the right panels (magnification, ×20).
DISCUSSION
The work described here highlights how a conditional tpi-TetON mutant helped identify conditions in which TPI is dispensable for growth of M. tuberculosis and provided genetic evidence for the capability of M. tuberculosis to cocatabolize two carbon substrates that feed opposite pathways in central carbon metabolism. TPI catalyzes an essential reaction in central carbon metabolism, and reports of bacterial tpi deletion mutants are scarce. They are limited to enteric bacteria, including E. coli, Klebsiella pneumoniae, and Salmonella enterica serovar Typhimurium, as well as the symbiotic soil bacterium Sinorhizobium meliloti (19, 20, 37, 38). Deletion of tpi in E. coli and in S. Typhimurium yielded viable bacteria on complex media containing multiple carbon substrates. Growth of these mutants with single carbon sources was only observed with those that bypass the TPI reaction, for example gluconate, which can be metabolized via the pentose phosphate (PP) or the Entner-Doudoroff (ED) pathway (38, 39). E. coli tpi mutants have also been shown to evolve to grow on glucose by routing carbon through the normally latent methylglyoxal bypass (17). K. pneumoniae contains two tpi genes, and only one of these could be successfully deleted (19). Sinorhizobium meliloti also expresses two TPI enzymes, and deletion of both prevented growth with gluconeogenic carbon sources, indicating that TPI is essential for gluconeogenesis (20). However, the double mutant replicated with glucose as the sole carbon source, presumably because in rhizobia, glucose is degraded primarily through the ED pathway (40–42).
M. tuberculosis lacks the ED pathway, and its genome contains a single tpi gene (21). TPI is central to glycolysis and gluconeogenesis, but in M. tuberculosis, which is capable of cocatabolizing multiple carbon substrates (2, 43, 44), the availability of a glycolytic and gluconeogenic carbon source should have made TPI dispensable. However, agar plates containing glucose, oleic acid, and glycerol were not permissive for growth of M. tuberculosis lacking TPI, as previously suggested by genome-wide transposon mutagenesis studies (22–24). The data presented here underscore the fact that gene essentiality is condition dependent and that essentiality predictions from genome wide transposon mutagenesis studies are contingent on the growth conditions used in the screen.
With the help of a conditional TPI mutant, we found that the presence of glycerol prevented growth on agar plates in the absence of TPI, possibly due to the intracellular accumulation of methylglyoxal (MG). Similarly, in liquid culture, the Δtpi strain displayed a significant growth defect in medium containing glycerol and acetate or glycerol and glutamate, while its growth was indistinguishable from that of the WT in the presence of glucose and acetate (Fig. 3). Most microorganisms possess a route for the detoxification of MG (31–33). M. tuberculosis might use one of two alternative pathways depending on an MG glyoxylase system or an MG reductase (34). Both pathways lead to the formation of pyruvate. However, we did not detect significant incorporation of 13C label derived from glycerol into pyruvate in the Δtpi strain (data not shown). MG reacts quickly with DNA and proteins, resulting in products that are difficult to quantify but likely also contribute to its toxicity. Thus, MG accumulation may have contributed to glycerol-mediated toxicity in the Δtpi strain, but this is challenging to prove experimentally, as we lack direct evidence of MG-derived adducts.
Carbon-tracing analysis of [U13-C]glucose and [U13-C]acetate demonstrated a metabolic block in the Δtpi strain characterized by the accumulation of the triose phosphates DHAP and G3P with predominant label incorporation from glucose, while accumulating metabolites downstream of the TPI-catalyzed reaction such as aspartate and malate (data not shown) contained acetate-derived label. Pentose-P and sedoheptulose-P also accumulated in the Δtpi strain, indicating that the PP pathway does not serve as a bypass for TPI in M. tuberculosis. The isotopomeric distribution of 13C-labeled triose-P and hexose-P in the Δtpi strain fed [U13-C]glycerol or [U13-C]acetate confirmed the lack of DHAP and G3P isomerization and established that the accumulating triose-P pool in the Δtpi strain consists predominantly of DHAP. Together, these metabolomic analyses demonstrate that M. tuberculosis contains only a single TPI enzyme that is required for both glycolytic and gluconeogenic carbon flow.
Deletion of tpi impaired replication of S. Typhimurium in mouse spleen and liver but did not fully attenuate S. Typhimurium, indicating that TPI-independent metabolic routes, including the PP and ED pathways, are used to metabolize in vivo available carbon substrates (38). In contrast, M. tuberculosis lacking TPI failed to replicate in mouse lungs, and the bacteria were rapidly killed (Fig. 7). This suggests that M. tuberculosis does not have access to sufficient quantities of a growth-permissive combination of glycolytic and gluconeogenic carbon sources during early stages of infection in mice. In culture, the Δtpi strain died in glycerol-containing media as well as in media containing a fatty acid as the sole carbon source. Glycerol is unlikely to be a dominant exogenous carbon source in vivo, because M. tuberculosis lacking glycerol kinase, while unable to utilize glycerol, was fully virulent in mice (34). The rapid death of the Δtpi strain in mouse lungs is thus consistent with previous suggestions that the host environment may consist of an abundance of fatty acids and a lack of available carbohydrates (8–11, 36). A number of metabolic enzymes have moonlighting activities, which are involved in virulence, including TPI from Staphylococcus aureus, which serves as an adhesin for the fungus Cryptococcus neoformans (45). The extent to which such secondary functions may also be true of and contribute to the impact of TPI deletion on virulence of M. tuberculosis remains to be determined.
Does TPI represent a potential drug target in M. tuberculosis? While it is essential for replication and establishment of infection in mice, several observations argue that TPI might not be an ideal target for tuberculosis chemotherapy. TPI is a highly conserved enzyme, and TPI from M. tuberculosis shares strong structural similarity with the human enzyme (26). TPI from parasites such as Plasmodium, Trypanosoma, and Giardia have been targets for rational drug design talking advantage of a cysteine residue that is required for stability of the TPI dimer but is replaced by a methionine in the human enzyme (46–48). However, like human TPI, the mycobacterial enzyme contains a methionine and not a cysteine in that position (25). A series of docking procedures identified additional residues in the dimer interface of TPI from Trypanosoma cruzi as sites for inhibiting the enzyme and provided potential for additional strategies of targeting TPI (49). Our data suggest that inhibition of TPI in M. tuberculosis will have to be substantial in order to abolish growth. Reducing TPI activity in the tpi-TetON mutant by 95% impaired but did not prevent replication (Table 1 and Fig. 2). Moreover, the tpi-TetON mutant replicated like the WT in mice even in the absence of inducer (data not shown), indicating that approximately 5% TPI activity is sufficient for growth of M. tuberculosis in mouse lungs. This also prevented us from investigating whether TPI is required to maintain a chronic infection in mice. Notwithstanding, the failure of the Δtpi strain to establish infection in mice and its rapid loss of viability support previous evidence that targeting metabolic enzymes holds promise for the development of inhibitors of vulnerable enzymes in M. tuberculosis central carbon metabolism.
MATERIALS AND METHODS
Strains, media, and growth conditions.
M. tuberculosis cultures were grown aerated in the presence of 5% CO2 at 37°C in Middlebrook 7H9 supplemented with 5% bovine albumin, 2% dextrose, 0.5% glycerol, 0.85% sodium chloride, and 0.05% Tween 80, except for the Δtpi strain, which was grown in 7H9 broth supplemented with 10% BBL, Middlebrook OADC enrichment (Becton, Dickinson), and 0.05% Tween 80. To measure growth and survival in carbon-defined media, we used either 7H9 broth supplemented with 5% bovine albumin, 0.85% sodium chloride, and 0.02% tyloxapol or a modified Sauton’s base medium (50) in which asparagine was replaced by 0.5 g/liter ammonium sulfate as the nitrogen source and 0.02% tyloxapol. Carbon substrates were used at 0.2% (wt/vol) unless otherwise indicated. Hygromycin B (50 µg/ml), kanamycin (20 µg/ml), and streptomycin (20 µg/ml) were included when selection was required. Anhydrotetracycline (Sigma) was used at 200 ng/ml. For CFU determinations, all strains were cultured on 7H9 base agar media (Difco 7H9 broth, 0.2% glycerol, 10% BBL, Middlebrook OADC enrichment, 1.5% Bacto agar), which supports growth of the Δtpi strain. For metabolite analysis, 5 × 108 bacteria from mid-log-phase cultures were seeded on nitrocellulose filters on top of 7H9 base agar with the carbon substrates as specified in Figs. 4 and 5 for 5 days and then transferred to freshly made 7H9 base agar plates with 0.2% (wt/vol) of U-13C-labeled carbon substrates. Metabolite extraction was done as previously described (2, 10).
Mutant construction.
Deletion of tpi in the merodiploid strain was achieved by allelic exchange using the specialized transducing phage phAE87 as previously described (10). Replacement transformations of the attL5 inserts were used to generate the tpi-TetON and Δtpi mutants. All attL5-integrating plasmids were generated using Gateway Cloning Technology (Invitrogen). The tpi gene and putative tpi promoter were amplified by PCR. Primer sequences are available upon request. The tpi-TetOn strain was generated using the Pmyc1 tetO promoter (27) and WT TetR (28). The complemented mutant expressed tpi from its putative promoter on a plasmid integrated in the attL5 site.
Immunoblot and TPI activity assay.
Protein extracts were prepared from bacterial pellets from 50-ml cultures in late exponential growth phase in the media as specified in the legend of Fig. 2. Briefly, cultures were washed with phosphate-buffered saline (PBS), 0.05% Tween 80 and resuspended in 1 ml 50 mM HEPES buffer (pH 7.4), 1× protease inhibitor cocktail (Roche). Cells were lysed by bead beating three times at 4,500 rpm for 30 s with 0.1-mm zirconia/silica beads. Beads and cell walls were removed through centrifugation (11,000 × g, 10 min, 4°C), and the supernatant was filtered through a 0.22 µm SpinX column (Corning). For immunoblots, 20- to 30-µg protein extracts were separated by SDS-PAGE, transferred to nitrocellulose membranes, and probed with rabbit antisera to TPI, enolase (ENO), or dihydrolipoamide acyltransferase (DLAT) (1:1,000 dilution in PBS, 0.05% Tween 20) (generated by Covance). As a secondary antibody, IRDye 800CW donkey anti-rabbit IgG(H+L) (heavy plus light chain) (LI-COR) was used. Proteins were detected using the Odyssey infrared imaging system (LI-COR Biosciences).
TPI activity was measured using G3P as the substrate in the presence of NADH (β-NADH; extinction coefficient at 340 nm = 6.2 mM−1 cm−1) in a coupled enzyme assay adapted from the work of Gracy (51). The reaction was initiated by addition of 0.2 mM G3P to a reaction mixture containing 20 mM HEPES (pH 7.4), 5 U α-glycerol 3-phosphate dehydrogenase (EC 1.1.1.8.), 0.1 mM β-NADH, and 25 to 100 µl of protein extract (50 µg total protein). Production of NAD+ was monitored at 340 nm in an Uvikon XL spectrophotometer. Protein concentrations were determined using the Bio-Rad DC protein assay following the manufacturer instructions.
Metabolomics using liquid chromatography-mass spectrometry (LC-MS).
M. tuberculosis metabolites were separated in an Agilent Accurate-Mass 6220 time-of-flight (TOF) mass spectrometer coupled to an Agilent 1200 liquid chromatography system using a Cogent Diamond hydride type C column (Microsolve Technologies) using solvents and configuration as described previously (52). Metabolite concentrations were normalized to bacterial biomasses of individual samples determined by measuring residual protein content (BCA protein assay kit; Pierce). Isotopomer data analysis was performed as previously described (52).
Determination of methylglyoxal levels.
MG levels were determined as described by Randell et al. (53) with some modifications. Briefly, M. tuberculosis was grown for 7 days on filters laid on 7H9 agar media containing the indicated carbon substrates in Fig. S4. At the time of harvest, filters were transferred to a 35-mm-diameter petri dish, and bacteria were metabolically quenched and harvested by the addition of precooled acetonitrilie-methanol-H2O (2:2:1) supplemented with 1% formic acid, 2.5 µM 2,3-hexanedione, and 125 µM O-phenylenediamine. Bacteria were immediately broken by bead beating and clarified by centrifugation. Metabolite samples were derivatized by incubation for 18 to 20 h at 4°C. The O-phenylenediamine derivatization products of methylglyoxal and 2,3-hexanedione, i.e., 2-methylquinoxaline and 1-methyl-2-propylquinoxaline, respectively, were analyzed by LC-MS using the standard method as described above. Derivatized metabolite identities were established by comparing with metabolite standards used to spike biological samples and derivatized with O-phenylenediamine as described for the samples.
Mouse and macrophage infections.
Female C57BL/6 mice (Jackson Laboratory) were infected by aerosol using an inhalation exposure system (Glas-Col) and early-log-phase M. tuberculosis cultures as single-cell suspensions in PBS to deliver 100 to 200 bacilli per mouse. At the indicated time points in Fig. 7, serial dilutions of lung and spleen homogenates were cultured on 7H9 agar base media (as described above) to determine CFU. The left lobe of each lung was fixed in 10% buffered formalin, further processed for histopathology, and stained with hematoxylin and eosin. We isolated and infected bone marrow-derived mouse macrophages as previously described (54). All procedures involving animals were reviewed and approved by the Institutional Animal Care and Use Committee of Weill Cornell Medical College.
SUPPLEMENTAL MATERIAL
Southern blot analysis of mutants generated in this study. Map of the tpi genomic region in WT M. tuberculosis (A) and tpi mutant (B) indicating probe location, PstI restriction sites, and hybridization product fragment size (gray bar). (C) Southern blot of PstI-digested genomic DNA of the WT (lane 1), Δtpi (lane 2), and tpi-TetON (lane 3) strains. (D to F) Map of the attL5 genomic region in the WT (D), tpi-TetON (E), and Δtpi (F) strains, indicating probe location, FspI and NheI restriction sites, and hybridization fragment size (gray bar). (G) Southern blot of FspI- and NheI-digested genomic DNA of the WT (lane 1), tpi-TetON (lane 2), and Δtpi (lane 3) strains. Download
Immunoblot analysis of the Δtpi strain. Immunoblot of M. tuberculosis protein extracts from the WT, Δtpi, and tpi-Comp strains probed with TPI (27 kDa)-specific rabbit antiserum. Dihydrolipoamide acyltransferase (DLAT; 57 kDa) served as the loading control. Download
Hydrogen peroxide susceptibility of the Δtpi strain. Survival of the WT, tpi-Comp, and Δtpi strains after exposure to 8 mM H2O2 for 4 h in medium containing 0.1% glucose and 0.1% acetate is shown. Strains were cultured on agar plates and CFU were determined. Data are means ± SD for triplicate cultures and are representative of 3 independent experiments. Download
Methylglyoxal accumulates in the Δtpi strain during growth in glycerol containing media. Methylglyoxal concentrations were determined by liquid chromatography-mass spectrometry in metabolite extracts from the indicated strains after growth for 7 days in the indicated carbon substrates as described in Materials and Methods. Data are means for triplicate cultures ± SD. *, P < 0.05 (Student’s t test). Data are representative of two independent experiments. Download
ACKNOWLEDGMENTS
We thank Luiz Pedro S. de Carvalho for insightful discussions and help with the enzymatic assay and Alexandra Dostal for technical help.
This work was supported by National Institutes of Health (NIH) grant AI63446 (to S.E.), grants 42848 and OPP1024065 (to D.S.) and OPP10249392 (to K.Y.R.) from the Bill and Melinda Gates Foundation, and a Burroughs Wellcome Career Award in the Biomedical Sciences (to K.Y.R.).
Footnotes
Citation Trujillo C, Blumenthal A, Marrero J, Rhee KY, Schnappinger D, Ehrt S. 2014. Triosephosphate isomerase is dispensable in vitro yet essential for Mycobacterium tuberculosis to establish infection. mBio 5(2):00085-14. doi:10.1128/mBio.00085-14.
REFERENCES
- 1. Wirth T, Hildebrand F, Allix-Béguec C, Wölbeling F, Kubica T, Kremer K, van Soolingen D, Rüsch-Gerdes S, Locht C, Brisse S, Meyer A, Supply P, Niemann S. 2008. Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog. 4:e1000160. 10.1371/journal.ppat.1000160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. de Carvalho LP, Fischer SM, Marrero J, Nathan C, Ehrt S, Rhee KY. 2010. Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem. Biol. 17:1122–1131. 10.1016/j.chembiol.2010.08.009 [DOI] [PubMed] [Google Scholar]
- 3. Kovárová-Kovar K, Egli T. 1998. Growth kinetics of suspended microbial cells: from single-substrate-controlled growth to mixed-substrate kinetics. Microbiol. Mol. Biol. Rev. 62:646–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Görke B, Stülke J. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6:613–624. 10.1038/nrmicro1932 [DOI] [PubMed] [Google Scholar]
- 5. Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, Schoolnik GK. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693–704. 10.1084/jem.20030846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Timm J, Post FA, Bekker LG, Walther GB, Wainwright HC, Manganelli R, Chan WT, Tsenova L, Gold B, Smith I, Kaplan G, McKinney JD. 2003. Differential expression of iron-, carbon-, and oxygen-responsive mycobacterial genes in the lungs of chronically infected mice and tuberculosis patients. Proc. Natl. Acad. Sci. U. S. A. 100:14321–14326. 10.1073/pnas.2436197100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bloch H, Segal W. 1956. Biochemical differentiation of Mycobacterium tuberculosis grown in vivo and in vitro. J. Bacteriol. 72:132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Muñoz-Elías EJ, McKinney JD. 2005. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 11:638–644. 10.1038/nm1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pandey AK, Sassetti CM. 2008. Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. U. S. A. 105:4376–4380. 10.1073/pnas.0711159105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marrero J, Rhee KY, Schnappinger D, Pethe K, Ehrt S. 2010. Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc. Natl. Acad. Sci. U. S. A. 107:9819–9824. 10.1073/pnas.1000715107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marrero J, Trujillo C, Rhee KY, Ehrt S. 2013. Glucose phosphorylation is required for Mycobacterium tuberculosis persistence in mice. PLoS Pathog. 9:e1003116. 10.1371/journal.ppat.1003116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Phong WY, Lin W, Rao SP, Dick T, Alonso S, Pethe K. 2013. Characterization of phosphofructokinase activity in Mycobacterium tuberculosis reveals that a functional glycolytic carbon flow is necessary to limit the accumulation of toxic metabolic intermediates under hypoxia. PLoS One 8:e56037. 10.1371/journal.pone.0056037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hasan MR, Rahman M, Jaques S, Purwantini E, Daniels L. 2010. Glucose 6-phosphate accumulation in mycobacteria: implications for a novel F420-dependent anti-oxidant defense system. J. Biol. Chem. 285:19135–19144. 10.1074/jbc.M109.074310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wierenga RK. 2001. The TIM-barrel fold: a versatile framework for efficient enzymes. FEBS Lett. 492:193–198. 10.1016/S0014-5793(01)02236-0 [DOI] [PubMed] [Google Scholar]
- 15. Knowles JR. 1991. Enzyme catalysis: not different, just better. Nature 350:121–124. 10.1038/350121a0 [DOI] [PubMed] [Google Scholar]
- 16. Wierenga RK, Kapetaniou EG, Venkatesan R. 2010. Triosephosphate isomerase: a highly evolved biocatalyst. Cell. Mol. Life Sci. 67:3961–3982. 10.1007/s00018-010-0473-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fong SS, Nanchen A, Palsson BO, Sauer U. 2006. Latent pathway activation and increased pathway capacity enable Escherichia coli adaptation to loss of key metabolic enzymes. J. Biol. Chem. 281:8024–8033. 10.1074/jbc.M510016200 [DOI] [PubMed] [Google Scholar]
- 18. Desai KK, Miller BG. 2008. A metabolic bypass of the triosephosphate isomerase reaction. Biochemistry 47:7983–7985. 10.1021/bi801054v [DOI] [PubMed] [Google Scholar]
- 19. Zheng P, Sun J, van den Heuvel J, Zeng AP. 2006. Discovery and investigation of a new, second triose phosphate isomerase in Klebsiella pneumoniae. J. Biotechnol. 125:462–473. 10.1016/j.jbiotec.2006.03.034 [DOI] [PubMed] [Google Scholar]
- 20. Poysti NJ, Oresnik IJ. 2007. Characterization of Sinorhizobium meliloti triose phosphate isomerase genes. J. Bacteriol. 189:3445–3451. 10.1128/JB.01707-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544. 10.1038/31159 [DOI] [PubMed] [Google Scholar]
- 22. Sassetti CM, Boyd DH, Rubin EJ. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77–84. 10.1046/j.1365-2958.2003.03425.x [DOI] [PubMed] [Google Scholar]
- 23. Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. 2011. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 7:e1002251. 10.1371/journal.ppat.1002251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang YJ, Ioerger TR, Huttenhower C, Long JE, Sassetti CM, Sacchettini JC, Rubin EJ. 2012. Global assessment of genomic regions required for growth in Mycobacterium tuberculosis. PLoS Pathog. 8:e1002946. 10.1371/journal.ppat.1002946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mathur D, Malik G, Garg LC. 2006. Biochemical and functional characterization of triosephosphate isomerase from Mycobacterium tuberculosis H37Rv. FEMS Microbiol. Lett. 263:229–235. 10.1111/j.1574-6968.2006.00420.x [DOI] [PubMed] [Google Scholar]
- 26. Connor SE, Capodagli GC, Deaton MK, Pegan SD. 2011. Structural and functional characterization of Mycobacterium tuberculosis triosephosphate isomerase. Acta Crystallogr. D Biol. Crystallogr. 67:1017–1022. 10.1107/S0907444911042971 [DOI] [PubMed] [Google Scholar]
- 27. Ehrt S, Guo XV, Hickey CM, Ryou M, Monteleone M, Riley LW, Schnappinger D. 2005. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 33:e21. 10.1093/nar/gni013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Klotzsche M, Ehrt S, Schnappinger D. 2009. Improved tetracycline repressors for gene silencing in mycobacteria. Nucleic Acids Res. 37:1778–1788. 10.1093/nar/gkp015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grüning NM, Rinnerthaler M, Bluemlein K, Mülleder M, Wamelink MM, Lehrach H, Jakobs C, Breitenbach M, Ralser M. 2011. Pyruvate kinase triggers a metabolic feedback loop that controls redox metabolism in respiring cells. Cell Metab. 14:415–427. 10.1016/j.cmet.2011.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cooper RA, Anderson A. 1970. The formation and catabolism of methylglyoxal during glycolysis in Escherichia coli. FEBS Lett. 11:273–276. 10.1016/0014-5793(70)80546-4 [DOI] [PubMed] [Google Scholar]
- 31. Inoue Y, Kimura A. 1995. Methylglyoxal and regulation of its metabolism in microorganisms. Adv. Microb. Physiol. 37:177–227. 10.1016/S0065-2911(08)60146-0 [DOI] [PubMed] [Google Scholar]
- 32. Ferguson GP, Tötemeyer S, MacLean MJ, Booth IR. 1998. Methylglyoxal production in bacteria: suicide or survival? Arch. Microbiol. 170:209–218. 10.1007/s002030050635 [DOI] [PubMed] [Google Scholar]
- 33. Booth IR, Ferguson GP, Miller S, Li C, Gunasekera B, Kinghorn S. 2003. Bacterial production of methylglyoxal: a survival strategy or death by misadventure? Biochem. Soc. Trans. 31:1406–1408. 10.1042/BST0311406 [DOI] [PubMed] [Google Scholar]
- 34. Pethe K, Sequeira PC, Agarwalla S, Rhee K, Kuhen K, Phong WY, Patel V, Beer D, Walker JR, Duraiswamy J, Jiricek J, Keller TH, Chatterjee A, Tan MP, Ujjini M, Rao SPS, Camacho L, Bifani P, Mak PA, Ma I, Barnes SW, Chen Z, Plouffe D, Thayalan P, Ng SH, Au M, Lee BH, Tan BH, Ravindran S, Nanjundappa M, Lin X, Goh A, Lakshminarayana SB, Shoen C, Cynamon M, Kreiswirth B, Dartois V, Peters EC, Glynne R, Brenner S, Dick T. 2010. A chemical genetic screen in Mycobacterium tuberculosis identifies carbon-source-dependent growth inhibitors devoid of in vivo efficacy. Nat. Commun. 1:1–8. 10.1038/ncomms1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Berney M, Weimar MR, Heikal A, Cook GM. 2012. Regulation of proline metabolism in mycobacteria and its role in carbon metabolism under hypoxia. Mol. Microbiol. 84:664–681. 10.1111/j.1365-2958.2012.08053.x [DOI] [PubMed] [Google Scholar]
- 36. McKinney JD, zu Bentrup K, Muñoz-Elías EJ, Miczak A, Chen B, Chan WT, Swenson D, Sacchettini JC, Jacobs WR, Russell DG. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735–738. 10.1038/35021074 [DOI] [PubMed] [Google Scholar]
- 37. Anderson A, Cooper RA. 1969. Gluconeogenesis in Escherichia coli. The role of triose phosphate isomerase. FEBS Lett. 4:19–20. 10.1016/0014-5793(69)80184-5 [DOI] [PubMed] [Google Scholar]
- 38. Paterson GK, Cone DB, Northen H, Peters SE, Maskell DJ. 2009. Deletion of the gene encoding the glycolytic enzyme triosephosphate isomerase (tpi) alters morphology of Salmonella enterica serovar Typhimurium and decreases fitness in mice. FEMS Microbiol. Lett. 294:45–51. 10.1111/j.1574-6968.2009.01553.x [DOI] [PubMed] [Google Scholar]
- 39. Eisenberg RC, Dobrogosz WJ. 1967. Gluconate metabolism in Escherichia coli. J. Bacteriol. 93:941–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Portais JC, Tavernier P, Gosselin I, Barbotin JN. 1999. Cyclic organization of the carbohydrate metabolism in Sinorhizobium meliloti. Eur. J. Biochem. 265:473–480. 10.1046/j.1432-1327.1999.00778.x [DOI] [PubMed] [Google Scholar]
- 41. Portais JC, Tavernier P, Gosselin I, Barbotin JN. 2000. Relevance and isotopic assessment of hexose-6-phosphate recycling in micro-organisms. J. Biotechnol. 77:49–64. 10.1016/S0168-1656(99)00207-2 [DOI] [PubMed] [Google Scholar]
- 42. Gosselin I, Wattraint O, Riboul D, Barbotin J, Portais J. 2001. A deeper investigation on carbohydrate cycling in Sinorhizobium meliloti. FEBS Lett. 499:45–49. 10.1016/S0014-5793(01)02518-2 [DOI] [PubMed] [Google Scholar]
- 43. Beste DJV, Bonde B, Hawkins N, Ward JL, Beale MH, Noack S, Nöh K, Kruger NJ, Ratcliffe RG, McFadden J. 2011. C metabolic flux analysis identifies an unusual route for pyruvate dissimilation in mycobacteria which requires isocitrate lyase and carbon dioxide fixation. PLoS Pathog. 7:e1002091. 10.1371/journal.ppat.1002091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Beste DJ, Nöh K, Niedenführ S, Mendum TA, Hawkins ND, Ward JL, Beale MH, Wiechert W, McFadden J. 2013. 13C-flux spectral analysis of host-pathogen metabolism reveals a mixed diet for intracellular Mycobacterium tuberculosis. Chem. Biol. 20:1–10. 10.1016/j.chembiol.2013.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Henderson B, Martin A. 2011. Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect. Immun. 79:3476–3491. 10.1128/IAI.00179-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maithal K, Ravindra G, Balaram H, Balaram P. 2002. Inhibition of Plasmodium falciparum triose-phosphate isomerase by chemical modification of an interface cysteine. Electrospray ionization mass spectrometric analysis of differential cysteine reactivities. J. Biol. Chem. 277:25106–25114. 10.1074/jbc.M202419200 [DOI] [PubMed] [Google Scholar]
- 47. Téllez-Valencia A, Olivares-Illana V, Hernández-Santoyo A, Pérez-Montfort R, Costas M, Rodríguez-Romero A, López-Calahorra F, Tuena De Gómez-Puyou M, Gómez-Puyou A. 2004. Inactivation of triosephosphate isomerase from Trypanosoma cruzi by an agent that perturbs its dimer interface. J. Mol. Biol. 341:1355–1365. 10.1016/j.jmb.2004.06.056 [DOI] [PubMed] [Google Scholar]
- 48. Hernández-Alcántara G, Torres-Larios A, Enríquez-Flores S, García-Torres I, Castillo-Villanueva A, Méndez ST, de la Mora-de la Mora I, Gómez-Manzo S, Torres-Arroyo A, López-Velázquez G, Reyes-Vivas H, Oria-Hernández J. 2013. Structural and functional perturbation of Giardia lamblia triosephosphate isomerase by modification of a non-catalytic, non-conserved region. PLoS One 8:e69031. 10.1371/journal.pone.0069031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Romo-Mancillas A, Téllez-Valencia A, Yépez-Mulia L, Hernández-Luis F, Hernández-Campos A, Castillo R. 2011. The design and inhibitory profile of new benzimidazole derivatives against triosephosphate isomerase from Trypanosoma cruzi: a problem of residue motility. J. Mol. Graph. Model. 30:90–99. 10.1016/j.jmgm.2011.06.009 [DOI] [PubMed] [Google Scholar]
- 50. Allen BW. 1998. Mycobacteria: general culture methodology and safety considerations. Methods Mol. Biol. 101:15–30 [DOI] [PubMed] [Google Scholar]
- 51. Gracy RW. 1975. Triosephosphate isomerase from human erythrocytes. Methods Enzymol. 41:442–447. 10.1016/S0076-6879(75)41096-5 [DOI] [PubMed] [Google Scholar]
- 52. Eoh H, Rhee KY. 2013. Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 110:6554–6559. 10.1073/pnas.1219375110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Randell EW, Vasdev S, Gill V. 2005. Measurement of methylglyoxal in rat tissues by electrospray ionization mass spectrometry and liquid chromatography. J. Pharmacol. Toxicol. Methods 51:153–157. 10.1016/j.vascn.2004.08.005 [DOI] [PubMed] [Google Scholar]
- 54. Vandal OH, Pierini LM, Schnappinger D, Nathan CF, Ehrt S. 2008. A membrane protein preserves intrabacterial pH in intraphagosomal Mycobacterium tuberculosis. Nat. Med. 14:849–854. 10.1038/nm.1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Southern blot analysis of mutants generated in this study. Map of the tpi genomic region in WT M. tuberculosis (A) and tpi mutant (B) indicating probe location, PstI restriction sites, and hybridization product fragment size (gray bar). (C) Southern blot of PstI-digested genomic DNA of the WT (lane 1), Δtpi (lane 2), and tpi-TetON (lane 3) strains. (D to F) Map of the attL5 genomic region in the WT (D), tpi-TetON (E), and Δtpi (F) strains, indicating probe location, FspI and NheI restriction sites, and hybridization fragment size (gray bar). (G) Southern blot of FspI- and NheI-digested genomic DNA of the WT (lane 1), tpi-TetON (lane 2), and Δtpi (lane 3) strains. Download
Immunoblot analysis of the Δtpi strain. Immunoblot of M. tuberculosis protein extracts from the WT, Δtpi, and tpi-Comp strains probed with TPI (27 kDa)-specific rabbit antiserum. Dihydrolipoamide acyltransferase (DLAT; 57 kDa) served as the loading control. Download
Hydrogen peroxide susceptibility of the Δtpi strain. Survival of the WT, tpi-Comp, and Δtpi strains after exposure to 8 mM H2O2 for 4 h in medium containing 0.1% glucose and 0.1% acetate is shown. Strains were cultured on agar plates and CFU were determined. Data are means ± SD for triplicate cultures and are representative of 3 independent experiments. Download
Methylglyoxal accumulates in the Δtpi strain during growth in glycerol containing media. Methylglyoxal concentrations were determined by liquid chromatography-mass spectrometry in metabolite extracts from the indicated strains after growth for 7 days in the indicated carbon substrates as described in Materials and Methods. Data are means for triplicate cultures ± SD. *, P < 0.05 (Student’s t test). Data are representative of two independent experiments. Download