Abstract
Polychlorinated biphenyls (PCBs) are persistent environmental contaminants, and exposure to PCBs and their hydroxylated metabolites (OHPCBs) has been associated with various adverse health effects. The mammalian cytosolic sulfotransferases (SULTs) catalyze the sulfation of OHPCBs, and the interaction of OHPCBs with both the SULT1 and SULT2 families of these enzymes has received attention both with respect to metabolic disposition of these molecules and the potential mechanisms for their roles in endocrine disruption. We have previously shown that OHPCBs interact with human hydroxysteroid sulfotransferase hSULT2A1, an enzyme that catalyzes the sulfation of dehydroepiandrosterone (DHEA), other alcohol-containing steroids, bile acids, and many xenobiotics. The objective of our current studies is to investigate the mechanism of inhibition of hSULT2A1 by OHPCBs by combining inhibition kinetics with determination of equilibrium binding constants and molecular modeling of potential interactions. Examination of the effects of fifteen OHPCBs on the sulfation of DHEA catalyzed by hSULT2A1 showed predominantly noncompetitive inhibition patterns. This was observed for OHPCBs that were substrates for sulfation reactions catalyzed by the enzyme as well as those that solely inhibited the sulfation of DHEA. Equilibrium binding experiments and molecular modeling studies indicated that the OHPCBs bind at the binding site for DHEA on the enzyme, and that the observed noncompetitive patterns of inhibition are consistent with binding in more than one orientation to more than one enzyme complex. These results have implications for the roles of SULTs in the toxicology of OHPCBs, while also providing molecular probes of the complexity of substrate/inhibitor interactions with hSULT2A1.
Keywords: hydroxylated polychlorinated biphenyl, OHPCB, PCB, sulfotransferase, hSULT2A1
1. Introduction
Polychlorinated biphenyls (PCBs) are worldwide pollutants that are implicated in a variety of adverse health effects. Although their production was banned over four decades ago, they remain of concern due to their persistence in the environment and their toxicities to humans and other animals [1–5]. One area of concern is that PCBs are present in paints, fluorescent light fixtures, caulk, and other materials contained in older buildings and schools that are still in use [6–9]. Since the PCBs with lower numbers of chlorine atoms are more volatile, there is increasing concern about their release into indoor air in older buildings [8, 10, 11]. In addition, these lower chlorinated PCB congeners are also being released into air from PCB-contaminated soils and water [9, 12–14].
The lower chlorinated PCBs can be oxidatively metabolized leading to formation of their hydroxylated derivatives (OHPCBs) [4–5]. Further metabolism of these OHPCBs may lead to formation of dihydroxylated PCBs, reactive quinones and semiquinones that can cause oxidative stress [15] and mutations that may lead to initiation of cancer [3]. The lower chlorinated OHPCBs have also been shown to interact with cytosolic sulfotransferases (SULTs) that are active in metabolism of steroid hormones [16] and thyroid hormones [17]. Moreover, OHPCBs [18, 19] as well as the products of sulfation of OHPCBs [20] have been shown to bind with high affinity to the thyroid hormone transport protein transthyretin. These interactions of OHPCBs with sulfotransferases involved in the metabolism of steroid hormones and with thyroid hormone carriers may contribute to the endocrine disruption observed with some PCBs [16,17].
Among the SULTs that catalyze sulfation of OHPCBs is the human hydroxysteroid sulfotransferase hSULT2A1 [21, 22], a family 2 SULT that is involved in the metabolism of various steroids, bile acids and xenobiotics [23–27]. The hSULT2A1 is highly expressed in the adrenal, liver, and intestine, but there is also evidence of its expression in the lung, kidney, and other tissues [28, 29].
Many endogenous and exogenous compounds inhibit the activity of sulfotransferases [16, 30–33]. Such inhibition could result from either a molecule serving solely as an inhibitor or as an alternate substrate that competes for the catalytic site of the enzyme. Inhibition of a SULT may be brought about either by binding to the sulfuryl acceptor site or by binding to the PAPS/PAP-site [32]. We have previously studied 15 OHPCBs as inhibitors of the sulfation of DHEA catalyzed by hSULT2A1 and observed that eight of these OHPCBs (i.e., 4-OHPCB 8, 4-OHPCB 11, 4-OHPCB 14, 4’-OHPCB 25, 4’-OHPCB 33, 4-OHPCB 34, 4-OHPCB 36, and 4’-OHPCB 68) served as alternate substrates for hSULT2A1 [21]. While we developed a predictive three-dimensional structure-activity relationship for OHPCBs as inhibitors of hSULT2A1 using comparative molecular field analysis (CoMFA), this did not address the molecular mechanism(s) of the interactions of these molecules as inhibitors. In the current study, our aim was to determine the molecular basis for inhibition of hSULT2A1 by these 15 OHPCBs.
2. Materials and Methods
2.1 Chemicals
All OHPCBs were synthesized and characterized by a previously described procedure [34]. 3’-Phosphoadenosine-5’-phosphosulfate (PAPS) was purchased from Sigma-Aldrich (St. Louis, MO) and further purified using a published method [35]. The final purity of the purified PAPS was greater than 98% as judged by HPLC.3H-DHEA (94.5 Ci/mmol) was purchased from Perkin Elmer Life and Analytical Sciences (Boston, MA). Econo-Safe biodegradable scintillation cocktail and Tris-HCl were purchased from RPI (Mt. Prospect, IL). Hydroxyapatite (Bio-Gel HT) was obtained from Bio-Rad Laboratories (Hercules, CA). The ammonium salt of 8-anilinonaphthalene-1-sulfonic acid (ANS) was from Fluka (Steinheim, Germany), and Tween 20 was obtained from J.T. Baker Chemicals (Phillipsburg, NJ). All other chemicals used were of the highest purity commercially available.
2.2 Protein Expression and Purification of recombinant hSULT2A1
Human sulfotransferase hSULT2A1 was expressed in recombinant Escherichia coli BL21 (DE3) cells using a previously described procedure [21]. Purification of hSULT2A1 was carried out as previously described [36], and the resulting enzyme preparation was homogeneous as judged by SDS-PAGE with Coomassie Blue staining. Protein concentrations were obtained using the modified Lowry procedure [37] with bovine serum albumin as standard. During the purification, catalytic activity of hSULT2A1 was determined by a standard paired ion extraction assay [38, 39].
2.3 Kinetic studies on the inhibition of hSULT2A1 by OHPCBs
Mechanistic studies of the kinetics on the inhibition of hSULT2A1 were carried out using a previously described radiochemical method for the sulfation of DHEA [40]. Assays contained fixed concentrations of [3H]DHEA (0.2, 0.4, 0.6 and 1 µM with final radioactive specific activity of 0.4, 0.8, 1.2 and 2 µCi/ nmol, respectively) in the presence and absence of variable concentrations of OHPCBs. Assays (total volume of 200 µL) also contained 200 µM PAPS, 0.25 M potassium phosphate at pH 7.0, and 7.5 mM 2-mercaptoethanol. OHPCBs were dissolved in ethanol for addition to assay mixtures, and the final concentration of ethanol in each reaction mixture was 2% (v/v). Sulfation reactions were started by the addition of 0.25 µg hSULT2A1 and carried out for 10 min at 37°C. Reactions were terminated by addition of 0.8 mL of 50 mM potassium hydroxide and 0.5 mL of chloroform, and the phases separated as described previously [40]. A 100 µL aliquot of the aqueous phase was added to 10 mL of liquid scintillation cocktail for determination of radioactivity using a Perkin Elmer TriCarb 2900TR liquid scintillation analyzer. The rate of sulfation was expressed as nmol of DHEA-sulfate formed per minute per mg of protein. Data were fit by non-linear regression analysis to equations for competitive, noncompetitive, mixed, and uncompetitive inhibition (Enzyme Kinetics Module 1.3; SigmaPlot v. 11.0; Systat software, Chicago, IL).
2.4. Ligand-binding studies on the interaction of OHPCBs with hSULT2A1
The binding of OHPCBs to hSULT2A1, to the enzyme-PAP complex, and to the enzyme-DHEA complex was evaluated by determining the change in fluorescence intensity of ANS upon its displacement from binding sites on the enzyme by higher affinity ligands. This method has been previously used for determination of Kd values for SULTs [41, 42]. The ligand-binding studies were carried out using a Perkin Elmer model LS-55 Luminescence spectrophotometer with a water-thermostated cell holder using a 10 mm path-length quartz cuvette. The fluorescence excitation and emission wavelengths were 380 nm and 465 nm, respectively, and slit widths were set at 5 nm for both emission and excitation beams. Ligand-binding was measured at 37°C in 0.25 M potassium phosphate buffer, pH 7.0, containing 7.5 mM 2-mercaptoethanol and 40 µM ANS in a total volume of 1 mL. All solutions were filtered before use with a Millex-GS 0.22 µm filter, and solutions of ANS were kept in the dark prior to use. The phosphate buffer solution was preincubated with 3 µg of enzyme for 2 min at 37°C before titrating with a range of concentrations of OHPCB(s) depending on the solubility limit of the given compound. The absolute value of the decrease in fluorescence (ΔF) upon displacement of ANS by OHPCB was used for analysis of titration experiments, and data were fit to a one-site saturable binding equation with an additional non-specific binding parameter (Sigma Plot, v.11; Systat Software, Chicago, IL).
2.5. Molecular modeling of the interactions of OHPCBs with hSULT2A1
In order to understand more clearly the molecular interactions between OHPCBs and hSULT2A1, docking studies were performed using Surflex-Dock in Sybyl 8.0 (Tripos International) on a Linux operating system. Two crystal structures of hSULT2A1 were used: hSULT2A1 in complex with DHEA (PDB file: 1J99) [43] and hSULT2A1 in complex with PAP (PDB file:1EFH) [44]. Amino acid residues appearing at a 5Å distance around DHEA and PAP were noted, and then DHEA and PAP were extracted from the two crystal structures before performing the docking experiments. Water molecules in the crystal structures were deleted, hydrogen atoms added, and the structures were then saved as mol2 files. Each of the ligands (ANS, 4’-OHPCB 9, 4-OHPCB 14, 4’-OHPCB 33 and 4-OHPCB 34) was constructed, hydrogen atoms were added, and their energies were minimized using the Powell method with the Tripos Force field. The termination gradient was set at 0.05 kcal/mol*Å, maximum iteration at 1000, dielectric constant of 1.0000, and RMS displacement at 0.001. The 3D structures of all ligands were placed in different molecular areas within the same file and saved in MOL2 format before docking with Surflex-Dock (Tripos Inc., St. Louis, MO). The protomol was then generated automatically (threshold of 0.3 with bloat of 1, and threshold of 0.5 with bloat of 0 for the crystal structures of hSULT2A1 in the DHEA-bound conformation and PAP-bound conformation, respectively). The ligands were docked, one at a time, into the binding site of the receptor, hSULT2A1, with additional starting conformation set at 100 per molecule and the default settings kept for all other parameters. After each run of Surflex-Dock, the ten best docked conformers or poses were sorted in a molecular spreadsheet, and binding affinities were represented as -log 10(Kd) based on the Surflex-dock scoring function (crash score (also pKd units), polar score, D-score, PMF-score, G-score, ChemSco and CScore). The best total score conformers of each of the docked ligands were selected based on a consensus score of 3 to 5. Amino acid residues appearing at a 5Å distance around each docked ligand (ANS, 4’-OHPCB 9, 4-OHPCB 14, 4’-OHPCB 33 and 4-OHPCB 34) were labeled and compared to the residues appearing at 5Å distance around DHEA and PAP (PDB code, 1J99 and 1EFH respectively). As additional control experiments, the extracted PAP and DHEA were docked in the two model structures as well. Images of the docking results were created with the PyMOL Molecular Graphics System (version 1.5.0.4; Schrödinger LLC, New York, NY).
3. Results
3.1. Initial Velocity Studies on the Inhibition of DHEA-sulfation by OHPCBs
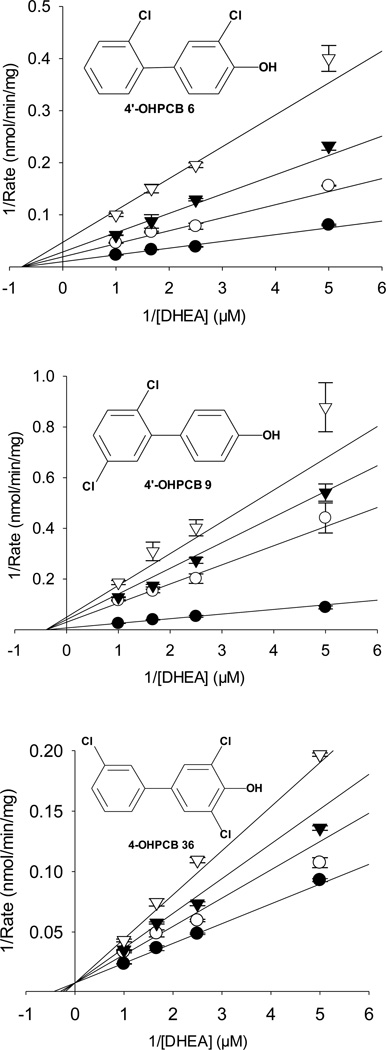
The mechanism for the inhibition of hSULT2A1-catalyzed sulfation of DHEA catalyzed by hSULT2A1 was investigated with 15 OHPCBs (structures in Figure 1) through the use of initial velocity studies at a fixed concentration of PAPS and varied concentrations of DHEA and inhibitor. Kinetic equations for competitive, noncompetitive, mixed, and uncompetitive inhibition were used to determine the most likely type of inhibition involved, and these kinetic equations are provided in Figure S1 (Supplementary Material). Plots of 1/reaction velocity vs. 1/[DHEA] at varied concentrations of each OHPCB displayed lines intersecting to the left of the y-axis for 14 of the 15 OHPCBs examined, thus indicating either a type of noncompetitive or mixed inhibition was occurring for most of the OHPCBs examined. Plots for the inhibition of the hSULT2A1-catalyzed sulfation of DHEA by 4’-OH PCB 6 and 4’-OHPCB 9 are shown in Figure 2 as representative of the data for those compounds that displayed noncompetitive and partial noncompetitive inhibition, respectively. As also seen in Figure 2, a plot of 1/reaction velocity vs. 1/[DHEA] for the hSULT2A1-catalyzed sulfation of DHEA at varied concentrations of 4-OHPCB 36 displayed lines intersecting on the y-axis, indicative of a form of competitive inhibition. In the case of 4-OHPCB 36, the kinetic data fit most closely to the equation for partial competitive inhibition. The values of R2 (conventional correlation) and AICc (corrected Akaike information criterion) obtained for all the inhibition models are listed in Table 1. The model having a high R2 with the lowest AICc value was chosen to be the most likely model for the kinetic mechanism of inhibition for each OHPCB.
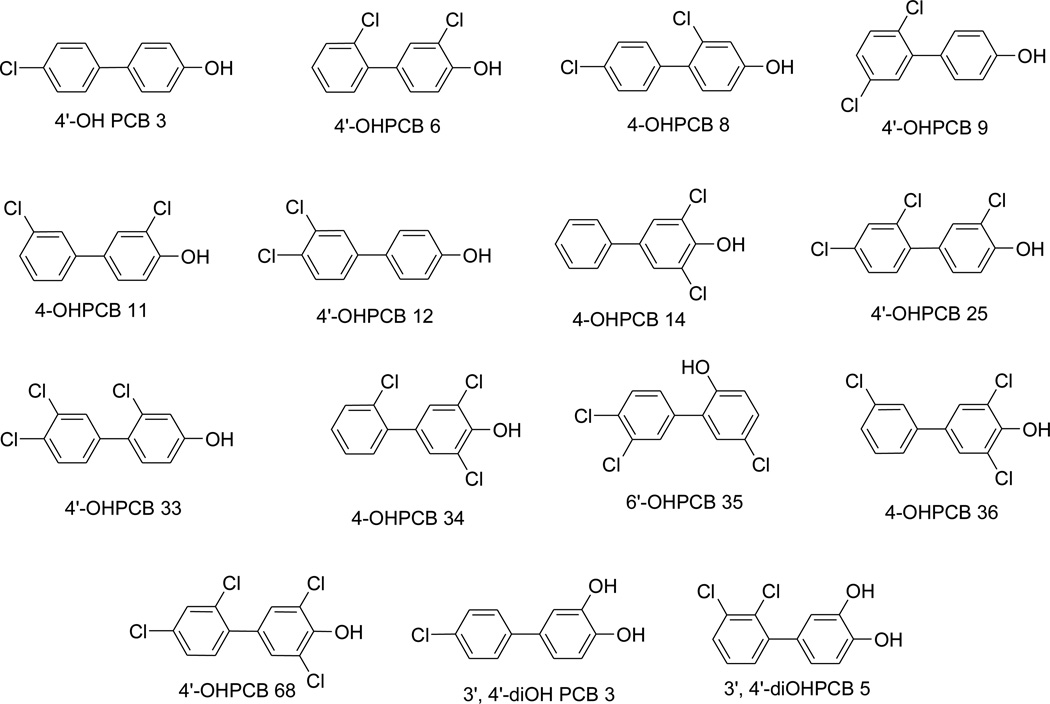
Fig 1.
Structures of the OHPCBs used in the study
Fig 2.
Kinetic analysis of OHPCBs as inhibitors of DHEA sulfation catalyzed by hSULT2A1. Nonlinear regression fits to noncompetitive (full), noncompetitive (partial), and competitive (partial) inhibition of the enzyme by 4’-OHPCB 6, 4’-OHPCB 9, and 4-OHPCB 36, respectively, are shown. Closed circles (●) represent the no inhibitor, open circle (○) represent 5 µM, 4 µM and 2.2 µM of 4’-OHPCB 6, 4’-OHPCB 9, and 4- OHPCB 36 respectively. Closed triangles (▼) represent 10 µM, 8 µM, and 4.4 µM of 4’-OHPCB 6, 4’-OHPCB 9, and 4-OHPCB 36 respectively. Open triangles (▽) represent 20 µM, 16 µM, and 8.8 µM of 4’-OHPCB 6, 4’-OHPCB 9, and 4-OHPCB 36 respectively. Data points represent the mean ± standard error of duplicate determinations at each combination of substrate and inhibitor. For each graph, the lines represent the calculated theoretical values derived from the kinetic model that best fit all of the data (as determined by the AICc and R2 values in Table 1) for inhibition of the hSULT2A1-catalyzed sulfation of DHEA by that OHPCB.
Table 1.
OHPCBs as inhibitors of the hSULT2A1-catalyzed sulfation of DHEA: comparison of statistical parameters for the fit of initial velocity data to various mechanisms of inhibition
| Noncompetitive | Mixed | Competitive | Uncompetitive | |||||
|---|---|---|---|---|---|---|---|---|
| OHPCBs | R2 | AICc | R2 | AICc | R2 | AICc | R2 | AICc |
| 4’-OHPCB 33 | 0.97c | 40.9 | 0.97 | 43.7 | 0.96 | 48.0 | 0.94 | 58.0 |
| 4’-OHPCB 68a | 0.97 | 6.4 | 0.97 | 9.9 | 0.97 | 8.4 | 0.95 | 23.4 |
| 4’-OHPCB 6 | 0.99 | 17.7 | 0.99 | 19.4 | 0.99 | 23.4 | 0.96 | 59.2 |
| 4-OHPCB 36b | 0.98 | 31.4 | 0.98 | 27.4 | 0.98 | 24.6 | 0.96 | 55.1 |
| 4-OHPCB 8 | 0.97 | 38.8 | 0.97 | 41.5 | 0.97 | 40.8 | 0.95 | 60.9 |
| 4-OHPCB 14 | 0.97 | 40.6 | 0.97 | 43.3 | 0.97 | 43.5 | 0.94 | 60.0 |
| 4’-OHPCB 9a | 0.99 | −1.1 | 0.99 | 0.8 | 0.99 | 4.1 | 0.98 | 42.5 |
| 4-OHPCB 11 | 0.97 | 53.2 | 0.97 | 54.1 | 0.96 | 60.3 | 0.95 | 64.4 |
| 4’-OHPCB 3 | 0.96 | 43.3 | 0.96 | 45.5 | 0.95 | 47.9 | 0.94 | 53.5 |
| 4’-OHPCB 12 | 0.98 | 20.6 | 0.98 | 23.3 | 0.98 | 23.0 | 0.97 | 44.6 |
| 4’-OHPCB 25a | 0.97 | 45.8 | 0.97 | 48.0 | 0.96 | 52.9 | 0.95 | 61.0 |
| 3’,4’-diOH PCB 3 | 0.98 | 33.9 | 0.98 | 35.6 | 0.97 | 45.0 | 0.97 | 43.8 |
| 3’,4’-diOH PCB 5 | 0.98 | 21.8 | 0.98 | 23.4 | 0.98 | 25.7 | 0.96 | 46.7 |
| 6’-OHPCB 35 | 0.98 | 10.8 | 0.99 | 11.4 | 0.97 | 25.0 | 0.97 | 33.9 |
| 4-OHPCB 34 | 0.97 | 37.1 | 0.97 | 38.7 | 0.96 | 47.2 | 0.96 | 46.9 |
fit to partial noncompetitive inhibition equation
fit to partial competitive inhibition equation
boldface type indicates the most likely model based on highest R2 446 and lowest AICc
The model that best fit the inhibition data for each OHPCB (Table 1) was used to determine inhibition constants for the enzyme with DHEA as substrate. The Ki values for all fifteen OHPCBs were obtained by nonlinear r egression fit to the appropriate kinetic equation, and these are listed in Table 2. Eleven OHPCBs displayed noncompetitive inhibition of the sulfation of DHEA, a type of mixed inhibition wherein an inhibitor has equal affinity for both the free enzyme (E) and enzyme-substrate (ES) complex (i.e., the value of α in the equation for mixed inhibition is equal to 1). Inhibition of hSULT2A1 by 4’-OHPCB 9, 4’-OHPCB 25, and 4’-OHPCB 68 was best described by an equation for partial noncompetitive inhibition, with β values of 0.11 ± 0.02, 0.19 ± 0.06, 0.23 ± 0.07, respectively. The β values signify formation of product (i.e., DHEA-sulfate) in the presence of the inhibitor. Of the fifteen OHPCBs examined, only 4-OHPCB 36 showed a form of competitive inhibition with respect to DHEA, and this was best modeled by a partial competitive inhibition equation with an α value of 4.2.
Table 2.
Dissociation constants for 15 OHPCBs as inhibitors of the sulfation of DHEA catalyzed by hSULT2A1
| OHPCB | Ki (µM)a |
|---|---|
| 4’-OHPCB 3 | 17 ± 1.5 |
| 4’-OHPCB 6 | 5.4 ± 0.2 |
| 4-OHPCB 8 | 5.0 ± 0.4 |
| 4’-OHPCB 9 | 0.7 ± 0.1 |
| 4-OHPCB 11 | 12 ± 0.9 |
| 4’-OHPCB 12 | 4.0 ± 0.3 |
| 4-OHPCB 14 | 13.5 ± 1.0 |
| 4’-OHPCB 25 | 3.0 ± 0.6 |
| 4’-OHPCB 33 | 3.0 ± 0.2 |
| 4-OHPCB 34 | 0.8 ± 0.1 |
| 4-OHPCB 36 | 3.4 ± 0.8 |
| 4’-OHPCB 68 | 0.6 ± 0.1 |
| 6’-OHPCB 35 | 9.0 ± 0.5 |
| 3’,4’-diOHPCB 3 | 32 ± 2.3 |
| 3’,4’-diOHPCB 5 | 14 ± 0.8 |
With the exception of 4’-OHPCB 36, values for Ki were calculated using a nonlinear regression fit to a noncompetitive inhibition equation. The Ki for 4’-OHPCB 36 was obtained from a fit of the data to an equation for competitive inhibition.
3.2. The binding of OHPCBs to hSULT2A1
Additional information about the binding of inhibitors to the enzyme was obtained by determining the equilibrium dissociation constants for interaction of the OHPCBs with the enzyme. These studies were carried out using the fluorescent probe, 8-anilino-1-naphthalene sulfonic acid (ANS). The non-covalent binding of ANS is a well established probe for binding of ligands to proteins, where binding of a ligand with high specificity for the protein displaces ANS yielding a change in fluorescence of ANS due to the change in the polarity of its environment [41, 42]. Preliminary experiments (data not shown) indicated that 40 µM ANS was sufficient to saturate the enzyme under the conditions of the assay, and this concentration was used in subsequent ligand-displacement experiments.
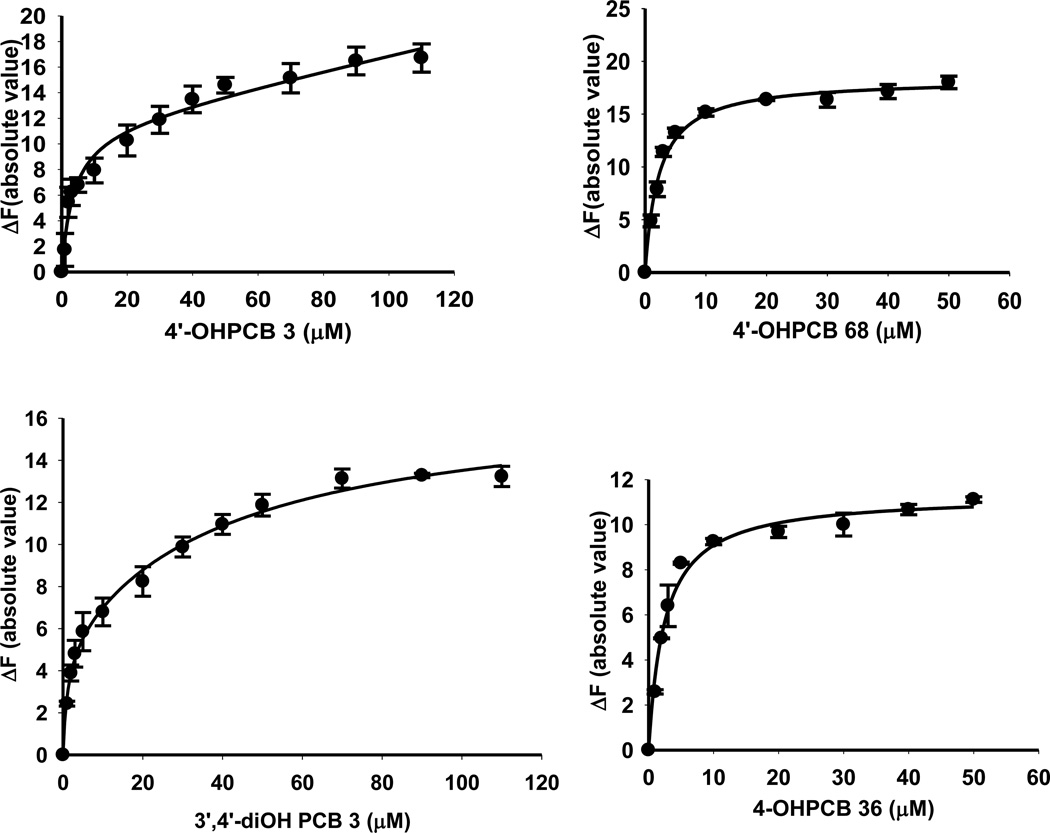
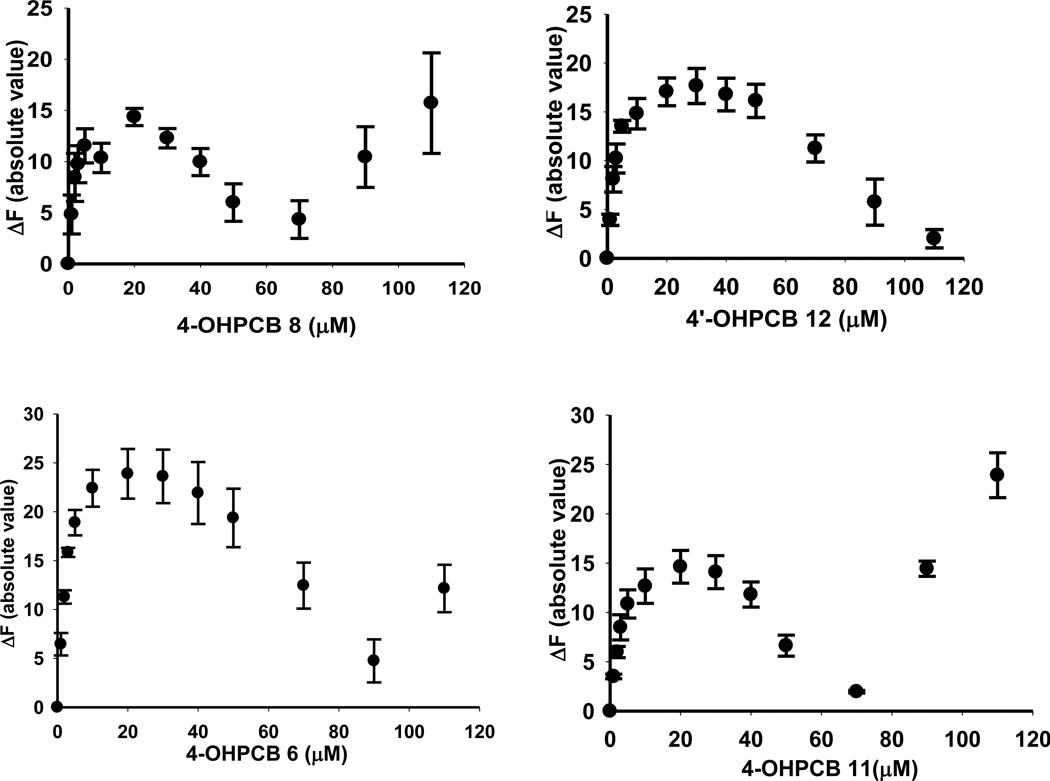
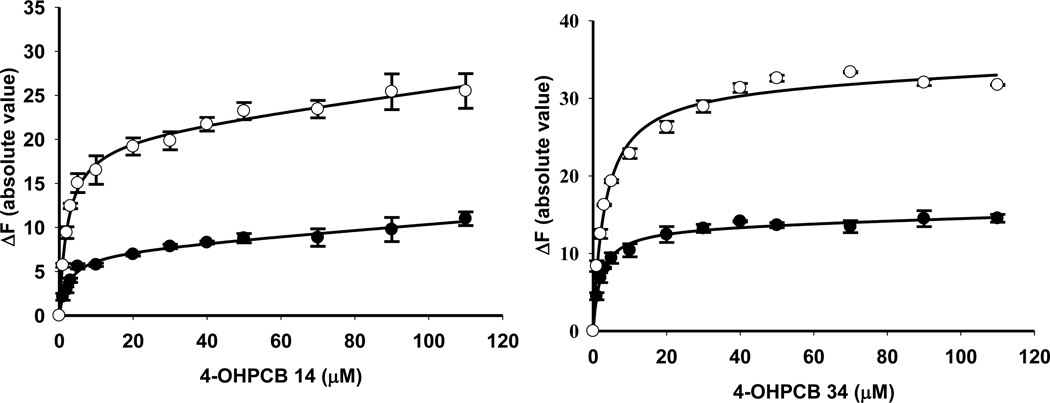
The binding of four representative OHPCBs, as determined by the absolute value of the change in fluorescence upon displacement of ANS from the active site of hSULT2A1, is shown in Figure 3, and the equilibrium dissociation constants (Kd values) for six OHPCBs are provided in Table 3. Several OHPCBs did not exhibit a classical binding pattern that could be easily fit to simple one-site or two-site binding equations, but, instead, these molecules displayed a multiphasic binding pattern (Figure 4). A control experiment indicated that for 6’-OHPCB 35 (one of the OHPCBs exhibiting multiphasic behavior), there was no direct interaction of the OHPCB with ANS in solution without the enzyme (data not shown). Thus, the observed changes in fluorescence intensity at higher concentrations of OHPCB were not due to any interactions of the OHPCB with ANS that had been displaced from binding sites on hSULT2A1. An explanation for the multiphasic binding pattern might be differential binding of these OHPCBs to enzyme-substrate or enzyme-product complexes. Experiments with 4-OHPCB 9 and 4-OHPCB33, however, did not show significant differences in the multiphasic binding pattern in the presence or absence of either DHEA or PAP (Supplementary Material, Figure S1). Moreover, as seen in Figure 5, the Kd value for binding of 4-OHPCB 14 to the E-PAP complex (2.21 ± 0.50 µM) was not significantly different from the Kd value for binding to free enzyme (2.55 ± 0.63 µM). Likewise, the Kd for the binding of 4-OHPCB 34 to the E-PAP complex (3.20 ± 0.31 µM) was similar to that observed for binding to the free enzyme (Kd = 2.07 ± 0.30 µM). It is of note, however, that the total change in fluorescence intensity was increased for the binding of both 4-OHPCB 14 and 4-OHPCB 34 to the E-PAP complex (Figure 5).
Fig 3.
Binding of 4-OHPCB 3, 4’-OHPCB 68, 3’4’-diOH PCB 3 and 4-OHPCB 36 to hSULT2A1. Plots of the absolute value of the fluorescence change upon titration with the indicated concentration of OHPCB are shown. Assays were carried out with a saturating concentration of ANS (40 µM). Data points are the mean ± standard error of triplicate determinations.
Table 3.
Kd values determined for six hydroxylated polychlorinated biphenyls (OHPCBs) binding to hSULT2A1 in the absence of substrates or products
| OHPCBs | Kd (µM) |
|---|---|
| 4’-OHPCB 3 | 3.48 ± 1.03 |
| 4-OHPCB 14 | 2.55 ± 0.63 |
| 4-OHPCB 36 | 2.46 ± 0.43 |
| 4-OHPCB 34 | 2.07 ± 0.30 |
| 4’-OHPCB 68 | 2.26 ± 0.33 |
| 3’,4’-diOHPCB 3 | 1.17 ± 1.11 (Kd1) |
| 41.9 ± 74.8 (Kd2) |
Note: The binding data for 3’, 4’-diOHPCB 3 fit to a two-site, nonspecific binding curve, others fit to a one-site nonspecific binding curve
Fig 4.
Binding of 4-OHPCB 8, 4’-OHPCB 12, 4’-OH PCB 6 and 4-OHPCB 11 to hSULT2A1. Plots of the absolute value of the fluorescence change upon titration with the indicated concentration of OHPCB are shown. Assays were carried out with a saturating concentration of ANS (40 µ M). Data points are the mean ± standard error of triplicate determinations.
Fig 5.
Binding of 4-OHPCB 14 and 4-OHPCB 34 to different forms of the enzyme (hSULT2A1). Assays done in the presence of (●) free enzyme, and (○) E-PAP complex. Plots of the absolute value of the fluorescence change versus OHPCBs concentration are shown. Assays were carried out with a saturating concentration of ANS (40 µM) and a saturating concentration of PAP (200 µM). Data points are the mean ± standard error of triplicate determinations.
3.3. Modeling of ligand-binding interactions with hSULT2A1
In order to gain additional insight as we interpreted our results from the ANS-displacement and inhibition studies, molecular docking experiments were carried out using two crystal structures: hSULT2A1 with bound DHEA (PDB file 1J99) [43] and hSULT2A1 with bound PAP (PDB file 1EFH) [44]. The bound DHEA and PAP molecules were removed from the crystal structures before docking studies were carried out. The appropriate OHPCBs were then docked into these structures and minimum energy docked structures were obtained. Control experiments were performed where DHEA, or PAP, was re-docked (cognate docking) into the models derived from crystal structures where the respective ligand had been removed. From among those with multiphasic binding to hSULT2A1, two OHPCBs (4’-OHPCB 9 and 4’-OHPCB 33) were selected for molecular docking, and two OHPCBs (4-OHPCB 14 and 4-OHPCB 34) that displayed classical single-site binding characteristics were also used in the docking experiments.
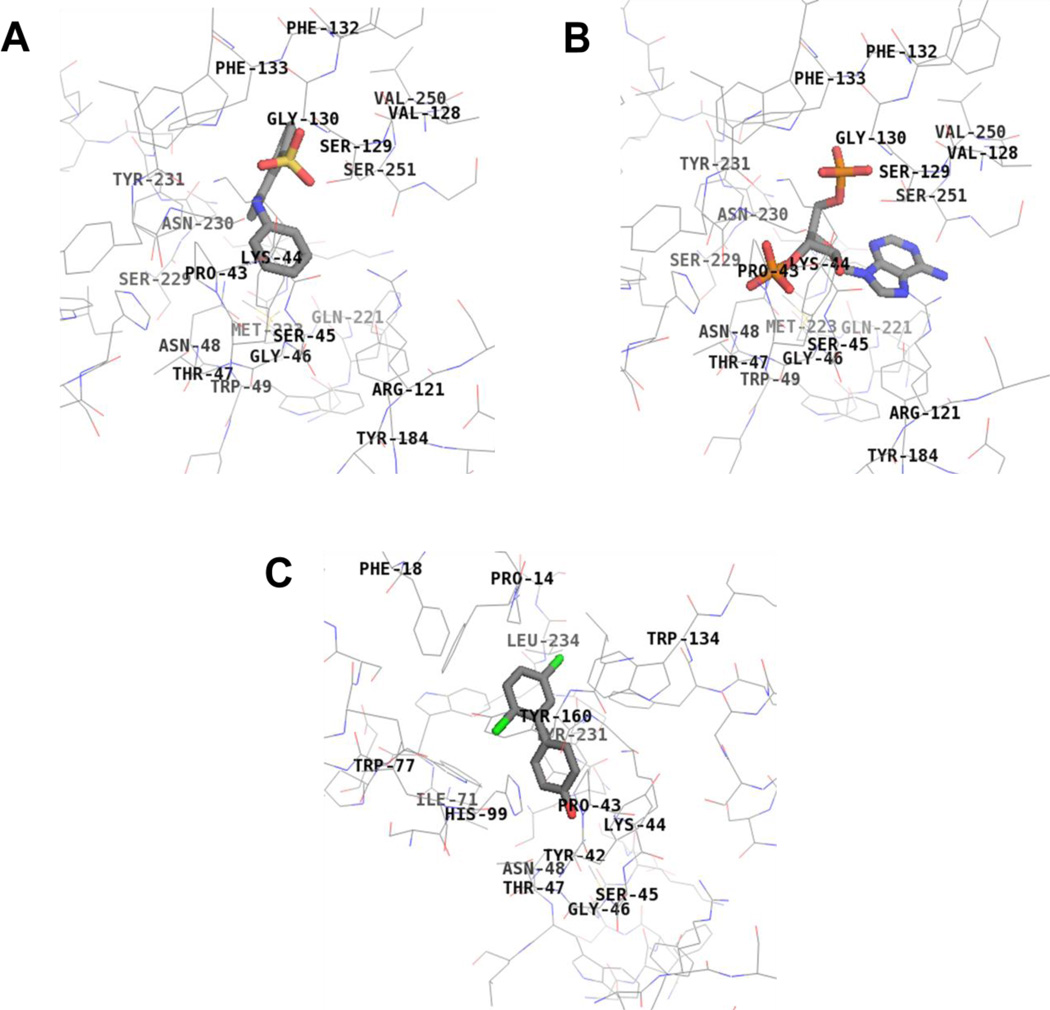
As seen in Figure 6, when ANS was docked into hSULT2A1 using the model derived from the crystal structure that originally had DHEA bound at the active site (PDB 1J99), it displayed binding at the PAP binding site. When docked into hSULT2A1 using the model derived from the crystal structure with PAP bound, the ANS displayed highest affinity binding at the DHEA-binding site (data not shown). Thus, the model predicts that ANS binds to both the DHEA site and the PAPS/PAP site.
Fig 6.
Binding interactions of ANS (Panel A), PAP (Panel B), and 4’-OHPCB 9 (Panel C) with hSULT2A1. ANS binds at the same site as PAP, and 4’-OHPCB 9 binds at the DHEA site. Key hSULT2A1 residues interacting with the ligands at the active site are shown. Total score of 3.78 based on a consensus score of 4 for binding of ANS, total score of 7.80 based on a consensus score of 5 for binding of PAP, and total score of 4.14 based on a consensus score of 5 for binding of 4’-OHPCB 9 were obtained. (crystal structure: hSULT2A1 in the conformation observed with DHEA bound (1J99)).
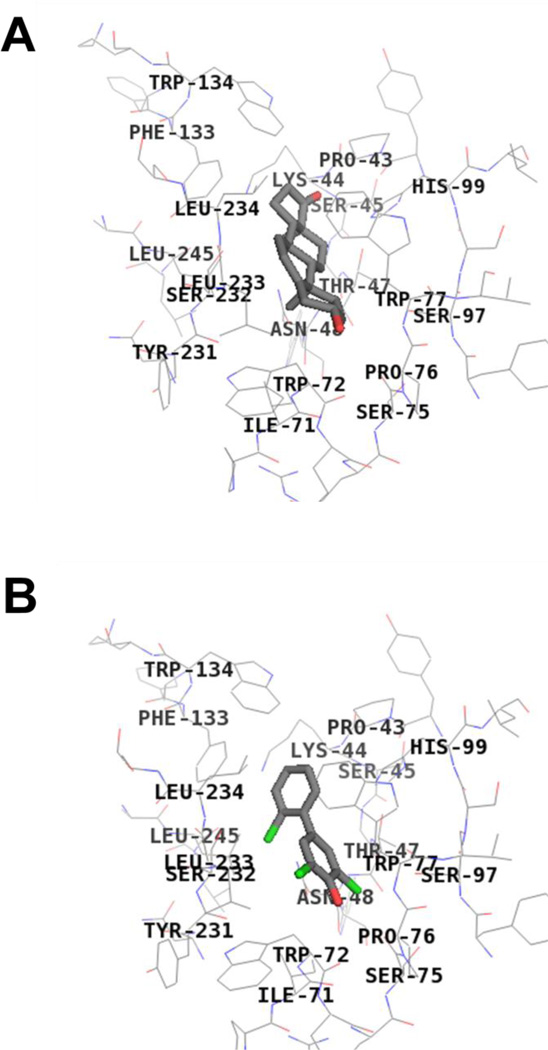
As exemplified by 4’-OHPCB 9 (Figure 6), the four OHPCBs (4’-OHPCB 9, 4’-OHPCB 33, 4-OHPCB 14, and 4-OHPCB 34) all docked at the DHEA binding site when using the model derived from the crystal with DHEA bound (PDB 1J99). Although there were differences in orientations of the docked molecules within the DHEA-binding site, all four OHPCBs yielded highest affinity docking results at this location within the model derived from the crystal structure with PAP bound (i.e., PDB 1EFH). An example of this docking at the DHEA-binding site is seen for 4-OHPCB 34 in Figure 7. Therefore, the docking experiments were consistent with the conclusion that all four OHPCBs are binding at the DHEA-binding site in both forms of the enzyme (i.e., the enzyme conformation m odeled by the crystal structure with DHEA bound and the conformation modeled by the crystal structure with PAP bound). Moreover, differences in the orientation of OHPCBs within the DHEA binding site in the model suggest that multiple modes of binding may occur within that binding cavity, and many of these may be catalytically non-productive.
Fig 7.
Binding interactions of DHEA and 4-OHPCB 34 with hSULT2A1 in the PAP-bound conformation. DHEA (Panel A) and 4-OHPCB 34 (Panel B) bind at the same site. Key hSULT2A1 residues interacting with the ligands at the active site are shown. Total score of 3.96 based on a consensus score of 3 for binding of DHEA, and total score of 3.63 based on a consensus score of 5 for binding of 4-OHPCB 34 were obtained. (crystal structure: hSULT2A1 in the conformation observed with PAP bound (1EFH)).
4. Discussion
Previous studies have shown that the fifteen OHPCBs investigated here inhibit the sulfation of DHEA catalyzed by hSULT2A1, and many of these were also substrates for the enzyme [21]. Our current results have indicated that most of the OHPCBs studied displayed noncompetitive inhibition patterns, and this included molecules that were known to be substrates for the enzyme as well as those for which no sulfation had been observed. The combination of equilibrium binding data and studies on inhibition kinetics for these OHPCBs has elucidated possible molecular interactions that influence specificity of the enzyme. Furthermore, molecular modeling studies have provided additional insight into the mechanisms for inhibition of the enzyme by these molecules.
The observation of a noncompetitive inhibition pattern for an enzyme indicates that a molecule binds to more than one form of the enzyme. While this sometimes means that an allosteric site is involved, it is common for two-substrate enzymes to display noncompetitive inhibition patterns due to the binding of the inhibitor to the same site as one substrate, but binding to different forms of the enzyme (e.g., free enzyme, enzyme bound to one substrate, and enzyme bound to one product) [45]. Indeed, an example of a sulfotransferase displaying this type of binding is illustrated in the recently reported crystal structure of hSULT1E1 with a noncompetitive inhibitor, 3,3’,5,5’-tetrabromo-diphenylether, located at the binding site for estradiol in the hSULT1E1-PAP-Inhibitor complex [46]. The combination of our studies on inhibition kinetics, equilibrium binding, and molecular modeling of interactions at the active site are consistent with a noncompetitive inhibition pattern derived from differences among binding interactions of the OHPCBs at the DHEA-binding site present in the free enzyme, the enzyme-PAPS complex, and/or the enzyme-PAP complex. Such an interpretation is also consistent with changes in conformation of the DHEA binding site that occur due to binding of PAPS to the enzyme. Recent simulations of structural changes in hSULT2A1 and other sulfotransferases indicate significant conformational changes in the DHEA-binding site that are linked to interactions with PAPS or PAP [47–49]. Such changes in the three dimensional structure of the DHEA-binding site would yield different enzyme forms that would provide noncompetitive inhibition patterns. Additionally, the multiple binding orientations of the OHPCBs within the active site of the enzyme that were observed through our modeling experiments may contribute to the observed inhibition patterns.
When the molecular docking studies are considered along with the binding of ANS to the PAP-site in a model derived from the crystal structure containing DHEA (i.e., PDB 1J99), the multiphasic nature of binding of some OHPCBs to the free enzyme may relate to differences in the ability of various OHPCBs to cause binding of ANS to the PAPS/PAP site. That is, the OHPCBs may differ in their ability to cause conformational changes in hSULT2A1 that mimic those elicited by DHEA, wherein ANS binds to the PAPS/PAP binding site. Further displacement of ANS from that site by the OHPCB might result in the multiphasic binding results observed for some of the OHPCBs.
Additional insight into the interactions of some OHPCBs with hSULT2A1 was obtained by examination of the binding of 4-OHPCB 14 and 4-OHPCB 34 to the free enzyme and to the enzyme-PAP complex (E-PAP). The equilibrium dissociation constants for the two molecules (Table 3) indicate that the binding to the free enzyme displays similar affinity. However, when binding to the free enzyme is compared to binding to the E-PAP complex (Figure 5), there is a difference in the change in fluorescence intensity due to displacement of ANS. This could be explained based on the structure of the enzyme wherein hSULT2A1, like other human cytosolic sulfotransferases, is a homodimer with two DHEA binding sites and two PAP binding sites. Therefore, under saturating concentrations of PAP there could be a conformational change that allows ANS to be displaced by OHPCBs from both subunits of the hSULT2A1 dimer resulting in the increase in the total change in fluorescence intensity observed. Moreover, in the first crystal structure solved for hSULT2A1 with bound PAP, the authors showed that hSULT2A1 activity may be regulated by conformational change that is induced by the dimer [44]. Studies on the binding of DHEA to hSULT2A1 have indicated negative cooperativity with two dissociation constants for interaction of DHEA with the enzyme [40]. Related differences in interaction with the DHEA-site in different enzyme forms may contribute to the noncompetitive inhibition pattern that was observed with most of the OHPCBs in the current study.
Thus, our results indicate that the OHPCBs interact with the DHEA-binding site of the hSULT2A1 in a manner that depends on the presence or absence of PAPS/PAP as well as the structure of the OHPCB. The precise role(s) that either the dimeric structure of hSULT2A1 or the conformational changes due to the presence or absence of PAPS or PAP may play in the structure-activity relationships for OHPCBs will require further extensive study with a broader range of both substrates and inhibitors. It is clear at this point, however, that in addition to their toxicological importance, OHPCBs serve as useful tools for probing the molecular mechanisms for catalysis and inhibition in hSULT2A1, an enzyme that functions in both the metabolism of endogenous steroids and bile acids as well as xenobiotics.
Supplementary Material
Highlights.
We studied the mechanism of inhibition of hSULT2A1 by hydroxylated PCBs (OHPCBs).
The OHPCBs studied bind at the sulfuryl-acceptor binding site of hSULT2A1.
This binding may occur with different orientations of the OHPCB.
The results suggest binding of OHPCBs to more than one form of hSULT2A1.
Acknowledgements
This study was supported by the National Institutes of Health through grants P42 ES013661 (to L.W.R., M.W.D. and H.J.L.) and R01 ES017425 (to H.J.L.) from the National Institute of Environmental Health Sciences, and research grant R01 CA038683 (to M.W.D.) from the National Cancer Institute. We also acknowledge programmatic support through the University of Iowa Environmental Health Sciences Research Center (NIEHS/NIH P30 ES05605). The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Abbreviations
- ANS
8-anilino-1-naphthalene sulfonic acid
- DTT
dithiothreitol
- OHPCB
hydroxylated polychlorinated biphenyl
- PAP
adenosine 3’,5’-diphosphate
- PAPS
adenosine 3’-phosphate 5’-phosphosulfate
- PCB
polychlorinated biphenyl
- 4’-OHPCB 3
4’-hydroxy-4-monochlorobiphenyl
- 4’-OHPCB 6
4’-hydroxy-2, 3’-dichlorobiphenyl
- 4-OHPCB 8
4-hydroxy-2, 4’-dichlorobiphenyl
- 4’-OHPCB 9
4’-hydroxy-2, 5-dichlorobiphenyl
- 4-OHPCB 11
4-hydroxy-3, 3’-dichlorobiphenyl
- 4’-OHPCB 12
4’-hydroxy-3, 4-dichlorobiphenyl
- 4-OHPCB 14
4-hydroxy-3, 5- dichlorobiphenyl
- 4’-OHPCB 25
4’-hydroxy-2, 3’, 4-trichlorobiphenyl
- 4’-OHPCB 33
4’-hydroxy-2’, 3, 4-trichlorobiphenyl
- 4-OHPCB 34
4-hydroxy-2’, 3, 5-trichlorobiphenyl
- 6’-OHPCB 35
6’-hydroxy-3, 3’-4 trichlorobiphenyl
- 4-OHPCB 36
4-hydroxy-3, 3’, 5-trichlorobiphenyl
- 4’-OHPCB 68
4’-hydroxy-2, 3’, 4, 5’-tetrachlorobiphenyl
- 3’
4’-diOHPCB 3, 3’, 4’-dihydroxy-4-monochlorobiphenyl
- 3’
4’-diOHPCB 5, 3’, 4’- dihydroxy-2, 3-dichlorobiphenyl
- SULT
sulfotransferase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare that there are no conflicts of interest.
References
- 1.Erickson MD, Kaley RG., 2nd Applications of polychlorinated biphenyls. Environ. Sci. Pollut. Res. Int. 2011;18:135–151. doi: 10.1007/s11356-010-0392-1. [DOI] [PubMed] [Google Scholar]
- 2.Martinez A, Wang K, Hornbuckle KC. Fate of PCB congeners in an industrial harbor of Lake Michigan. Environ. Sci. Technol. 2010;44:2803–2808. doi: 10.1021/es902911a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ludewig G, Lehmann L, Esch H, Robertson LW. Metabolic Activation of PCBs to Carcinogens in Vivo - A Review. Environ. Toxicol. Pharmacol. 2008;25:241–246. doi: 10.1016/j.etap.2007.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robertson LW, Hansen LG. PCBs: Recent Advances in Environmental Toxicology and Health Effects. Lexington: University of Kentucky Press; 2001. [Google Scholar]
- 5.Safe SH. Polychlorinated biphenyls (PCBs): environmental impact, biochemical and toxic responses, and implications for risk assessment. Crit. Rev. Toxicol. 1994;24:87–149. doi: 10.3109/10408449409049308. [DOI] [PubMed] [Google Scholar]
- 6.Currado GM, Harrad S. Comparison of polychlorinated biphenyl concentrations in indoor and outdoor air and the potential significance of inhalation as a human exposure pathway. Environ. Sci. Technol. 1998;32:3043–3047. [Google Scholar]
- 7.Harrad S, Ren J, Hazrati S, Robson M. Chiral signatures of PCB#s 95 and 149 in indoor air, grass, duplicate diets and human faeces. Chemosphere. 2006;63:1368–1376. doi: 10.1016/j.chemosphere.2005.09.027. [DOI] [PubMed] [Google Scholar]
- 8.Herrick RF, McClean MD, Meeker JD, Baxter LK, Weymouth GA. An unrecognized source of PCB contamination in schools and other buildings. Environ. Health Perspect. 2004;112:1051–1053. doi: 10.1289/ehp.6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Correa PA, Lin L, Just CL, Hu D, Hornbuckle KC, Schnoor JL, Van Aken B. The effects of individual PCB congeners on the soil bacterial community structure and the abundance of biphenyl dioxygenase genes. Environ. Int. 2010;36:901–906. doi: 10.1016/j.envint.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gabrio T, Piechotowski I, W allenhorst T, Klett M, Cott L, Friebel P, Link B, Schwenk M. PCB-blood levels in teachers, working in PCB-contaminated schools. Chemosphere. 2000;40:1055–1062. doi: 10.1016/s0045-6535(99)00353-7. [DOI] [PubMed] [Google Scholar]
- 11.Herrick RF, Meeker JD, Altshul L. Serum PCB levels and congener profiles among teachers in PCB-containing schools: a pilot study. Environ. Health. 2011;10:56. doi: 10.1186/1476-069X-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez A, Norstrom K, Wang K, Hornbuckle KC. Polychlorinated biphenyls in the surficial sediment of Indiana Harbor and Ship Canal, Lake Michigan. Environ. Int. 2010;36:849–854. doi: 10.1016/j.envint.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Norstrom K, Czub G, McLachlan MS, Hu D, Thorne PS, Hornbuckle KC. External exposure and bioaccumulation of PCBs in humans living in a contaminated urban environment. Environ. Int. 2010;36:855–861. doi: 10.1016/j.envint.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Persoon C, Peters TM, Kumar N, Hornbuckle KC. Spatial distribution of airborne polychlorinated biphenyls in Cleveland, Ohio and Chicago, Illinois. Environ. Sci. Technol. 2010;44:2797–2802. doi: 10.1021/es901691s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oakley GG, Devanaboyina U, Robertson LW, Gupta RC. Oxidative DNA damage induced by activation of polychlorinated biphenyls (PCBs): implications for PCB-induced oxidative stress in breast cancer. Chem. Res. Toxicol. 1996;9:1285–1292. doi: 10.1021/tx960103o. [DOI] [PubMed] [Google Scholar]
- 16.Kester MH, Bulduk S, Tibboel D, Meinl W, Glatt H, Falany CN, Coughtrie MW, Bergman A, Safe SH, Kuiper GG, Schuur AG, Brouwer A, Visser TJ. Potent inhibition of estrogen sulfotransferase by hydroxylated PCB metabolites: a novel pathway explaining the estrogenic activity of PCBs Endocrinology. 2000;141:1897–1900. doi: 10.1210/endo.141.5.7530. [DOI] [PubMed] [Google Scholar]
- 17.Schuur AG, Brouwer A, Bergman A, Coughtrie MW, Visser TJ. Inhibition of thyroid hormone sulfation by hydroxylated metabolites of polychlorinated biphenyls. Chem. Biol. Interact. 1998;109:293–297. doi: 10.1016/s0009-2797(97)00140-3. [DOI] [PubMed] [Google Scholar]
- 18.Lans MC, Klasson-Wehler E, Willemsen M, Meussen E, Safe S, Brouwer A. Structure-dependent, competitive interaction of hydroxy-polychlorobiphenyls, -dibenzo-p-dioxins and -dibenzo-furans with human transthyretin. Chem. Biol Interact. 1993;88:7–21. doi: 10.1016/0009-2797(93)90081-9. [DOI] [PubMed] [Google Scholar]
- 19.Purkey HE, Palaninathan SK, Kent KC, Smith C, Safe SH, Sacchettini JC, Kelly JW. Hydroxylated polychlorinated biphenyls selectively bind transthyretin in blood and inhibit amyloidogenesis: rationalizing rodent PCB toxicity. Chem. Biol. 2004;11:1719–1728. doi: 10.1016/j.chembiol.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 20.Grimm FA, Lehmler HJ, He X, Robertson LW, Duffel MW. Sulfated metabolites of polychlorinated biphenyls are high-affinity ligands for the thyroid hormone transport protein transthyretin. Environ. Health Perspect. 2013;121:657–662. doi: 10.1289/ehp.1206198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ekuase EJ, Liu Y, Lehmler HJ, Robertson LW, Duffel MW. Structure-activity relationships for hydroxylated polychlorinated biphenyls as inhibitors of the sulfation of dehydroepiandrosterone catalyzed by human hydroxysteroid sulfotransferase SULT2A1. Chem. Res. Toxicol. 2011;24:1720–1728. doi: 10.1021/tx200260h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y, Apak TI, Lehmler HJ, Robertson LW, Duffel MW. Hydroxylated polychlorinated biphenyls are substrates and inhibitors of human hydroxysteroid sulfotransferase SULT2A1. Chem. Res. Toxicol. 2006;19:1420–1425. doi: 10.1021/tx060160+. [DOI] [PubMed] [Google Scholar]
- 23.Duffel MW. Sulfotransferases. In: McQueen CA, Guengerich FP, editors. Comprehensive Toxicology, Vol. 4 Biotransformation. Oxford: Elsevier; 2010. pp. 367–384. Vol. Ed. [Google Scholar]
- 24.Falany CN. Enzymology of human cytosolic sulfotransferases. FASEB J. 1997;11:206–216. doi: 10.1096/fasebj.11.4.9068609. [DOI] [PubMed] [Google Scholar]
- 25.Gamage N, Barnett A, Hempel N, Duggleby RG, Windmill KF, Martin JL, McManus ME. Human sulfotransferases and their role in chemical metabolism. Toxicol. Sci. 2006;90:5–22. doi: 10.1093/toxsci/kfj061. [DOI] [PubMed] [Google Scholar]
- 26.Radominska A, Comer KA, Zimniak P, Falany J, Iscan M, Falany CN. Human liver steroid sulphotransferase sulphates bile acids. Biochem. J. 1990;272:597–604. doi: 10.1042/bj2720597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strott CA. Hydroxysteroid sulfotransferases SULT2A1, SULT2B1a, and SULT2B1b. In: Pacifici GM, Coughtrie MWH, editors. Human cytosolic sulfotransferases. Boca Raton, FL: CRC Press; 2005. pp. 231–253. [Google Scholar]
- 28.Barker EV, Hume R, Hallas A, Coughtrie WH. Dehydroepiandrosterone sulfotransferase in the developing human fetus: quantitative biochemical and immunological characterization of the hepatic, renal, and adrenal enzymes. Endocrinology. 1994;134:982–989. doi: 10.1210/endo.134.2.8299591. [DOI] [PubMed] [Google Scholar]
- 29.Riches Z, Stanley EL, Bloomer JC, Coughtrie MW. Quantitative evaluation of the expression and activity of five major sulfotransferases (SULTs) in human tissues: the SULT "pie". Drug Metab. Dispos. 2009;37:2255–2261. doi: 10.1124/dmd.109.028399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mesia-Vela S, Sanchez RI, Estrada-Muniz E, Alavez-Solano D, Torres-Sosa C, Jimenez M, Estrada, Reyes-Chilpa R, Kauffman FC. Natural products isolated from Mexican medicinal plants: novel inhibitors of sulfotransferases, SULT1A1 and SULT2A1. Phytomedicine. 2001;8:481–488. doi: 10.1078/s0944-7113(04)70070-7. [DOI] [PubMed] [Google Scholar]
- 31.Pacifici GM, Temellini A, Castiglioni M, D'Alessandro C, Ducci A, Giuliani L. Interindividual variability of the human hepatic sulphotransferases. Chem. Biol. Interact. 1994;92:219–231. doi: 10.1016/0009-2797(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 32.Wang LQ, James MO. Inhibition of sulfotransferases by xenobiotics. Curr. Drug Metab. 2006;7:83–104. doi: 10.2174/138920006774832596. [DOI] [PubMed] [Google Scholar]
- 33.Waring RH, Ayers S, Gescher AJ, Glatt HR, Meinl W, Jarratt P, Kirk CJ, Pettitt T, Rea D, Harris RM. Phytoestrogens and xenoestrogens: the contribution of diet and environment to endocrine disruption. J. Steroid Biochem. Mol. Biol. 2008;108:213–220. doi: 10.1016/j.jsbmb.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 34.Lehmler HJ, Robertson LW. Synthesis of hydroxylated PCB metabolites with the Suzuki coupling. Chemosphere. 2001;45:1119–1127. doi: 10.1016/s0045-6535(01)00052-2. [DOI] [PubMed] [Google Scholar]
- 35.Sekura RD. Adenosine 3'-phosphate 5'-phosphosulfate. Methods Enzymol. 1981;77:413–415. [Google Scholar]
- 36.Gulcan HO, Liu Y, Duffel MW. Pentachlorophenol and other chlorinated phenols are substrates for human hydroxysteroid sulfotransferase hSULT2A1. Chem. Res. Toxicol. 2008;21:1503–1508. doi: 10.1021/tx800133d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bensadoun A, Weinstein D. Assay of proteins in the presence of interfering materials. Anal. Biochem. 1976;70:241–250. doi: 10.1016/s0003-2697(76)80064-4. [DOI] [PubMed] [Google Scholar]
- 38.Sekura RD, Jakoby WB. Phenol Sulfotransferases. J. Biol. Chem. 1979;254:5658–5663. [PubMed] [Google Scholar]
- 39.Sheng J, Sharma V, Duffel MW. Measurement of aryl and alcohol sulfotransferase activity, Current Protocols in Toxicology. New York: John Wiley & Sons, Inc; 2001. pp. 4.5.1–4.5.9. [DOI] [PubMed] [Google Scholar]
- 40.Gulcan HO, Duffel MW. Substrate inhibition in human hydroxysteroid sulfotransferase SULT2A1: studies on the formation of catalytically non-productive enzyme complexes. Arch. Biochem. Biophys. 2011;507:232–240. doi: 10.1016/j.abb.2010.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Y, Lehmler HJ, Robertson LW, Duffel MW. Physicochemical properties of hydroxylated polychlorinated biphenyls aid in predicting their interactions with rat sulfotransferase 1A1 (rSULT1A1) Chem. Biol. Interact. 2011;189:153–160. doi: 10.1016/j.cbi.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marshall AD, Darbyshire JF, Hunter AP, McPhie P, Jakoby WB. Control of activity through oxidative modification at the conserved residue Cys66 of aryl sulfotransferase IV. J. Biol. Chem. 1997;272:9153–9160. doi: 10.1074/jbc.272.14.9153. [DOI] [PubMed] [Google Scholar]
- 43.Rehse PH, Zhou M, Lin SX. Crystal structure of human dehydroepiandrosterone sulphotransferase in complex with substrate. [erratum appears in Biochem J 2002 Jun 15;364(Pt 3):888.] Biochem. J. 2002;364:165–171. doi: 10.1042/bj3640165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pedersen LC, Petrotchenko EV, Negishi M. Crystal structure of SULT2A3, human hydroxysteroid sulfotransferase. FEBS Lett. 2000;475:61–64. doi: 10.1016/s0014-5793(00)01479-4. [DOI] [PubMed] [Google Scholar]
- 45.Blat Y. Non-competitive inhibition by active site binders. Chem Biol Drug Des. 2010;75:535–540. doi: 10.1111/j.1747-0285.2010.00972.x. [DOI] [PubMed] [Google Scholar]
- 46.Gosavi RA, Knudsen GA, Birnbaum LS, Pedersen LC. Mimicking of estradiol binding by flame retardants and their metabolites: a crystallographic analysis. Environ. Health Perspect. 2013;121:1194–1199. doi: 10.1289/ehp.1306902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cook I, Wang T, Almo SC, Kim J, Falany CN, Leyh TS. The gate that governs sulfotransferase selectivity. Biochemistry. 2013;52:415–424. doi: 10.1021/bi301492j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cook I, Wang T, Almo SC, Kim J, Falany CN, Leyh TS. Testing the sulfotransferase molecular pore hypothesis. J. Biol. Chem. 2013;288:8619–8626. doi: 10.1074/jbc.M112.445015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cook I, Wang T, Falany CN, Leyh TS. A nucleotide-gated molecular pore selects sulfotransferase substrates. Biochemistry. 2012;51:5674–5683. doi: 10.1021/bi300631g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.