Abstract
Background
Studies have suggested that selective microbial targets prevail in the fecal microbiota of infants with eczema. This study evaluated the composition of fecal microbiota of infants who developed eczema in the first 5 years of life and compared these with those of healthy controls.
Findings
Children who developed eczema in the first 2 years, those with eczema at 5 years of age and healthy controls were selected from the placebo arm of a birth cohort of at-risk infants participating in a randomized double-blind trial on the protective effects of supplemental probiotics in early life on allergic outcomes. Molecular evaluation of fecal microbiota were conducted using Fluorescence In Situ Hybridization-Flow Cytometry (FISH-FC) for fecal samples collected. Longitudinal analysis of fecal microbiota composition at three days, one and three months and one year of life revealed higher abundance of Enterobacteriaceae [coefficient (B): 1.081, 95% CI: 0.229-1.933, adj p = 0.014] and Clostridium perfringens [coefficient (B): 0.521, 95% CI: 0.556-0.988, adj p = 0.03] in those who developed eczema in the first 2 years life. In those with eczema at 5 years of age, a lower abundance of Bifidobacterium was observed [coefficient (B): -27.635, 95% CI: -50.040 - -5.231, adj p = 0.018].
Conclusions
The differences in infant fecal microbiota observed in eczema subjects in this study support the notion that relative abundance of selective microbial targets may contribute to the subsequent development of eczema in childhood.
Findings
Background
Allergic diseases, such as eczema, are chronic inflammatory disorders with increasing global trends [1]. Intestinal microbiota play a role in the regulation of innate and adaptive immunity [2] and has been implicated in the development of allergy diseases [3]. The composition of fecal microbiota of infants and young children with eczema differ from healthy children [4]. This study aims to evaluate and monitor the composition, maturation, development of fecal microbiota at 4 times points of an at risk birth cohort in the first year of life, and compare these findings in relation to the development of eczema at 2 and 5 years of age with those of healthy controls.
Methods
Subjects with eczema and healthy controls were selected from the placebo arm of a birth cohort of at-risk infants participating in a randomized double-blind trial on the protective effects of supplemental probiotics in early life on allergic outcomes [5]. Subjects who developed eczema in the first 2 years (eczema, n = 28; healthy, n = 32), and those with eczema at 5 years of age (eczema, n = 15 [persistent from 2 years: n = 11; new cases: n = 4]; healthy, n = 19) were studied. Thirteen controls were excluded in the 5 year analysis because they developed allergen sensitization, rhinitis and wheeze at 5 years of age (Figure 1). Informed consent was obtained from all families. The study was approved by the hospital’s institutional ethical review board (Ref Code: 2006/00008). Stool samples were collected at 4 time points (3 days, 1, 3 and 12 months of age) as previously described [6] There were missing stool samples in 2% to 25% of subjects at different time points, but these were not different between cases and controls. Subjects with eczema in the first 2 years and at 5 years of age were subclassified into atopic (positive skin prick test to common allergens) eczema (2 yrs: n = 13; 5 yrs: n = 12) and non-atopic eczema (2 yrs: n = 15; 5 yrs: n = 3).
Figure 1.
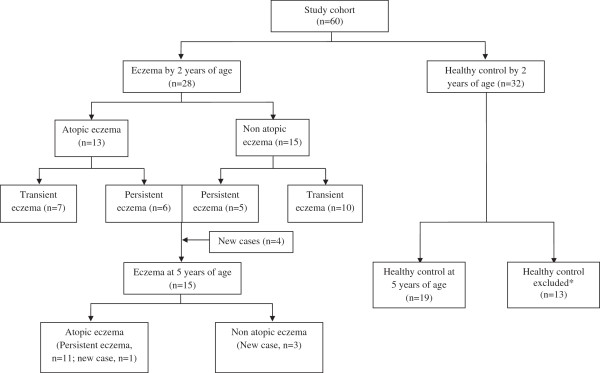
Flowchart of study subjects showing the progress of eczema development in the first 2 years and at 5 years of age. *Thirteen controls were excluded in the 5 year analysis because they developed allergen sensitization, rhinitis and wheeze at 5 years of age.
Molecular evaluation of fecal microbiota was conducted using/harnessing whole-cell-based detection approach based on Fluorescence In Situ Hybridization combined with Flow Cytometry (FISH-FC) for fecal samples collected at 3 days, 1, 3 and 12 months. 16S rRNA probes were selected to target and quantify the Eubacterium rectale-Clostridium coccoides group (Erec 482), Clostridium leptum subgroup (Clep 866 and the corresponding competitor probes), Bacteroides-Prevotella group (Bac 303), Bifidobacterium genus (Bif 164), Atopobium group (Ato 291), Lactobacilli-Enterococci group (Lab 158), Enterobacteriaceae family (Enter 1432), Clostridium perfringens (Cperf191), Clostridium difficile (Cdif198) and Escherichia coli (Eco 1531) as previously described [7].
Linear mixed model was used to evaluate the longitudinal differences (i.e. 4 time points) of bacterial targets with adjustments for gender, mode of delivery, breastfeeding up to 6 months and comorbidities (rhinitis and wheeze) [8].
Results
The demographic characteristics and data showing the relative abundance of fecal bacterial groups for children who developed eczema by 2 and at 5 years of age and their healthy controls are summarized in Tables 1, 2 and 3 respectively. Longitudinal monitoring of the dynamics of intestinal bacterial colonization over 4 time points (3 days, one month, three months and one year of life) in the first year of life showed higher relative abundance of Enterobacteriaceae [coefficient (B): 1.081, 95% CI: 0.229-1.933, adj p = 0.014] in children with eczema by 2 years of age, and Clostridium perfringens [coefficient (B): 0.521, 95% CI: 0.556-0.988, adj p = 0.03] in eczema n = 28) by 2 years of age compared to healthy controls (n = 32) (Table 2 and Figure 2a).
Table 1.
Demographic characteristics of children with eczema by 2 years and at 5 years of age and their healthy controls
| |
By 2 years of age |
|
At 5 years of age |
|
||
|---|---|---|---|---|---|---|
| Eczema (n = 28) | Healthy control (n = 32) | p value | Eczema (n = 15) | Healthy control (n = 19) | p value | |
|
Gender (%) |
|
|
|
|
|
|
| Male |
14 (50) |
15 (46.9) |
0.809 |
6 (40) |
9 (47.4) |
0.667 |
| Female |
14 (50) |
17 (53.1) |
|
9 (60) |
10 (52.6) |
|
|
Mode of delivery (%) |
|
|
|
|
|
|
| Lower segment caesarean section |
7 (25) |
9 (28.1) |
0.785 |
4 (26.7) |
4 (21.1) |
1 |
| Vaginal delivery |
21 (75) |
23 (71.9) |
|
11 (73.3) |
15 (78.9) |
|
|
Feeding history from birth to month 6 (%) |
|
|
|
|
|
|
| Breastfeeding and formula feeding |
21 (75) |
26 (81.3) |
0.558 |
11 (73.3) |
16 (84.2) |
0.672 |
| Total formula feeding | 7 (25) | 6 (18.7) | 4 (26.7) | 3 (15.8) | ||
Table 2.
Relative abundance of fecal bacterial groups for children with eczema by 2 years of age and their healthy controls
| |
|
Healthy |
Eczema by 2 years of age |
||
|---|---|---|---|---|---|
| Time points | Bacterial group | n | Mean (SD) | n | Mean (SD) |
| 3 day |
E.rectale - C. coccoides |
21 |
0.27 (1.139) |
19 |
ND |
| |
Clostridium leptum |
21 |
3.08 (9.155) |
19 |
ND |
| |
Bacteriodes-Prevotella |
21 |
5.56 (11.654) |
19 |
6.88 (19.386) |
| |
Bifidobacterium |
21 |
29.35 (36.872) |
19 |
15.88 (30.175) |
| |
Atopobium |
21 |
0.44 (0.801) |
19 |
0.86 (3.555) |
| |
Lactobacilli–Enterococci |
21 |
3.22 (8.631) |
19 |
0.49 (1.370) |
| |
Enterobacteriaceae |
21 |
26.22 (35.55) |
19 |
41.64 (37.819) |
| |
Clostridium perfringens |
12 |
2.05 (7.093) |
16 |
2.99 (8.137) |
| |
Clostridium difficile |
12 |
ND |
16 |
0.66 (2.365) |
| |
E.coli |
11 |
14.17 (29.862) |
16 |
34.98 (40.363) |
| 1 month |
E.rectale–C. coccoides |
23 |
0.29 (0.589) |
19 |
6.65 (18.510) |
| |
Clostridium leptum |
23 |
ND |
19 |
2.47 (9.339) |
| |
Bacteriodes-Prevotella |
23 |
1.65 (3.309) |
19 |
6.01 (16.408) |
| |
Bifidobacterium |
23 |
41.27 (38.514) |
19 |
28.38 (33.945) |
| |
Atopobium |
23 |
6.52 (12.635) |
19 |
2.32 (6.41) |
| |
Lactobacilli–Enterococci |
23 |
2.74 (7.265) |
19 |
7.98 (15.762) |
| |
Enterobacteriaceae |
23 |
15.63 (23.726) |
19 |
20.1 (23.837) |
| |
Clostridium perfringens |
15 |
0.19 (0.751) |
18 |
0.55 (1.482) |
| |
Clostridium difficile |
15 |
ND |
18 |
ND |
| |
E.coli |
15 |
9.01 (18.952) |
18 |
3.62 (4.631) |
| 3 month |
E.rectale–C. coccoides |
28 |
5.45 (10.551) |
24 |
7.43 (13.389) |
| |
Clostridium leptum |
28 |
0.46 (1.590) |
24 |
0.01 (0.059) |
| |
Bacteriodes-Prevotella |
28 |
3.53 (7.845) |
24 |
3.32 (5.126) |
| |
Bifidobacterium |
28 |
55.86 (32.344) |
24 |
54.21 (33.827) |
| |
Atopobium |
28 |
10.19 (16.06) |
24 |
3.72 (7.037) |
| |
Lactobacilli–Enterococci |
28 |
2.77 (3.121) |
24 |
2.62 (3.404) |
| |
Enterobacteriaceae |
28 |
3.8 (7.222) |
24 |
6.51 (10.024) |
| |
Clostridium perfringens |
17 |
0.72 (1.547) |
20 |
2.26 (6.246) |
| |
Clostridium difficile |
17 |
ND |
20 |
ND |
| |
E.coli |
17 |
1.16 (2.725) |
20 |
4.26 (10.987) |
| 1 year |
E.rectale–C. coccoides |
27 |
30.40 (18.445) |
24 |
31.25 (16.0.42) |
| |
Clostridium leptum |
27 |
4.22 (7.022) |
24 |
1.33 (3.047) |
| |
Bacteriodes–Prevotella |
27 |
10.05 (10.082) |
24 |
4.175 (6.049) |
| |
Bifidobacterium |
27 |
28.88 (23.227) |
24 |
27.92 (25.196) |
| |
Atopobium |
27 |
5.91 (8.639) |
24 |
5.05 (9.337) |
| |
Lactobacilli–Enterococci |
27 |
1.57 (3.437) |
24 |
1.45 (4.99) |
| |
Enterobacteriaceae |
27 |
0.10 (0.3) |
24 |
1.65 (3.162) |
| |
Clostridium perfringens |
18 |
ND |
24 |
0.30 (1.457) |
| |
Clostridium difficile |
18 |
ND |
22 |
0.07 (0.328) |
| E.coli | 18 | 0.05 (0.129) | 23 | 0.83 (2.22) | |
ND, not detected.
Table 3.
Relative abundance of fecal bacterial groups for children with eczema at 5 years of age and their healthy controls
| |
|
Healthy |
Eczema at 5 years of age |
||
|---|---|---|---|---|---|
| Time points | Bacterial group | n | Mean (SD) | n | Mean (SD) |
| 3 day |
E.rectale–C. coccoides |
10 |
ND |
9 |
ND |
| |
Clostridium leptum |
10 |
2.7 (8.526) |
9 |
3.74 (11.217) |
| |
Bacteriodes-Prevotella |
10 |
10.32 (15.729) |
9 |
7.49 (19.590) |
| |
Bifidobacterium |
10 |
21.65 (33.2) |
9 |
24.45 (34.194) |
| |
Atopobium |
10 |
0.21 (0.439) |
9 |
0.12 (0.229) |
| |
Lactobacilli–Enterococci |
10 |
5.48 (12.272) |
9 |
0.64 (1.307) |
| |
Enterobacteriaceae |
10 |
47.73 (41.03) |
9 |
30.89 (39.958) |
| |
Clostridium perfringens |
7 |
ND |
5 |
ND |
| |
Clostridium difficile |
7 |
ND |
5 |
ND |
| |
E.coli |
7 |
21.87 (35.99) |
5 |
20.36 (39.873) |
| 1 month |
E.rectale–C. coccoides |
13 |
0.32 (0.670) |
10 |
0.74 (1.687) |
| |
Clostridium leptum |
13 |
ND |
10 |
4.06 (12.829) |
| |
Bacteriodes-Prevotella |
13 |
2.92 (4.008) |
10 |
6.56 (20.824) |
| |
Bifidobacterium |
13 |
48.29 (36.270) |
10 |
21.28 (28.332) |
| |
Atopobium |
13 |
5.15 (9.403) |
10 |
6.01 (12.463) |
| |
Lactobacilli–Enterococci |
13 |
1.91 (3.987) |
10 |
5.77 (18.246) |
| |
Enterobacteriaceae |
13 |
19.54 (28.826) |
10 |
18.19 (20.289) |
| |
Clostridium perfringens |
10 |
0.29 (0.92) |
6 |
ND |
| |
Clostridium difficile |
10 |
ND |
6 |
ND |
| |
E.coli |
10 |
11.62 (22.528) |
6 |
5.27 (7.354) |
| 3 month |
E.rectale–C. coccoides |
16 |
4.83 (10.187) |
12 |
7.67 (14.804) |
| |
Clostridium leptum |
16 |
0.17 (0.688) |
12 |
0.2 (0.678) |
| |
Bacteriodes-Prevotella |
16 |
2.83 (5.904) |
12 |
6.42 (9.913) |
| |
Bifidobacterium |
16 |
55.21 (33.0) |
12 |
42.557 (35.960) |
| |
Atopobium |
16 |
12.73 (19.887) |
12 |
5.96 (9.282) |
| |
Lactobacilli–Enterococci |
16 |
2.79 (3.085) |
12 |
3.07 (3.726) |
| |
Enterobacteriaceae |
16 |
4.28 (6.634) |
12 |
10.94 (14.169) |
| |
Clostridium perfringens |
12 |
0.98 (1.791) |
7 |
6.50 (9.55) |
| |
Clostridium difficile |
12 |
ND |
7 |
ND |
| |
E.coli |
12 |
1.64 (3.152) |
7 |
6.83 (13.998) |
| 1 year |
E.rectale–C. coccoides |
15 |
28.20 (19.361) |
12 |
29.78 (15.534) |
| |
Clostridium leptum |
15 |
3.54 (7.102) |
12 |
1.84 (2.359) |
| |
Bacteriodes-Prevotella |
15 |
9.22 (10.450) |
12 |
4.86 (8.232) |
| |
Bifidobacterium |
15 |
37.97 (23.939) |
12 |
18.88 (24.0) |
| |
Atopobium |
15 |
6.45 (9.575) |
12 |
4.40 (5.199) |
| |
Lactobacilli–Enterococci |
15 |
1.59 (3.724) |
12 |
3.29 (6.987) |
| |
Enterobacteriaceae |
15 |
0.04 (0.152) |
12 |
1.59 (3.133) |
| |
Clostridium perfringens |
12 |
ND |
9 |
ND |
| |
Clostridium difficile |
12 |
ND |
7 |
0.22 (0.582) |
| E.coli | 12 | 0.07 (0.155) | 8 | 1.78 (3.521) | |
ND, not detected.
Figure 2.
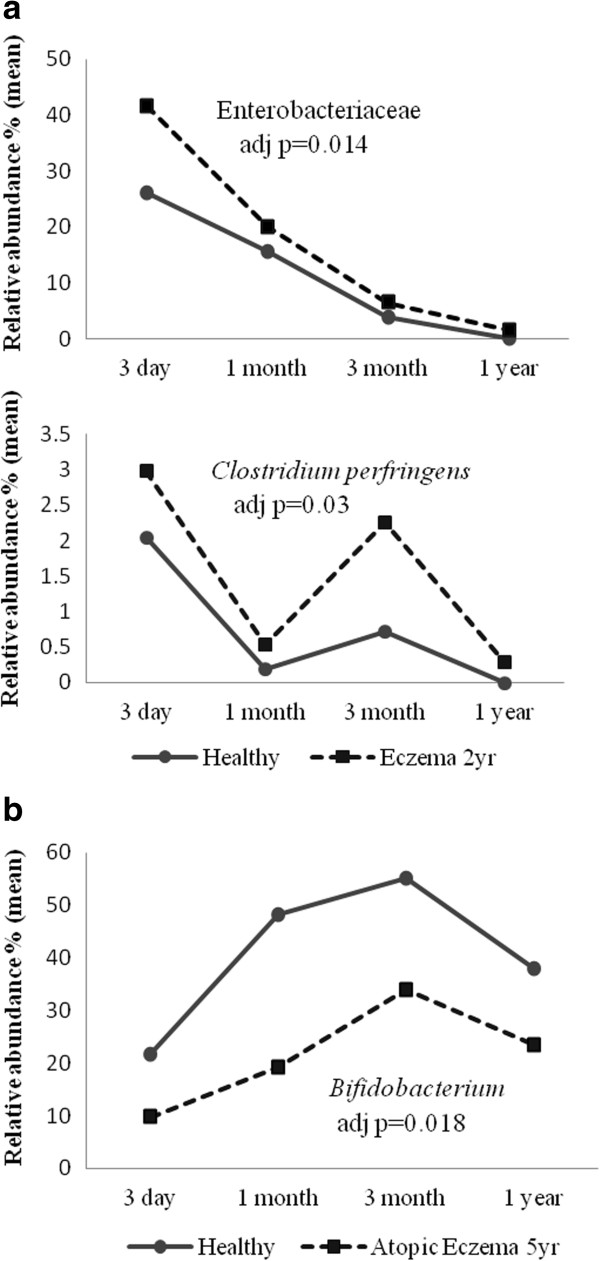
Longitudinal comparison of the fecal microbiota of children with and without eczema. Linear mixed model analysis of fecal microbiota of children with eczema in (a) the first 2 years of life and (b) at 5 years of age, compared to healthy controls. Only microbial targets with statistically significant difference between eczema and controls are shown.
Sub-group analysis of atopic eczema (n = 13) and non-atopic eczema (n = 15) cases with onset in first 2 years of life yielded similar results, with higher abundance of Enterobacteriaceae compared to controls [atopic eczema: coefficient (B): 0.949, 95% CI: 0.214-1.683, adj p = 0.013; non atopic eczema: coefficient (B): 1.119, 95% CI: 0.136-2.102, adj p = 0.027], and Clostridium perfringens for non-atopic eczema only [coefficient (B): 0.713, 95% CI: 0.124-1.303, adj p = 0.022]. This analysis was also carried out with controls (n = 19) who were healthy all the way till 5 years of age (that is did not develop eczema after the age of 2 years), and similar findings were observed (data not shown).
For atopic eczema at 5 years old, a lower relative abundance of Bifidobacterium was found in cases (n = 15) compared to healthy controls (n = 19) [coefficient (B): -27.635, 95% CI: -50.040 - -5.231, adj p = 0.018] (Table 3 and Figure 2b).
Discussion
In this study significant differences in fecal microbiota signatures analyzed at 4 times points in the first year of life were observed in infants who developed eczema in the first 5 years of life. These data suggests that specific microbial signatures appearing in early life might be predictive of eczema. Previous studies have also compared infant microbiota at more than one time point in infancy [3,9]. Sjögren et al. reported lower prevalence of lactobacilli group I and Bifidobacterium adolescentis at 1 week after birth for atopic dermatitis compared to non-allergic controls, but no significant differences were found for the other time points i.e. 1 month and 2 months [3]. In another study, reduced microbial diversity at 1 month and 12 months were associated with the presence of serum specific IgE, while allergic rhinitis group at school age showed less microbial diversity at 1 month [9]. Instead of making comparisons between single time points, our study utilized linear mixed model analysis to enabled comparisons over 4 time points in the first year of life. Our findings indicate that a consistent pattern in profile of fecal microbiota over the first year was associated with development of childhood eczema.
Our findings support the single time point cross sectional studies, where increased numbers of bacterium from Clostridium cluster IV and XIVa have been reported. These clusters of Clostridium are normally found in abundance in the adult intestine [10,11]. Although we did not find any difference for Clostridium cluster IV and XIVa in our study but we did observe an increase in Clostridium perfringens in children with eczema. This findings was supported by a recent study where Clostridium cluster I was associated with higher risk of developing atopic dermatitis [12].
An increased abundance of Enterobacteriaceae has also been observed in atopic subjects [13]. We have also previously shown by 16S rRNA pyrosequencing in this same cohort that fecal Enterobacteriaceae was more abundant at the age of 1 month in eczema compared to match controls [14]. Higher abundance of Enterobacteriaceae has been reported to be associated with intestinal inflammation (dextran sodium sulfate induced colitis), suggesting that Enterobacteriaceae may be responsible for promoting immune dysregulation [15].
The reduced abundance of Bifidobacterium in eczema compared to healthy controls has been observed in several other studies [3,7], and is associated with an increased risk of atopic dermatitis [16]. We did not observe reduced abundance of Bifidobacterium for eczema in the first 2 years of life compared to healthy controls. However, in subjects whose onset of eczema occurred by the age of 2 years (n = 28), we found that in those with eczema persistent till 5 years (n = 11) had lower abundances of Bifidobacterium compared with those with transient eczema (resolved before 5 years) (n = 17) as well as healthy controls, but these differences did reach statistical significance (p = 0.099) (data not shown). These observed differences, occurring in early life, in the maturation and development of the infant gut microbiota between subjects with atopic dermatitis and healthy controls suggests that the gut microbiota may influence immune system development [17] and contribute to disease severity [2].
In conclusion, this prospective study which profiled the dynamics of intestinal bacterial colonization over infancy supports the notion that relative abundance of selective microbial targets may contribute to the subsequent development of eczema in childhood.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
GCY performed the experiments, data analysis and statistical analysis. EXLL drafted the manuscript. GCY and BWL helped to revise the manuscript. QSL participated in collation of clinical data. MA, LPCS and BWL participated in the study design and helped in coordination of sample and clinical data collection. All authors read and approved the final manuscript.
Contributor Information
Gaik Chin Yap, Email: paeyapgc@nus.edu.sg.
Evelyn Xiu Ling Loo, Email: paelxle@nus.edu.sg.
Marion Aw, Email: paeawm@nus.edu.sg.
Qingshu Lu, Email: qingshu.lu@scri.edu.sg.
Lynette Pei-Chi Shek, Email: paeshekl@nus.edu.sg.
Bee Wah Lee, Email: paeleebw@nus.edu.sg.
Acknowledgements
The authors would like to thank Drs Dawn Lim, Irvin Gerez and Genevieve Llanora, Ms Hor Chuen Yee, Corinne Kwek Poh Lian and Judy Anthony who assisted with follow-up of the subjects. The voluntary participation of all subjects in this study is sincerely appreciated. This study was supported by MOH’s National Medical Research Council (NMRC/EDG/1057/2011).
References
- Ngoc PL, Gold DR, Tzianabos AO, Weiss ST, Celedon JC. Cytokines, allergy, and asthma. Curr Opin Allergy Clin Immunol. 2005;5(2):161–166. doi: 10.1097/01.all.0000162309.97480.45. [DOI] [PubMed] [Google Scholar]
- Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474(7351):327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjogren YM, Jenmalm MC, Bottcher MF, Bjorksten B, Sverremark-Ekstrom E. Altered early infant gut microbiota in children developing allergy up to 5 years of age. Clin Exp Allergy. 2009;39(4):518–526. doi: 10.1111/j.1365-2222.2008.03156.x. [DOI] [PubMed] [Google Scholar]
- Forno E, Onderdonk AB, McCracken J, Litonjua AA, Laskey D, Delaney ML, Dubois AM, Gold DR, Ryan LM, Weiss ST, Celedon JC. Diversity of the gut microbiota and eczema in early life. Clin Mol Allergy: CMA. 2008;6:11. doi: 10.1186/1476-7961-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh SE, Aw M, Gerez I, Chong YS, Rauff M, Ng YP, Wong HB, Pai N, Lee BW, Shek LP. Probiotic supplementation in the first 6 months of life in at risk Asian infants–effects on eczema and atopic sensitization at the age of 1 year. Clin Exp Allergy. 2009;39(4):571–578. doi: 10.1111/j.1365-2222.2008.03133.x. [DOI] [PubMed] [Google Scholar]
- Mah KW, Chin VI, Wong WS, Lay C, Tannock GW, Shek LP, Aw MM, Chua KY, Wong HB, Panchalingham A, Lee BW. Effect of a milk formula containing probiotics on the fecal microbiota of asian infants at risk of atopic diseases. Pediatr Res. 2007;62(6):674–679. doi: 10.1203/PDR.0b013e31815991d5. [DOI] [PubMed] [Google Scholar]
- Mah KW, Bjorksten B, Lee BW, van Bever HP, Shek LP, Tan TN, Lee YK, Chua KY. Distinct pattern of commensal gut microbiota in toddlers with eczema. Int Arch Allergy Immunol. 2006;140(2):157–163. doi: 10.1159/000092555. [DOI] [PubMed] [Google Scholar]
- Chan YH. Biostatistics 301A: repeated measurement analysis (mixed models) Singapore Med J. 2004;45(10):456–461. [PubMed] [Google Scholar]
- Bisgaard H, Li N, Bonnelykke K, Chawes BL, Skov T, Paludan-Muller G, Stokholm J, Smith B, Krogfelt KA. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J Allergy Clin Immunol. 2011;128(3):646–652. doi: 10.1016/j.jaci.2011.04.060. e641-645. [DOI] [PubMed] [Google Scholar]
- Nylund L, Satokari R, Nikkila J, Rajilic-Stojanovic M, Kalliomaki M, Isolauri E, Salminen S, de Vos WM. Microarray analysis reveals marked intestinal microbiota aberrancy in infants having eczema compared to healthy children in at-risk for atopic disease. BMC Microbiol. 2013;13:12. doi: 10.1186/1471-2180-13-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penders J, Gerhold K, Stobberingh EE, Thijs C, Zimmermann K, Lau S, Hamelmann E. Establishment of the intestinal microbiota and its role for atopic dermatitis in early childhood. J Allergy Clin Immunol. 2013;132(3):601–607. doi: 10.1016/j.jaci.2013.05.043. [DOI] [PubMed] [Google Scholar]
- Candela M, Rampelli S, Turroni S, Severgnini M, Consolandi C, De Bellis G, Masetti R, Ricci G, Pession A, Brigidi P. Unbalance of intestinal microbiota in atopic children. BMC Microbiol. 2012;12:95. doi: 10.1186/1471-2180-12-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong PY, Lee BW, Aw M, Shek LP, Yap GC, Chua KY, Liu WT. Comparative analysis of fecal microbiota in infants with and without eczema. PLoS One. 2010;5(4):e9964. doi: 10.1371/journal.pone.0009964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2(2):119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Narisawa Y, Arase S, Okamatsu H, Ikenaga T, Tajiri Y, Kumemura M. Differences in fecal microflora between patients with atopic dermatitis and healthy control subjects. J Allergy Clin Immunol. 2003;111(3):587–591. doi: 10.1067/mai.2003.105. [DOI] [PubMed] [Google Scholar]
- Brandtzaeg PE. Current understanding of gastrointestinal immunoregulation and its relation to food allergy. Ann N Y Acad Sci. 2002;964:13–45. doi: 10.1111/j.1749-6632.2002.tb04131.x. [DOI] [PubMed] [Google Scholar]


