A wide diversity of pathogens, mutualists and pests of plant and animal hosts, including bacteria, fungi, oomycetes, apicomplexan parasites, nematodes, and insects produce effector proteins that enter the cytoplasm of host cells in order to modify the physiology of those cells, usually to favor of colonization (Torto-Alalibo et al., 2010). Two basic mechanisms for introducing those proteins into host cells have been observed, entry mechanisms dependent on microbe-derived machinery, and entry mechanisms that utilize only host machinery (Tyler, 2011). Examples of microbe-derived machinery include the type III, type IV and type VI secretion machineries of gram negative bacteria (Tseng et al., 2009), and rhoptries of apicomplexan parasites (Boothroyd and Dubremetz, 2008). Examples of microbe-independent cell entry include type II secreted toxin proteins of bacteria that enter human cells by endocytosis following binding to glycosphingolipid receptors (Sandvig et al., 2010), and protein toxins secreted by necrotrophic plant pathogenic fungi, such as ToxA (Manning and Ciuffetti, 2005). While large numbers of effector proteins have been identified from oomycete and fungal plant pathogens, the mechanisms by which these effector proteins enter plant cells continues to be a very active area of research. A major question has been whether or not entry by these effectors can occur independently of the microbe or requires machinery provided by the microbe (Ellis et al., 2006).
Much of the research on entry has centered around oomycete effectors that carry the N-terminal motifs, RxLR (arginine, anything, leucine, arginine) and dEER (a string of acidic amino acids followed by arginine) (Kale and Tyler, 2011; Tyler, 2011). These motifs have been demonstrated to be required for efficient effector delivery during infection by Phytophthora sojae and P. infestans (Whisson et al., 2007; Dou et al., 2008; Grouffaud et al., 2008). Two classes of experiments have provided information that these effectors do not require the microbe for entry. The first are plant transient expression experiments, by bombardment or agroinfiltration, in which the effector in question is secreted from the plant and then must re-enter to trigger a measured response (usually cell death in the presence of a cognate R gene) (Dou et al., 2008; Kale et al., 2010; Kale and Tyler, 2010; Rafiqi et al., 2010; Gu et al., 2011; Anderson et al., 2012; Sun et al., 2013). The second is to expose root tips or leaf tissue to purified effectors and then, to measure entry microscopically using antibodies, or by the presence of a chemical or protein fluorescent label attached to the effector (Dou et al., 2008; Kale et al., 2010; Plett et al., 2011; Wawra et al., 2012). In the case of P. sojae effector Avr1b, the same RxLR and dEER motif mutations that abolished entry by the effectors from the P. sojae transformants also abolished entry as measured by the transient expression and purified protein assays (Dou et al., 2008; Kale and Tyler, 2010), supporting the conclusion that these assays are relevant to infection in vivo. Similar experiments have also identified fungal effectors that appear to enter plant cells in the absence of the microbe, via RxLR-like motifs ([R, K, H]×[L, M, I, F, W, Y]); with no dEER-like motifs) (Kale and Tyler, 2010; Gu et al., 2011; Plett et al., 2011). The demonstration that those oomycete and fungal effectors can bind host cell surface phosphatidyinositol-3-phosphate (PI-3-P) via the RxLR motifs, and that PI-3-P binding is required for entry has provided a mechanistic basis for RxLR-dependent entry (Kale et al., 2010; Sun et al., 2013). A surprising finding to emerge from these studies was that many of these oomycete and fungal effectors could enter not only plant cells but also human lung epithelial cells (Kale et al., 2010; Sun et al., 2013).
Recently, Wawra et al. (2013) presented a re-examination of whether the RxLR domain of oomycete RxLR effectors is sufficient for microbe-independent entry into host cells. The authors, from the van West, Kahmann and Nuernberger labs, presented data in support of the conclusion that the RxLR domains of P. infestans Avr3a and of P. sojae Avr1b alone are NOT sufficient to enable microbe-independent entry of proteins into host and non-host plant and animal cells. More specifically, they reported that they were unsuccessful in their attempts to reproduce key experiments previously reported by Dou et al. (2008) and Kale et al. (2010) in which GFP fusions to Avr1b and its RxLR domain were observed to enter soybean root cells and human lung epithelial cells. Instead, they reported that any fluorescent protein can be observed to enter soybean roots if exposed to the roots for an extended period. Data on non-specific uptake into soybean roots were contributed by the Kahmann lab, negative uptake data for human and fish cells were contributed by the van West lab, and negative uptake data for Arabidopsis and Nicotiana cells were contributed by the Nuernberger lab.
In this letter, we summarize the wide diversity of data from different experimental systems that support the conclusion that Avr1b and its N-terminal domain can in fact enter host cells independently of the microbe. These include new, more detailed data that unambiguously demonstrate that the RxLR domain of Avr1b does show efficient and specific entry into soybean root cells, and also into wheat leaf cells, at levels well above background non-specific entry. We also summarize host cell entry experiments with a wide diversity of oomycete and fungal effectors with RxLR or RxLR-like motifs that have been independently carried out by the seven different labs that co-authored this letter. Finally we discuss possible reasons why Wawra et al. (2013) may have failed to observe specific cell entry.
The principal means for establishing whether entry of Avr1b into host cells is relevant to infection is based on mutagenesis experiments published by Dou et al. (2008). Avr1b has two RxLR motifs, RSLR at position 20 of the mature protein (called RXLR1) and RFLR at position 31 (called RXLR2). Furthermore, it has a dEER motif, EEDDAGER at position 37. Mutations in RXLR2 and dEER, but not RXLR1, abolished the ability of the Avr1b-1 gene to confer avirulence on P. sojae when infecting soybean plants containing the cognate resistance gene Rps1b. Transient expression of Avr1b proteins in soybean leaf cells by particle bombardment demonstrated that, when the proteins were expressed in the host cytoplasm, the RXLR2 and dEER mutants did not impair the ability of the protein to trigger cell death in the presence of Rps1b; thus the mutations did not interfere with recognition of the protein by Rps1b. Further bombardment experiments in the presence of the Avr1b secretory leader revealed that, when the proteins were secreted from the host cells, the RXLR2 and dEER mutations did abolish Rps1b-mediated cell death; thus the RXLR2 and dEER motifs were inferred to be required for re-entry. In order to demonstrate that the N-terminal domain of Avr1b could deliver a C-terminal domain into soybean cells from an unambiguously external location, a purified GFP fusion protein carrying the Avr1b N-terminus (1–48) was exposed to soybean root tips. Entry of the protein into the root cells was observed microscopically only when the RXLR2 and dEER motifs were intact. Plasmolysis of the root cells confirmed that the protein was inside the cells. Kale et al. (2010) confirmed the root cell entry experiments of Dou et al (2008) using full length Avr1b protein and using more specific mutations of RXLR2 (qFLR) that agreed with results of bombardment experiments of Dou et al. (2008). Furthermore, validating an observation of Shan et al. (2004), Kale et al. (2010) showed that full length Avr1b protein could trigger cell death on leaves containing the Rps1b resistance gene, but not on leaves lacking Rps1b. The ability to trigger Rps1b-mediated cell death was abolished by the qFLR mutation and a dEER motif mutation, consistent with RxLR-dependent entry into soybean cells. The strong concordance among the four experiments - P. sojae transformants, particle bombardment transient expression, GFP fusion root entry, and leaf cell death – with respect to the effects of mutations in the RXLR2 and dEER motifs formed the basis for concluding that Avr1b could enter host cells in the absence of the microbe, as a result of the action of the RxLR and dEER motifs. The further demonstration that PI-3-P binding also depended on the RxLR and dEER motifs suggested that the mechanism of entry was lipid receptor-mediated endocytosis.
More detailed biophysical experiments with the Avr1b paralog Avh5 (Sun et al., 2013), clarified that PI-3-P binding and entry into soybean root cells and human lung cells depended not only on the N-terminal RxLR domain, but also on residues in the C-terminus. These root entry experiments were done using Dylight488-labeled Avh5 and shorter incubation times (2 hr.) ruling out artifacts resulting from the use of GFP fusions or long incubation times.
The original description of microbe-independent cell entry made with Avr1b led to the subsequent observation of microbe-independent entry by a wide diversity of oomycete and fungal effectors. The subsequent reports used the same methodologies pioneered for Avr1b, including bombardment assays and uptake of fluorescent fusion proteins into soybean root and leaf cells and into human lung epithelial cells. These subsequent discoveries thus provide further diverse and independent validation of the initial discovery of microbe-independent entry by Avr1b. Here, we summarize the experience of seven different laboratories and sixteen researchers in those laboratories in documenting microbe-independent entry by oomycete and fungal effectors into plant and animal cells, using the approaches developed using Avr1b.
B.M. Tyler, Virginia Tech. Subsequent to the Avr1b experiments, soybean and human cell entry experiments were done with Avr1k (P. sojae Avh331), Avr2 (Fusarium oxysporum), AvrLm6 (Leptosphaeria maculans), AvrL567 (Melampsora lini) (Kale et al., 2010), MiSSP7 (Plett et al., 2011), AvrLm4–7 (Leptosphaeria maculans) (unpublished), Af2 (Aspergillus fumigatus) (unpublished), Af3 (Aspergillus fumigatus) (unpublished), and a secreted allergen (Alternaria alternata) (unpublished). Root and/or human cell entry experiments were conducted by B. Gu, V. Antignani and Q. Chen in addition to S. Kale, as indicated in Table 1. In each case, specific entry was established by use of mutations in RxLR(-like) motifs and inhibition by PI3P-binding proteins. The demonstration that AvrL567 enters plant cells independently of the microbe, dependent on the RXLR-like motif RFYR, was independently validated by (Rafiqi et al., 2010).
Table 1.
Selected host cell entry experiments that have been reproduced by individual researchers within and among different labs.
| Proteina | Tyler Lab, VTb | Additional labs with same or similar resultsc | ||
|---|---|---|---|---|
| Avr1b (soybean) | D. Dou, S.D. Kale, Q. Chen, V. Antignani, B. Gu, W. Shand | Kale Lab: H.R. Clarke, K. Drewse, S.D. Kalee (also wheat) | Tyler Lab, OSU: Q. Wang, K. Tao | Shan lab: B. Gu |
| MiSSP7 (poplar) | S.D. Kale, V. Antignani (soybean) | Martin Lab: J. Plette | ||
| AvrLm4/7 (oilseed rape) | S.D. Kalee (soybean) | Rouxel LabI. Fudale | ||
| Af2 (human) | V. Antignani | Kale Lab: H.R. Clark, K. Drews, S.D. Kale | Lawrence Lab: A. Rumore, T. Hayes | |
| Af3 (human) | S.D. Kale, V. Antignani | Keller Lab: Affeldt, Fischer, Berthier | ||
| Allergen (human) | S.D. Kale | Lawrence Lab: A. Rumore, T. Hayes | ||
Protein assayed and, in parentheses, the native host target species of the protein.
Researchers in the Tyler Lab at Virginia Tech. Target species is noted in parentheses if it is not the natural target listed in column 1
Researchers in other labs who documented cell entry either under the same conditions, or with a different target species, as noted in parentheses, or labeling procedure. In the case of MiSSP7 and the Alternaria allergen, entry was first observed by the Martin and Lawrence labs, respectively, then repeated in the other labs.
W. Shan conducted the first Avr1b cell entry experiments into soybean leaves while in B. Tyler’s lab at the University of California, Davis.
These experiments used proteins that were labeled chemically with a dye; otherwise fusions to fluorescent proteins were used.
B.M. Tyler, Oregon State. In early 2012 the Tyler lab moved to Oregon State, while S. Kale remained at Virginia Tech with his own lab. Subsequently, the root cell entry assays and PI-3-P binding assays with the Avr1b RxLR domain were both re-established after some period of trouble shooting. For example, the growth conditions and age of the soybean plants, the quality of the protein preparations, and buffer and pH of the protein solution, and the time and temperature of incubation of the roots all were important for successful root uptake experiments. Q. Wang and K. Tao conducted the root cell entry assays. In particular they conducted the experiment shown in Fig. 1, in which GFP alone, Avr1b RxLR domain fused to GFP, or the Avr1b RxLR domain with the mutation RFLR -> AAAA, were exposed to soybean root tips. mCherry protein was mixed with each GFP protein in order to observe the level of non-specific entry into the same cells that were being observed for specific entry. The results demonstrate that the intact Avr1b-GFP fusion, but not GFP itself or the Avr1b RxLR mutant, accumulate in soybean roots to a substantially higher level than mCherry.
Fig. 1.
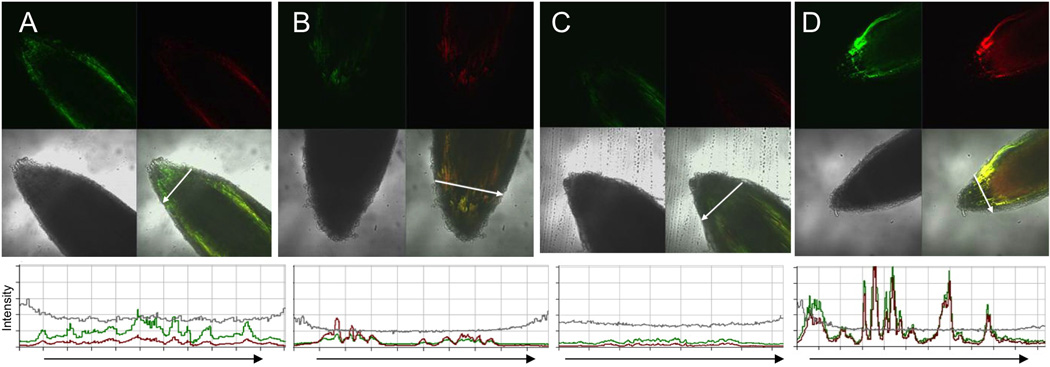
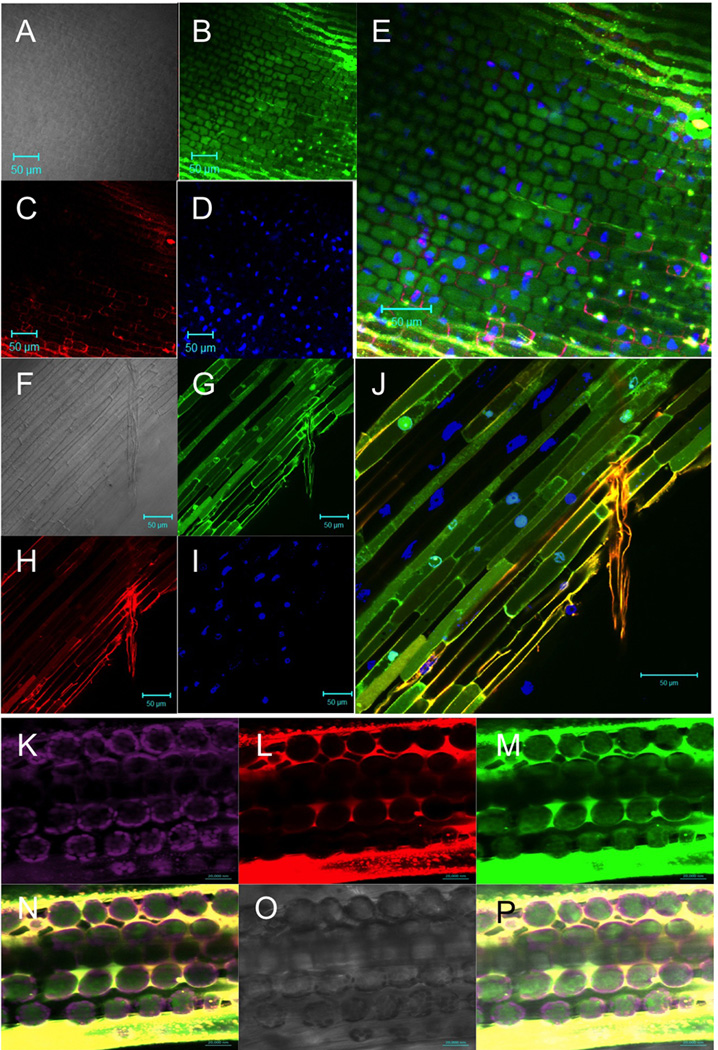
Specific microbe-independent entry of Avr1bNt-GFP fusions into soybean root cells. A, Avr1b N-terminus (residues 1–50 of the mature protein) fused to GFP mixed with mCherry protein. B, Avr1b N-terminus (residues 1–50) with RSLR->AAAA and RFLR->AAAA mutations, fused to GFP mixed with mCherry protein. C and D, GFP protein (no fusions) mixed with mCherry protein. Purified fusion proteins (0.5 mg/ml each) were incubated with soybean (Williams) root tips, for 12 h in the dark at 23°C in phosphate-buffered saline (10 mM Na phosphate, 138 mM NaCl, 2.7 mM KCl, adjusted to pH 6.8 with HCl; Sigma P5368), then washed four times for 1 h each in PBS pH 6.8 at 23°C. Proteins all carried C-terminal his6 tags and were purified as described (Kale et al., 2010). The roots were imaged with a Zeiss LSM 510 confocal microscope, with an excitation wavelength of 488 nm and emission window of 505–530 nm for GFP or an excitation wavelength of 543 nm and emission window of >560 nm for mCherry. Master gain was set to 500 for A-C and to 650 for D. In each figure section, the upper left panel shows the GFP image, the upper right the mCherry image, the middle left, the light image, the middle right the overlay of GFP, mCherry and light images, AND the bottom panel shows an intensity scan of the GFP (green), mCherry (red) and light (gray) channels along the transect shown by the white arrow in the overlay image. The intensity scale for the scans are all on the same scale. The experiments shown in this figure were conducted at Oregon State University.
S. Kale, Virginia Tech. S. Kale opened his own lab at Virginia Tech in summer 2011. Subsequently he and his lab members, H. Clark and K. Drews, conducted soybean and human cell entry experiments with Avr1b, Avh5, AvrLm4–7, Af2, Af3, and an Alternaria allergen. In particular, his lab conducted the experiments shown in Fig. 2 that demonstrate that the Avr1b RxLR domain labeled with Dylight488, but not the Avr1b RxLR mutant protein labeled with Dylight550, could enter soybean root cells and wheat leaf cells after only 2 hr when the two proteins were incubated together within the same tissues.
Fig. 2.
Specific microbe-independent entry of Avr1bNt proteins into soybean root cells and wheat leaf cells. A to E, Soybean root entry by Avr1b N-terminus (residues 1–50) labeled by Dylight488, and counter stained with propidium iodide and 4',6-diamidino-2-phenylindole (DAPI). F to J, Soybean root entry by Avr1b N-terminus (residues 1–50) labeled by Dylight488, mixed with Avr1b N-terminus with RFLR->qFLR mutation labeled with DyLight550, and counter stained with DAPI. A,F, light image; B,G, Dylight488 image; C, propidium image; H, Dylight550 image; D and I, DAPI image; E and J, overlay of the three fluorescent images. In A to J, purified fusion proteins (0.4 mg/ml each) were incubated with soybean (Williams) root tips, for 2 hours at 25°C in PBS buffer adjusted to pH 7.2, then washed for 15 min in formalin (PBS + 10% formaldehyde) containing 0.2 µg/ml propidium iodide (A to E only) and 0.4 µg/ml DAPI. K to P, Wheat leaf cell entry by Avr1b N-terminus (residues 1–50) labeled by Dylight488 mixed with Avr1b N-terminus (residues 1–50) with RFLR->qFLR mutation labeled by Dylight550. Purified fusion proteins (0.4 mg/ml each) in PBS buffer adjusted to pH 7.2 were infiltrated with a blunt syringe into ~9 day old wheat seedling leaves. Leaves were imaged after 6 h without washing. K, chloroplast fluorescence (excitation 488 nm; emission meta filter 675–715 nm); L, Dylight550 image (excitation 543 nm; emission window 585–615 nm; gain 600 to 654; digital offset −0.1 to 0); M, Dylight488 image (excitation 488 nm; emission window 505–530 nm; gain 580 to 610; digital offset −0.1 to 0); N, overlay of K to M; O, light image; overlay of N and O. Labeling of proteins with Dylight dyes was as described (Sun et al., 2013). The experiments shown in this figure were conducted at Virginia Tech using a Zeiss LSM 510 Meta confocal microscope.
C.B. Lawrence, Virginia Tech. The Lawrence lab conducts research on the interactions of human lung cells with fungal proteins. Lab members A. Rumore and T. Hayes independently carried out the initial human cell entry experiments with Af2 and the Alternaria allergen, including validation of specific entry using mutations in RxLR-like motifs and inhibition by PI3P-binding proteins. Proteins were either directly labeled with DyLight488 or fused with reporters GFP and mCherry. Moreover, they have confirmed PI3P-binding to these proteins and the importance of the RxLR-like motif using lipid blots and isothermal titration calorimetry. (unpublished).
W. Shan, Northwest A & F University, China. The Shan lab studies interactions of plants with oomycete and fungal plant pathogens. W. Shan conducted the first Avr1b cell entry experiments into soybean leaves while in B. Tyler’s lab at the University of California, Davis (Shan et al. 2004) that were later replicated in Kale and associates (2010). Lab member B. Gu visited the Tyler lab at Virginia Tech where he carried out numerous root cell entry experiments including replicates of those described in Kale and associates (2010), and particularly experiments with Avr1bNt-GFP. After returning to the Shan lab he independently replicated the cell entry experiments with Avr1bNt-GFP and carried out new experiments documenting entry of a candidate wheat rust effector, Ps87, into soybean root cells, dependent on an RxLR-like motif. Both those results are illustrated in Figure 4 of Gu and associates (2011).
F. Martin, INRA, Nancy. The Martin lab studies interactions between trees and the mutualistic ectomycorrhizal fungus Laccaria bicolor. They demonstrated that MYCORRHIZAL iNDUCED SMALL SECRETED PROTEIN 7 (MiSSP7), the most highly symbiosis-upregulated gene from L. bicolor (Martin et al., 2008), encodes an effector protein indispensible for the establishment of mutualism (Plett et al., 2011). Jonathan Plett showed that 5,6-carboxyfluorescein (FAM)- tagged MiSSP7 protein is actively taken up into roots of host (Populus trichocarpa) and non-host plants (e.g. Arabidopsis thaliana, Glycine max) and poplar suspension cells in less than 20 min in the absence of the fungus (Plett et al., 2011). Then, synthetic and recombinant MiSSP7 proteins were both found to enter host cells. Numerous controls were performed in the Martin and Tyler labs to ensure that MiSSP7 entry was not artifactual, including the use of deletion and substitution mutations in the RALG (RxLR-like) motif and inhibition by PI3P-binding proteins and by many other inhibitors. Under the same experimental conditions, other MiSSPs, such as MiSSP8 and hydrophobins, did not enter poplar root cells (J. Plett, S. Kale, unpublished). To ensure that MiSSP7 entry was not due to passive diffusion due to the small size of the protein, heat killed poplar roots were shown to be unable to take up the protein.
T. Rouxel, INRA, Thiverval-Grignon. The Rouxel lab studies effectors produced by Leptosphaeria maculans, a hemi-biotrophic fungal pathogen of oilseed rape (Brassica napus). Cell entry experiments into oilseed rape roots were performed by I. Fudal using 70 µg/ml of full length AvrLm4–7 protein coupled to 5,6-FAM, with an incubation time of 3 hours. The Kale lab did similar experiments with soybean roots and DyLight488-labeled proteins. In both cases, rapid entry of the proteins was observed within 2–3 hr and in the case of experiments on soybean roots specific entry was established using mutations in RxLR-like and C-terminal motifs (unpublished).
N. Keller, University of Wisconsin, Madison. The Keller lab studies the interaction of Aspergillus fumigatus with animals. They identified the putative effector Af3 bioinformatically and shared the sequence with the Tyler lab. Specific entry of Af3-GFP fusions into human lung epithelial cells was demonstrated first in the Tyler lab, and then validated in the Keller lab (unpublished).
There are several possible technical explanations why Wawra and associates (2013) may have obtained the negative results that they report.
i) The Avr1b-GFP protein used for the experiment may have been inactive or degraded. It appears that just a single preparation of Avr1b-GFP protein, prepared in the van West lab and distributed to the other collaborators, was used for all the studies. If this protein preparation was inactive then all experiments in the study would be negative. The isolation procedure used by the authors is quite different than that used by Dou and associates (2008) and Kale and associates (2010), so it is possible the differences in isolation led to differences in activity. It is the experience of the Tyler, Kale, Lawrence and Rouxel labs that effector preparations stored for any length of time soon lose activity, so it is our routine practice to use recently prepared proteins for all assays. Protein degradation can be a problem also. Although Wawra and associates (2013) showed mass spectrometry data to validate the quality of their initial preparation of protein, they did not use an assay such as western blots to check the intactness of the protein immediately prior to and during the cell entry assays. Thus it remains possible the protein was already degraded prior to the start of the experiments or was rapidly degraded following exposure to the target cells. Kale and associates (2010) and Plett and associates (2011) did use western blots to check the intactness of the proteins before and after their entry assays.
ii) In the case of soybean, the roots may not have been prepared in a way that left them competent for specific cell entry. The wheat germ agglutinin positive control used for the human cell experiments failed to enter the soybean roots. Furthermore, Wawra and associates (2013) did not utilize the Arg9-GFP control developed by Chang and associates (2005) and used by and associates (2008) and Kale and associates. (2010). The experience of the Tyler lab (at both Virginia Tech and Oregon State) is that batches of roots are occasionally encountered that fail to exhibit specific RxLR-dependent entry activity and instead exhibit only the weak non-specific entry reported by Wawra and associates (2013). As documented in our publications, entry by the numerous oomycete and fungal proteins we have studied has in every case been validated to be specific by use of proteins with RxLR mutations, and by the use of PI3P-blocking inhibitors. In our published figures we have routinely used stringent confocal microscope settings that exclude non-specific background entry. Furthermore, in recent work (Fig. 2; Sun et al., 2013) we have introduced the use of propidium iodide to identify cells which may have died and hence may take up protein non-specifically.
iii) Wawra and associates (2013) observed no entry into human A549 lung epithelial cells. However, our experience (Tyler, Kale, Lawrence and Keller labs) is that entry into these cells is straightforward to observe. Even if their effector protein preparations were not inactive, it is likely that the presence of 10% foetal bovine serum (FBS) in the medium during their uptake assays would have interfered with entry. We routinely incubate our cells in serum free medium for at least two hours before adding fluorescent proteins for entry assays. It seems likely that the high concentration of proteins and lipids in the FBS could interfere with the binding and entry of lipid-binding proteins.
iv) We (Tyler lab) have occasionally encountered the GFP-related precipitation problem mentioned by Wawra and associates (2013). However as noted by the authors, there is no correlation between precipitation and entry. GFP-mediated precipitation certainly cannot explain entry that is abolished by the presence of RxLR mutations or the presence of PI3P-binding proteins (Dou et al., 2008; Kale et al., 2010; Gu et al., 2011). Furthermore, we have observed strong, specific entry within 2–3 hr. when using chemically-labeled Avr1b (Fig. 2), Avh5 (Sun et al., 2013) or MiSSP7 (Plett et al., 2011), clearly ruling out GFP-mediated precipitation as a cause of non-specific entry. Moreover, we have shown in multiple repeated experiments with several proteins that the RxLR-dependent cell entry phenomenon occurs within minutes in multiple lung epithelial cells types, namely immortalized cell lines (A549 and BEAS-2b) and primary cells derived directly from human (NHBE) (Rumore, Hayes and Lawrence, unpublished).
v) Wawra et al. (2013) reported being unable to observe specific entry into Arabidopsis root cells or N. benthamiana leaf cells. We also have never observed entry into N. benthamiana cells and have only observed entry in Arabidopsis cells by MiSSP7 (Plett et al., 2011). Either the correct way to prepare these tissues to preserve their specific uptake activity has not yet been found, or else the cells of these two species simply never exhibit a high enough activity to detect entry using fluorescent proteins. On the other hand, we could readily observe RxLR-dependent entry into poplar root cells (Plett et al., 2011), into rapeseed root cells (Fudal and Rouxel, unpublished) and into wheat leaf cells (Fig. 2), indicating that a high level of specific uptake activity is not restricted to soybean. We have not tested RxLR-dependent entry into fish cells.
In conclusion, it is the overwhelming experience of the sixteen researchers and seven labs that are co-authors of this letter that microbe-independent entry of oomycete and fungal effectors into both plant and animal cells is specific and independently reproducible. We welcome a vigorous, ongoing and constructive debate on the question of how effectors enter host cells, as a means for stimulating research into this extremely important topic. We renew our invitation, expressed many times in the past, for researchers from the van West, Kahmann and Nuernberger labs to visit our labs and learn the technical details of how we assay specific entry.
ACKNOWLEDGEMENTS
This work was supported by the National Science Foundation Grant IOS-0924861 (to B. M. Tyler), the NIH NIAID grant 1R21A1094071-01 (to C. B. Lawrence), and by funds from the Virginia Bioinformatics Institute and Oregon State University. NPK, KJA, EB and GF acknowledge support from NSF IOS-0965649 and the American Asthma Foundation.
LITERATURE CITED
- Anderson RG, Casady MS, Fee RA, Vaughan MM, Deb D, Fedkenheuer K, Huffaker A, Schmelz EA, Tyler BM, McDowell JM. Homologous RXLR effectors from Hyaloperonospora arabidopsidis and Phytophthora sojae suppress immunity in distantly related plants. Plant J. 2012;72:882–893. doi: 10.1111/j.1365-313X.2012.05079.x. [DOI] [PubMed] [Google Scholar]
- Boothroyd JC, Dubremetz JF. Kiss and spit: the dual roles of Toxoplasma rhoptries. Nat Rev Microbiol. 2008;6:79–88. doi: 10.1038/nrmicro1800. [DOI] [PubMed] [Google Scholar]
- Chang M, Chou JC, Lee HJ. Cellular internalization of fluorescent proteins via arginine-rich intracellular delivery peptide in plant cells. Plant & cell physiology. 2005;46:482–488. doi: 10.1093/pcp/pci046. [DOI] [PubMed] [Google Scholar]
- Dou D, Kale SD, Wang X, Jiang RHY, Bruce NA, Arredondo FD, Zhang X, Tyler BM. RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. Plant Cell. 2008;20:1930–1947. doi: 10.1105/tpc.107.056093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J, Catanzariti AM, Dodds P. The problem of how fungal and oomycete avirulence proteins enter plant cells. Trends Plant Sci. 2006;11:61–63. doi: 10.1016/j.tplants.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Grouffaud S, van West P, Avrova AO, Birch PR, Whisson SC. Plasmodium falciparum and Hyaloperonospora parasitica effector translocation motifs are functional in Phytophthora infestans. Microbiology (Reading, England) 2008;154:3743–3751. doi: 10.1099/mic.0.2008/021964-0. [DOI] [PubMed] [Google Scholar]
- Gu B, Kale SD, Wang Q, Pan Q, Cao H, Meng Y, Kang Z, Tyler BM, Shan W. Rust secreted protein Ps87 is conserved in diverse fungal pathogens and contains an RXLR-like motif sufficient for translocation into plant cells. PLoS ONE. 2011;6:e27217. doi: 10.1371/journal.pone.0027217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale SD, Tyler BM. Assaying effector function in planta using double-barreled particle bombardment. In: McDowell JM, editor. Methods in Molecular Biology. The Plant Immune Response. Totowa, NJ: Humana; 2010. Page in press. [DOI] [PubMed] [Google Scholar]
- Kale SD, Tyler BM. Entry of oomycete and fungal effectors into plant and animal cells. Cellular Microbiology. 2011;13:1839–1848. doi: 10.1111/j.1462-5822.2011.01659.x. [DOI] [PubMed] [Google Scholar]
- Kale SD, Gu B, Capelluto DGS, Dou D-L, Feldman E, Rumore A, Arredondo FD, Hanlon R, Fudal I, Rouxel T, Lawrence CB, Shan W-X, Tyler BM. External lipid PI-3-P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell. 2010;142:284–295. doi: 10.1016/j.cell.2010.06.008. [DOI] [PubMed] [Google Scholar]
- Manning VA, Ciuffetti LM. Localization of Ptr ToxA produced by Pyrenophora tritici-repentis reveals protein import into wheat mesophyll cells. Plant Cell. 2005;17:3203–3212. doi: 10.1105/tpc.105.035063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Aerts A, Ahren D, Brun A, Danchin EG, Duchaussoy F, Gibon J, Kohler A, Lindquist E, Pereda V, Salamov A, Shapiro HJ, Wuyts J, Blaudez D, Buee M, Brokstein P, Canback B, Cohen D, Courty PE, Coutinho PM, Delaruelle C, Detter JC, Deveau A, DiFazio S, Duplessis S, Fraissinet-Tachet L, Lucic E, Frey-Klett P, Fourrey C, Feussner I, Gay G, Grimwood J, Hoegger PJ, Jain P, Kilaru S, Labbe J, Lin YC, Legue V, Le Tacon F, Marmeisse R, Melayah D, Montanini B, Muratet M, Nehls U, Niculita-Hirzel H, Oudot-Le Secq MP, Peter M, Quesneville H, Rajashekar B, Reich M, Rouhier N, Schmutz J, Yin T, Chalot M, Henrissat B, Kues U, Lucas S, Van de Peer Y, Podila GK, Polle A, Pukkila PJ, Richardson PM, Rouze P, Sanders IR, Stajich JE, Tunlid A, Tuskan G, Grigoriev IV. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008;452:88–92. doi: 10.1038/nature06556. [DOI] [PubMed] [Google Scholar]
- Plett JM, Kemppainen M, Kale SD, Kohler A, Legué V, Brun A, Tyler BM, Pardo AG, Martin F. A secreted effector protein of Laccaria bicolor is required for symbiosis development. Current Biology. 2011;21:1197–1203. doi: 10.1016/j.cub.2011.05.033. [DOI] [PubMed] [Google Scholar]
- Rafiqi M, Gan PH, Ravensdale M, Lawrence GJ, Ellis JG, Jones DA, Hardham AR, Dodds PN. Internalization of flax rust avirulence proteins into flax and tobacco cells can occur in the absence of the pathogen. Plant Cell. 2010;22:2017–2032. doi: 10.1105/tpc.109.072983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandvig K, Torgersen ML, Engedal N, Skotland T, Iversen TG. Protein toxins from plants and bacteria: probes for intracellular transport and tools in medicine. FEBS Lett. 2010;584:2626–2634. doi: 10.1016/j.febslet.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Shan W, Cao M, Leung D, Tyler BM. The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps 1b. Mol. Plant Microbe Interact. 2004;17:394–403. doi: 10.1094/MPMI.2004.17.4.394. [DOI] [PubMed] [Google Scholar]
- Sun F, Kale SD, Azurmendi HF, Li D, Tyler BM, Capelluto DGS. Structural basis for interactions of the Phytophthora sojae RXLR effector Avh5 with phosphatidylinositol 3-phosphate and for host cell entry. Mol. Plant-Microbe Interact. 2013;26:330–344. doi: 10.1094/MPMI-07-12-0184-R. [DOI] [PubMed] [Google Scholar]
- Torto-Alalibo T, Collmer CW, Gwinn-Giglio M, Lindeberg M, Meng S-W, Chibucos MC, Tseng T-T, Lomax J, Biehl B, Ireland A, Bird D, Dean RA, Glasner JD, Perna N, Setubal JC, Collmer A, Tyler BM. Unifying themes in microbial associations with animal and plant hosts described using the Gene Ontology. Microbiol. Molec. Biol. Rev. 2010;74:479–503. doi: 10.1128/MMBR.00017-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng TT, Tyler BM, Setubal JC. Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol. 2009;9(Suppl 1):S2. doi: 10.1186/1471-2180-9-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler BM. Entry of oomycete and fungal effectors into host cells. In: Martin F, Kamoun S, editors. Effectors in Plant-Microbe Interactions. Oxford: Wiley-Blackwell; 2011. pp. 243–278. [Google Scholar]
- Wawra S, Djamei A, Kuefner I, Nuernberger T, Kahmann R, van West P. In vitro translocation experiments with RxLR-reporter fusion proteins of Avr1b from Phytophthora sojae and AVR3a from Phytophthora infestans fail to demonstrate autonomous uptake in plant and animal cells. Mol. Plant-Microbe Interact. 2013;26(5):528–536. doi: 10.1094/MPMI-08-12-0200-R. [DOI] [PubMed] [Google Scholar]
- Wawra S, Bain J, Durward E, Bruijn Id, Minor KL, Matena A, Löbach L, Whisson SC, Bayer P, Porter AJ, Birch PRJ, Secombes CJ, West Pv. Host-targeting protein 1 (SpHtp1) from the oomycete Saprolegnia parasitica translocates specifically into fish cells in a tyrosine-O-sulphate–dependent manner. Proc. Natl. Acad. Sci. USA. 2012;109(6):2096–2101. doi: 10.1073/pnas.1113775109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, Gilroy EM, Armstrong MR, Grouffaud S, West Pv, Chapman S, Hein I, Toth IK, Pritchard L, Birch PRJ. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450:115–119. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]