Abstract
Objective
To extract disorder-associated genes from the scientific literature in PubMed with greater sensitivity for literature-based support than existing methods.
Methods
We developed a PubMed query to retrieve disorder-related, original research articles. Then we applied a rule-based text-mining algorithm with keyword matching to extract target disorders, genes with significant results, and the type of study described by the article.
Results
We compared our resulting candidate disorder genes and supporting references with existing databases. We demonstrated that our candidate gene set covers nearly all genes in manually curated databases, and that the references supporting the disorder–gene link are more extensive and accurate than other general purpose gene-to-disorder association databases.
Conclusions
We implemented a novel publication search tool to find target articles, specifically focused on links between disorders and genotypes. Through comparison against gold-standard manually updated gene–disorder databases and comparison with automated databases of similar functionality we show that our tool can search through the entirety of PubMed to extract the main gene findings for human diseases rapidly and accurately.
Keywords: PubMed Search Tool, Disorder Genes, Gene-Disease Relationships, Literature Mining, Bibliome
Background
With the advance of genotyping and sequencing technologies, a rising number of studies have reported genetic association with various disorders in the past decade. As hundreds of genes may be involved in one complex disorder, a thorough literature review is a fundamental starting point to understand genetic risk factors of any given human disorder. For example, if we are interested in the genetic etiology of schizophrenia, we would first like to know which genes have been reported for association with the most important literature evidence to justify the association. However, few applications have been available to help search and keep track of up-to-date, gene–disease associations. Many sites provide detailed gene data, including GeneCards,1 PharmGKB,2 WikiGenes,3 and iHOP,4 but it is not easy to find the disorders associated with a given gene using these sites. Another group of tools, including LitInspector,5 MuGeX,6 Quertle (http://quertle.info) and NEXTBIO genetic markers (http://nextbio.com) provide supporting references and snippets of text from abstracts highlighting the target gene or disorder, but their genetic and/or disease coverage is limited and the methods, for gene–disorder association (such as simple co-occurrence of terms), often yield high rates of false positives. Disorder-oriented sites like SFARI gene7 and SZgene8 provide candidate genes for a target disorder with or without references, but cover only a single disorder. Previously we built a meta-search tool that integrates results from several of these sites.9 Using this tool, we discovered significant discrepancies between databases, with few providing adequate references to supporting literature.
Provided this context, our goal in the present study was to build a novel PubMed extraction tool that focuses on identifying target disorders and associated human genes from all original research articles, and to compare the results from this tool with the existing databases that provide disorder candidate genes and supporting references, including Online Mendelian Inheritance in Man (OMIM),10 HuGE Navigator,11 and Genetic Association Database (GAD).12 Various text-mining algorithms have been proposed to address entity recognition in scientific literature13 14 and infer novel gene–disorder relationships.15–20 We set our scope in this work, however, precisely to extract gene–disease linkages reported in the research articles, rather than to discover new associations based on literature information.
Methods
Retrieving disorder-related articles
For each target disorder name, we built a comprehensive PubMed query to retrieve disorder-specific research or review articles. First, we mapped the given disorder name to the corresponding medical subject headings (MeSH),21 and obtained target disorder aliases from MeSH, MedlinePlus,22 and Genetics Home Reference.23 After expanding names for plurals (eg, disorder → disorders) and synonyms (eg, syndrome ↔ disorder/disease), we required in the query that these names and aliases appear in the title or MeSH entries sections of papers. We did not search in any field or in abstracts as we observed false-positive findings (ie, articles that are not directly related to the target disorder) when we allowed these fields. Second, we filtered out articles with publication types that were not relevant to research or reviews, for example bibliographies, comments, and editorials. We also limited results to publication dates after 1990, as we were interested in retrieving recent, genetics-oriented research articles. This was a conservative publication date filter, considering that the human genome project began in 1990 and the pilot phase of sequencing was done in 1999.24 An example of the expanded query targeted to autism spectrum disorder (ASD) is shown in figure 1. We used E-utilities25 to execute this query, extracting PubMed article identifiers and details for further steps.
Figure 1.

An example of expanded query for a user input, ‘autism spectrum disorders’. This is a translated query so query terms without matching documents are not displayed. Colors are added for illustration purposes. Green texts are disorder aliases to appear in titles; red texts are the mapped MeSH entries, including sub-tree terms; blue texts specify publication dates; purple texts are publication types that should be excluded from this search; and orange texts are added to exclude comments, erratum and retracted articles when publication types are not available.
Screening genetics-related articles
In the first step, we retrieved articles related to the target disorder; we then performed further steps to reduce this set to include only papers related to genetics. First, if an article has MeSH annotations, we examined whether they include terms under ‘genetic techniques’ sub-tree; terms under ‘genetic phenomena’ sub-tree; or ‘genetics’ subheading. Second, if the article has no MeSH annotation, we examined if the title or abstract include genetics-related keywords. We obtained these keywords by comparing two training sets of abstracts. In particular, we selected 32 disorders from the genome-wide association studies (GWAS) catalog26 and downloaded two sets of abstracts from PubMed: those that include MeSH terms fitting in above conditions (158 745 articles); and those that have MeSH terms, but do not include terms of the first set (385 383 articles). After removing common frequent words based on Corpus of Contemporary American English,27 we measured the word frequency and compared the top 5000 words from each set in order to find out keywords that uniquely or dominantly (ie, top 1% after Wilcoxon signed-rank test) appear in the first set only. Supplementary table 1 (available online only) shows the top 20 keywords sorted by word frequency. We also used this keyword extraction method to identify abstract structures and study types that we explain below.
Analyzing structure of abstracts, study types and negations
As described in the background section, many existing tools still use simple co-occurrence to show gene–disorder associations. This is not reliable when we want to learn the exact study and reference in which specific associations are tested and reported. For example, sentences like ‘neuroligin genes have been associated with autism’ can occur in the introduction section of an abstract, but the main topic of the paper may not be relevant to neuroligin genes at all. Therefore we decided to use the structure of abstract28 to address this issue. We assume that the main findings of a research article must be reported in the result/conclusion sections of the abstract, or in the title, and in these locations only. This assumption enables us to separate tested genes in the background or methods section (eg, ‘We tested A, B, and C genes’) from associated genes (eg, ‘Only C gene was highly expressed’), and introductory statements in the background section (eg, ‘We previously showed that gene A is associated’). For abstracts without designated structure, we built a set of keywords and rules to identify them, using the available structured abstracts as training data. To extract the study type, we used publication types (eg, ‘reviews’ or ‘case report’) and MeSH terms (eg, ‘disease models, animal’ or ‘genome-wide association study’) when such information is available, or used keywords if MeSH terms or publication types are not annotated. We derived another set of rules to find negated statements in either the title or abstracts; for this we used example sentences obtained from BioNOT.29
Gene representation
Finding gene symbols and their names in the literature and mapping them to unique identifiers is one of the major topics in biological literature mining,30 31 and a large number of algorithms exist to address this issue within various contexts.16 32–40 While following up our previous study,9 we recognized that many genetic–disorder-related articles only use gene symbols or protein symbols, rather than using full names. We tested two of the widely used entity recognition tools trained for human genes (ABNER41 and BANNER),42 but they did not show high precision for this type of task. To address this, we implemented a precision-based gene recognition procedure, which is similar to the protein name extraction algorithm of Fukuda et al43 or LitInspector.5 For each article, we first scanned the title and abstract to identify symbols that match up with gene patterns. For example, official human gene symbols can be identified with these regular expressions: /[A-Z][A-Z0-9\-]+/ or /C(X|0–9+)orf0–9+/. When such a symbol was found, we determined the semantics of the symbol based on the context in which the symbol is located. For example, symbol ‘CGH’ may be used as an alias term of HTC2 gene, but may also mean array-CGH. We checked whether the immediate previous/following text around this symbol includes (1) in a list of gene symbols (eg, ‘X chromosome genes like DMD, MAOA, CGH, and FMR1’); (2) full (official) gene name (eg, ‘hypertrichosis 2 (CGH)’); (3) genetic keywords defined in the previous section (eg, ‘CGH deletion’); (4) other full names for the same pattern (eg, ‘comparative genome hybridization (CGH)’); or (5) other (dis)qualifier for the same pattern (eg, ‘array CGH’). We kept the track of found symbols per article, assuming that a symbol can only have one meaning in the same article. The scan was performed twice, because the meaning of a symbol may not be decided in the current position, but in the later part of the title or abstract. The comparison output of our algorithm with ABNER and BANNER, including test input sentences and tagged words, is shown in the supplementary files (available online only).
Assessing the significance of articles and genes
Ranking of articles and terms based on the strength of publication and the structure of the article has been thoroughly studied by Demner-Fushman and Lin.44 We combined the temporal significance of articles with positive/negative findings of genes in order to assess the significance of an association between a gene and a target disorder. We defined the significance of an article based on the impact factor of the published journal and the publication year. The score of an article a for a given disorder d was defined as follows:
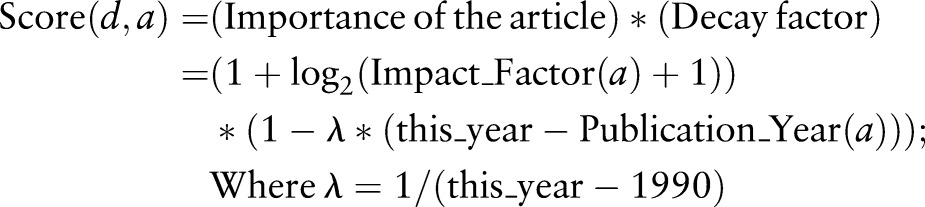 |
For example, if an article is published this year in a journal without a known impact factor, the score for the article is 1.0, and will decrease every year after publication. We used the decay factor to put priority on more recent findings. All other things being equal, recent articles will have slightly higher scores. The significance of a gene g for a given disorder d was defined by the sum of scores of articles reporting an association of gene g and disorder d.
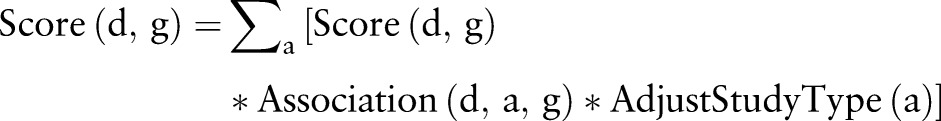 |
Where Association(d, a, g) is (1) 0, if g is not one of the main findings of this article; (2) 1.0, if gene g is reported in the title/results/conclusion; or (3) −1.0 if g is one of the main findings, but the statement is negative. AdjustStudyType(a) is defined as (1) 0, if the article is a review or hypothesis; (2) 0.5, if the article is a case study or examines blood/serum protein levels. The collective gene score can have a negative value when there is a preponderance of evidence against the association according to our scoring algorithm. The overall procedure of our search tool is summarized in figure 2.
Figure 2.
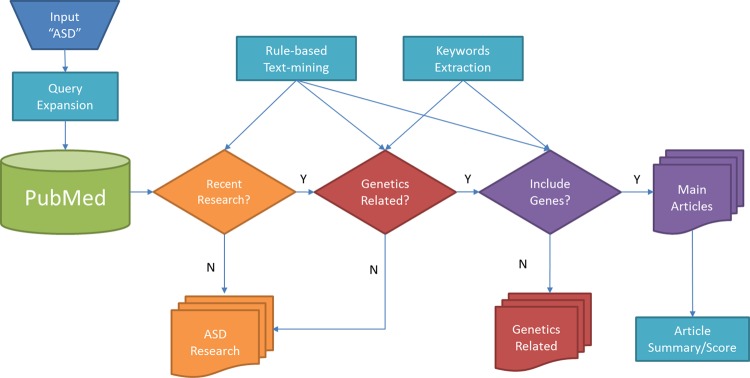
The overall workflow of our search tool. Using an expanded query per given disorder, we retrieve target-disorder-specific, research article information from PubMed. Then we examine whether the given article is genetics-related, or has gene-related terms in the title or abstract, by applying extracted keywords and rule-based text-mining approaches. Finally, we score each document based on the impact factor of the published journal, and score genes using collective scores of articles associated with the target gene.
Results
Reference coverage comparison with existing databases
We tested our implementation with 10 complex disorders and genetic syndromes selected from the GWAS Catalog (attention deficit with hyperactivity disorder, ankylosing spondylitis, ASD, bipolar disorder, multiple sclerosis, and schizophrenia) and genetics home reference (Angelman syndrome, Down syndrome, Huntington disorder, and Lynch syndrome). Table 1 shows the number of gene–disorder association references we found and the number of such references from the union of HuGE Navigator, OMIM, and GAD. For each target disorder, our tool covered more references than the union of results from these sites and we confirmed that all of these articles are specific to the target disorder by manual inspection. We examined all the articles our tool did not retrieve, and found that a majority of them was not related to the target disorder. For example, the target disorder name may be stated in the abstract, but the main topic is for a different disorder. Other causes for exclusion included: the PubMed ID was not available for the article, the article was a commentary article, or the article was published before 1990. We also show high-profile reference samples that were not included in the compared repositories in table 1 and supplementary table 1 (available online only).
Table 1 .
A comparison summary of gene–disorder association references per target disorder
| Target disorder | #Ref. gene-disorder association | #Ref. from other DBs | #Ref. missed in our result | Example ref. in our result only* |
|---|---|---|---|---|
| ADHD | 847 | 463 | 3 | 45 |
| Angelman syndrome | 319 | 11 | 6 | 46 |
| Ankylosing spondylitis | 680 | 210 | 1 | 47 |
| ASD | 1158 | 279 | 3 | 48 |
| Bipolar disorder | 1480 | 935 | 182 | 49 |
| Down syndrome | 1402 | 119 | 21 | 50 |
| Huntington disorder | 1045 | 108 | 9 | 51 |
| Lynch syndrome | 1264 | 161 | 1 | 52 |
| Multiple sclerosis | 2774 | 878 | 95 | 53 |
| Schizophrenia | 4419 | 2691 | 384 | 54 |
*Single reference per disorder is shown. Full references are available in supplementary table 1 (available online only).
ADHD, attention deficit hyperactivity disorder; ASD, autism spectrum disorder.
Tested, positive result, or negative result genes
As shown in the Methods section, we separately identified tested (or simply mentioned) genes, gene with positive findings, and genes with negative findings. By analyzing negating expressions, gene symbols, and disorder names that appear within the same sentence of the title, result, or conclusion sections, we found that a significant number of articles report negative associations or null findings, in which targeted genes showed no difference in case–control experiments, or genetic variants were not found in patient groups. Currently GAD is the only external resource that provides such references for multiple disorders, so we compared our result with those of GAD. As shown in table 2, our result covers more references than GAD for all target disorders. We examined articles shown only in GAD and found that some of the tested genes without positive association findings were reported as negative associations, while we only count genes combined with explicit negating statements in either title or abstract. We also show reference samples that are not included in GAD in table 2 and supplementary table 2 (available online only).
Table 2 .
References with negative results from our tool and GAD
| Target disorder | #Ref. with negative result | #Ref. with negative result in GAD | #Ref. missed in our result | Example ref. in our result only* |
|---|---|---|---|---|
| ADHD | 135 | 15 | 1 | 55 |
| Angelman syndrome | 21 | 0 | 0 | 56 |
| Ankylosing spondylitis | 114 | 13 | 0 | 57 |
| ASD | 168 | 31 | 3 | 58 |
| Bipolar disorder | 341 | 75 | 6 | 59 |
| Down syndrome | 185 | 3 | 0 | 60 |
| Huntington disorder | 82 | 3 | 2 | 61 |
| Lynch syndrome | 203 | 0 | 0 | 62 |
| Multiple sclerosis | 496 | 73 | 6 | 63 |
| Schizophrenia | 1029 | 104 | 5 | 64 |
*Single reference per disorder is shown. Full references are available in supplementary table 2 (available online only).
ADHD, attention deficit hyperactivity disorder; ASD, autism spectrum disorder; GAD, genetic association database.
An example of a single disorder result: ASD
Identifying association study articles and categories
Next, we report a detailed result of single disorder case, ASD. As of October 2012, we identified 23 661 ASD-related articles in PubMed by querying disorder name and aliases plus MeSH terms without applying any filter. Our expanded query with filters, shown in figure 1, returned about half of them (12 984) as ASD-specific original research or review articles. Second, we identified 1581 genetics-related publications among them by scanning them for targeted MeSH terms or keywords. Third, we categorized these articles in order to weight the scores by their publication or study types. We found 232 reviews (will not be included in scoring) and 135 case reports (50% of the original score will be applied) by publication types. We also found 37 animal models, 272 common variant-focused articles including GWAS, 146 rare variant-focused publications including copy-number variation (CNV) and exome sequencing studies, 52 gene expression-related articles, and 23 blood/serum protein level articles (50% of the original score will be applied). In table 3 and supplementary table 3 (available online only), we summarize our categorization result with references, in order to demonstrate that our tool can effectively retrieve high-impact research articles related to a target disorder in different study types.
Table 3 .
Categorization of ASD-specific articles
| Category | # Ref. | Example ref.* | Category | # Ref. | Example ref.* |
|---|---|---|---|---|---|
| Reviews | 232 | 65 | Common/GWAS | 272 | 66 |
| Case reports | 135 | 67 | Rare/CNV | 139 | 68 |
| Animal models | 37 | 69 | Exome sequencing | 5 | 70 |
| Gene expression | 52 | 71 | Blood/serum | 23 | 72 |
*Single reference per category is shown. Full references are available in supplementary table 3 (available online only).
ASD, autism spectrum disorder; CNV, copy-number variation; GWAS, genome-wide association studies.
Assessment of candidate genes and references
We identified 597 gene symbols; 437 of them have their collective score greater than 1.0. Although the fragile X (FMR1) and methyl CpG binding (MECP2) genes have the largest number of associated articles, our result shows that CNTNAP2 (contactin- associated protein-like 2) is the highest score gene with more recent, high-profile publications. Table 4 and supplementary table 4 (available online only) show our top 10 candidate genes and supporting reference examples. To examine whether we found proper articles without missing a significant portion, we selected two external resources to compare our result with. First, HuGE Navigator maintains genetics-related publications using an algorithmic search, and we obtained 256 articles with a disease term of ‘autistic disorder’. Second, SFARI gene is a manually curated, ASD-specific database, and we obtained 278 articles that are associated with category 1 (confident) to 5 (minimal evidence) genes, according to their classification method. Compared with the set of genes from HuGE Navigator, our reference set missed one article primarily describing schizophrenia73 and not ASD. Similar cases were found for SFARI set; our result missed 48 articles; however, the main topic of such articles is not ASD specific but on comorbid disorders including epilepsy,84 85 intellectual disability,86 87 and attention-deficit hyperactivity disorder.88 89
Table 4 .
Top 10 candidate genes, ordered by collective article score for each gene
| Symbol | Name | Score | #Ref. | Example references* |
|---|---|---|---|---|
| CNTNAP2 | Contactin-associated protein-like 2 | 56.23 | 34 | 74 |
| FMR1 | Fragile X mental retardation 1 | 50.10 | 68 | 75 |
| SHANK3 | SH3 and multiple ankyrin repeat domains 3 | 48.89 | 32 | 76 |
| MET | Met proto-oncogene | 47.18 | 19 | 77 |
| SLC6A4 | Neurotransmitter transporter, serotonin | 42.87 | 36 | 78 |
| GABRB3 | γ-Aminobutyric acid A receptor, subunit β 3 | 40.08 | 30 | 79 |
| MECP2 | Methyl CpG binding protein 2 | 39.64 | 47 | 80 |
| PTEN | Phosphatase and tensin homolog | 38.09 | 27 | 81 |
| NRXN1 | Neurexin 1 | 32.52 | 23 | 82 |
| EN2 | Engrailed 2 | 31.72 | 17 | 83 |
*Single reference per gene is shown. Full references are available in supplementary table 4 (available online only).
Next, we compared our ASD candidate gene sets with those from external databases. Our set of 597 genes included (1) all candidate genes of GeneCards (31 genes) and PharmGkb (four genes); (2) 121/133 genes of category 1 to 4 in SFARI gene; (3) 21/22 syndromic genes in SFARI; and (4) 231/426 genes in HuGE Navigator. For all genes we missed in the SFARI set, gene names or symbols were not actually listed in the title or abstract. While we missed about a half of candidate genes in the HuGE set, this set included tested genes, not genes with significant findings, as associated genes (eg, 129 genes were associated with a single article,90 according to HuGE Navigator).
Discussion
The main motivation for this study was that although there are many excellent sites91–93 providing detailed data on the human genome, it is cumbersome or not possible to retrieve the disease-associated genes together with supporting references from these sites. There are a few resources that provide disorder-targeted genes with references,14 94 but we found some issues related to references in such sites that cannot be addressed easily by the user as exemplified in the results section.
The innovation of our approach over existing tools is in the increased precision and that it is directly applicable within the context of statistical genetics and human genetic disorder research. Our method includes three formal steps—(1) extended query, (2) keyword filter, and (3) evaluation of abstract structure—to retrieve target disorder-specific, genetics-related articles. When compared to the mainstream data repositories such as HuGE Navigator, GAD, and OMIM, our three steps appear consistently to avoid the inclusion of non-genetics/non-research references and avoid mis-tagging genes/disorders.
Despite the encouraging results shown here, there are a number of limitations of our approach. First, as it focuses on extracting human genes and disorders, it will not accurately extract genes from model animal studies, for example zebrafish as an animal model for human fetal brain development.95 96 Second, because our tool uses a precision-based algorithm to extract genes, non-authoritative gene names/symbols that are not included in NCBI genes or HGNC may not be properly matched to the correct gene symbols. Finally, our method searches titles and abstracts and therefore will not recover a gene association that is only mentioned in the main text. This could impact the sensitivity with GWAS that report many genes or loci in one article as a list in the main text.
We plan to expand our approach to the full text of the articles as future work. As expected from the BioCreative II task,31 36 our rule-based algorithm successfully worked within the focused set of human genome research articles, and within concise data of titles and abstracts. However, statistical or hybrid entity recognition approaches may perform better in full text analysis, as shown in the BioCreative III task.30 We will examine this hypothesis with conditional random field-based models.41 42 97 98
Conclusion
In this work, we introduced a novel PubMed extraction tool that can find and summarize research articles presenting evidence of gene–disorder associations. Comparison with existing resources demonstrated that our tool can cover more references in general and extract candidate genes with an accuracy comparable to manually curated sites. This application provides a fundamental basis for conducting cross-disorder analysis among related disorders, including solid evidence in the literature for every gene–disorder association. The overall candidate gene sets and supporting reference information are available at http://genehawk.hms.harvard.edu, and we plan to update result sets periodically as new publications come out.
Supplementary Material
Footnotes
Contributors: DPW conceived the study. JYJ, DPW and TFD designed the study. JYJ, TFD and THN participated in implementation. All authors contributed to writing and approval of the final manuscript.
Funding: This work was supported by the National Science Foundation grant nos 0543480 and 0640809 to DPW; and the National Institutes of Health grant no. LM009261 to DPW.
Competing interests: None.
Patient consent: Obtained.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: The overall candidate gene sets and supporting reference information are available online: http://genehawk.hms.harvard.edu.
References
- 1.Safran M, Dalah I, Alexander J, et al. GeneCards Version 3: the human gene integrator. Database (Oxford) 2010;2010:baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thorn CF, Klein TE, Altman RB. Pharmacogenomics and bioinformatics: PharmGKB. Pharmacogenomics 2010;11:501–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann R. A wiki for the life sciences where authorship matters. Nat Genet 2008;40:1047–51 [DOI] [PubMed] [Google Scholar]
- 4.Hoffmann R, Valencia A. A gene network for navigating the literature. Nat Genet 2004;36:664. [DOI] [PubMed] [Google Scholar]
- 5.Frisch M, Klocke B, Haltmeier M, et al. LitInspector: literature and signal transduction pathway mining in PubMed abstracts. Nucleic Acids Res 2009;37(Web Server issue):W135–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erdogmus M, Sezerman OU. Application of automatic mutation-gene pair extraction to diseases. J Bioinform Comput Biol 2007;5:1261–75 [DOI] [PubMed] [Google Scholar]
- 7.Basu SN, Kollu R, Banerjee-Basu S. AutDB: a gene reference resource for autism research. Nucleic Acids Res 2009;37(Database issue):D832–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen NC, Bagade S, McQueen MB, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet 2008;40:827–34 [DOI] [PubMed] [Google Scholar]
- 9.Wall DP, Pivovarov R, Tong M, et al. Genotator: a disease-agnostic tool for genetic annotation of disease. BMC Med Genomics 2010;3:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Online Mendelian Inheritance in Man, OMIM®. [Internet]. McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University. http://omim.org/
- 11.Yu W, Gwinn M, Clyne M, et al. A navigator for human genome epidemiology. Nat Genet 2008;40:124–5 [DOI] [PubMed] [Google Scholar]
- 12. Genetic Association Database [Internet]. http://geneticassociationdb.nih.gov/
- 13.Jensen LJ, Saric J, Bork P. Literature mining for the biologist: from information retrieval to biological discovery. Nat Rev Genet 2006;7:119–29 [DOI] [PubMed] [Google Scholar]
- 14.Rebholz-Schuhmann D, Oellrich A, Hoehndorf R. Text-mining solutions for biomedical research: enabling integrative biology. Nat Rev Genet 2012;13:829–39 [DOI] [PubMed] [Google Scholar]
- 15.Karic A, Karic A. Using the BITOLA system to identify candidate genes for Parkinson's disease. Bosn J Basic Med Sci 2011;11:185–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kastrin A, Hristovski D. A fast document classification algorithm for gene symbol disambiguation in the BITOLA literature-based discovery support system. AMIA Annu Symp Proc 2008:358–62 [PMC free article] [PubMed] [Google Scholar]
- 17.Perez-Iratxeta C, Bork P, Andrade MA. Association of genes to genetically inherited diseases using data mining. Nat Genet 2002;31:316–19 [DOI] [PubMed] [Google Scholar]
- 18.Cheung WA, Ouellette BF, Wasserman WW. Quantitative biomedical annotation using medical subject heading over-representation profiles (MeSHOPs). BMC Bioinformatics 2012;13:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fleuren WW, Verhoeven S, Frijters R, et al. CoPub update: CoPub 5.0 a text mining system to answer biological questions. Nucleic Acids Res 2011;39(Web Server issue):W450–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frijters R, Heupers B, van Beek P, et al. CoPub: a literature-based keyword enrichment tool for microarray data analysis. Nucleic Acids Res 2008;36(Web Server issue):W406–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Medical Subject Headings (MeSH) [Internet]. Bethesda (MD): National Library of Medicine (US). [cited October 2012]. http://www.ncbi.nlm.nih.gov/mesh.
- 22.MedlinePlus [Internet]. Bethesda (MD): National Library of Medicine (US) (cited October 2012). http://www.nlm.nih.gov/medlineplus/
- 23.Genetics Home Reference [Internet]. Bethesda (MD): National Library of Medicine (US) (cited October 2012). http://ghr.nlm.nih.gov/
- 24.Dunham I, Shimizu N, Roe BA, et al. The DNA sequence of human chromosome 22. Nature 1999;402:489–95 [DOI] [PubMed] [Google Scholar]
- 25.Sayers E. E-utilities Quick Start: Bethesda (MD): National Center for Biotechnology Information (US); 2008. http://www.ncbi.nlm.nih.gov/books/NBK25500/
- 26.Hindorff A MJ, Morales J, Junkins HA, et al. A Catalog of Published Genome-Wide Association Studies. October 2012. http://www.genome.gov/gwastudies/
- 27. Word frequency and dictionary [Internet]. [cited October 2012]. http://www.wordfrequency.info/sample.asp.
- 28.Ripple AM, Mork JG, Knecht LS, et al. A retrospective cohort study of structured abstracts in MEDLINE, 1992–2006. J Med Libr Assoc 2011;99:160–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agarwal S, Yu H, Kohane I. BioNOT: a searchable database of biomedical negated sentences. BMC Bioinformatics 2011;12:420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Z, Kao HY, Wei CH, et al. The gene normalization task in BioCreative III. BMC Bioinformatics 2011;12(Suppl. 8):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgan AA, Lu Z, Wang X, et al. Overview of BioCreative II gene normalization. Genome Biol 2008;9(Suppl. 2):S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai HJ, Chang YC, Tsai RT, et al. Integration of gene normalization stages and co-reference resolution using a Markov logic network. Bioinformatics 2011;27:2586–94 [DOI] [PubMed] [Google Scholar]
- 33.Huang M, Liu J, Zhu X. GeneTUKit: a software for document-level gene normalization. Bioinformatics 2011;27:1032–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wermter J, Tomanek K, Hahn U. High-performance gene name normalization with GeNo. Bioinformatics 2009;25:815–21 [DOI] [PubMed] [Google Scholar]
- 35.Hur J, Schuyler AD, States DJ, et al. SciMiner: web-based literature mining tool for target identification and functional enrichment analysis. Bioinformatics 2009;25:838–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hakenberg J, Plake C, Royer L, et al. Gene mention normalization and interaction extraction with context models and sentence motifs. Genome Biol 2008;9(Suppl. 2):S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alex B, Grover C, Haddow B, et al. Automating curation using a natural language processing pipeline. Genome Biol 2008;9(Suppl.2):S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu H, Fan JW, Hripcsak G, et al. Gene symbol disambiguation using knowledge-based profiles. Bioinformatics 2007;23:1015–22 [DOI] [PubMed] [Google Scholar]
- 39.Podowski RM, Cleary JG, Goncharoff NT, et al. Suregene, a scalable system for automated term disambiguation of gene and protein names. J Bioinform Comput Biol 2005;3:743–70 [DOI] [PubMed] [Google Scholar]
- 40.Pahikkala T, Ginter F, Boberg J, et al. Contextual weighting for Support Vector Machines in literature mining: an application to gene versus protein name disambiguation. BMC Bioinformatics 2005;6:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Settles B. ABNER: an open source tool for automatically tagging genes, proteins and other entity names in text. Bioinformatics 2005;21:3191–2 [DOI] [PubMed] [Google Scholar]
- 42.Leaman R, Gonzalez G. BANNER: an executable survey of advances in biomedical named entity recognition. Pac Symp Biocomput 2008:652–63 [PubMed] [Google Scholar]
- 43.Fukuda K, Tamura A, Tsunoda T, et al. Toward information extraction: identifying protein names from biological papers. Pac Symp Biocomput 1998:707–18 [PubMed] [Google Scholar]
- 44.Demner-Fushman D, Lin J. Answering clinical questions with knowledge-based and statistical techniques. Computational Linguistics 2007;33:63–103 [Google Scholar]
- 45.Sakrikar D, Mazei-Robison MS, Mergy MA, et al. Attention deficit/hyperactivity disorder-derived coding variation in the dopamine transporter disrupts microdomain targeting and trafficking regulation. J Neurosci 2012;32:5385–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chamberlain SJ, Chen PF, Ng KY, et al. Induced pluripotent stem cell models of the genomic imprinting disorders Angelman and Prader–Willi syndromes. Proc Natl Acad Sci USA 2010;107:17668–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doyle GA, Lai AT, Chou AD, et al. Re-sequencing of ankyrin 3 exon 48 and case-control association analysis of rare variants in bipolar disorder type I. Bipolar Disord 2012;14:809–21 [DOI] [PubMed] [Google Scholar]
- 48.Novarino G, El-Fishawy P, Kayserili H, et al. Mutations in BCKD-kinase lead to a potentially treatable form of autism with epilepsy. Science 2012;338:394–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rueckert EH, Barker D, Ruderfer D, et al. Cis-acting regulation of brain-specific ANK3 gene expression by a genetic variant associated with bipolar disorder. Mol Psychiatry 2013;18:922–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roy A, Cowan G, Mead AJ, et al. Perturbation of fetal liver hematopoietic stem and progenitor cell development by trisomy 21. Proc Natl Acad Sci USA 2012;109:17579–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu D, Pendergraff H, Liu J, et al. Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant huntingtin expression. Cell 2012;150:895–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Engel C, Loeffler M, Steinke V, et al. Risks of less common cancers in proven mutation carriers with Lynch syndrome. J Clin Oncol 2012;30:4409–15. [DOI] [PubMed] [Google Scholar]
- 53.Bai L, Lennon DP, Caplan AI, et al. Hepatocyte growth factor mediates mesenchymal stem cell-induced recovery in multiple sclerosis models. Nat Neurosci 2012;15:862–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu B, Ionita-Laza I, Roos JL, et al. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat Genet 2012;44:1365–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stevenson J, Sonuga-Barke E, McCann D, et al. The role of histamine degradation gene polymorphisms in moderating the effects of food additives on children's ADHD symptoms. Am J Psychiatry 2010;167:1108–15 [DOI] [PubMed] [Google Scholar]
- 56.Bressler J, Tsai TF, Wu MY, et al. The SNRPN promoter is not required for genomic imprinting of the Prader–Willi/Angelman domain in mice. Nat Genet 2001;28:232–40 [DOI] [PubMed] [Google Scholar]
- 57.Davidson SI, Liu Y, Danoy PA, et al. Association of STAT3 and TNFRSF1A with ankylosing spondylitis in Han Chinese. Ann Rheum Dis 2011;70:289–92 [DOI] [PubMed] [Google Scholar]
- 58.Palmieri L, Papaleo V, Porcelli V, et al. Altered calcium homeostasis in autism-spectrum disorders: evidence from biochemical and genetic studies of the mitochondrial aspartate/glutamate carrier AGC1. Mol Psychiatry 2010;15:38–52 [DOI] [PubMed] [Google Scholar]
- 59.Liu C, Shi J, Badner JA, et al. No association of trace amine receptor genes with bipolar disorder. Mol Psychiatry 2007;12:979–81 [DOI] [PubMed] [Google Scholar]
- 60.Heywood W, Wang D, Madgett TE, et al. The development of a peptide SRM-based tandem mass spectrometry assay for prenatal screening of Down syndrome. J Proteomics 2012;75:3248–57 [DOI] [PubMed] [Google Scholar]
- 61.Ramos EM, Latourelle JC, Lee JH, et al. Population stratification may bias analysis of PGC-1alpha as a modifier of age at Huntington disease motor onset. Hum Genet 2012;131:1833–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goel A, Xicola RM, Nguyen TP, et al. Aberrant DNA methylation in hereditary nonpolyposis colorectal cancer without mismatch repair deficiency. Gastroenterology 2010;138:1854–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuhle J, Pohl C, Mehling M, et al. Lack of association between antimyelin antibodies and progression to multiple sclerosis. N Engl J Med 2007;356:371–8 [DOI] [PubMed] [Google Scholar]
- 64.Mathieson I, Munafo MR, Flint J. Meta-analysis indicates that common variants at the DISC1 locus are not associated with schizophrenia. Mol Psychiatry 2012;17:634–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lewis S. Synaptic physiology: meeting point for autism and fragile X syndrome. Nat Rev Neurosci 2012;13:74023034480 [Google Scholar]
- 66.Anney R, Klei L, Pinto D, et al. Individual common variants exert weak effects on the risk for autism spectrum disorderspi. Hum Mol Genet 2012;21:4781–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Newbury DF, Mari F, Sadighi Akha E, et al. Dual copy number variants involving 16p11 and 6q22 in a case of childhood apraxia of speech and pervasive developmental disorder. Eur J Hum Genet 2013;21:361–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moreno-De-Luca D, Sanders SJ, Willsey AJ, et al. Using large clinical data sets to infer pathogenicity for rare copy number variants in autism cohorts. Mol Psychiatry [Epub ahead of print 9 Oct 2012] doi: 10.1038/mp.2012.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hsiao EY, McBride SW, Chow J, et al. Modeling an autism risk factor in mice leads to permanent immune dysregulation. Proc Natl Acad Sci USA 2012;109:12776–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanders SJ, Murtha MT, Gupta AR, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 2012;485:237–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Konopka G, Wexler E, Rosen E, et al. Modeling the functional genomics of autism using human neurons. Mol Psychiatry 2012;17:202–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bressler JP, Gillin PK, O'Driscoll C, et al. Maternal antibody reactivity to lymphocytes of offspring with autism. Pediatr Neurol 2012;47:337–40 [DOI] [PubMed] [Google Scholar]
- 73.Akahane A, Kunugi H, Tanaka H, et al. Association analysis of polymorphic CGG repeat in 5′ UTR of the reelin and VLDLR genes with schizophrenia. Schizophr Res 2002;58:37–41 [DOI] [PubMed] [Google Scholar]
- 74.Anderson GR, Galfin T, Xu W, et al. Candidate autism gene screen identifies critical role for cell-adhesion molecule CASPR2 in dendritic arborization and spine development. Proc Natl Acad Sci USA 2012;109:18120–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hatton DD, Sideris J, Skinner M, et al. Autistic behavior in children with fragile X syndrome: prevalence, stability, and the impact of FMRP. Am J Med Genet A 2006;140A:1804–13 [DOI] [PubMed] [Google Scholar]
- 76.Moessner R, Marshall CR, Sutcliffe JS, et al. Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet 2007;81:1289–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Campbell DB, Sutcliffe JS, Ebert PJ, et al. A genetic variant that disrupts MET transcription is associated with autism. Proc Natl Acad Sci USA 2006;103:16834–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ma DQ, Rabionet R, Konidari I, et al. Association and gene-gene interaction of SLC6A4 and ITGB3 in autism. Am J Med Genet B Neuropsychiatr Genet 2010;153B:477–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thanseem I, Anitha A, Nakamura K, et al. Elevated transcription factor specificity protein 1 in autistic brains alters the expression of autism candidate genes. Biol Psychiatry 2012;71:410–18 [DOI] [PubMed] [Google Scholar]
- 80.Cukier HN, Lee JM, Ma D, et al. The Expanding Role of MBD Genes in Autism: Identification of a MECP2 Duplication and Novel Alterations in MBD5, MBD6, and SETDB1. Autism Res 2012;5:385–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee TL, Raygada MJ, Rennert OM. Integrative gene network analysis provides novel regulatory relationships, genetic contributions and susceptible targets in autism spectrum disorders. Gene 2012;496:88–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu Y, Hu Z, Xun G, et al. Mutation analysis of the NRXN1 gene in a Chinese autism cohort. J Psychiatr Res 2012;46:630–4 [DOI] [PubMed] [Google Scholar]
- 83.Lin PI, Chien YL, Wu YY, et al. The WNT2 gene polymorphism associated with speech delay inherent to autism. Res Dev Disabil 2012;33:1533–40 [DOI] [PubMed] [Google Scholar]
- 84.Mefford HC, Muhle H, Ostertag P, et al. Genome-wide copy number variation in epilepsy: novel susceptibility loci in idiopathic generalized and focal epilepsies. PLoS Genet 2010;6:e1000962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu Y, Feng Y, Pang JR, et al. Study on expression of laminin in patients with intractable epilepsy. Int J Neurosci 2009;119:2219–27 [DOI] [PubMed] [Google Scholar]
- 86.Pagan C, Botros HG, Poirier K, et al. Mutation screening of ASMT, the last enzyme of the melatonin pathway, in a large sample of patients with intellectual disability. BMC Med Genet 2011;12:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zweier C, de Jong EK, Zweier M, et al. CNTNAP2 and NRXN1 are mutated in autosomal-recessive Pitt-Hopkins-like mental retardation and determine the level of a common synaptic protein in Drosophila. Am J Hum Genet 2009;85: 655–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Elia J, Gai X, Hakonarson H, et al. Structural variations in attention-deficit hyperactivity disorder. Lancet 2011;377:377–8; author reply 8 [DOI] [PubMed] [Google Scholar]
- 89.Park J, Willmott M, Vetuz G, et al. Evidence that genetic variation in the oxytocin receptor (OXTR) gene influences social cognition in ADHD. Prog Neuropsychopharmacol Biol Psychiatry 2010;34:697–702 [DOI] [PubMed] [Google Scholar]
- 90.de Krom M, Staal WG, Ophoff RA, et al. A common variant in DRD3 receptor is associated with autism spectrum disorder. Biol Psychiatry 2009;65: 625–30 [DOI] [PubMed] [Google Scholar]
- 91.Fernandez-Suarez XM, Galperin MY. The 2013 Nucleic Acids Research Database Issue and the online molecular biology database collection. Nucleic Acids Res 2013;41(Database issue):D1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Frodsham AJ, Higgins JP. Online genetic databases informing human genome epidemiology. BMC Med Res Methodol 2007;7:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thorisson GA, Muilu J, Brookes AJ. Genotype-phenotype databases: challenges and solutions for the post-genomic era. Nat Rev Genet 2009;10:9–18 [DOI] [PubMed] [Google Scholar]
- 94.Krallinger M, Leitner F, Valencia A. Analysis of biological processes and diseases using text mining approaches. Methods Mol Biol 2010;593:341–82 [DOI] [PubMed] [Google Scholar]
- 95.Howe K, Clark MD, Torroja CF, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013;496:498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fetcho JR. Neuroscience: crystal-clear brains. Nature 2012;485:453–5 [DOI] [PubMed] [Google Scholar]
- 97.Hsu CN, Chang YM, Kuo CJ, et al. Integrating high dimensional bi-directional parsing models for gene mention tagging. Bioinformatics 2008;24:i286–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tsai RT, Sung CL, Dai HJ, et al. NERBio: using selected word conjunctions, term normalization, and global patterns to improve biomedical named entity recognition. BMC Bioinformatics 2006;7(Suppl. 5):S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


