Abstract
Objective
To research computational methods for discovering body site and severity modifiers in clinical texts.
Methods
We cast the task of discovering body site and severity modifiers as a relation extraction problem in the context of a supervised machine learning framework. We utilize rich linguistic features to represent the pairs of relation arguments and delegate the decision about the nature of the relationship between them to a support vector machine model. We evaluate our models using two corpora that annotate body site and severity modifiers. We also compare the model performance to a number of rule-based baselines. We conduct cross-domain portability experiments. In addition, we carry out feature ablation experiments to determine the contribution of various feature groups. Finally, we perform error analysis and report the sources of errors.
Results
The performance of our method for discovering body site modifiers achieves F1 of 0.740–0.908 and our method for discovering severity modifiers achieves F1 of 0.905–0.929.
Discussion
Results indicate that both methods perform well on both in-domain and out-domain data, approaching the performance of human annotators. The most salient features are token and named entity features, although syntactic dependency features also contribute to the overall performance. The dominant sources of errors are infrequent patterns in the data and inability of the system to discern deeper semantic structures.
Conclusions
We investigated computational methods for discovering body site and severity modifiers in clinical texts. Our best system is released open source as part of the clinical Text Analysis and Knowledge Extraction System (cTAKES).
Keywords: natural language processing, relation extraction, biomedical informatics, information extraction
Background and significance
It is widely accepted that the clinical narrative within electronic health records contains a substantial part of the patient's health information, but in its raw form does not represent computable data structures suitable for biomedical applications. Increasingly over the last decade the field of clinical natural language processing (NLP) has focused on developing methods for the semantic processing of clinical text that are use case and disease agnostic, and can thus be incorporated into a variety of clinical applications. The clinical NLP community has been converging around the use of conventions and standards for semantic processing to foster intra and inter-operability such as the unified medical language system1 (UMLS),2 Penn Treebank,3 PropBank,4 TimeML5 and Health Level 7. This shift from use case-specific applications to more general purpose and standards-based tools is characteristic of the last few years of clinical NLP efforts especially within the environment of meaningful use stage 2.6
The transformation of free text into a structured computable representation model is known as Information Extraction.7 In the general NLP domain, such representation models have been defined by the NIST-sponsored Automatic Content Extraction (ACE)8 and Text Analysis Conference (TAC)9 shared tasks, which included templates for person and organization and template slots such as employee_of and city_of_residence. However, these representations are of little relevance to the clinical domain. Instead, representations such as the Consolidated Clinical Document Architecture (CCDA) for Meaningful Use Stage 2, the Clinical Element Model10 (CEM) or the College of American Pathologists (CAP) protocols are more relevant. CCDA provides clinical and functional context for practical implementations of the Health Level 7 balloted standards6 and can be thought of as the normalization target for electronic health records information.
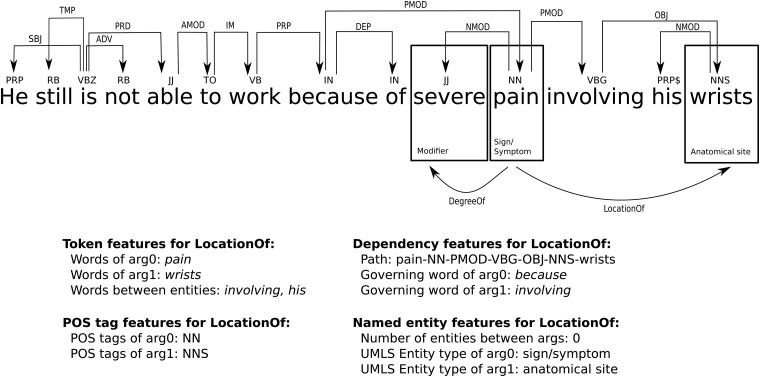
Body site and severity modifiers are two of the attributes (or template slots) associated with healthcare representation models such as CCDA, CEM and the CAP. These modifiers are usually attached to a disease/disorder, sign/symptom or procedure. Consider a sentence from a clinical record of a rheumatoid arthritis patient: He still is not able to work because of severe pain involving his wrists. In this sentence we would like to discover two facts: (1) that the body site of pain is the patient's wrists, and (2) that the severity level of pain is severe.
There is earlier work on discovering tumor body sites from pathology notes. MedKAT/P11 employs hand-built rules to populate a colon cancer template in which the body location of the primary tumor is one of the attributes. caTIES12 identifies all tumor site mentions in pathology reports using regular expressions. Martinez and Li13 explore a machine learning methodology for populating a colorectal cancer template with six attributes including the tumor site. They report an F score of 58.1, for a model whose most predictive features are based on UMLS and SNOMED-CT. Jouhet et al14 work with pathology notes from the French Poitou-Charentes Cancer Registry automatically to discover the primary tumor site and code to the International Classification of Diseases—Oncology (ICD-O)15 codes using machine learning techniques. Kuvuluru et al16 focus on extracting the generic ICD-O code for primary cancers reported in pathology reports. The body site of interest is the one of the primary tumor. MedLEE17 18 ‘has an integrated syntactic and semantic component which is realized in the form of its grammar. The MedLEE grammar consists of a specification of semantic (and sometimes syntactic) components and is used to interpret the semantic properties of the individual terms and of their relations with other terms, and to generate a target output. The semantic grammar rules were developed based on co-occurrence patterns observed in clinical text.’ MedLEE's scope includes processing radiology notes, discharge summaries and clinical reports.
In this paper we demonstrate that the problem of body site and severity modifier discovery can be successfully treated as a relation extraction task, a well-established semantic processing task. Relation extraction focuses on determining the relationships between entities in text. We use the UMLS definitions to type the relations and the entities. In our sample sentence, the entities pain and wrists are the participants of the LocationOf relation and can be succinctly captured as LocationOf(wrists, pain). The relationship between the entity pain and the modifier severe can be expressed as DegreeOf(pain, severe). The first argument of the LocationOf relation is an anatomical site, while the second argument is a sign/symptom, disease/disorder, or procedure. The first argument of the DegreeOf relation is either a sign/symptom or a procedure, while the second argument is a modifier (eg, significant, severe, marked).
In general, semantic processing of language aims to capture the meaning behind the many surface forms that written language can assume. For example, the relationship we represented earlier as LocationOf (wrists, pain) is often also expressed in clinical notes as pain in his wrists, pains involving his wrists, wrist pain, or his main complaints of joint pain are presently at the wrists bilaterally. Because of this diversity of clinical language, a rule-based approach is hard to implement. Instead, we adopt a supervised machine learning approach, in which we pair up candidate clinical entities and delegate the decision about whether they participate in a relation to a supervised classifier.
Supervised learning has been applied for relation extraction in the general domain. Feature-based methods19 20 represent relation instances using carefully engineered sets of features. Kernel-based methods21 22 make it possible to explore large (in some cases infinite) feature spaces automatically. In this work, we attempt both approaches and demonstrate that the feature-based approach is more promising for our task. In the clinical domain, relation extraction was the focus of the 2010 integrating the biology and the bedside (i2b2)/VA shared task,23 although the targeted relations were very different from ours. A recent work24 applied supervised learning for identifying anatomical locations of a small number of manually selected actionable findings in appendicitis-related radiology reports. Unlike their work, we do not limit the input of our system to a set of predefined findings; instead our system is potentially capable of identifying the anatomical sites for any sign/symptom, disease/disorder, or procedure that exists in UMLS. Open information extraction25–27 offers an alternative to supervised learning via the use of lightly supervised methods for extracting relations and their arguments from large collections of text. However, this work is not directly applicable to our task due to the difficulty of mapping the open set of relations to our relations of interest.
Our main contributions are:
We design and develop a machine learning system for discovering intra-sentential body site and severity modifiers from the clinical narrative, modeling the problem as a relation extraction task.
We conduct feature ablation experiments to determine the most salient features for the task.
We experiment with tree kernels, which have not been used in the past for relation extraction from the clinical narrative.
We demonstrate that our models are highly portable across different types of notes.
To allow result replication we make the gold standard corpus we used in our experiments available to the research community, and release our best-performing methods open source as part of the Apache clinical Text Analysis and Knowledge Extraction System28 (cTAKES)29 allowing replication of experiments as well as adoption and improvements thus strengthening the clinical NLP ecosystem.
Materials and methods
Corpus
In our experiments, we utilize two annotated corpora that have been in development for the past 3 years and that are now made available to the community through data use agreements with the contributing institution (to initiate the process, contact the last author)—the Strategic Health Advanced Research Project: area 4 (SHARP)30 and the shared annotated resource (ShARe).31 Table 1 provides the high-level characteristics of the corpora and box 1 gives a few example annotations.
Table 1 .
Description and statistics of the SHARP and ShARe corpora
| Corpus | SHARP | ShARe |
|---|---|---|
| Type of notes | Radiology, pathology, oncology | ICU notes, discharge summaries |
| Tokens | 70 704 | 104 918 |
| Sentences | 4801 | 8058 |
| Entity mentions | 11 781 | 5541 |
| Entity mention pairs | 36 865 | 6441 |
| LocationOf relations | 5025 | 2190 |
| DegreeOf relations | 729 | 702 |
| LocationOf agreement | 0.74 | 0.80 |
| DegreeOf agreement | 0.87 | 0.66 |
ShARe, Shared Annotated Resource; SHARP, Strategic Health Advanced Research Project.
Box 1 . Example annotations.
The [common femoral] had [moderate] [disease] without [stenosis].
LocationOf(common femoral, disease); LocationOf (common femoral, stenosis); DegreeOf(disease, moderate)
The patient had a [[skin] tumor] removed from [behind his left ear].
LocationOf(skin, skin tumor); LocationOf(behind his left ear, skin tumor)
The SHARP corpus provides several layers of annotations—syntax and semantics based on Treebank, PropBank and UMLS,32 and normalization targets based on CEM.33 The corpus consists of an equal amount of radiology notes, from Mayo Clinic peripheral arterial disease patients, and breast cancer oncology and pathology notes, from Seattle Group Health. The SHARP corpus is annotated for such clinical entities as drugs, diseases/disorders, signs/symptoms, procedures and anatomical sites. Diseases/disorders, sign/symptoms and procedures have body site modifiers expressed as a relation between the anchor and an anatomical site. Diseases/disorders and sign/symptoms have a severity modifier, expressed as a relation between the anchor and a severity indicator normalized to none, slight, moderate or severe. At the time of our experiments, the ‘seed’ part of the SHARP corpus, consisting of 18 batches (subsections) and a total of 183 notes, was fully completed including double annotation and adjudication. We split this corpus into training (140 notes: batches 2–9, 13–16, 18–19), development (21 notes: batches 10, 17), and test (22 notes: batches 11, 12) sets.
The ShARe corpus consists of MIMIC intensive care unit notes and discharge summaries as part of the PhysioNet project.34 It annotates parts of speech (POS) and phrasal chunks consistent with the SHARP corpus. Annotated named entities are a subset of the SHARP types: anatomical sites and diseases/disorders. The latter have body site and severity modifiers also consistent with the SHARP corpus. At the time of our experiments, the first 13 batches of the ShARe corpus were fully annotated and adjudicated. We used these 13 batches (130 notes) for our experiments. We split the set of notes into a training set (80 notes), development set (25 notes), and test set (25 notes). The full details of the ShARe annotations will be described in a separate paper; here we focus only on the relevant relation annotations.
Inter-annotator agreement on these corpora is computed with F1 score. Human agreement typically suggests the upper bound of system performance but is not necessarily the ceiling.
Classification task
We view the problems of body site and severity modifier discovery as relation extraction tasks. Formally, we define a relation extraction task as: given two sets of entities, E and F, and a relation, R⊆E×F, find all pairs (e, f)∈R. Essentially, a relation extraction task requires us to search over all pairs of entities in E and F, and identify the ones that participate in the relation R. The set E will contain entities like symptoms and diseases for both the body site (LocationOf) and severity (DegreeOf) relations, while the set F will contain anatomical sites for the Location-Of relation, and severity expressions for the DegreeOf relation.
We cast this relation extraction task as a supervised learning problem. Given a pair of entities (e, f), we train a classifier to decide whether or not (e, f)∈R. Thus, the classification task is binary and the classifier must assign each pair (e, f) one of the classes {R, No-R}. In particular, we focus on a sentence-level task, in which the classifier must look at all pairs of entities within a sentence, and learn to predict the class R if a relation was annotated between those two entities, and the class No-R if a relation was not annotated. We train two relation extraction classifiers, one for R=LocationOf and one for R=DegreeOf.
In this paper, we train support vector machines (SVM) classifiers for these tasks. SVM perform well on a variety of NLP tasks.35
Classifier features
To train a classifier, we must characterize each (e, f) pair with a set of features that provide clues as to whether or not this pair of entities participates in the relation R. We utilize rich linguistic features including lexical, syntactic, and semantic features. Figure 1 illustrates the features. Many of our features are based on Zhou et al20 and the best-performing systems36 37 from 2011 i2b2 challenge.23 Below, we briefly summarize our features and refer the reader to these publications for details:
Token: the first and the last word of each entity, all words of the entity as a bag, the preceding and the following three words, and the number of words between the two entities
POS: the POS tags of each entity as a bag
Chunking: the head words of the syntactic base phrase chunks between the two entities
Dependency tree: the governing word and its POS tag for each entity's head word
Dependency path: the length of the path through the dependency from each entity to their common ancestor, and the path between the two entities as a string
Named entity: the number of entities between the two entities, UMLS types of both entities, and whether the first entity is enclosed in the second one (or vice versa).
Figure 1.
Some of the features used to predict the LocationOf relation in an example sentence.
We also experimented with tree kernel features, which have been used successfully for relation extraction and semantic role labeling in the general domain.38 39 Tree kernels offer a generalized approach to representing syntactic features. An instance is represented by some phrase structure context, and the similarity between two instance structures is computed by taking a weighted sum of similar substructures (see Collins and Duffy40 for details). In this work, we use a representation called path-enclosed tree,39 which, starting from a complete automatic parse of a sentence, represents each potential relation instance with the smallest sub-tree in the sentence containing both arguments. In addition, new nodes labeled ARG1-{type} and ARG2-{type} are inserted into the tree above the lowest node that dominates the respective arguments, where {type} represents the UMLS semantic type of the argument. All features are generated automatically by cTAKES, which includes a POS tagger, a UMLS dictionary lookup, a phrase-chunker and the dependency parser from Albright et al.41
Classifier parameters
In addition to a set of features, most supervised classifiers have a set of parameters that are not set during the learning process, and must be separately specified. SVMs have several such parameters, including the cost of misclassification (SVMC), the kernel type (SVMkernel, eg, linear vs radial basis function), and additional kernel-specific parameters (eg, SVMgamma in the radial basis function kernel). To address specific issues associated with entity-relation data, our models include several additional classifier parameters beyond the standard SVM parameters.
Learning from imbalanced data is a central challenge in training relation extraction systems. Recall that we generate training instances with classes {R, No-R} for all pairs of entities within each sentence. As most entities and modifiers in a sentence are unrelated, we typically end up with significantly more negative than positive examples. Without additional guidance, most classifiers learn to favor the more dominant class. Thus, our models include a down-sampling parameter, Pneg, to address this imbalance. During training, this parameter is used randomly to discard negative (ie, dominant class) examples with probability (1—Pneg).
We also consider, as a classifier parameter, a variation to the classification paradigm. Note that in the standard binary classifier approach described above, if there is any overlap between sets E and F we may have to classify two entities e and f twice: once for the pair (e, f) and once for the pair (f, e). An alternative to this approach is to train a three-way classifier. We first order all of the entities by their location in the clinical text, and then pair up entities only with other entities that are later in the text. This means that we will see only (e, f) or (f, e), but not both. Now, we train our classifier to assign each pair (x, y) one of three classes: {R, R−1, No-R}, where the class R indicates that the relation R(x, y) is present, the class R−1 indicates that the relation R(y, x) is present, and No-R indicates that there is no relation for either ordering of the entities. Thus in our set-up, we have a strategy parameter that is set to one of {2-class, 3-class}.
The tree kernel requires the setting of a parameter λ, which represents a discount of larger tree structures. In addition, tree kernels can be used on their own or incorporated with other features in a composite kernel, which takes a weighted linear combination of a traditional feature kernel with a tree kernel.
Experimental set-up
Models are evaluated on these corpora using measures commonly employed in NLP—namely precision, recall and F1 score.7 42
To set the various model parameters (SVMC, SVMkernel, SVMgamma, Pneg, strategy), models are trained on the training set and evaluated on the development set. We explore the space of possible parameter settings using a grid search, training one model for each set of parameters. The parameter settings for the model with the highest F1 on the development set are used to train a model on the combination of the training and development data. This final model is then evaluated on the testing data. Note that this parameter tuning is performed separately for the DegreeOf and LocationOf models, so the two models may have different parameter settings chosen by their respective grid searches. For the tree kernel parameters, we set λ=0.4, and use a composite kernel (combining the grid search-optimized feature kernel and the tree kernel), normalizing both kernels and giving them equal weight. These tree kernel parameters can be optimized using a grid search, but it is computationally quite expensive to train tree kernels, so we set the parameters based on values found to perform well in previous work.
We implement five rule-based baselines to which we compare the performance of our system. The first four baselines only link pairs of entities that have appropriate entity types for their respective relations (DegreeOf or LocationOf). The first baseline predicts relations only in sentences with exactly two entities. The second baseline searches for sentences with one or more modifiers (anatomical site for LocationOf, severity for DegreeOf) and exactly one other entity, and predicts a relation between the entity and the closest modifier. The third baseline associates each modifier with the nearest entity, as long as there is no intervening modifier. The fourth baseline predicts a relation only between entities that are enclosed in the same noun phrase. The fifth baseline approximates a grammar/rule-based system. It trains an SVM model using only the dependency path feature (with words on both ends replaced with their UMLS semantic types), essentially allowing the SVM to memorize dependency paths between clinical entities that are likely indicators of LocationOf or DegreeOf relations. We train a model using only this feature, tuning the model parameters on the development set.
Results
Model tuning
As described earlier, a grid search over possible parameter settings was performed using the training and development data. This search determined that for the SHARP corpus, the best parameters for the LocationOf classifier were SVMC=100.0, SVMkernel=radial basis function, SVMgamma=0.01, Pneg=0.5 and strategy=3-class; the best parameters for the DegreeOf classifier were SVMC=0.05, SVMkernel=linear, Pneg=1.0 and strategy=3-class. For the ShARe corpus, the best parameters for the LocationOf classifier were SVMC=100.0, SVMkernel=radial basis function, SVMgamma=0.001, Pneg=1.0 and strategy=3-class; the best parameters for the DegreeOf classifier were SVMC=0.5, SVMkernel=linear, Pneg=1.0 and strategy=3-class.
So, for most models, the 3-class strategy was most effective and downsampling was not necessary.
Model evaluation
In this section we conduct an evaluation on a held-out test set, which provides an estimate of the system performance that can be achieved in practice. For each corpus, we train the models for the LocationOf and DegreeOf relations on the combination of the training and development data, and using the parameters determined in the previous section. We then evaluate the models against the test set. We also evaluate the baseline models on the same test set. To assess the portability of our models we also evaluate the models trained on the SHARP training and development sets against the ShARe test set. Results are shown in table 2. We do not test a ShARe-trained model on the SHARP test set because ShARe annotates only a subset of the SHARP entity types (see the Corpus section). So for example, a ShARe model will never see a procedure mention in the ShARe training data, but would be asked to find relations for procedure mentions in the SHARP test set.
Table 2 .
Model performance for on the SHARP and ShARe test sets
| Relation | Test corpus | Model | Precision | Recall | F1 |
|---|---|---|---|---|---|
| LocationOf | SHARP | Baseline 1 | 0.900 | 0.096 | 0.174 |
| Baseline 2 | 0.910 | 0.198 | 0.325 | ||
| Baseline 3 | 0.858 | 0.431 | 0.574 | ||
| Baseline 4 | 0.551 | 0.522 | 0.536 | ||
| Baseline 5 | 0.758 | 0.340 | 0.470 | ||
| SVM trained on SHARP | 0.786 | 0.699 | 0.740 | ||
| Composite (TK+features) | 0.828 | 0.661 | 0.735 | ||
| Human agreement | – | – | 0.744 | ||
| ShARe | Baseline 1 | 1.000 | 0.356 | 0.525 | |
| Baseline 2 | 1.000 | 0.381 | 0.552 | ||
| Baseline 3 | 0.971 | 0.777 | 0.863 | ||
| Baseline 4 | 0.521 | 0.700 | 0.598 | ||
| Baseline 5 | 0.941 | 0.556 | 0.699 | ||
| SVM trained on ShARe | 0.953 | 0.867 | 0.908 | ||
| SVM trained on SHARP | 0.916 | 0.883 | 0.899 | ||
| Human agreement | – | – | 0.800 | ||
| DegreeOf | SHARP | Baseline 1 | 1.000 | 0.044 | 0.084 |
| Baseline 2 | 1.000 | 0.044 | 0.084 | ||
| Baseline 3 | 0.907 | 0.857 | 0.881 | ||
| Baseline 4 | 0.896 | 0.758 | 0.821 | ||
| Baseline 5 | 0.860 | 0.473 | 0.610 | ||
| SVM trained on SHARP | 0.869 | 0.945 | 0.905 | ||
| Composite (TK+features) | 0.840 | 0.923 | 0.880 | ||
| Human agreement | – | – | 0.871 | ||
| ShARe | Baseline 1 | 0.944 | 0.121 | 0.214 | |
| Baseline 2 | 0.947 | 0.128 | 0.225 | ||
| Baseline 3 | 0.977 | 0.887 | 0.929 | ||
| Baseline 4 | 0.929 | 0.745 | 0.827 | ||
| Baseline 5 | 0.404 | 0.979 | 0.571 | ||
| SVM trained on ShARe | 0.929 | 0.929 | 0.929 | ||
| SVM trained on SHARP | 0.926 | 0.887 | 0.906 | ||
| Human agreement | – | – | 0.664 |
ShARe, Shared Annotated Resource; SHARP, Strategic Health Advanced Research Project.
Feature ablation experiments
To quantify the utility of each feature group, we performed all-but-one feature ablation experiments on the development set. That is, we left out each feature group, retrained the model, and evaluated it on the development set. We report the results for the SHARP corpus in table 3.
Table 3 .
Performance of models with various features removed on the SHARP development set
| Included features | LocationOf | DegreeOf | ||
|---|---|---|---|---|
| F1 | ΔF1 | F1 | ΔF1 | |
| All | 0.776 | 0.972 | ||
| No token features | 0.742 | −0.034 | 0.909 | −0.063 |
| No POS features | 0.768 | −0.008 | 0.963 | −0.009 |
| No chunking features | 0.766 | −0.010 | 0.972 | 0 |
| No named entity features | 0.712 | −0.064 | 0.904 | −0.068 |
| No dependency tree features | 0.757 | −0.019 | 0.944 | −0.028 |
| No dependency path features | 0.755 | −0.021 | 0.954 | −0.018 |
SHARP, Strategic Health Advanced Research Project.
Discussion
The results of our evaluation indicate that for both LocationOf and DegreeOf, model performance is high—typically achieving the same level as the human agreement. The performance of the DegreeOf model is consistently higher than that of the LocationOf model, probably because the task of discovering DegreeOf relations is easier: on average, the arguments of a DegreeOf relation are 0.61 tokens apart, while the arguments of a LocationOf relation are 3.22 tokens apart, and for the DegreeOf relation, the classifier had to consider only 2643 candidate entity pairs (28% of which were true relations), but for LocationOf it had to consider 36 865 pairs (14% of which were relations).
On the SHARP corpus, the SVM models outperformed all five rule-based baselines. On the ShARe corpus, the SVM LocationOf model outperformed all five baselines, but for DegreeOf, baseline 3 performed as well as the SVM. Baseline 3, which added relations for adjacent modifiers and entities, generally had good performance on DegreeOf, in which the arguments were on average only 0.61 tokens apart. However, for LocationOf, the baseline did not perform as well as the SVM models, which could handle better the more distant and complex relations. This was especially true on the SHARP corpus, in which the SVM model outperformed baseline 3 by 0.166 F1 (0.740 vs 0.574).
Across different corpora, the results are consistently better when the evaluation is conducted on the ShARe corpus. The difference is probably due to the fact that the ShARe project annotated fewer entity types than SHARP, making the task of discovering body site and severity modifiers simpler. But we also found that when evaluating on the ShARe test data, a model trained on the SHARP data performs almost as well as a model trained on the ShARe data, indicating that the SHARP model is fairly portable to other domains.
Our feature ablation experiments indicate that most features contribute to the overall system performance. Across both relation types, the most important feature group is the named entity type features, followed by the token features, which is consistent with the findings in the general domain.20 Unlike in the general domain, where chunking features appear to be among the largest contributors, in our experiments the chunking features did not improve the performance by much. Similarly, tree kernel features did not improve performance, contrary to several studies in the general domain. Finally, similar to the general domain, the dependency features provided only a modest boost to the system performance.
To analyze the sources of errors, we manually reviewed 50 LocationOf errors the system made on the SHARP data. Out of those 50, 22 instances were due to an error in the human annotations and 28 instances were actual system errors. It appears that the system errors could be attributed to one of three sources:
Sentence segmentation errors (one instance)
Infrequent patterns in training data (eight instances)
Inability of the system to discern more complex semantic patterns (19 instances).
An example of (2) is that the system mistakenly discovered LocationOf(abdominal aorta, aortogram) in Aortogram: Patent abdominal aorta that tapers from approximately 20 m… probably due to the frequent appearance of a similar pattern in the data, for example, Lungs: Equal AE bilaterally, no rales, no rhonchi in which Lungs (anatomical site) appears in a similar position as Aortogram (procedure). An example of (3) is that the system erroneously identified LocationOf(feet, femoropopliteal disease) in Non-invasive studies suggest significant femoropopliteal disease with monophasic Doppler signals in the feet probably due to incorrectly attaching the PP with monophasic Doppler signals to femoropopliteal disease even though such attachment does not make sense semantically.
Conclusion
We presented a methodology for the discovery of two key attributes from the clinical narrative—body site and severity. We showed that the task can be successfully cast as a supervised machine learning relation extraction problem, and that key features include the surrounding tokens and UMLS named entities. The best-performing methods identify LocationOf relations with F1 of 0.740–0.908 and DegreeOf relations with F1 of 0.905–0.929. These models are implemented as modules within cTAKES, thus providing an open source end-to-end system to the community for research and direct use purposes. In addition, the developed framework represents a general purpose utility for the semantic task of relation extraction thus contributing to the clinical NLP ecosystem.
This work focused on the discovery of body site and severity modifiers of clinical entities within the same sentence. Extending this work to inter-sentential relations will probably require leveraging sophisticated discourse processing including coreference resolution and in some cases textual entailment. Another challenge is relation discovery with underspecified, omitted or implicit information. For example, a mass mentioned in a breast cancer pathology report without an explicit anatomical site implies that the location is highly likely to be the breast.
The work described here is a step towards building a classification framework for relation discovery from the clinical narrative. Although in this work, we focused on DegreeOf and LocationOf relations, our system is easily extendable to many other relation types. In fact, to include new relations, no software changes are required; it is sufficient simply to include the examples of new relation types in the training data. The SHARP corpus currently includes several other UMLS relation types such as manages/treats and causes/brings_about. We are planning to retrain our system to include these relations in the near future. Our next steps will also include the implementation of the best methods in translational science applications such as phenotyping for the electronic medical record and genomics, informatics for i2b2, automatic disease activity classification43 as part of the pharmacogenomics research network, and clinical question answering as part of the multi-source integrated platform for answering clinical questions.44
Footnotes
Contributors: All authors contributed to the design, experiments, analysis, and writing the manuscript.
Funding: The project described was supported by awards from the Office of the National Coordinator of Healthcare Technologies 90TR002 (SHARP), from NIH R01GM090187 (ShARe), R01LM10090 (THYME), and U54LM008748 (i2b2).
Competing interests: GKS is on the Advisory Board of Wired Informatics, LLC, which provides services and products for clinical NLP applications.
Ethics approval: Research was conducted under an approved institutional board review protocol.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: Annotations from the Mayo Clinic and Seattle Group Health notes are available on an individual basis through a data use agreement. Annotations for the MIMIC corpus are distributed after a data use agreement with PhysioNet has been approved (http://www.physionet.org/).
References
- 1.Unified Medical Language System. http://www.nlm.nih.gov/research/umls/ (accessed 2/8/2013).
- 2.Bodenreider O, McCray AT. Exploring semantic groups through visual approaches. J Biomed Inform. 2003;36:414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marcus MP, Marcinkiewicz MA, Santorini B. Building a large annotated corpus of English: the Penn treebank. Comput Linguist 1993;19:313–30 [Google Scholar]
- 4.Palmer M, Gildea D, Kingsbury P. The proposition bank: an annotated corpus of semantic roles . Comput Linguist 2005;31:71–106 [Google Scholar]
- 5.Pustejovsky J, Castaño J, Ingria R, et al. TimeML: robust specification of event and temporal expressions in text. Proceedings of Fifth International Workshop on Computational Semantics (IWCS-5) 2003 [Google Scholar]
- 6.Standards & Interoperability Framework. http://wiki.siframework.org/file/view/Companion_Guide_Draft+3+for+Consensus_10012012.pdf (accessed 1/3/2013).
- 7.Jurafsky D, Martin JH. Speech and language processing: an introduction to natural language processing, computational linguistics, and speech recognition. Prentice Hall, 2009 [Google Scholar]
- 8.Automatic Content Extraction. http://www.itl.nist.gov/iad/mig/tests/ace/ (accessed 2/8/2013).
- 9.Text Analysis Conferences. http://www.nist.gov/tac/ (accessed 2/8/2013).
- 10.Clinical Element Model. http://www.clinicalelement.com/ (accessed 2/8/2013).
- 11.Coden A, Savova G, Sominsky I, et al. Automatically extracting cancer disease characteristics from pathology reports into a disease knowledge representation model. J Biomed Inform 2009;42:937. [DOI] [PubMed] [Google Scholar]
- 12.Crowley RS, Castine M, Mitchell K, et al. caTIES: a grid based system for coding and retrieval of surgical pathology reports and tissue specimens in support of translational research. J Am Med Inform Assoc 2010;17:253–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez D, Li Y. Information extraction from pathology reports in a hospital setting. Proceedings of the 20th ACM international conference on Information and knowledge management 2011 [Google Scholar]
- 14.Jouhet V, Defossez G, Burgun A, et al. Automated classification of free-text pathology reports for registration of incident cases of cancer. Methods Inf Med 2012;51:242. [DOI] [PubMed] [Google Scholar]
- 15.Fritz AG. International classification of diseases for oncology: ICD-O. World Health Organization, 2000 [Google Scholar]
- 16.Kavuluru R, Hands I, Durbin E, et al. Automatic extraction of ICD-O-3 primary sites from cancer pathology reports. Clinical Research Informatics AMIA symposium (forthcoming) 2013 [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman C. A broad-coverage natural language processing system. Proceedings of the AMIA Symposium 2000 [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman C. Semantic text parsing for patient records. Medical informatics. Springer, 2005 [Google Scholar]
- 19.Kambhatla N. Combining lexical, syntactic, and semantic features with maximum entropy models for extracting relations. Proceedings of the ACL 2004 on Interactive poster and demonstration sessions 2004 [Google Scholar]
- 20.Zhou G, Su J, Zhang J, et al. Exploring various knowledge in relation extraction. Proceedings of the 43rd Annual Meeting on Association for Computational Linguistics 2005 [Google Scholar]
- 21.Zelenko D, Aone C, Richardella A. Kernel methods for relation extraction. J Mach Learn Res 2003;3:1083–106 [Google Scholar]
- 22.Bunescu RC, Mooney RJ. A shortest path dependency kernel for relation extraction. Proceedings of the conference on Empirical Methods in Natural Language Processing 2005 [Google Scholar]
- 23.Uzuner Ö, South BR, Shen S, et al. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011;18:552–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts K, Rink B, Harabagiu SM, et al. A machine learning approach for identifying anatomical locations of actionable findings in radiology reports. AMIA Annual Symposium Proceedings 2012 [PMC free article] [PubMed] [Google Scholar]
- 25.Etzioni O, Banko M, Soderland S, et al. Open information extraction from the web. Commun ACM 2008;51:68–74 [Google Scholar]
- 26.Wu F, Weld DS. Open information extraction using Wikipedia. Proceedings of the 48th Annual Meeting of the Association for Computational Linguistics 2010 [Google Scholar]
- 27.Zhu J, Nie Z, Liu X, et al. StatSnowball: a statistical approach to extracting entity relationships. Proceedings of the 18th International Conference on World Wide Web 2009 [Google Scholar]
- 28.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Infor Assoc 2010;17:507–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clinical Text Analysis and Knowledge Extraction System (cTAKES). http://incubator.apache.org/ctakes/ (accessed 2/8/2013). [DOI] [PMC free article] [PubMed]
- 30.Strategic Health Advanced Research Project (SHARP). http://www.sharpn.org (accessed 2/8/2013).
- 31.Shared Annotated Resource (ShARe). https://www.clinicalnlpannotation.org (accessed 2/8/2013).
- 32.Albright D, Lanfranchi A, Fredriksen A, et al. Towards syntactic and semantic annotations of the clinical narrative. J Am Med Inform Assoc 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rea S, Pathak J, Savova G, et al. Building a robust, scalable and standards-driven infrastructure for secondary use of EHR data: the SHARPn project. J Biomed Inform 2012;45:763–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.PhysioNet. http://www.physionet.org (accessed 2/8/2013).
- 35.Joachims T. Learning to classify text using support vector machines: methods, theory and algorithms. Kluwer Academic Publishers, 2002 [Google Scholar]
- 36.de Bruijn B, Cherry C, Kiritchenko S, et al. Machine-learned solutions for three stages of clinical information extraction: the state of the art at i2b2 2010. J Am Med Inform Assoc 2011;18:557–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rink B, Harabagiu S, Roberts K. Automatic extraction of relations between medical concepts in clinical texts. J Am Med Inform Assoc 2011;18:594–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moschitti A. A study on convolution kernels for shallow semantic parsing. Proceedings of ACL 2004 [Google Scholar]
- 39.Zhang M, Zhang J, Su J. Exploring syntactic features for relation extraction using a convolution tree kernel. Proceedings of the Human Language Technology Conference of the North American Chapter of the ACL 2006 [Google Scholar]
- 40.Collins M, Duffy N. Convolution kernels for natural language. Proceedings of Neural Information Processing Systems (NIPS 14) 2001 [Google Scholar]
- 41.Albright D, Lanfranchi A, Fredriksen A, et al. Towards comprehensive syntactic and semantic annotations of the clinical narrative. J Am Med Inform Assoc 2013;20:922–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hripcsak G, Rothschild AS. Agreement, the f-measure, and reliability in information retrieval. J Am Med Inform Assoc 2005;12:296–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin C, Canhao H, Miller T, et al. Feature engineering and selection for rheumatoid arthritis disease activity classification using electronic medical records. ICML Workshop on Machine Learning for Clinical Data Analysis 2012 [Google Scholar]
- 44.Cairns BL, Nielsen RD, Masanz JJ, et al. The MiPACQ clinical question answering system. AMIA Annual Symposium Proceedings 2011 [PMC free article] [PubMed] [Google Scholar]