Abstract
The environment's contribution to health has been conceptualized as the exposome. Biomedical research interest in environmental exposures as a determinant of physiopathological processes is rising as such data increasingly become available. The panoply of miniaturized sensing devices now accessible and affordable for individuals to use to monitor a widening range of parameters opens up a new world of research data. Biomedical informatics (BMI) must provide a coherent framework for dealing with multi-scale population data including the phenome, the genome, the exposome, and their interconnections. The combination of these more continuous, comprehensive, and personalized data sources requires new research and development approaches to data management, analysis, and visualization. This article analyzes the implications of a new paradigm for the discipline of BMI, one that recognizes genome, phenome, and exposome data and their intricate interactions as the basis for biomedical research now and for clinical care in the near future.
Keywords: Informatics, Environmental Exposure, Biomedical Research, Gene-environment interaction, Phenotype
The opportunities
The phenotype of an individual results from the interplay between the genome (the complete set of genetic information) and the external/environmental elements to which it is exposed.1 The environment's contribution to health has been conceptualized as the exposome, defined as ‘every exposure to which an individual is subjected from conception to death, requiring consideration of the nature of the exposures and their changes and can be considered as internal, specific external and general external.’2
Biomedical research interest in environmental exposures as a determinant of physiopathological processes is rising as such data become increasingly available. The collection of new types of data on microbiomes,3 epigenomics,4 and physiological changes5 is proving very valuable in exposure assessment. Moreover, the panoply of miniaturized sensing devices now accessible and affordable for individuals to use to monitor a widening range of parameters—from clinical parameters such as blood pressure or glucose levels, to environmental parameters such as physical activity, food intake, the ambient temperature, or the presence of pollutants6—opens up a new world of research data. All of these data can be considered relevant for understanding the exposome; their integration and combined analysis looks very promising for advancing biomedical research.7
This situation presents new opportunities for biomedical informatics (BMI) to evolve as a discipline. For most of the 20th century, BMI mainly studied, represented, and analyzed phenotypic information related to health and disease states. In the last 20 years, due to advances in molecular medicine, BMI has started to deal significantly with ‘-omics’ information, and this has had a profound impact on BMI as a discipline.8 Many studies combining phenomic and genomic data, including genome-wide association studies (GWAS), have yielded important results. However, these approaches have also been criticized for their limited capability to explain the mechanisms underlying complex diseases.9 There is also increasing evidence that major determinants of common disease are based on exposure and behaviors.10 11 Now advances in exposome data collection12 13 and processing may be extending BMI again, probably pushing it towards another substantial revision.
A new paradigm for BMI is demanded by the increasing need to deal with inter-related exposome, genome, and phenome data or, as it has been termed, exposure science information.14 Five examples illustrate this point. First, continuous collection of real-time, highly dynamic environmental, genetic, and physiological data is now possible, using the new sensors.15 This is also closely related to the concept of ‘reality mining,’ which refers to the analysis of behavioral and self-reported data extracted from social networks and handheld devices such as mobile phones and applications.16 Second, genetic phenomena such as mosaicism and chimerism (eg, gene therapy, allogenic organ transplant, or intra-tumor cell genome heterogeneity17) reveal that a single individual might be composed of different genomes, adding a dynamic dimension to our previously static view of genomes. Third, epigenetic changes in response to environmental factors involve new probabilistic and multidimensional elements in health and disease.18 Fourth, advances in nanotechnology and its applications in medicine require the consideration of data on nanomaterials and their effects on living cells, as another aspect to be included in exposome informatics.19 20 Fifth, data from the human microbiome21 project sit at the intersection of genome, exposome, and phenome information. Definitions for key concepts are provided in table 1.
Table 1.
Definition of key concepts
| Concept | Definition | Source |
|---|---|---|
| Mosaicism | Condition in which cells within the same person have a different genetic makeup | Medline Plus http://www.nlm.nih.gov/medlineplus/ency/article/001317.htm |
| Epigenetics | Concerns the mechanisms that make organisms or parts of organisms look different, despite the fact they have the same genes and are in the same environment | The Conversation http://theconversation.com/explainer-what-is-epigenetics-13877 |
| Nanomaterial | Materials with at least one external dimension in the size range from approximately 1–100 nanometers | Centers for Disease Control and Prevention http://www.cdc.gov/niosh/docs/2009-125/ |
| Microbiome | Collective genomes of the microbes (composed of bacteria, bacteriophage, fungi, protozoa, and viruses) that live inside and on the human body | National Human Genome Research Institute http://www.genome.gov/27549400 |
These are examples of how the equation ‘Phenotype=Genotype×Environment’ poses enormous challenges to current biomedical research information systems. Current systems show something like a snapshot of the information available at certain stages. In comparison, future information systems for research will have to use new methods to process the flow and mix of data that will generate the coming wave of biomedical information and insights.
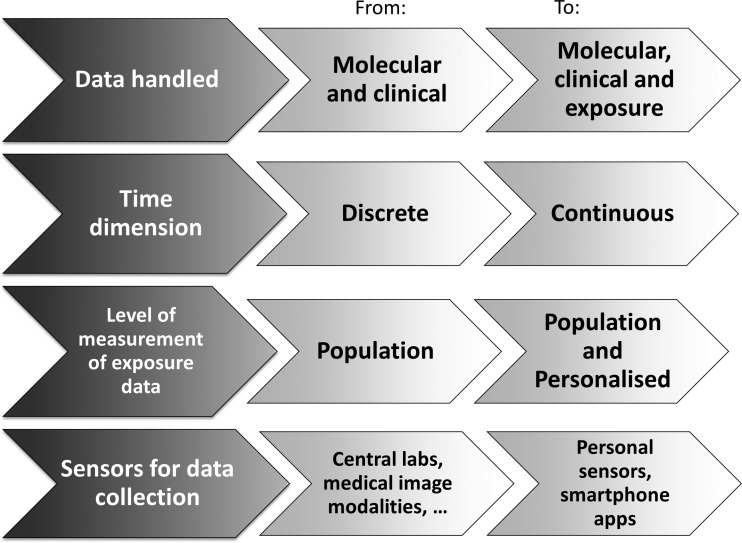
This new paradigm for BMI will bring a change in focus as well as in methods, insofar as it realizes the vision of more personalized biomedical research. Traditionally, most available exposure data have been captured through population studies. However, with the new sensors each individual can monitor their own exposures autonomously. Furthermore, new approaches to data integration can support individuals to combine such data with geospatial and behavioral tracking data.22 We have moved into an era when complex data monitoring and handling processes can be driven not only through large formal health research infrastructures, but also by individuals who wish to build their personal understanding of their own health (figure 1).
Figure 1.
Evolution of data collection methods.
The challenges
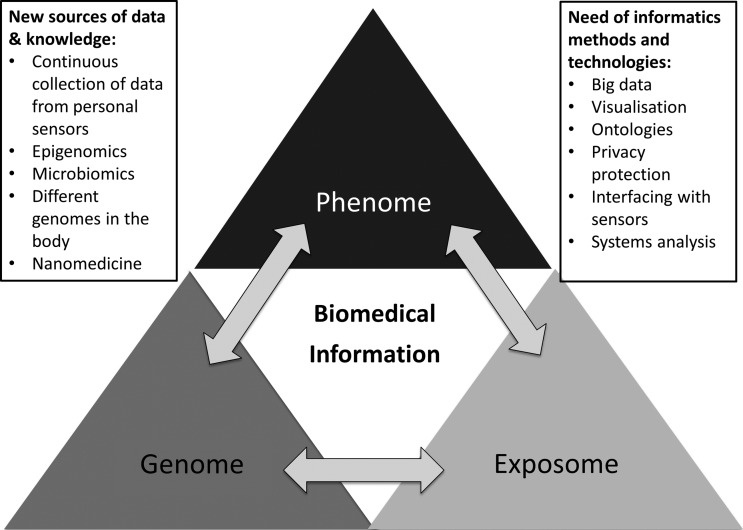
The combination of these more continuous, comprehensive, and personalized data sources requires new BMI research and development approaches to data management, analysis, and visualization. BMI must provide a coherent framework for dealing with multi-scale population data including the phenome, the genome, the exposome, and their interconnections (figure 2). The work involves defining an informatics infrastructure able to handle all of these types of data with a three-fold goal: (i) to perform population-based analysis that improves our knowledge of basic human health behaviors and determinants of common diseases; (ii) to provide data for basic and clinical research that combines phenotype, genotype, and exposure data at the level of the individual; and (iii) to build an augmented, data-rich personal health record which produces personal research results, tracking a person's exposome and giving him or her highly individualized, multi-faceted, disease risk profiles. A number of technical, organizational, and societal challenges have to be faced in implementing this BMI infrastructure to support both institutional and personal research.
Figure 2.
New research data types will require changes in biomedical informatics methods.
Let us consider what is involved in dealing with the ‘general external exposome’ (GEE).2 GEE data are generated routinely by everyone who engages in the information society through our communications using mobile phones, our movements using transit passes and recorded by security cameras, our purchases on bank cards, our utility consumption metered in the household, and our lifestyle choices reflected in social media, complemented by fixed and wearable sensors for sporting activity, ambulatory care monitoring, and ambient assisted living in smart homes. They are heterogeneous and selective (variety), there is a huge amount of data (volume), and their speed of processing needs to be high for optimal use (velocity). An additional crucial dimension of GEE data is time, characterized by multiple granularities: the GEE may include signals, for example collected by sensors (on a time scale of seconds, minutes, or hours), lifestyle data, such as information on food and nutrition (on a time scale of days or months), and finally long-term exposure data, such as the presence of pollutants (on a time scale of years or decades). In other words, GEE are not simply ‘big data,’23 but time series of big data.
Therefore, their very nature requires BMI implementation studies of novel informatics architectures that integrate recent data warehousing efforts, such as i2b224 and tranSMART25 which are aimed at managing phenotypes and molecular data, with NoSQL (Not only SQL) frameworks26 such as CouchDB27 and Cassandra,28 which are naturally scalable and can be implemented in a distributed environment, storing petabytes of data.
BMI also has a critical contribution to make in organizing these data conceptually, relying on a knowledge representation layer, based on suitable domain ontologies. For instance, the unstructured nature of GEE data requires extra effort in cataloging the information sources and the type of queries that can be performed in NoSQL repositories, making metadata essential to assess the quality of evidence that can be extracted from such data by suitable analytics.29
The types of analytical methods that are suited to cope with distributed, heterogeneous data is another area that needs particular attention from BMI, both in terms of scope—including information-based correlation analysis, detection of emergent phenomena, visualization, trends, and temporal abstractions—and in terms of computational efficiency.30 Pioneering efforts have been already made in the area of association studies with environmental/genomic/phenomic data,31–36 comprehensive molecular self-monitoring,37 the data collection surveys carried out by some direct-to-consumer genomic-testing companies,38 and previous epidemiological studies. However, those approaches lack the comprehensive treatment of data that is proposed here, namely coverage of individual exposure data facilitated by new technologies and sensors.
Last but not least, the design and implementation of a global BMI infrastructure for GEE data raises fundamental issues of security, privacy, and national and international legal compliance. These issues are related to the three-fold goal that a GEE-enabled biomedical research information system may pursue.
In the first case, of population-based analysis, the main concern is the implementation of a secure and reliable system for data gathering and data anonymization, that is, permanently and completely removing personal identifiers from data so that they can no longer be re-associated with an individual in any manner. This is a true challenge given the nature of GEE data, but could be achieved by providing aggregated data as advocated by the European Union eHealth Taskforce under the theme ‘Liberate the data.’39
A second, more complex issue is also one whose resolution is potentially much more valuable. This entails the definition of up-to-date strategies and policies for managing GEE data for clinical research at the individual level, even if de-identified, within the proper biomedical research governance infrastructure, including careful management of informed consent and risk management.40
Lastly, a cornerstone of a GEE-enabled biomedical research information system is the issue of building and maintaining a personal health record capable of including all clinical, genetic, and exposome data in a virtual repository. This must be under the ultimate control of ‘participatory biocitizens,’41 who may grant access for clinical care, clinical research, or epidemiological studies on a ‘my data my decision’ basis.39
Ways forward
In this article we have focused only on GEE, the first of Wild's2 three categories of exposures, but the complexity and volume of data exponentially increase when we incorporate the other two categories (table 2).
Table 2.
Examples of the data of interest for future information systems
| Group | Subgroup | Measure |
|---|---|---|
| Exposome | General external | Climate |
| Education | ||
| Socio-economical aspects | ||
| Natural and built environment | ||
| Specific external | Noise, humidity, CO, NOx, temperature, O3, radiation, particulate matter | |
| Medication, nanomaterials, medical procedures | ||
| Sedentary behaviors, physical activity | ||
| Smoking, diet, sleep, alcohol consumption | ||
| Infectious agents | ||
| Internal | Metabolites, hormones, oxidative stress, inflammation | |
| Phenome | Molecular traits | Gene expression, proteomics |
| Lipids, HDL, triglycerides | ||
| Cellular traits | Signaling pathways | |
| Cell cycle, apoptosis | ||
| Cell migration | ||
| Tissue/organ traits | Organ malformations, morphology, medical imaging | |
| Blood pressure | ||
| Organismal traits | Body mass index, weight, height | |
| Disease phenotypes | Pathologies | |
| Behavior | Stress, mood | |
| Endophenotypes | Cholesterol, immunoglobulins | |
| Genome | Sequence information | Whole genome, exome |
| Genomic variation | Single nucleotide variants (SNPs, mutations, …), structural variants (CNVs, In/Dels, …). | |
| Haplotypes | Blocks of variants | |
| Epigenomics | Methylation profiles |
Moreover, the internal exposome category (eg, metabolism, hormones, oxidative stress) can be measured using molecular biomarkers, reinforcing the points this article makes about data. Furthermore, these data too can be collected not only through sophisticated equipment available in institutions, but also through personalized, real time, continuous input from affordable devices and DIY services.
As already mentioned, it is worthwhile noticing that Wild's classification looks at the problem mainly from the data collection angle. As a matter of fact, BMI may not only provide instruments for data analysis but also tools for data representation and memorization, which may allow a clear description of the information and its consequent integration into an information system. For example, well-known disease nosology systems that include behavior and exposures, like SNOMED, provide clean, albeit orthogonal to Wild's view, ways to describe exposure factors, by giving different axes (ie, Living organisms; Physical agents, activities, and forces; Chemical, drugs and biological products) of classification. Such axes may be then properly exploited when the exposome is fitted into, for example, an electronic medical record.
What are the implications of a new paradigm for the discipline of BMI, one that recognizes genome, phenome, and exposome data, and their intricate interactions as the basis for biomedical research now and for clinical care in the near future?
The new generation of researchers in BMI should be familiar with the main methods and technological solutions required for the management of these new types of data (including big data, sensors, privacy and security, ontologies, systems analysis, and advanced visualization including geospatial systems). The new data types and sources may complement other studies and provide insights that are useful to understand the risks and the causes of the development of disease phenotypes. This has important consequences for the way we design BMI training programs and for the way we structure and specify the underlying competencies of experts in the discipline. In connection with this, the organization of BMI forums for professional development and knowledge exchange may need review to ensure sufficient scope for both established and new topics and themes.
The development of new information systems capable of linking these new data types and sources with personal health records could entrench recognition of the role of BMI expertise within other areas of biomedical research and development. And BMI has all the potentials, and tools, including a collection of ontologies, terminologies, and standards, to deal with such a challenge. There will be growing expectation that biomedical research routinely will include the design, implementation, and evaluation of comprehensive data-rich environments, in which to investigate the causative elements associated with pathologies to improve risk profiling, and so to contribute to advancing preventive medicine. To our knowledge no-one yet is fully engaged in realizing the vision proposed in this article, although recent initiatives probably will require many of the elements described herein.42 43
Lastly, the way we think about the contribution of BMI as a discipline will need to have regard for new insights that the exposome will bring, into the connections between human health and the health of the biosphere. BMI may increasingly support shared decision making in settings beyond traditional health sciences.
Footnotes
Contributors: FMS designed the general structure of the paper, prepared the figures and table, contributed to writing the ‘Ways forward’ section, and reviewed the paper. KG critically revised the paper and edited it. RB wrote the ‘The challenges’ section, provided technical oversight, and reviewed the paper. GLC contributed to writing the ‘The opportunities’ section and provided scientific oversight.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Weatherall D. From genotype to phenotype: genetics and medical practice in the new millennium. Philos Trans R Soc Lond B Biol Sci 1999;354:1995–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wild CP. The exposome: from concept to utility. Int J Epidemiol 2012;41:24–32 [DOI] [PubMed] [Google Scholar]
- 3.Human Microbiome Project Consortium. A framework for human microbiome research. Nature 2012;486:215–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirst M. Epigenomics: sequencing the methylome. Methods Mol Biol 2013;973:39–54 [DOI] [PubMed] [Google Scholar]
- 5.Gohlke JM, Thomas R, Zhang Y, et al. Genetic and environmental pathways to complex diseases. BMC Syst Biol 2009;3:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Tongeren M, Cherrie JW. An integrated approach to the exposome. Environ Health Perspect 2012;120:A103–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buck Louis GM, Sundaram R. Exposome: time for transformative research. Stat Med 2012;31:2569–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Athey BD, Cavalcoli JD, Jagadish HV, et al. The NIH National Center for Integrative Biomedical Informatics (NCIBI). J Am Med Inform Assoc 2012;19:166–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McClellan J, King MC. Genetic heterogeneity in human disease. Cell 2010;141:210–17 [DOI] [PubMed] [Google Scholar]
- 10.Bickham DS, Blood EA, Walls CE, et al. Characteristics of screen media use associated with higher BMI in young adolescents. Pediatrics 2013;131:935–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Selikoff J, Hammond EC, Churg J. Asbestos exposure, smoking and neoplasia. JAMA 1968;204:106–12 [PubMed] [Google Scholar]
- 12.Callaway E. Daily dose of toxics to be tracked. Nature 2012;49:647 [DOI] [PubMed] [Google Scholar]
- 13. The Human Exposome Project. http://humanexposomeproject.com (accessed 5 Aug 2013)
- 14.Committee on Human And Environmental Exposure Science in the 21st Century, Board on Environmental Studies and Toxicology, National Research Council of The Academies. Exposure science in the 21st century: a vision and a strategy. Washington, DC: National Academies Press, 2012 [PubMed] [Google Scholar]
- 15.Pentland A, Lazer D, Brewer D, et al. Using reality mining to improve public health and medicine. Stud Health Technol Inform 2009;149:93–102 [PubMed] [Google Scholar]
- 16.Komatireddy R, Topol EJ. Medicine unplugged: the future of laboratory medicine. Clin Chem 2012;58:1644–7 [DOI] [PubMed] [Google Scholar]
- 17.Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochim Biophy Acta 2010;1805:105–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langevin SM, Kelsey KT. The fate is not always written in the genes: epigenomics in epidemiologic studies. Environ Mol Mutagen 2013;54:533–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen Y, Rallo R, Liu R, et al. In silico analysis of nanomaterials hazard and risk. Acc Chem Res 2012;46:802–12 [DOI] [PubMed] [Google Scholar]
- 20.Thomas DG, Klaessig F, Harper SL, et al. Informatics and standards for nanomedicine technology. Wiley interdisciplinary reviews. Nanomedicine Nanobiotechnol 2011;3:511–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smarr L. Quantifying your body: a how-to guide from a systems biology perspective. Biotechnol J 2012;7:980–91 [DOI] [PubMed] [Google Scholar]
- 22.Boulos MNK, Resch B, Crowley DN, et al. Crowdsourcing, citizen sensing and sensor web technologies for public and environmental health surveillance and crisis management: trends, OGC standards and application examples. Int J Health Geogr 2011;10:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eaton C, DeRoos D, Deutsch T, et al. Understanding Big Data. McGraw Hill, 2012 [Google Scholar]
- 24.Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc 2010;17:124–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szalma S, Koka V, Khasanova T, et al. Effective knowledge management in translational medicine. J Transl Med 2010;8:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee KK, Tang WC, Choi KS. Alternatives to relational database: comparison of NoSQL and XML approaches for clinical data storage. Comput Methods Programs Biomed 2013;110:99–109 [DOI] [PubMed] [Google Scholar]
- 27.Manyam G, Payton MA, Roth JA, et al. Relax with CouchDB—into the non-relational DBMS era of bioinformatics. Genomics 2012;100:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hewitt E. Cassandra: the definitive guide. 1st edn O'Reilly Media, 2010 [Google Scholar]
- 29.Megler VM, Maier D. When Big Data leads to lost data. Proceedings of the 5th PhD Workshop on Information and Knowledge 2012:1–8 [Google Scholar]
- 30.Cherian A, Sra S, Banerjee A, et al. Jensen-Bregman LogDet divergence with application to efficient similarity search for covariance matrices. IEEE Trans Pattern Anal Mach Intell 2013;35:2161–74 [DOI] [PubMed] [Google Scholar]
- 31.Patel CJ, Bhattacharya J, Butte AJ. An environment-wide association study (EWAS) on type 2 diabetes mellitus. PloS One 2010;5:e10746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patel CJ, Chen R, Butte AJ. Data-driven integration of epidemiological and toxicological data to select candidate interacting genes and environmental factors in association with disease. Bioinformatics 2012;28:i121–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel CJ, Chen R, Kodama K, et al. Systematic identification of interaction effects between genome- and environment-wide associations in type 2 diabetes mellitus. Hum Genet 2013;132:495–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tzoulaki I, Patel CJ, Okamura T, et al. A nutrient-wide association study on blood pressure. Circulation 2012;126:2456–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel CJ, Cullen MR, Ioannidis JP, et al. Systematic evaluation of environmental factors: persistent pollutants and nutrients correlated with serum lipid levels. Int J Epidemiol 2012;41:828–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lind PM, Risérus U, Salihovic S, et al. An environmental wide association study (EWAS) approach to the metabolic syndrome. Environ Int 2013;55:1–8 [DOI] [PubMed] [Google Scholar]
- 37.Chen R, Mias GI, Li-Pook-Than J, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 2012;148:1293–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eriksson N, Macpherson JM, Tung JY, et al. Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genet 2010;6:e1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.e-Health task force report. Redesigning Health In Europe for 2020. Luxembourg: Publications Office of the European Union, 2012. http://ec.europa.eu/digital-agenda/en/news/eu-task-force-ehealth-redesigning-health-europe-2020 (accessed 25 Jan 2013) [Google Scholar]
- 40.Prainsack B. Voting with their mice: personal genome testing and the participatory turn in disease research. Account Res 2011;18:132–47 [DOI] [PubMed] [Google Scholar]
- 41.Swan M. Health 2050: The realization of personalized medicine through crowdsourcing, the quantified self, and the participatory biocitizen. J Pers Med 2012;2:93–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Health eHeart https://www.health-eheartstudy.org (accessed 12 Apr 2013)
- 43. CancerCommons http://www.cancercommons.org (accessed 12 Apr 2013)