Abstract
Pharmacogenetics (PG) examines gene variations for drug disposition, response, or toxicity. At the National Institutes of Health Clinical Center (NIH CC), a multidepartment Pharmacogenetics Testing Implementation Committee (PGTIC) was established to develop clinical decision support (CDS) algorithms for abacavir, carbamazepine, and allopurinol, medications for which human leukocyte antigen (HLA) variants predict severe hypersensitivity reactions. Providing PG CDS in the electronic health record (EHR) during order entry could prevent adverse drug events. Medical Logic Module (MLM) programming was used to implement PG CDS in our EHR. The MLM checks to see if an HLA sequence-based gene test is ordered. A message regarding test status (result present, absent, pending, or test not ordered) is displayed on the order form, and the MLM determines if the prescriber can place the order, place it but require an over-ride reason, or be blocked from placing the order. Since implementation, more than 725 medication orders have been placed for over 230 patients by 154 different prescribers for the three drugs included in our PG program. Prescribers commonly used an over-ride reason when placing the order mainly because patients had been receiving the drug without reaction before implementation of the CDS program. Successful incorporation of PG CDS into the NIH CC EHR required a coordinated, interdisciplinary effort to ensure smooth activation and a positive effect on patient care. Prescribers have adapted to using the CDS and have ordered PG testing as a direct result of the implementation.
Keywords: Pharmacogenetics, Informatics, Clinical Decision Support, Computerized Prescriber Order Entry, Patient Safety
Background
Pharmacogenetics (PG) examines inherited or acquired variations in genes that dictate drug response, disposition, or toxicity and explores how these variations can be used to optimize medication therapy.1 Over 100 medications approved by the Food and Drug Administration contain PG information in their product labels.2 Integrating PG into clinical care has been challenging for many reasons, including the inability to define drug disposition/response/toxicity phenotypes, the lack of cost-effective PG tests, and the lack of readily available software that can integrate this information into an electronic health record (EHR).3 However, progress is being made on defining both gene variation–drug response information and clinical standards for PG information.4 5 The Clinical Pharmacogenetics Implementation Consortium (CPIC) has defined a methodology for developing clinical practice standards for PG information.6
The objective of clinical decision support (CDS) is to provide the necessary information, to the right clinician, in the right intervention format, in the right channel, at the right time in the workflow to help make healthcare decisions.7 8 To integrate PG information into patient care, organizations need to identify where it can definitively identify cases where this information can improve patient outcomes. Although using PG information in CDS would optimize the EHR, only a few institutions have been able to accomplish this integration.9–12
At the National Institutes of Health Clinical Center (NIH CC), we have implemented PG CDS to prevent severe hypersensitivity reactions associated with three medications: abacavir, allopurinol, and carbamazepine. This paper describes the clinical and technical approach to integrating PG information and CDS into the EHR.
Approach
We formed a multidisciplinary team to evaluate the PG information. The team was charged by the hospital director to determine the feasibility of, and develop a plan for, implementing PG testing. The team reported to the Pharmacy and Therapeutics Committee, which reports to the Medical Executive Committee. This team defined the PG testing and reporting methodology, vetted information through institutional processes for approving medication-related decisions, developed the technical solution within the EHR, and educated NIH CC staff about the program.
Pharmacogenetics Testing Implementation Committee (PGTIC)
The PGTIC comprised individuals with a wide range of backgrounds and expertise. The PGTIC made recommendations from published PG literature, including strong evidence that human leukocyte antigen (HLA) variants are associated with drug toxicity, such as severe hypersensitivity reactions, and evidence that drug metabolizing enzymes and transporter (DMET) variants affect the pharmacokinetics and/or pharmacodynamics of many drugs. Because HLA genotyping capabilities existed in our Department of Transfusion Medicine (DTM), the PGTIC recommended that HLA testing be implemented first. Based on the literature and CPIC guidelines, the PTGIC selected three drugs—abacavir (HLA-B*57:01), allopurinol (HLA-B*58:01), and carbamazepine (HLA-A*31:01, HLA-B*15:02)—as the initial drugs and variants in the program.13–20
Laboratory component
The committee determined that sequence-based typing (SBT) methodology was optimal for high-resolution HLA genotyping (HLA-A and HLA-B allele SEQR Typing Kits (Atria Genetics, Hayward, California, USA) on a 3730xL DNA analyzer (Applied Biosystems, Carlsbad, California, USA)).21 Sequence data were compared with reference sequences for known HLA alleles (IMGT/HLA database).22 The HLA SBT testing is compliant with the regulations under the Clinical Laboratory Improvement Amendments for ‘tests of high complexity’. Results are entered into the SCC Soft Computer Laboratory Information System and transmitted through an HL7 message to the EHR. The availability of the result can be emailed to the requesting prescriber. To avoid the need for secure email to send personally identifiable information, the order number is sent to the prescriber. The prescriber then retrieves the test results from our EHR using the order number. The desired turnaround time was discussed and communicated with the prescribers. HLA testing for the PG program is given a high priority, and the laboratory performs the test on the basis of priority.
Organizational approval process
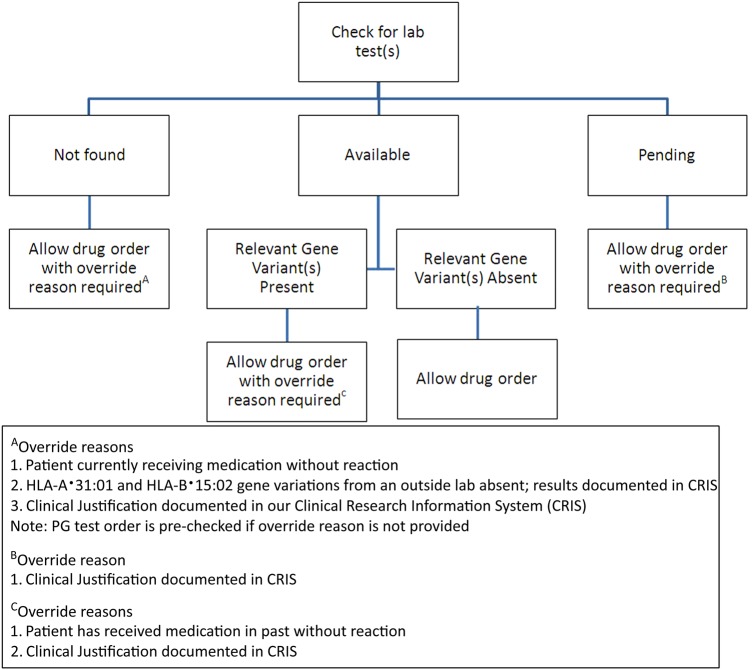
Medication use guidelines, PG testing infrastructure, and prescriber clinical decision algorithms were established for the three drugs (abacavir, allopurinol, and carbamazepine). An example for carbamazepine is shown in figure 1. Four scenarios are possible for test status and results depending on whether the PG test has:
not been ordered
been ordered but result is not yet available
been ordered but result shows risk of toxicity
been ordered and result shows low risk of toxicity.
Figure 1.
The clinical decision support data flow algorithm used for carbamazepine is shown. The medical logic module checks for the laboratory (lab) test first. If laboratory test results are found, a message appropriate to the results is displayed to the prescriber. If results are not found, then a different message is displayed recommending that the pharmacogenetics (PG) test is carried out. In this case, the PG Testing Implementation Committee (PGTIC) decided that an order can be placed if the gene variation is found, the test results are pending, or to proceed without ordering the test as long an over-ride reason is provided. In contrast, the PGTIC decided to block the order for abacavir if the relevant gene variation is detected.
For each of these cases, the following questions were answered.
What message is displayed to the user?
Should an order be permitted?
If an over-ride is required, what are the allowable parameters?
Should ordering the test be facilitated by preselecting the laboratory order on the form?
These answers resulted in a control table used to program the CDS (table 1). The control table is customized for each drug by creating specific entries for each of the four cases for the message provided to the user, whether the order can be placed, for the over-ride reasons, and to facilitate ordering the PG test through the order set form.
Table 1.
Example of a PG CDS control table template for carbamazepine
| Case number | Situation | Message to user | Allow order to be entered | Over-ride reasons | Facilitate laboratory order |
|---|---|---|---|---|---|
| 1 | PG test has not been ordered | Serious dermatologic reactions, including toxic epidermal necrolysis (TEN) and Stevens–Johnson syndrome (SJS), have been reported with carbamazepine. >90% of reactions occur within 2–3 months of treatment. Testing for HLA-A*31:01 and HLA-B*15:02 is recommended to assess risk | Yes with over-ride reason required |
|
Yes—if not over-ridden No—if over-ridden |
| 2 | PG test has been ordered but result has not been provided | An HLA genotype test has been ordered but not yet resulted | Yes with over-ride reason required |
|
No—prescriber blocked from entering an additional HLA/gene test |
| 3 | PG test has been ordered but result shows risk of toxicity | The patient carries either or both of the HLA-A*31:01 and HLA-B*15:02 alleles, indicating the patient is at risk of a serious reaction, including TEN or SJS. Alternative therapy is highly recommended Carbamazepine should only be used if the benefit clearly outweighs the risk | Yes with over-ride reason required | 1. Patient has received medication in past without reaction 2. Clinical justification documented in the EHR |
No |
| 4 | PG test has been ordered and result shows low risk of toxicity | HLA-A*31:01 and HLA-B*15:02 tests are both negative. Patient is at low risk of serious HLA-A*31:01- and HLA-B*15:02-related reactions to carbamazepine | Yes | Not applicable |
CDS, clinical decision support; EHR, electronic health record; PG, pharmacogenetics.
The control table was reviewed and approved by the Pharmacy and Therapeutics Committee. The use and content of this CDS template will be evaluated at least annually for clinical content validity and outcome assessment. Recently, we established a PG subcommittee comprising all members of the PGTIC, an additional physician, and a nurse. The PG subcommittee will perform periodic content review for medications in our PG algorithms and consider additional medications.
Technical solution
The NIH CC Clinical Research Information System uses Allscripts Sunrise Clinical Manager, V.5.5, Service Pack 1 Rollup 7 as the central component of our EHR. The CDS program is completely integrated into the medication order pathway in our EHR and was developed to support the following ordering and resulting PG testing processes.
HLA and genetics tests are ordered from the EHR.
HLA and genetic test results are stored in the EHR for both user review and usage within CDS.
Physicians order medications included in our PG program through ‘order set forms.’
CDS within the order determines if the HLA SBT has been ordered and/or resulted. A message regarding test status and results is presented to the prescriber.
The control table information and the data flow are used to complete the CDS programming within the EHR. The CDS is programmed using Medical Logic Modules (MLMs) with Arden Syntax programming language, which allows complex logic to create rules and actions working from the EHR patient and order context and for querying the EHR database. To handle data retrieval from the EHR database, stored procedures were written in Structured Query Language to retrieve EHR content, results, and logic from site-defined tables, which makes EHR queries very easy to develop. The control table approach simplifies managing the logic defined in the template, allows changes on demand when new medications or tests need to be considered, and provides an area to store the PG test results. The MLMs have two purposes: (1) to interpret results as they are received from the ancillary laboratory computer system; and (2) to process clinical rules for medication orders.
From the technical perspective, a primary goal was to simplify the prescriber order entry process by presenting the relevant clinical information on a single order entry screen. To meet this goal, a structured results table with actionable data was necessary for gathering the historical and ongoing test results. Each HLA result is evaluated with the control table. For example, with the HLA-B test results, each laboratory result text was searched for the specific allele values in the text for each drug. An interesting design issue was handling the recording method when an allele nomenclature changed (ie, HLA B 5701 vs HLA B 57:01). The control table allowed for this expansion and could handle variants for each test and drug combination. For example, both 5701 and 57:01 were searched in the case of abacavir.
Allele names are controlled by the WHO, and such standard conventions were used. Complexity was brought into the algorithm when historical data needed to be viewed. Since PG CDS has been implemented, there no longer exists an issue in retrieving and analyzing historical data.
The next step was to write an MLM that would run in the background when each new result is received via the interface. This MLM has three steps to process.
Step 1: retrieve the order and patient identifier of the laboratory test and send to a stored procedure to analyze the result against the control table. For each laboratory component with a corresponding medication from the control table, the allele search term is checked against the results value and comments of the test. The output of this stored procedure includes the name of the medication, the test component name, the allele search term, and whether a positive match occurred. This information is sent to the structured results table. These interpreted results are called by another MLM during the ordering process.
Step 2: notify one or more prescribers about HLA test results. The MLM pulls prescriber names and email addresses and sends an email with the order number noting the new results. The email provides instructions to access the results and patient information.
Step 3: log all events. The MLM firing event is tracked in the EHR with our defined messages. Additionally, if an email is requested, a separate tracking table records the email recipients, the date and time it was sent, and the associated order number.
System performance was considered in the design when building the MLMs. In a clinical research organization with primarily longitudinal studies of patients, laboratory results can accumulate over many years.23 Minimization of the retrieval time for results required pre-interpretation of the historical data at the time of implementation. The result table contains the following information.
Identification numbers for the patient, laboratory test, and medication order
Table logic parameters (drug name, laboratory test name, allele searched)
Dates (laboratory test requested date, results received date, and date added to table)
The result for the variant
A textual description of the results value (eg, ‘No Matches with 57:01 for SEQ HLA B*’).
The retrieval of stored procedures used in MLMs was further tuned by optimizing the results table indexes to ensure a sub-second response time.
Multiple variations of ordering screen methodologies were considered in the planning and early design phase. A structured order set, which supports the use of MLMs to drive clinical rules and logic needed for ordering medications, was created. A structured order set uses a custom-designed order form to present various fields for the relevant information or data entry and grids to present the laboratory test and medications that may be ordered. When a structured order set is selected and opened, an MLM can be used to drive clinical rules on the underlining fields and grids.
The MLM was written to apply the following workflow steps when the structured order set form opens.
Step 1: retrieve patient and medication information and pass to a stored procedure. This stored procedure returns the actionable laboratory results, if available, and information if the test was previously ordered and if resulted.
Step 2: determine the case level from the control table on the basis of the data from the clinical rules logic.
Step 3: populate the informational fields on the form to show when and if a test has been ordered, the drug-specific message, and allowable over-ride reasons, if applicable.
- Step 4: apply logic to the ordering grids related to facilitating the PG test order and ordering the medication.
- 4a: PG test: if there is no history of an HLA test, the order will be preselected. If the patient has results or an order with pending results, the MLM blocks the laboratory test from being orderable, preventing unnecessary duplicate orders.
- 4b: medication order: the MLM can allow or block the medication order on the basis of the medication and laboratory test result—for example, if there are clinical situations where patients may need allopurinol despite the presence of the HLA B*15:02 variation; an over-ride reason is required here. However, an abacavir order entry should be blocked if the HLA B*57:01 variation is detected, because of the nature of the hypersensitivity reaction.
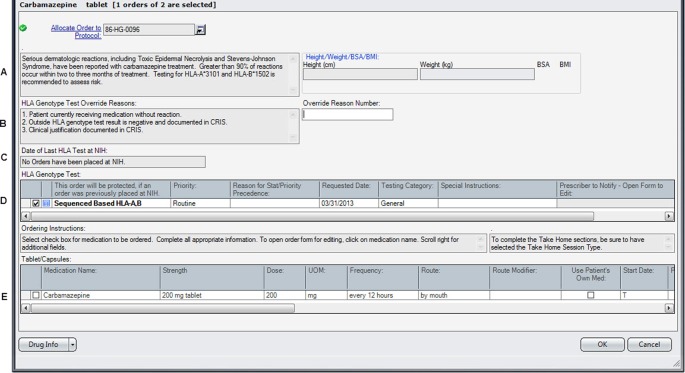
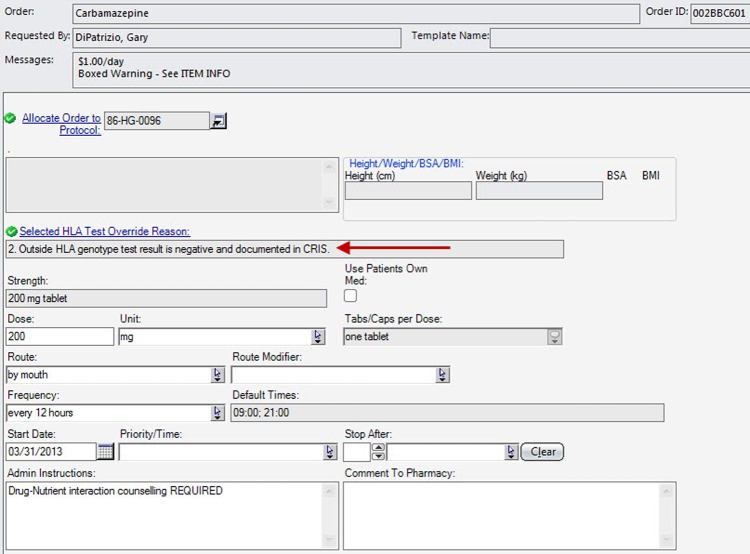
After the form opens, the MLM processes the information. The prescriber can then make a clinically informed decision. If an over-ride reason is allowed and selected, that information is carried down to the medication order tracking fields. This is essential information for other care providers to review when reordering the medication or for pharmacists and nurses who will be verifying, dispensing, and administering the medication. Since these medications were already present in existing order sets, each occurrence was replaced with a reference to the structured order set to ensure that the process was uniformly followed. The ordering process met the CDS objectives by providing the correct information at the time of order entry. An example of what the prescriber sees when placing an order for carbamazepine in a patient with the HLA B*15:02 variant is shown in figure 2. The resulting order placed from this order form, which includes the PG information, is shown in figure 3.
Figure 2.
The design for the order set form for drugs included in the pharmacogenetics (PG) program is shown. In this case, the information displayed to the prescriber is based on the case where the HLA test has not yet been ordered. The displayed messages and order actions are determined by the control table. The standard design for the order set form includes the following areas. (A) Message box where the clinical information message is displayed. (B) Message box where over-ride reasons are displayed, if appropriate for the case and test result. The ‘over-ride reason number’ field becomes a required entry if an over-ride reason is allowed. (C) Message box where PG test information is displayed. (D) Grid where PG tests can be ordered or are automatically preselected depending on the case. (E) Grid where medications can be ordered. CRIS, Clinical Research Information System.
Figure 3.
The resulting order placed through one of the order set forms is shown. A field has been added to our standard order form that provides the over-ride reason, if appropriate, selected by the prescriber. BMI, body mass index; BSA, body surface area.
Before activation, all possible scenarios for each medication were validated in our test system by defining test cases and expected responses for each algorithm. Orders were placed to accommodate all of the scenarios. The displayed messages, laboratory results, prechecking the order, requirement for an over-ride, and blocking or allowing the drug order were checked for each case. Design and testing was performed in collaboration with all stakeholders and was activated after successful validation.
Educational efforts before activation
Implementing PG testing was a new initiative for the NIH CC. While the responsibility of ensuring appropriate HLA testing belongs to prescribers, education was provided to several healthcare disciplines, such as prescribers, pharmacists, and nurses. The goal was to inform these healthcare professionals about how the CDS would be incorporated into our EHR.
A two-page competency document summarizing the clinical information regarding severe hypersensitivity reactions with the three drugs in relation to the gene variants was developed. The document contained a five-question quiz. All pharmacists and nurses were required to complete this competency. In addition, we educated our clinical pharmacy specialists, nurse clinical specialists, and nursing leadership groups so that they could educate their healthcare team. Prescribers were educated by the clinical pharmacy specialists and through education documents distributed via email.
Clinical implementation
Data were collected from October 1, 2012 to August 31, 2013 (11 months) to analyze prescriber reaction to the system and to determine if the CDS facilitated incorporation of PG information into drug therapy management (table 2).
Table 2.
Analysis of pharmacogenetic clinical decision support
| Abacavir | Allopurinol | Carbamazepine | |
|---|---|---|---|
| Number of patients | 56 | 133 | 45 |
| Number of patients where a PG test was performed onsite and the relevant variation was not found (no over-ride reason required) | 0 | 21 | 3 |
| Number of patients where an over-ride reason was entered | 56 | 112 | 42 |
| Over-ride reason entered* | |||
| Negative HLA test from an outside laboratory | 7 | 3 | 0 |
| Clinical justification documented in the EHR | 13 | 61 | 7 |
| Patient previously received medication without reaction | 47 | 73 | 37 |
*The total number of over-ride reasons for each drug exceeds the total number of patients with an over-ride reason because some patients had different over-ride reasons entered for the same drug.
EHR, electronic health record; PG, pharmacogenetics.
Over 725 medication orders have been placed for 234 patients by 154 different prescribers since implementation of the program. A total of 24 PG test results were available to prescribers through the CDS program to assist their clinical decision making including 12 tests ordered directly from the order set form. The CDS is configured to retrieve the test result regardless of the order source. In all cases where a laboratory result was available, the relevant gene variation(s) was not detected and therefore an over-ride reason was not required to be entered. In all other cases, an over-ride reason was required. Review of the over-ride reasons selected identified that prescribers sometimes used a different reason when placing orders for the same medication for the same patient. For example, ‘Clinical justification documented in the EHR’ and ‘Patient currently receiving medication without reaction’ were used to indicate the same clinical circumstance. When searching through the patient documentation, we found that the specific clinical reason was difficult to locate because various documentation note types were used. Prescribers often selected ‘Clinical justification documented in the EHR’ and then entered ‘Patient has received this drug in the past’ in the clinical documentation note instead of selecting ‘Patient previously received medication without reaction’ from the preconfigured over-ride reasons. To encourage the proper use of the over-ride reasons, additional prescriber education will be completed. The committee will also review the possibility of creating a specific PG note type in the EHR.
For many years before initiation of the CDS program, HLA testing before the start of abacavir therapy was considered the standard of care. Because testing was routine and it is not common to initiate abacavir as an early treatment for HIV infection, all patients had either been tested by an outside laboratory or had been receiving abacavir for a long time. Thus, it is not surprising that all of the orders placed included an over-ride reason.
The clinical situation for allopurinol and carbamazepine is different than for abacavir. The incidence of severe allergic reactions in a patient with the relevant gene variation is low. For this reason, the HLA test is recommended rather than required. It was interesting to discover that a high number of HLA tests had been performed for patients receiving allopurinol. Upon investigation, it was determined that this group consisted mainly of patients who had received bone marrow or stem cell transplants, in whom HLA tests for immune function determination are performed as part of the routine care.
The number of times an over-ride reason was selected for allopurinol and carbamazepine was not unexpected. Many of these patients have safely received these medications for seizure prevention or neuropathic pain in the case of carbamazepine, and for treatment of gout in the case of allopurinol, and are thought to be at low risk of a severe allergic reaction. ‘Clinical justification’ was a very common over-ride reason when allopurinol was prescribed for prevention of tumor lysis syndrome. For prevention of tumor lysis syndrome, there is an urgent need to start the medication before the HLA test results are available. This is a predictable problem with current PG tests, where the turnaround time is longer than for routine tests.
Discussion
At the NIH CC, CDS for PG testing was implemented successfully starting in September 2012. An interdisciplinary team facilitated integration and seamless activation of PG CDS directly into the medication order pathway. Prescribers have adapted to using the CDS and have ordered PG testing as a direct result of the implementation.
Initial implementation required that the ‘reorder/copy’ function for medication ordering be deactivated for these three medications to ensure that all orders passed through the CDS logic. Several months after implementation, feedback from prescribers identified the need to redesign the CDS tool to allow reorders to facilitate workflow. The CDS tool was redesigned to ensure that each prescription was reassessed with each order, yet facilitated reordering so that key orders would not be omitted.
The application of the program to just three drugs and HLA molecular variants provided the opportunity to validate the algorithm and explore its usefulness and robustness during routine clinical care. The MLM components, data tables, and stored procedures can be easily adapted to other PG tests and medications. Future plans include incorporating low-resolution HLA testing, which is commonly requested at the NIH CC to assess the immune system. These tests are also performed on site by the DTM laboratory, which allows control of the results formatting.
In the second implementation phase, PG testing will be incorporated for drugs affected by polymorphisms in DMET. Mercaptopurine and azathioprine are two examples of drugs affected by polymorphisms in the thiopurine methyltransferase (TPMT) gene. A CPIC guideline has been published that recommends testing for TPMT gene variations to guide dosing for these drugs.24 25
Although the technical implementation for DMET testing is anticipated to be straightforward in that the current CDS algorithm is adaptable to any PG–drug pair, the turnaround time for the test and incorporation of outside PG test results will be challenging. For example, mercaptopurine is used for induction therapy for leukemia; it is therefore important that the PG test results are either already available from prior PG testing or available within a few hours to use this information for drug dosing.
Unless a PG test panel can be found where results are available with a very quick turnaround time or pre-emptive PG testing is readily accepted, clinicians will not be able to take advantage of the additional PG information, and therapy will be based on current clinical decision parameters (eg, adjustment of the international normalization ratio for warfarin). Institutions will then be challenged to decide whether pre-emptive PG testing is cost-effective and contributes significantly to improved drug therapy. However, common DMET testing arrays provide results for many drug–gene variation pairs, so after a patient is genotyped, results could be available for future drug therapy decisions.
Conclusion
At the NIH CC, CDS for PG testing was successfully incorporated into the EHR. Implementation required a coordinated, interdisciplinary effort to ensure smooth activation and a positive effect on patient care. Prescribers have adapted to using the CDS and have ordered PG testing as a direct result of the implementation.
Footnotes
Contributors: BRG coordinated writing the drafts of the paper. WAF, SDA, and TAF developed the laboratory components of the CDS and provided information about testing the algorithms. GD constructed the CDS programming and Medical Logic Modules. BRG, TS, SP, LGB, WDF, and JJLL performed literature searches to develop the CDS algorithms. BRG, GD, JJP DH, and JWM developed the order entry screens. BRG, GD, and JJP tested the programming before implementation. All authors contributed to the development and implementation of this CDS as a task force and all made critical revisions to the paper.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Hamburg MA, Collins FS. The path to personalized medicine. N Engl J Med 2010;363:301–4 [DOI] [PubMed] [Google Scholar]
- 2.Table of Pharmacogenomic Biomarkers in Drug Labels. US Food and Drug Administration. http://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.htm (accessed 1 Apr 2013). [Google Scholar]
- 3.Roden DM, Altman RB, Benowitz NL, et al. Pharmacogenetics Research Network. Pharmacogenomics: challenges and opportunities. Ann Intern Med 2006;145:749–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manolio TA, Chisholm RL, Ozenberger B, et al. Implementing genomic medicine in the clinic: the future is here. Genet Med 2013;15:258–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crews KR, Hicks JK, Pui CH, et al. Pharmacogenomics and individualized medicine: translating science into practice. Clin Pharmacol Ther 2012;92:467–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther 2011;89:464–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Approaching clinical decision support in medication management. In: Osheroff J. ed Improving medication use and outcomes with clinical decision support: a step-by-step guide. Chicago: Healthcare Information and Management Systems Society, 2009:1–10 [Google Scholar]
- 8.Osheroff JA, Teich JM, Middleton B, et al. A roadmap for national action on clinical decision support. J Am Med Inform Assoc 2007;14:141–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hicks JK, Crews KR, Hoffman JM, et al. A clinician-driven automated system for integration of pharmacogenetic interpretations into an electronic medical record. Clin Pharmacol Ther 2012;92:563–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Welch BM, Kawamoto K. Clinical decision support for genetically guided personalized medicine: a systematic review. J Am Med Inform Assoc 2013;20:388–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peterson JF, Bowton E, Field JRet al. Electronic health record design and implementation for pharmacogenomics: a local perspective. Genet Med 2013;15:833–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bell GC, Crews KR, Wilkinson MRet al. Development and use of active clinical decision support for preemptive pharmacogenomics. J Am Med Inform Assoc 2014;21:e93–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mallal S, Phillips E, Carosi G, et al. ; PREDICT-1 Study Team. HLA-B*5701 screening for hypersensitivity to abacavir. N Engl J Med 2008;358:568–79 [DOI] [PubMed] [Google Scholar]
- 14.Hughes CA, Foisy MM, Dewhurst N, et al. Abacavir hypersensitivity reaction: an update. Ann Pharmacother 2008;42:387–96 [DOI] [PubMed] [Google Scholar]
- 15.Martin MA, Klein TE, Dong BJ, et al. Clinical pharmacogenetics implementation consortium. Clinical pharmacogenetics implementation consortium guidelines for HLA-B genotype and abacavir dosing. Clin Pharmacol Ther 2012;91:734–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCormack M, Alfirevic A, Bourgeois S, et al. HLA-A*3101 and carbamazepine-induced hypersensitivity reactions in Europeans. N Engl J Med 2011;364:1134–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen P, Lin J-J, Lu C-S, et al. Carbamazepine-induced toxic effects and HLA-B*1502 screening in Taiwan. N Engl J Med 2011;364:1126–33 [DOI] [PubMed] [Google Scholar]
- 18.Leckband SG, Kelsoe JR, Dunnenberger HMet al. Clinical pharmacogenetics implementation consortium guidelines for hla-B genotype and carbamazepine dosing. Clin Pharmacol Ther 2013;94:324–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zineh I, Mummaneni P, Lyndly J, et al. Allopurinol pharmacogenetics: assessment of potential clinical usefulness. Pharmacogenomics 2011;12:1741–9 [DOI] [PubMed] [Google Scholar]
- 20.Hershfield MS, Callaghan JT, Tassaneeyakul W, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for human leukocyte antigen-B genotype and allopurinol dosing. Clin Pharmacol Ther 2013;93:153–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams S, Robbins FM, Chen D, et al. HLA class I and II genotype of the NCI-60 cell lines. J Transl Med 2005;3:11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson J, Halliwell JA, McWilliam H, et al. The IMGT/HLA database. Nucleic Acids Res 2013;41:D1222–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ioannidis JPA. To replicate or not to replicate: the case of pharmacogenetic studies. Circ Cardiovasc Genet 2013;6:413–18 [DOI] [PubMed] [Google Scholar]
- 24.Relling MV, Gardner EE, Sandborn WJ, et al. Clinical pharmacogenetics implementation consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing: 2013 update. Clin Pharmacol Ther 2013;93:324–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Relling MV, Gardner EE, Sandborn WJ, et al. ; Clinical Pharmacogenetics Implementation Consortium. Clinical Pharmacogenetics Implementation Consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing. Clin Pharmacol Ther 2011;89:387–91 Erratum in: Clin Pharmacol Ther. 2011;90:894 [DOI] [PMC free article] [PubMed] [Google Scholar]