Summary
The Arp2/3 complex is an actin filament nucleator involved in cell motility and vesicle trafficking. Owing to the role the complex plays in important and fundamental cell biological processes, the purified complex is used in biochemical assays, reconstituted motility assays and structural biology. As this is a eukaryotic complex assembled from seven polypeptides, the complex is purified from eukaryotic sources. Described here is a detailed method for purification of the complex from a mammalian tissue, bovine thymus.
Keywords: Actin nucleation factor, Arp2/3 complex, biochemical purification, endogenous source, detailed protocol
1. Introduction
Dynamic rearrangements of the actin cytoskeleton are used by eukaryotic cells to generate protrusive and pinching forces, at specific places and times (1). These rearrangements underlie aspects of cell motility (2), endocytosis (3,4), cell division (5), and bacterial and viral pathogenesis (6,7). The nucleation of new actin filaments (8) is an essential component of each of these processes. Spontaneous nucleation of actin filaments is prevented in cells by a variety of factors including the proteins profilin and thymosin, but this can be overcome by the action of regulated actin nucleation factors (2). One prominent nucleation factor is the Arp2/3 complex.
The Arp2/3 complex is formed from seven polypeptides, including two actin related proteins, Arp2 and Arp3 (9–11). The Arp2/3 complex nucleates new filaments from the side of existing filaments, and anchors the nucleated filament in a branched structure (12–15). In this process, Arp2, Arp3 and additional actin monomers serve as an actin nucleus, from which the new filament grows (16,15). Arp2/3 complex is basally inhibited, and can be stimulated to nucleate new filaments by the action of nucleation promoting factors, in particular those of the WASP family (8). Together, nucleation promoting factors and Arp2/3 complex control the assembly of cellular structures such as lamellipodia (2) and the pinching off of nascent vesicles (17,4). Pathogens, such as Listeria, override the cellular control of Arp2/3 complex by providing their own highly potent activators of the complex (6,18).
Purified Arp2/3 complex is used in two biochemical contexts. The first is the reconstitution of actin polymerization (19). In this system, purified actin is present as a monomer that slowly polymerizes into filaments. Polymerization can be tracked through the use of a pyrene labeled actin, whose fluorescence intensity increases upon incorporation into an actin filament (20,21). Inclusion of Arp2/3 complex and an activator, such as the VCA domain of the WASP family member N-WASP, accelerates nucleation of new filaments and overall actin polymerization is much faster (22,8,23). Alternatively, a variety of imaging based assays have been used to directly visualize Arp2/3 complex dependent branch formation using actin labeled with a variety of fluorophores (24,12,13). A second context in which purified Arp2/3 complex is used is in reconstituted bead motility assays. In these assays, a mixture of purified proteins can trigger bacteria (25) or functionalized beads (26) to move through solution on a ‘comet tail’ of polymerized actin. This reconstituted system has opened a window into how larger actin structures assemble and interact with surfaces (27–30).
For both of these assay systems, purified Arp2/3 complex is needed. Owing to the fact that this is an assembly of seven polypeptides, several of which are anticipated to need eukaryotic chaperones, overexpression in bacteria is not a viable option. Instead, purification from endogenous sources or recombinant expression in a eukaryotic host has been used. Purification of Arp2/3 complex from endogenous sources can be accomplished using one of two methods. A classical biochemical approach has been used to purify the complex from a variety of sources (31,32,18,33,34), and we provide a time-tested version of one protocol here. Alternatively, engineered affinity columns can be used (35–37,10,38) including columns derived from the VCA domain of N-WASP (36). This approach has been adapted to the purification of Arp2/3 complex from a variety of sources (35–38); purification of Arp2/3 complex from commercially available baker’s yeast (S. cerevisiae) by this method is described in an accompanying chapter of this book (39). Recombinant methods include the simultaneous overexpression of all the subunits of the human complex in insect cell lines (40), and the recombinant fusion of affinity tags to selected subunits in yeast (41–46), allowing purification by affinity methods. These two recombinant methods allow the production of mutant Arp2/3 complexes, and the use of readily available affinity chromatography methods. The primary difficulties of the recombinant systems are that eukaryotic expression systems are needed, and that the yields are low. These practical concerns have prevented the recombinant sources from displacing the endogenous sources for studies not requiring modified Arp2/3 complex.
Here we describe a method for purifying endogenous Arp2/3 complex from bovine thymus, derived from a method originally developed for purification from human leukocytes (31). The method relies on classical biochemical methods including ammonium sulfate precipitation, ion exchange chromatography and gel filtration chromatography. The described protocol takes three days to perform, not including stock solution preparation. While a single person can perform the entire protocol, two people working as a team (particularly on the first day) simplify the procedure. Typical yields from the protocol range between 5 and 8 mg of highly purified complex. It should be noted that the protocol does not discriminate between the different subunit isoforms, for example ArpC1 runs as a multiplet on the final gel (47), owing to multiple isoforms. This has also been reported for ArpC1 of human origin (18). It is also expected that there will be a degree of heterogeneity at the level of post-translational modification. One characterized modification, phosphorylation of Arp2 at several sites (48), is expected to be present, but may not be uniformly present. The protocol can be scaled up and down. Given the columns described here, between one and four calf thymuses may be used.
2. Materials
All buffer and salt stocks are prepared using ultrapure water (>18 MOhm, using Millipore brand Milli-Q water purification system). Except where noted otherwise, all solutions are filtered through a 0.22 μm cellulose acetate membrane. Working buffers are prepared by dilution of buffer stocks into prechilled ultrapure water (except where noted). Where specific sources are recommended, the manufacturer and part numbers are indicated.
2.1. Stock Materials and Solutions
Bovine calf thymus: This protocol is described for two bovine calf thymuses, roughly 400 g total weight. Frozen thymus can be obtained from Pel-Freez Biologicals (Rogers, AR). Note the appearance of the thymus in a given lot, and track the lot number (see Note 1). Store thymus at −80°C for less than 2 years.
Leupeptin hydrochloride (Bachem #N-1000.0100): 1 mg/mL stock in ultrapure water, filtered and stored at −20°C in 250 μL aliquots.
Calpain Inhibitor II 10 mM stock (ALLM, Calbiochem #208721): 25 mg in 6.2 mL 100% ethanol and stored as a single volume at −20°C until needed (use within 6 months).
Cathespin B Inhibitor I 10 mM stock (Caspase inhibitor, Calbiochem #342000): 5 mg in 1.3 mL 100% ethanol and stored at −20°C until needed (use within 6 months).
PMSF (Phenylmethanesulphonyl fluoride, Sigma #P7626): 100 mM stock in isopropanol, filtered and stored in 1 mL aliquots at −20°C (see Note 2).
Benzamidine dihydrochloride (Sigma #B6506-100g): 1 M stock in ultrapure water, filtered and stored at −20°C in 250 μL aliquots.
Antipain dihydrochloride (Sigma #A6191-100mg): 1 mg/mL stock in ultrapure water, filtered and stored at −20°C in 250 μL aliquots.
Dithiothreitol (DTT): 1 mM stock in ultrapure water, filtered and stored at −20°C in 1 mL aliquots. Thaw in cool water until liquid then transfer to ice until needed. Once added to buffers, assume DTT is no longer functional after 48 hours when stored at 4°C.
1 M Tris-HCl pH 8.0.
0.5 M PIPES pH 6.8: Prepared using PIPES acid (piperazine-N,N′-bis(2-ethanesulfonic acid), and titrated with sodium hydroxide to pH 6.8 at room temperature (see Note 3).
1 M Imidazole pH 7.0: Solution is titrated with hydrochloric acid to pH 7.0 at room temperature.
2 M KCl.
5 M NaCl.
0.5 M EGTA: Solution is titrated to pH 8.0 with sodium hydroxide. Initially, EGTA is insoluble, but comes into solution as the pH is adjusted.
1 M MgCl2.
2.2. Working Buffers
See Note 4 for recommended buffer preparation schedule.
Buffer EB: 40 mM Tris-HCl pH 8, 10 mM EGTA, 1 mM MgCl2, 1mM DTT. Make with room temperature ultrapure water, not 4°C water (see Note 5). Also, place 100 mL of EB on ice to be used for rinsing the blender jar.
Buffer QA: 20 mM Tris-HCl pH 8, 75 mM NaCl, 1 mM EGTA, 1 mM MgCl2, 1 mM DTT, 10 μM Calpain Inhibitor II, 20 μg/mL leupeptin, 1 mM benzamidine, 2 μg/mL antipain.
Buffer DB: 2 mM Tris-HCl pH 8, 0.5 mM EGTA, 0.5 mM MgCl2, 1 mM DTT.
Buffer QC: 20 mM Tris-HCl pH 8, 1 mM EGTA, 1 mM MgCl2, 1 mM DTT.
Buffer QD: 20 mM Tris-HCl pH 8, 1 mM EGTA, 1 mM MgCl2, 1 mM DTT, 1 M NaCl.
Buffer SA: 10 mM PIPES pH 6.8, 1 mM EGTA, 1 mM MgCl2, 1 mM DTT.
Buffer SB: 10 mM PIPES pH 6.8, 1 mM EGTA, 1 mM MgCl2, 1 mM DTT, 2 M NaCl.
Buffer KMEId: 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10 mM Imidazole pH 7, 1 mM DTT.
Buffer FB: 50 mM KCl, 10 mM Imidazole pH 7.0, 1 mM MgCl2 1 mM EGTA, 0.5 mM DTT, 100 μM ATP and 60% (weight to volume) sucrose. This is prepared by adding appropriate volumes of salt and buffer stocks for 10 mL of buffer to 6 g of dry sucrose, and adding water to ~9 mL. The solution is mixed until most of the sucrose has come into solution, and the volume is corrected to 10 mL. The solution is then allowed to come completely into solution. At that point it is passed through a 0.22 μm PES filter. Prior to adding to the protein solution it is chilled on ice for ~10 minutes. This is enough time to reduce the temperature, but will not result in sucrose crystallizing out of solution. Use buffer FB the same day that it is prepared.
Sodium chloride (5 M).
Q Cleaning buffer: 6 M guanidine hydrochloride, 200 mM acetic acid (see Note 6).
2.3. Chromatography Columns
Recommended flow rates and operating pressures are typical for a column in good condition. The operating pressures for the Q Sepharose FF column and SOURCE 15Q column refer to the pressure difference between the top of the column and the fractionation or waste outlet. If it is difficult to achieve the recommended flow rates without exceeding the operating pressure for the column, make sure that there is not a back pressure device after the column, that tubing bores are reasonable, and that you account for pressure build-up prior to the column.
150 mL Q Sepharose FF: 150 mL Q Sepharose FF (GE #17-0510-01) is packed into an XK 26/40 (GE #28-9889-49) column. The maximum flow rate and pressure for the column is 26.5 mL/min and 0.5 MPa, respectively. We run this column at a flow rate of 8–15 mL/min at less than 0.5 MPa.
50 mL SOURCE 15Q: Bulk SOURCE (GE #17-0947-05) is packed in an XK 26/20 column (GE #28-9889-48). The maximum flow rate is 26.5 mL/min at 0.6 MPa maximum back-pressure. We typically run this column at a flow rate of 6–10 mL/min at less than 0.6 MPa.
MonoS GL 10/100 column (GE #17-5169-01): The maximum pressure and flow rate for this column is 6 mL/min and 4 MPa, respectively. We run this column at 2.5 – 4 mL/min and at less than 4 MPa.
Superdex 200pg column: This is a prepacked HiLoad 26/600 Superdex 200 pg column, with 320 mL of resin in an XK 26/600 column hardware (GE# 28-9893-36). The maximum flow rate is 4.25 mL/min with a maximum pressure of 0.5 MPa. We run this column run at 1 – 2.5 mL/min at less than 0.5 MPa back-pressure.
2.4. Hardware and Equipment
Liquid chromatography system(s) capable of delivering linear gradients and operating columns at the described flow rates and pressures. A system capable of applying linear gradients to columns at 1–5 mL/min and at 2–5 MPa is needed for the Mono S column. A system capable of applying the >500 mL of clarified lysate at low pressure is needed for the Q Sepharose column.
Three centrifuges are needed: (1) A low speed refrigerated centrifuge with swinging bucket rotor, capable of spinning 1 L bottles at 1,600 x g. (2) An ultracentrifuge (Beckman Optima or equivalent) with Type 45 Ti rotor. (3) A refrigerated high-speed centrifuge (Beckman Coulter Avanti Centrifuge), equipped with JA-10 and JA-25.50 rotors. Centrifuges equipped with a Beckman JA-20 or Sorvall SLA-3000 and SS-34 are equivalent for this purpose. All centrifugation steps are at 4°C.
Warring blender for homogenization, with 1–2 L stainless steel jar.
Three each large stir plates, magnetic stir bars and 4 L beakers.
Cold room or cold box, with space for large stir-plates.
Conductivity meter.
Dialysis tubing (50 kDa cut-off dialysis membrane (Spectra/Por 6, #132544)) and clamps.
AmiconUltra-15 30kD cut-off (Fisher #UFC903024) centrifugal concentrator.
3. Methods
The overall protocol takes three days. A single person can complete the entire protocol, but working in a team of two greatly eases the process. We break each day up into a separate sub-protocol, but they are meant to occur on consecutive days.
3.1. Day 1: lysis, clarification and ammonium sulfate cut
On the first day of the preparation, it is very helpful to have a second set of hands to keep up with the dishes that are generated during the process without needing to slow down other aspects of the preparation. Generally, assembly of materials and lysis can be completed within an hour and a half, clarification takes three hours, the Q Sepharose column takes an hour, and the ammonium sulfate cut takes roughly three and a half hours, including setting up the dialysis afterwards. This timing doesn’t account for cleanup, which can generally occur during subsequent steps. Thus, with two people, the protocol can be completed in about nine hours.
Chill the three centrifuges; 1 L capacity swinging bucket, JA-10 and Type 45 Ti rotors, and bottles (see Note 7).
Weigh the thymuses individually, aiming for a total weight of 200 to 450 grams. Note their weight, and then place them on ice. The scale limiting steps are the high-speed centrifugation step and the first two columns.
Use a lead brick to smash one frozen thymus (at −80°C) into small pieces (see Note 8). Place the thymus pieces into blender jar and add 2 to 3 mL of room temperature EB per gram of thymus (as weighed prior to breaking into pieces). Blend to homogenize, making sure that the thymus-buffer mix does not freeze. Add additional room temperature buffer if the homogenate appears slushy. After approximately one minute of blending, stop the blender and add 10 μM calpain inhibitor II, 10 μM cathespin B Inhibitor I, 20 μg/mL leupeptin, 2 μg/mL antipain, 1 mM PMSF, and 1 mM benzamidine (according to volume of EB used). Then, continue blending until the final consistency and appearance is roughly that of a strawberry banana smoothie, typically after 3 – 5 minutes of total blending time (see Note 9). Split the homogenate across two 1 L bottles, rinse the blender with about 50 mL of EB and add to the homogenate in the 1 L bottles.
Repeat step 3 for each thymus.
Centrifuge homogenate in 1 L bottles at ~1,600 x g (2,600 rpm in the Sorvall H6000A rotor) for 30 minutes.
Inspect the centrifuged homogenate, there should be a small pellet, and a whitish, fatty disc at the top (see Note 1). Scoop out the fatty disc and decant the supernatant through four layers of cheesecloth into a 2 L beaker.
Split the supernatant from step 6 into 500 mL bottles and spin at ~4,000 x g (6,000 rpm in the Beckman JA-10 rotor) for 15 minutes.
Decant the supernatant through four layers of cheesecloth into a 2 L beaker.
Split the supernatant from step 8 into multiple 70 mL polycarbonate bottle assemblies. Centrifuge the bottles at 138,000 x g for one hour (42,000 rpm in the Type 45 Ti rotor). Note that the bottles must be full. If there is less than 65 mL of supernatant that does not fit into the bottles, discard the excess. It is common for the supernatant from step 8 to fill more bottles than fit in a single Type 45 Ti rotor. If this is the case, keep the excess on ice until repeating the spin with additional bottles.
While the centrifuge for step 9 is running, equilibrate the 150 mL Q Sepharose FF column with approximately 500 mL of buffer QA. Note the conductivity at the end of equilibration.
Decant the supernatant from step 9 through four layers of cheesecloth into a 2 L beaker. Supernatant should be clear, but red in color.
Check the conductivity of QA using conductivity meter. QA at 4°C should be ~5–6 mS/cm. With the beaker containing the pooled supernatant on wet ice, adjust the conductivity of the supernatant to be 0.2–0.5 mS/cm greater than QA by adding 5 M sodium chloride. Note the initial, final and QA reference conductivities.
Inject the conductivity corrected, pooled supernatant from step 12 onto the 150 mL Q Sepharose column equilibrated with QA and collect flow through. Rinse column with 1 column volume of QA and also collect flow through. To minimize volume, begin collection when the color of the flow through changes to red, and stop collecting flow through when the conductivity shifts back down to that seen for QA alone. Clean Q Sepharose column (see Note 10).
Measure volume of the combined flow through. Begin stirring the supernatant in a 4°C cold room or packed into wet ice using a large magnetic stir plate and a large stir bar. Stirring should be aggressive enough to create a vortex with a depression which reaches at least halfway down the height of the liquid. The depression should not contact the stir bar, which would introduce air.
Weigh out enough solid ammonium sulfate to bring the solution to 35% saturated (200 g of solid ammonium sulfate per L). Very slowly add the ammonium sulfate (see Note 11) while stirring.
Let solution stir for 30 minutes at 4°C.
Transfer the ammonium sulfate solution into 500 mL centrifuge bottles, filling the bottles approximately half full. Centrifuge at ~9,000 x g for 30 minutes (9,000 rpm in the JA-10 rotor and bottles).
Carefully decant the supernatant into a glass beaker in wet ice. Discard the pellets.
Measure the volume of the supernatant with a cold graduated cylinder.
Weigh out enough ammonium sulfate to bring this 35% saturated solution to 60% saturated (an additional 150 grams of ammonium sulfate solid per L of supernatant). Very slowly add the ammonium sulfate (see Note 11) while stirring.
Let solution stir for 30 minutes at 4°C.
Transfer the ammonium sulfate solution into 500 mL centrifuge bottles, filling the bottles approximately half full. Centrifuge at ~9,000 x g for 30 minutes (9,000 rpm min in the JA-10 rotor and bottles).
Carefully decant off the supernatant. Place the bottles at an angle in a large ice bucket with the pellets in contact with the ice, but not at the lowest point. Let remaining liquid drain from the walls to the lowest point of the bottle for a few minutes, then use a pipette to remove it. Discard the supernatant.
Resuspend the pellet in 20–40 mL of buffer DB by pipetting up and down with a powered pipette aid, minimizing air introduction. If pellets do not resuspend quickly, they will ‘soften’ somewhat as they are in contact with the DB.
Cut and rinse lengths of 50 kDa cut-off dialysis membrane with ultrapure water. Enough dialysis tubing should be prepared to hold roughly twice the resuspended volume. For this dialysis tubing cut a total of 50 cm per 60 mL, split across at least two pieces. Transfer the resuspended pellets into the dialysis tubing, leaving at least 30% of the length as slack, after affixing dialysis clips to the ends. The resuspended pellets are of high salt content and thus will increase in volume during dialysis owing to osmotic effects. Dialyze the pellets against 8 L of DB overnight (greater than 6 hours) at 4°C, stirring slowly. It is usually necessary to use two medium or large dialysis clips at either end of the tubing to give the suspension/tubing sufficient buoyancy to prevent it from hitting the stir bar.
3.2. Day 2: SOURCE 15Q column
The second day begins with the resuspended and dialyzed pellets from step 25 of Section 3.1. Typically, this day can be performed by one person. Clarifying and correcting the salt concentration in the dialysate typically takes about an hour. The SOURCE 15Q column takes 2 – 5 hours to run, depending on the volume to be loaded and the flow rate that is used. SDS-PAGE analysis takes roughly an hour. Setting up the dialysis and cleaning the SOURCE 15Q column typically takes about an hour. All told the second day typically can be completed in less than eight hours.
Transfer the dialysate from step 25 of Section 3.1 to centrifuge tubes and spin at ~ 7,000 x g (9,000 rpm in a JA-25.50 rotor) for 30 minutes.
Decant the supernatant into a beaker. Dilute the supernatant with 2 mM Tris-HCl, pH 8.0, until the conductivity is lower than QC. Cold QC buffer has a conductivity of ~1 mS/cm. Note the initial, final and reference conductivities.
Filter the conductivity corrected dialysate through a 0.45 μm filter (see Note 11).
Purify the filtered, conductivity corrected dialysate using SOURCE 15Q ion exchange chromatography (see Note 13). Equilibrate the SOURCE 15Q column in buffer QC. Inject the filtered dialysate onto the column at 5 mL/min, wash out unbound proteins using 2 column volumes of QC. Elute proteins in 14 mL fractions, including Arp2/3 complex, from the column using a linear buffer gradient of 100% QC to 90% QC/10% QD developed over 20 column volumes (1 L of buffer volume).
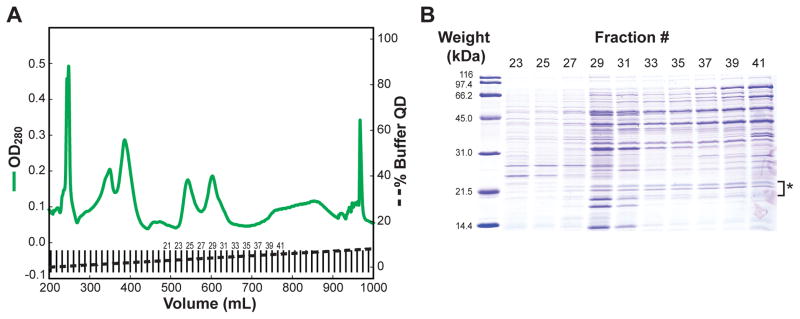
Identify fractions to pool using SDS-PAGE (see Note 14). See example in Fig. 1.
Clean SOURCE 15Q column (see Note 15).
Place SOURCE 15Q pool into 50 kDa dialysis tubing, dialyze against 12 L of buffer SA overnight. Follow the protocol in step 25 of Section 3.1, but allow less space for expansion as the osmotic differences will be lower in this case. Leaving 25% of the tubing length as slack is sufficient.
Figure 1. Purification of bovine Arp2/3 complex using SOURCE 15Q ion exchange chromatography.
(A) An example of the UV absorbance detected chromatogram for bovine Arp2/3 complex purification on a 50 mL SOURCE 15Q column. UV absorbance is shown with a thick green line, fraction of QD (with buffer QC making up the remaining volume) is shown with a dashed line. Fractions collected are shown with short vertical lines. Relevant fraction numbers are shown. (B) SDS-PAGE analysis of relevant fractions was performed a using 15% acrylamide gel, and stained using coomassie brilliant blue dye. At this stage the complex is only marginally pure, and fraction assessment is accomplished by looking for ArpC3, ArpC4 and ArpC5, which run as a triplet between 19 kDa and 23 kDa. This region of the gel is indicated with a bracket and an asterisk. Fraction numbers from the chromatogram are indicated at the top of the gel lanes, and molecular weights of standards are shown at the left. For this run fractions 28 through 35 were pooled and further purified.
3.3. Day 3: Mono S and Superdex 200 columns
The third day begins with the dialyzed pool from step 7 of Section 3.2, and typically requires only a single person to complete. Preparing and running the Mono S column typically takes 3 – 4 hours. SDS-PAGE analysis takes about an hour, but may be performed while equilibrating the gel filtration column. Running the gel filtration column (once equilibrated) takes about 2 hours. SDS-PAGE analysis takes about an hour, and is typically begun immediately after the primary peak elutes. Running the two columns can be completed in about eight hours, if the SDS-PAGE steps are performed when described. If fractions are pooled, concentrated, measured and frozen on the same day (this may be delayed until the next day), this adds an additional 2 – 3 hours to day 3.
Collect the dialysate from step 7 of Section 3.2. Filter through a 0.45 μm filter (see Note 12).
Measure the conductivity of the dialysate and dilute with 10 mM PIPES pH 6.8 until the conductivity is lower than buffer SA. Buffer SA has a conductivity of about ~1 mS/cm. Note the initial, final and reference conductivity.
Purify Arp2/3 complex using MonoS cation exchange chromatography. Equilibrate the MonoS column with buffer SA. Inject onto the column at 2–4 mL/min. Wash unbound materials from the column with 3 column volumes of buffer SA. Elute proteins in 5 mL fractions, including the Arp2/3 complex, from the column with a linear buffer gradient from 100% SA to 90% SA/10% SB, developed over 30 column volumes (240 mL of buffer volume).
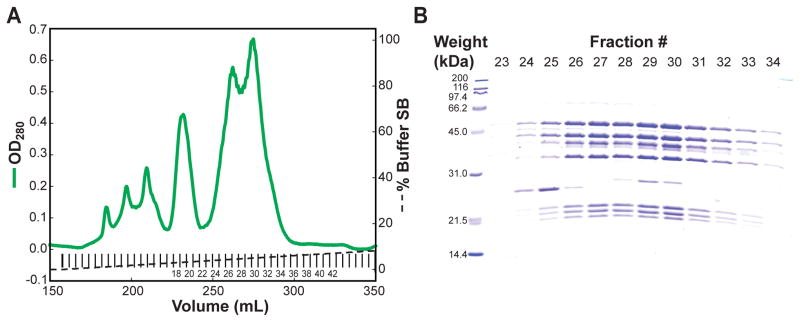
Pool fractions containing Arp2/3 complex, as assessed by SDS PAGE (see Fig. 2 and Note 16).
Concentrate pooled fractions from step 4 to roughly 11 mL using an AmiconUltra-15 30kD cut-off centrifugal concentrator. Spin at ~2,000 x g in a swinging bucket rotor (3,500 rpm in a Sorvall Legend with swinging bucket rotor). The concentrate should be mixed thoroughly and topped up after every 3 to 5 minutes of centrifugation time. If any precipitate is observed, filter using a 0.22 μm syringe filter (not usually needed).
Purify Arp2/3 complex using Superdex 200 gel filtration chromatography. Equilibrate a 320 mL Superdex 200pg column with at least 300 mL KMEId (see Note 17). Inject the concentrated Arp2/3 complex pool onto the column at 1 mL/min. Elute the Arp2/3 complex by flowing an additional column volume of buffer over the column, collecting 4 to 5 mL fractions beginning after 0.3 column volumes of buffer. Arp2/3 complex typically elutes as a single peak, with a maxima occurring when approximately 0.5 – 0.6 column volumes of buffer have been applied to the column.
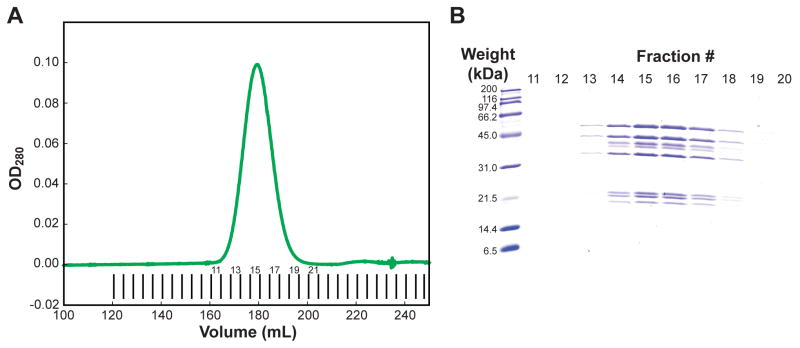
Identify fractions to keep by SDS-PAGE analysis (see Fig. 3). Pool or keep fractions separate according to subsequent needs.
Quantify Arp2/3 complex concentration using ultraviolet absorption (see Note 18). If necessary, the complex may be concentrated as in step 5.
Either store fractions away from light at 4°C for less than four weeks, or freeze the fractions (see Note 19). Beyond SDS-PAGE and gel filtration assessment of purity, typical quality control steps are to assay activity using pyrene actin polymerization assays (19). 10 nM of the bovine Arp2/3 complex should not show observable activity in the absence of nucleation promotion factor, and should trigger rapid actin nucleation in the presence of monomeric 250 nM N-WASP VCA.
Figure 2. Purification of bovine Arp2/3 complex using MonoS ion exchange chromatography.
(A) An example of the UV absorbance detected chromatogram for bovine Arp2/3 complex purification on an 8 mL MonoS column. UV absorbance is shown with a thick green line, fraction of SB (with buffer SA making up the remaining volume) is shown with a dashed line. Fractions collected are shown with short vertical lines. Relevant fraction numbers are shown. (B) SDS-PAGE analysis of relevant fractions was performed a using 15% acrylamide gel, and stained using coomassie brilliant blue dye. At this stage the complex is mostly pure, with contaminants at 26 kDa, 28 kDa and 74 kDa. Fraction numbers from the chromatogram are indicated at the top of the gel lanes, and molecular weights of standards are shown at the left. For this run fractions 24 through 32 were pooled and further purified.
Figure 3. Purification of bovine Arp2/3 complex using Superdex 200 gel filtration chromatography.
(A) An example of the UV absorbance detected chromatogram for bovine Arp2/3 complex purification on a 320 mL Superdex 200 pg column. UV absorbance is shown with a thick green line. Fractions collected are shown with short vertical lines. Relevant fraction numbers are shown. (B) SDS-PAGE analysis of relevant fractions was performed a using 15% acrylamide gel, and stained using coomassie brilliant blue dye. At this stage the complex is pure, with no contaminants observed. Multiple isoforms of ArpC1 are present, and appears as a multiplet. Fraction numbers from the chromatogram are indicated at the top of the gel lanes, and molecular weights of standards are shown at the left. Depending on intended use, Arp2/3 complex may be pooled (e.g., fractions 14 – 17) or kept separate.
Acknowledgments
We thank Dr. Ayman Ismail for assembling early versions of this protocol.
Footnotes
Bovine thymus can be obtained in a few ways. Traditionally, they are obtained from a local slaughterhouse. This can work, but the relationship between the slaughterhouse and the laboratory is critical. Here, calf thymuses of approximately 200 – 300 g are necessary and they must be cooled promptly after collection. Larger thymus or those that have been allowed to sit at room temperature for an hour or more will yield Arp2/3 complex of poor purity and activity. Due to the variability in slaughterhouse events, and a lack of universal slaughterhouse availability, we recommend obtaining frozen calf thymus from Pel-Freez.
This has improved our reliability substantially, with one exception. One lot arrived with particularly small thymuses, which were of somewhat different color. At the first centrifugation step it was noted that there was a general absence of the ‘fat-pad’ normally observed at the top of the 1 L centrifugation bottles. This has correlated with low activity. The problem persisted through the entire lot of 10 thymuses and cleared up once a new lot was obtained. If poor results are obtained and a fat pad was not observed at the first centrifugation step, consider obtaining a new lot of thymus.
PMSF is toxic. Care should be used when preparing this stock. In particular, use of gloves, lab coats, protective eye wear and dust masks will reduce inhalation and cross contamination. One effective strategy is to purchase <100 g bottles, and prepare the entire bottle at one time trusting the manufacturer’s provided weight. The stock should be stored as 1 mL aliquots in 1.5 mL microfuge tubes at −20°C. Once the stocks have been frozen PMSF will crystallize out of solution. Warming in a room temperature beaker of water for a few minutes is usually sufficient to bring the PMSF back into solution. Agitation by inversion may be needed. When adding PMSF to a buffer, avoid splashing the stock and buffers onto gloves, skin and eyes.
In the case of the PIPES buffer, you must use the free acid (Sigma #P6757), not the disodium salt. This is necessary to reduce overall ionic strength in later steps.
Recommended buffer preparation schedule and volumes for the provided protocol scale. On the first day you will need 1 L of EB, 1 L QA and 8 L of DB. The DB buffer can be prepared shortly before use, but make sure that you have a sufficient quantity of cold water at hand. We accomplish this by keeping 12 L carboys of ultrapure water in the cold room for several hours. On the second day, you will need 1.5 L QC, 0.5 L QD, and 12 L of SA. As on day 1, ensure that you have sufficient cold water prepared to make 12 L of SA at the end of the day. On the third day you will need 1 L of SA, 0.5 L SB, and 1 L KMEId.
If you are using thymus stored at −80°C, keep them as cold as possible until homogenizing. Since the thymuses are homogenized in a moderate excess of buffer, their heat capacity matters. While keeping the homogenate as cold as possible is desirable in general, when homogenizing thymus at near −80°C with cold buffer, the entire homogenate can freeze solid and damage the blender. If you are using fresh (ice cold) thymus, cold buffer should be used.
Q Cleaning buffer is 6 M guanidine hydrochloride acidified with 200 mM acetic acid. This is generally useful for cleaning anion exchange columns. Technical or 98% pure guanidine hydrochloride can be used for this buffer (analytic quality is prohibitively expensive). If some particulate matter remains insoluble, it can be removed by centrifugation at low speed (e.g., 3,000 x g for 20 minutes) prior to 0.22 μm filtration.
We use a Sorvall RC 3C plus centrifuge equipped with a H6000A rotor (swinging bucket rotor capable of holding six 1 L bottles), a Beckman Avanti J-26XPI centrifuge equipped with a JA-10 rotor and a Beckman Optima L-90X ultracentrifuge equipped with a Type 45 Ti rotor. The first two rotors are used with polypropylene copolymer bottles with threaded closures, and the Type 45 Ti rotor uses 70 mL thick wall polycarbonate bottles with two part aluminum closures. Check centrifuge availability and temperature. For the ultracentrifugation step, allow sufficient time for the centrifuge to cool to 4°C. For most Beckman ultracentrifuges, the vacuum must be pulled for cooling to occur.
Here is a specific setup useable for breaking up and homogenizing calf thymus. A 38 quart, heavy-duty, stainless steel stock pot is placed against a cabinet on the floor, with eight layers of paper towels to protect the cabinet. A lead brick is wrapped with 5 to 8 layers of clean heavy-duty aluminum foil. The ‘lead brick’ is a piece of lead the size and shape of a construction brick. The purpose of the brick is that the mass is sufficient to shatter the thymus when hitting it.
A frozen (−80°C) thymus is placed, still in its packaging from Pel-Freez, into the pot. The thymus is then smashed into pieces with the lead brick. With a little practice one can break up a thymus in 3 to 5 minutes, which minimizes thawing. A good method is to first smash the thymus into 3 to 6 pieces, push them to one side and remove the plastic bag. Then individual pieces can be smashed into smaller pieces that are less than 2 cm in their longest dimension. Once a large piece is completely broken down, use the brick to push the small pieces to one side. After one thymus is broken into pieces, it gets homogenized, and the homogenate goes on ice. Then the process is repeated for additional thymuses and the homogenates are combined.
Ideally, the blender jar will be sized such that one thymus plus buffer will fill it 40–80% full. We use a 1 L blender jar to homogenize one thymus. A balance needs to be struck between blending to complete homogeneity and heating of the homogenate by the blender. A good working method is to blend in pulses of 5 to 10 seconds, counting the number of pulses. Initially, larger chunks can be heard hitting the bottom of the blender when a pulse is stopped. When this can no longer be heard, note how many pulses were used to reach this point, and continue with the same number of additional pulses. Total blending time will be 3 – 5 minutes. Once this point is reached, allow the blender to sit for ~20 seconds prior to opening to allow mists to condense and settle. Transfer to pre-chilled 1 L centrifuge bottles, rinse blender with cold EB and check for any large chunks remaining. These can be added to the next homogenization run or discarded after the final run.
Performance of the Q Sepharose column is substantially degraded during use. It is common for observable fat to accumulate at the top of the column during the run. This can be largely removed by cleaning the column on the same day that it is used. Cleaning is accomplished by flowing 3 column volumes (450 mL) of 5 M sodium chloride over the column, followed by 3 column volumes (450 mL) of 6 M guanidine hydrochloride/200 mM acetic acid and then 5 column volumes (750 mL) of water with 0.5 mM sodium azide. The column is stored in the latter buffer. Even with this cleaning procedure, we reserve this column for Arp2/3 complex purification only, and completely repack it with fresh media every third preparation.
A common mistake during the ammonium sulfate cut is to add the solid ammonium sulfate too quickly or all at once. This results in locally high concentrations of ammonium sulfate, which can cause undesired proteins to precipitate. A few precautions can minimize this problem.
First, inspect the ammonium sulfate as it is weighed out. Often there are clumps of crystals present, some of which may be larger than 5 mm. If this is a problem, break them up by mashing them with a mortar and pestle. The goal is not to smash the crystals into powder (which does help, but not enough to warrant routinely doing) but just to break up any large crystals present.
Second, the addition of ammonium sulfate should occur over 20 to 25 minutes for the amounts described here (time can be scaled down somewhat for smaller volumes). The stirred solution should be checked periodically while the solid is added. Look at the bottom of the beaker, make sure solid ammonium sulfate is not accumulating. If it does, the ammonium sulfate is being added too quickly. Wait a few minutes for the accumulated solid to disperse, then continue to add, but more slowly. A practical way to add the solid is to put half to one third of it in a plastic weigh boat, and to tap it with a spatula. By varying the frequency of tapping a reasonably controllable and uniform addition rate may be found.
For historical reasons we use a Whatman ZapCap-S 0.45 μm (VWR # 28152-182) with one Whatman #30 glass membrane on the top, pouring the dialysate through a portion of the membrane only. The supernatant typically still has some particulate remaining after centrifugation, and the addition of a glass prefilter speeds filtration.
Choice of SOURCE 15Q hardware and column size. The recommended linear flow rates for SOURCE 15 resins (>50 mL/min for a 26 mm wide column) produces column pressures well in excess of the tolerance of the XK26 hardware. Thus, flow rates are dictated by the column pressure tolerance, and flow rates may be a low as 5 mL/min. If the user is scales down the preparation to one ~150 gram thymus, then performance may be improved by packing 20 mL of SOURCE 15Q into HR16/10 hardware (which is designed for operation at higher pressures). Reduced flow rates (<8 mL/min) are still a good idea in this context, as our experience with SOURCE 15Q resins indicates that increased flow rate decreases resolution.
Pooling fractions at the SOURCE 15Q stage. Arp2/3 complex elutes roughly in the middle of the 0–10% buffer QD gradient. At this stage however, the mixture is still complex enough that the appearance of a well defined peak cannot be reliably used to identify the fractions containing Arp2/3 complex. Further, each preparation is a little different, and the exact contaminants vary somewhat. Fractions containing Arp2/3 complex to pool are identified using SDS-PAGE analysis. 15% acrylamide gels are run until the bromophenol blue loading dye runs off the gel. The gels are stained using coomassie brilliant blue. Fractions to pool are identified by the presence of the ArpC3/ArpC4/ArpC5 triplet near the bottom of the gel (see Fig. 1). Typically, two 15-lane gels are run, analyzing every second 14 mL fraction through the middle of the gradient.
The SOURCE 15Q column should be cleaned the same day it is used. It is cleaned by passing three column volumes (150 mL) of 5 M sodium chloride over the column, followed by three column volumes (150 mL) of 6 M guanidine hydrochloride/200 mM acetic acid. Finally, five column volumes (250 mL) of 0.5 mM sodium azide are passed over the column and the column is stored in the latter buffer. The entire SOURCE 15Q column is repacked with fresh resin after four Arp2/3 complex preparations, and the column is reserved for only Arp2/3 complex preparation. As SOURCE15Q resin is quite expensive when purchased at the 10 mL lab pack scale, we reduce costs by purchasing Bioprocess scale packages (200 mL and larger).
Arp2/3 complex elutes from the MonoS column as two peaks, both of which are active (31). All Arp2/3 complex containing fractions are customarily pooled at this point. Arp2/3 complex elutes between 3% and 6% buffer SB. There should be pronounced peaks for the complex, guiding choice of fractions to analyze by SDS-PAGE. See Fig. 2 for an example of the chromatogram and gel. Fractions are chosen to contain Arp2/3 complex and to reject contaminants. At this point the complex should be pure enough that Arp3, Arp2, ArpC2 and a multiplet for ArpC1/ArpC1B can be easily seen. Contaminants at 26 kDa, 28 kDa and 74 kDa are expected and tolerable, they are removed during gel filtration.
Typically we use the KMEId for the final gel filtration step, but this buffer is largely chosen for compatibility with our standard pyrene actin polymerization assay conditions. This buffer may be varied subtly at the user’s discretion. Changes that we have not seen to affect the purification: substitution of 5 or 10 mM HEPES pH 7.0 for the imidazole, using pH 7.5 instead of pH 7.0, inclusion of 100 μM ATP, omission of the DTT. When experiments require substantially different buffers from this, we typically purify the complex under the standard conditions, then concentrate the complex and use a further gel filtration step to exchange into the desired buffer.
Our method of quantifying Arp2/3 complex is based on UV absorption. An extinction coefficient at 280 nm of 233320 M−1 cm−1 was estimated from the number of tryptophan and tyrosine residues (24 and 68, respectively) in a complex composed of the reference sequences for Arp3, Arp2, ArpC1B, ArpC2, ArpC3, ArpC4 and ArpC5 (accession numbers: NP_776651.1, AAI51357.1, NP_001014844.1, NP_001029885.1, NP_001029443.1, NP_001069631.1, and NP_001030524.1). As the complex may bring an ATP or ADP nucleotide along with it during purification, we routinely measure absorption at 290 nm, and correct for any scatter by using the absorption at 314 nm. By measuring the relative absorbance of a sample judged to be devoid of nucleotide at 280 nm and 290 nm, we found the A290/A280 ratio to be 0.6, and thus routinely measure the concentration using an extinction coefficient at 290 nm of 140000 M−1 cm−1.
Directly freezing small aliquots of Arp2/3 complex in KMEId in liquid nitrogen results in a slight degree of aggregation, and measurable loss of activity. This can be corrected for by including ATP and a cryoprotectant in the freezing buffer. For routine assay at 10 nM of Arp2/3 complex in pyrene actin polymerization assays, we now supplement 600 nM Arp2/3 complex in KMEId with one half of a part freezing buffer (i.e., 5 mL of buffer FB are added to 10 mL of Arp2/3 complex solution). While glycerol can also be used, our typical freezing buffer uses sucrose as a cryoprotectant (see recipe for buffer FB). The concentration of Arp2/3 complex should be measured prior to dilution, and some care should be used to carefully measure and dispense all of the viscous, sucrose-containing, buffer FB solution. At that point the concentration is inferred from the know dilution factor. Small volumes (e.g., 80 μL) can then be placed into small tubes (200 μL thin wall PCR tubes are convenient for this) and snap frozen in liquid nitrogen. Aliquots can then be stored at −80°C until needed; complex maintains activity for at least one year under these conditions. Aliquots should be used soon after thawing.
We have also had good results storing Arp2/3 complex at 4°C, with DTT present for up to four weeks. More prolonged storage typically results in loss of activity, perhaps due to loss of phosphorylation at critical residues (48).
For experiments that require a buffer incompatible with freezing, we routinely concentrate Arp2/3 complex to >5 mg/mL, quantify the concentration as in Note 18, supplement with freezing buffer as above and freeze in ~500 μL aliquots. These can be thawed, spun briefly and passed over a Superdex200 10/300 analytic gel filtration column into a desired buffer and quantified by UV absorption.
References
- 1.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326 (5957):1208–1212. doi: 10.1126/science.1175862. doi: 326/5957/1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112 (4):453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 3.Mooren OL, Galletta BJ, Cooper JA. Roles for actin assembly in endocytosis. Annu Rev Biochem. 2012;81:661–686. doi: 10.1146/annurev-biochem-060910-094416. [DOI] [PubMed] [Google Scholar]
- 4.Weinberg J, Drubin DG. Clathrin-mediated endocytosis in budding yeast. Trends Cell Biol. 2012;22 (1):1–13. doi: 10.1016/j.tcb.2011.09.001. doi: S0962-8924(11)00175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pollard TD. Mechanics of cytokinesis in eukaryotes. Curr Opin Cell Biol. 2010;22 (1):50–56. doi: 10.1016/j.ceb.2009.11.010. doi: S0955-0674(09)00216-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haglund CM, Welch MD. Pathogens and polymers: microbe-host interactions illuminate the cytoskeleton. J Cell Biol. 2011;195 (1):7–17. doi: 10.1083/jcb.201103148. doi: jcb.201103148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munter S, Way M, Frischknecht F. Signaling during pathogen infection. Sci STKE. 2006;2006(335):re5. doi: 10.1126/stke.3352006re5. [DOI] [PubMed] [Google Scholar]
- 8.Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- 9.Goode BL, Rodal AA, Barnes G, Drubin DG. Activation of the Arp2/3 complex by the actin filament binding protein Abp1p. J Cell Biol. 2001;153 (3):627–634. doi: 10.1083/jcb.153.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Machesky LM, Atkinson SJ, Ampe C, Vandekerckhove J, Pollard TD. Purification of a cortical complex containing two unconventional actins from Acanthamoeba by affinity chromatography on profilin-agarose. J Cell Biol. 1994;127 (1):107–115. doi: 10.1083/jcb.127.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mullins RD, Stafford WF, Pollard TD. Structure, subunit topology, and actin-binding activity of the Arp2/3 complex from Acanthamoeba. J Cell Biol. 1997;136 (2):331–343. doi: 10.1083/jcb.136.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amann KJ, Pollard TD. The Arp2/3 complex nucleates actin filament branches from the sides of pre-existing filaments. Nat Cell Biol. 2001;3 (3):306–310. doi: 10.1038/35060104. [DOI] [PubMed] [Google Scholar]
- 13.Blanchoin L, Amann KJ, Higgs HN, Marchand JB, Kaiser DA, Pollard TD. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 2000;404 (6781):1007–1011. doi: 10.1038/35010008. [DOI] [PubMed] [Google Scholar]
- 14.Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci U S A. 1998;95 (11):6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rouiller I, Xu XP, Amann KJ, Egile C, Nickell S, Nicastro D, Li R, Pollard TD, Volkmann N, Hanein D. The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol. 2008;180 (5):887–895. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelleher JF, Atkinson SJ, Pollard TD. Sequences, structural models, and cellular localization of the actin-related proteins Arp2 and Arp3 from Acanthamoeba. J Cell Biol. 1995;131 (2):385–397. doi: 10.1083/jcb.131.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galletta BJ, Mooren OL, Cooper JA. Actin dynamics and endocytosis in yeast and mammals. Curr Opin Biotechnol. 2010;21 (5):604–610. doi: 10.1016/j.copbio.2010.06.006. doi: S0958-1669(10)00113-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welch MD, Iwamatsu A, Mitchison TJ. Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature. 1997;385 (6613):265–269. doi: 10.1038/385265a0. [DOI] [PubMed] [Google Scholar]
- 19.Doolittle LK, Rosen MK, Padrick SB. Measurement and Analysis of in vitro Actin Polymerization. Methods in Molecular Biology in progress. doi: 10.1007/978-1-62703-538-5_16. (In progress) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper JA, Walker SB, Pollard TD. Pyrene actin: documentation of the validity of a sensitive assay for actin polymerization. J Muscle Res Cell Motil. 1983;4 (2):253–262. doi: 10.1007/BF00712034. [DOI] [PubMed] [Google Scholar]
- 21.Kouyama T, Mihashi K. Fluorimetry study of N-(1-pyrenyl)iodoacetamide-labelled F-actin. Local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur J Biochem. 1981;114 (1):33–38. [PubMed] [Google Scholar]
- 22.Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci U S A. 1999;96 (7):3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welch MD, Rosenblatt J, Skoble J, Portnoy DA, Mitchison TJ. Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science. 1998;281 (5373):105–108. doi: 10.1126/science.281.5373.105. [DOI] [PubMed] [Google Scholar]
- 24.Zuchero In vitro actin purification and assembly assay using TIRF to visualise filament dynamics. Methods in Molecular Biology (in progress) [Google Scholar]
- 25.Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401 (6753):613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- 26.Bernheim-Groswasser A, Wiesner S, Golsteyn RM, Carlier MF, Sykes C. The dynamics of actin-based motility depend on surface parameters. Nature. 2002;417 (6886):308–311. doi: 10.1038/417308a. [DOI] [PubMed] [Google Scholar]
- 27.Achard V, Martiel JL, Michelot A, Guerin C, Reymann AC, Blanchoin L, Boujemaa-Paterski R. A “primer”-based mechanism underlies branched actin filament network formation and motility. Curr Biol. 2010;20 (5):423–428. doi: 10.1016/j.cub.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 28.Akin O, Mullins RD. Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell. 2008;133 (5):841–851. doi: 10.1016/j.cell.2008.04.011. doi: S0092-8674(08)00498-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Co C, Wong DT, Gierke S, Chang V, Taunton J. Mechanism of actin network attachment to moving membranes: barbed end capture by N-WASP WH2 domains. Cell. 2007;128 (5):901–913. doi: 10.1016/j.cell.2006.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dayel MJ, Akin O, Landeryou M, Risca V, Mogilner A, Mullins RD. In silico reconstitution of actin-based symmetry breaking and motility. PLoS Biol. 2009;7 (9):e1000201. doi: 10.1371/journal.pbio.1000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higgs HN, Blanchoin L, Pollard TD. Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry. 1999;38 (46):15212–15222. doi: 10.1021/bi991843+. doi: bi991843+ [DOI] [PubMed] [Google Scholar]
- 32.Insall R, Muller-Taubenberger A, Machesky L, Kohler J, Simmeth E, Atkinson SJ, Weber I, Gerisch G. Dynamics of the Dictyostelium Arp2/3 complex in endocytosis, cytokinesis, and chemotaxis. Cell Motil Cytoskeleton. 2001;50 (3):115–128. doi: 10.1002/cm.10005. [DOI] [PubMed] [Google Scholar]
- 33.Welch MD, Mitchison TJ. Purification and assay of the platelet Arp2/3 complex. Methods Enzymol. 1998;298:52–61. doi: 10.1016/s0076-6879(98)98008-9. [DOI] [PubMed] [Google Scholar]
- 34.Ma L, Rohatgi R, Kirschner MW. The Arp2/3 complex mediates actin polymerization induced by the small GTP-binding protein Cdc42. Proc Natl Acad Sci U S A. 1998;95 (26):15362–15367. doi: 10.1073/pnas.95.26.15362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beltzner CC, Pollard TD. Pathway of actin filament branch formation by Arp2/3 complex. J Biol Chem. 2008;283 (11):7135–7144. doi: 10.1074/jbc.M705894200. doi: M705894200. [DOI] [PubMed] [Google Scholar]
- 36.Egile C, Loisel TP, Laurent V, Li R, Pantaloni D, Sansonetti PJ, Carlier MF. Activation of the CDC42 effector N-WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin-based motility. J Cell Biol. 1999;146 (6):1319–1332. doi: 10.1083/jcb.146.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lechler T, Jonsdottir GA, Klee SK, Pellman D, Li R. A two-tiered mechanism by which Cdc42 controls the localization and activation of an Arp2/3-activating motor complex in yeast. J Cell Biol. 2001;155 (2):261–270. doi: 10.1083/jcb.200104094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zalevsky J, Lempert L, Kranitz H, Mullins RD. Different WASP family proteins stimulate different Arp2/3 complex-dependent actin-nucleating activities. Curr Biol. 2001;11 (24):1903–1913. doi: 10.1016/s0960-9822(01)00603-0. doi: S0960-9822(01)00603-0. [DOI] [PubMed] [Google Scholar]
- 39.Doolittle LK, Rosen MK, Padrick SB. Purification of Arp2/3 complex from Saccharomyces cerevisiae. Methods in Molecular Biology in progress. doi: 10.1007/978-1-62703-538-5_15. (In progress) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gournier H, Goley ED, Niederstrasser H, Trinh T, Welch MD. Reconstitution of human Arp2/3 complex reveals critical roles of individual subunits in complex structure and activity. Mol Cell. 2001;8 (5):1041–1052. doi: 10.1016/s1097-2765(01)00393-8. doi: S1097-2765(01)00393-8. [DOI] [PubMed] [Google Scholar]
- 41.Balcer HI, Daugherty-Clarke K, Goode BL. The p40/ARPC1 subunit of Arp2/3 complex performs multiple essential roles in WASp-regulated actin nucleation. J Biol Chem. 2010;285 (11):8481–8491. doi: 10.1074/jbc.M109.054957. doi: M109.054957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daugherty KM, Goode BL. Functional surfaces on the p35/ARPC2 subunit of Arp2/3 complex required for cell growth, actin nucleation, and endocytosis. J Biol Chem. 2008;283 (24):16950–16959. doi: 10.1074/jbc.M800783200. doi: M800783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin AC, Xu XP, Rouiller I, Kaksonen M, Sun Y, Belmont L, Volkmann N, Hanein D, Welch M, Drubin DG. Effects of Arp2 and Arp3 nucleotide-binding pocket mutations on Arp2/3 complex function. J Cell Biol. 2005;168 (2):315–328. doi: 10.1083/jcb.200408177. doi: jcb.200408177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pan F, Egile C, Lipkin T, Li R. ARPC1/Arc40 mediates the interaction of the actin-related protein 2 and 3 complex with Wiskott-Aldrich syndrome protein family activators. J Biol Chem. 2004;279 (52):54629–54636. doi: 10.1074/jbc.M402357200. doi: M402357200. [DOI] [PubMed] [Google Scholar]
- 45.Rodal AA, Sokolova O, Robins DB, Daugherty KM, Hippenmeyer S, Riezman H, Grigorieff N, Goode BL. Conformational changes in the Arp2/3 complex leading to actin nucleation. Nat Struct Mol Biol. 2005;12 (1):26–31. doi: 10.1038/nsmb870. doi: nsmb870. [DOI] [PubMed] [Google Scholar]
- 46.Winter DC, Choe EY, Li R. Genetic dissection of the budding yeast Arp2/3 complex: a comparison of the in vivo and structural roles of individual subunits. Proc Natl Acad Sci U S A. 1999;96 (13):7288–7293. doi: 10.1073/pnas.96.13.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK. Arp2/3 complex is bound and activated by two WASP proteins. Proc Natl Acad Sci U S A. 2011;108 (33):E472–479. doi: 10.1073/pnas.1100236108. doi: 1100236108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.LeClaire LL, 3rd, Baumgartner M, Iwasa JH, Mullins RD, Barber DL. Phosphorylation of the Arp2/3 complex is necessary to nucleate actin filaments. J Cell Biol. 2008;182 (4):647–654. doi: 10.1083/jcb.200802145. doi: jcb.200802145. [DOI] [PMC free article] [PubMed] [Google Scholar]