Abstract
Pancreatic β-cell dysfunction is a common feature of type 2 diabetes. Earlier, we had cloned IG20 cDNA from a human insulinoma and had shown that IG20/MADD can encode six different splice isoforms that are differentially expressed and have unique functions, but its role in β-cell function was unexplored. To investigate the role of IG20/MADD in β-cell function, we generated conditional knockout (KMA1ko) mice. Deletion of IG20/MADD in β-cells resulted in hyperglycemia and glucose intolerance associated with reduced and delayed glucose-induced insulin production. KMA1ko β-cells were able to process insulin normally but had increased insulin accumulation and showed a severe defect in glucose-induced insulin release. These findings indicated that IG20/MADD plays a critical role in glucose-induced insulin release from β-cells and that its functional disruption can cause type 2 diabetes. The clinical relevance of these findings is highlighted by recent reports of very strong association of the rs7944584 single nucleotide polymorphism (SNP) of IG20/MADD with fasting hyperglycemia/diabetes. Thus, IG20/MADD could be a therapeutic target for type 2 diabetes, particularly in those with the rs7944584 SNP.
Introduction
Type 2 diabetes affects ∼8% of all Americans and 366 million people worldwide with significant resulting morbidity and mortality. Peripheral insulin resistance and insulin secretion deficit characterize type 2 diabetes. In an attempt to identify genes that are involved in pancreatic β-cell function, we cloned a number of genes, including the IG20 (clone number 20 and hence IG20), that were differentially expressed in a human insulinoma through subtractive hybridization with a human glucagonoma (1–3). Subsequently, IG20 and MADD/DENN cDNAs, which were independently cloned, were found to be nearly identical to each other and were splice isoforms of the same gene (4,5). Alternative splicing allows a single gene to encode several proteins and thus enhance proteome diversity. IG20/MADD is one such gene that can undergo cell-specific alternate splicing and give rise to six different protein isoforms with unique functions (3–17).
Although the IG20pa and IG20-SV2 isoforms are not always expressed, the MADD and DENN-SV isoforms are constitutively expressed in all cells and tissues and are overexpressed in many human tumors and cancer cell lines (3,14,15,17,18). The function of IG20-SV2 is not yet known; however, the MADD isoform is an Akt substrate and plays an important role in cancer cell survival and confers resistance to TRAIL (tumor necrosis factor [TNF]-related apoptosis-inducing ligand)- and TNF-α–induced apoptosis (6,14,17,19). The IG20pa isoform is proapoptotic and functions like a “tumor suppressor”; in contrast, the DENN-SV isoform acts as an “oncogene” and promotes tumor cell proliferation (9,10).
The other two isoforms, KIAA0358 and IG20-SV4, are expressed only in certain neuronal and neuroendocrine tissues (7) and are important in regulating neurotransmission and neuronal cell survival (7,20–22). The KIAA0358 isoform of IG20/MADD is a human ortholog of the rat Rab3A guanosine diphosphate–guanosine triphosphate exchange protein (12) that regulates neurotransmitter release at the neuromuscular junction (23,24) during the postdocking process of postsynaptic exocytosis (23).
Whereas the above studies revealed the function of IG20/MADD in neurotransmission and cell survival, human genome-wide association studies have shown a strong association of the rs7944584 single nucleotide polymorphism (SNP) in IG20/MADD with fasting hyperglycemia in European cohorts (25) and in type 2 diabetes in Han Chinese (26). This suggested that IG20/MADD might play a role in glucose homeostasis by an unknown mechanism. This finding, combined with the fact that IG20 cDNA was cloned from a human insulinoma and that the β-cells are of neuroendocrine origin, led us to hypothesize that IG20/MADD might be important in β-cell function. We therefore explored the role of IG20/MADD in pancreatic β-cells by generating mice in which IG20/MADD can be selectively deleted in β-cells and studied the consequent effects on glucose homeostasis, including β-cell function.
Research Design and Methods
Generation of Ig20/Madd-knockout Mice
All animal experiments were according to the protocol approved by the University of Illinois at Chicago Institutional Animal Care and Use Committee. The mouse IG20/MADD homologous sequence was isolated from a 129Sv/Pas bacterial artificial chromosome library using the PCR primers listed in Supplementary Table 1. The targeting vector contained exons 3 and 4 flanked by a distal LoxP site at the 3′ end and a LoxP–flippase recognition target sites (FRT)–neomycin (Neo)-FRT–positive selection cassette at the 5′ end. The Neo cassette flanking the FRT sites allowed for its deletion by crossing with an Flp recombinase-expressing mouse. The linearized targeting vector (Fig. 1A) was electroporated into mouse E14 embryonic stem (ES) cells, and G418-resistant clones were selected. The primers listed in Supplementary Tables 2 and 4 were used under the conditions reported in Supplementary Tables 3 and 5, respectively, for PCR amplification to detect the homologous recombinants at the 5′ end (2.6-kb fragment; Supplementary Fig. 1) and 3′ end (5.4-kb fragment; Supplementary Fig. 2). Bgl II and AvrII digested genomic DNA was subjected to Southern blotting with probes generated using PCR primers listed in Supplementary Tables 6 and 7 for detecting the 5′ (8.4-kb fragment) and 3′ (7.0-kb fragment) fragments, respectively (Supplementary Figs. 3 and 4).
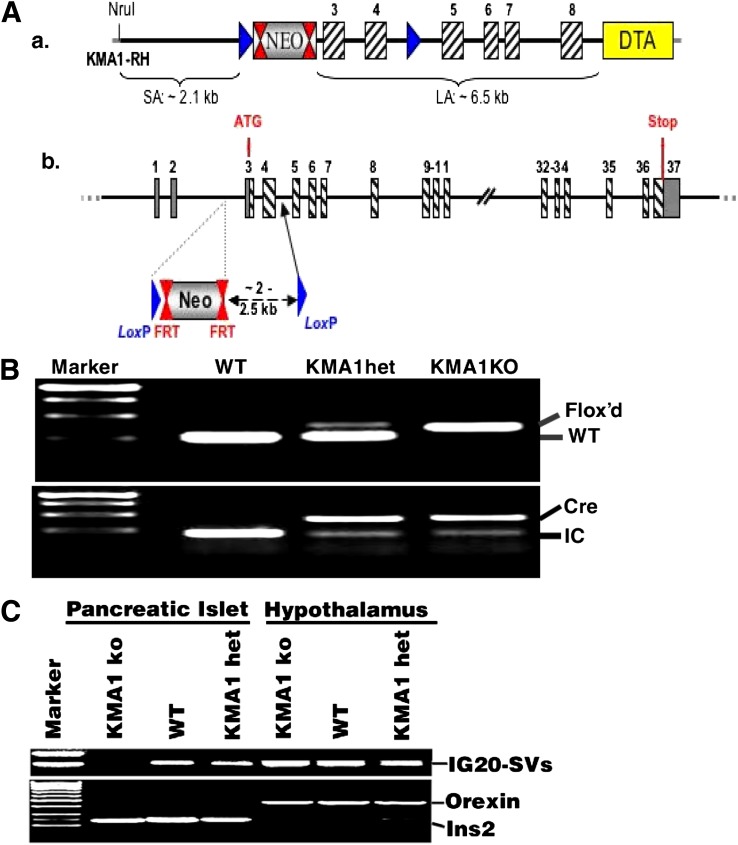
Figure 1.
Generation of mice with conditional KO of IG20/MADD in β-cells. A: Shows schematic representation of the strategy used for generating floxed KMA1 mice. B: Shows genotyping results from offspring of KMA1het (KMA1flox+/−: Cre) mice intercrossing. Primers used for PCR are listed in Supplementary Table 8. Representative figure from 98 mice is shown. C: Results from RT-PCR using RNA isolated from hypothalamus and islets from WT, KMA1het, and KMA1ko mice. Orexin and insulin-2 served as specific markers of hypothalamus and pancreatic islets, respectively. Representative data from 3 mice per group are shown.
To generate chimeric mice, targeted ES cells were injected into C57BL/6 blastocysts and reimplanted into pseudopregnant females. KMA1 Flox+/− mice were obtained by mating chimeric males with C57BL/6 females (Supplementary Fig. 5). These mice, generated by GenOway, Lyon, France, were intercrossed to generate KMA1 (Flox+/+) mice. KMA1 Flox+/− Cre mice were obtained by crossing KMA1 Flox+/+ mice with heterozygous (het; Ins2-cre/Esr1)1Dam/J (The Jackson Laboratory). The KMA1 Flox+/− Cre mice were intercrossed to produce KMA1 Flox+/+Cre mice (Ig20/Madd-knockout [KMA1ko]). Mice were genotyped using PCR primers listed in Supplementary Table 8. Expected size of DNA fragments from wild-type (WT) and KMA1ko was 245 bp and 363 bp, respectively (Fig. 1B), the Cre fragment was 410 bp, and the internal control (IC) MGSCv37 (NT_039606.7) fragment was 200 bp (Fig. 1B). Mice were maintained on a C57BL/6 background.
Genotyping
Tail DNA from 3- to- 4-week-old mice was extracted using the DNeasy Blood & Tissue Kit (Qiagen, Germantown, MD). The PCR primer sets (Supplementary Table 8) were used for genotyping. The Flox'd allele and the Cre allele were detected by PCR.
Induction of KMA1 Deletion
Mice were housed in a pathogen-free facility on a 12:12-h light-dark cycle and fed ad libitum with a standard mouse chow diet. To delete KMA1, 40 mg/kg/day tamoxifen in 100 μL peanut oil was injected intraperitoneally into the peritoneum of 4- to 5-week-old mice for 5 days (27). Blood glucose was measured between 9:00 and 9:30 a.m. using a glucometer (Bayer HealthCare LLC, Whippany, NJ). Plasma collected during the same time was used to test normal-feeding insulin levels.
RNA Extraction and RT-PCR
Total cellular RNA from the islets and hypothalamus of WT, KMA1het, and KMA1ko mice was extracted using TRIzol reagent (Life Technologies, Grand Island, NY) and was subjected to RT-PCR using the SuperScript III One-Step RT-PCR System with PlatinumTaq reagent (Life Technologies). To detect deletion of the IG20/MADD gene, primers flanking exon 4 (forward: 5′-TCTTCAGGTGCTAACCTGCATCCT-3′) and 5 (reverse: 5′-AACGCCATCACAGACATGGAGAGT-3′) were used. Orexin primers (forward: 5′-ACGGCCTCAGACTTCTTGGGTATT-3′ and reverse: 5′-TGCTAAAGCG-GTGGTAGTTACGGT-3′) and insulin-2 (Ins2) primers (forward: 5′-ACCATCAGCAAGCAGGAAGGTACT-3′ and reverse: 5′-GAACCACAAAGGTGCTGCTTGACA-3′) were used to amplify orexin and Ins2 as markers for hypothalamus and pancreatic islets, respectively.
Glucose and Insulin Tolerance Tests
Mice (n = 10 per group) fasted for 16 h were injected intraperitoneally with 2 g/kg glucose (Sigma-Aldrich, St. Louis, MO) in saline. Blood from the tail vein was obtained, and blood glucose levels were measured using a Bayer Contour Blood Glucose Meter (Bayer HealthCare LLC). Bovine insulin (0.75 units/kg; Sigma-Aldrich) was injected intraperitoneally. In both tolerance tests, blood glucose levels were measured at 0, 15, 30, 45, 60, and 120 min.
Proinsulin, Insulin, and C-Peptide Measurement
Plasma insulin, proinsulin, and C-peptide levels in mice fasted for 16 h were determined using an ultrasensitive mouse insulin kit (Crystal Chem Inc., Downers Grove, IL) and mouse proinsulin and C-peptide ELISA kits (ALPCO Diagnostics, Salem, NH), respectively.
Homeostasis Model Assessment for Insulin Resistance and β-Cell Function
The homeostasis model assessment (HOMA) method was used to calculate insulin resistance (HOMA-IR) and β-cell function (HOMA-B), as described before (28,29). The fasting plasma glucose and insulin level was used to determine the values of HOMA-IR and HOMA-B according the following equations: HOMA-IR = (FPI × FPG)/22.5, and HOMA-B = (20 × FPI)/(FPG − 3.5) (30), where FPI is the fasting plasma insulin concentration (mU/L) and FPG is the fasting plasma glucose level (mmol/L).
Islet Isolation and Culture
Pancreatic islets were isolated as previously described (31) and purified using discontinuous Ficoll (Mediatech, Herndon, VA) gradients (32). The islets were cultured in RPMI-1640 supplemented with 10% FBS (Hyclone Inc., Waltham, MA), 11.2 mmol/L glucose, and antibiotics at 37°C in an incubator containing 5% CO2 in air.
Assay for Insulin Content
Insulin extracted using an acid-ethanol method (33,34) from 10 islets per group or total pancreas was measured using an ultrasensitive ELISA kit (Crystal Chem Inc.).
Pancreas Staining
Tissue sections were prepared from pancreas fixed in 10% buffered formalin and stained with hematoxylin and eosin or subjected to antigen retrieval procedure (Vector Laboratories, Burlingame, CA). Guinea pig anti-insulin antibody (1:1,000; Invitrogen, Cat#: 18-0067) and TRITC-conjugated anti-guinea pig antibody (1:1,000; Sigma-Aldrich, Cat#: T7153) were used to detect insulin.
Apoptosis Assay
Paraffin was removed from the tissue sections, and pancreatic β-cell apoptosis was assessed using the ApopTag Plus Fluorescein In Situ Apoptosis Detection Kit (Chemicon International Inc., Billerica, MA). Pancreatic β-cells were stained for insulin as above, and DAPI was used to label the nucleus.
Islet Perifusion
Simultaneous islet perifusion and fluorescence imaging were performed as described before (31). Briefly, 10 islets per sample were incubated with 5 μmol/L Fura-2/AM (Molecular Probes, Grand Island, NY) and/or 2.5 μmol/L rhodamine 123 (Rh123, Sigma-Aldrich) for 30 min at 37°C in Krebs-Ringer buffer (KRB) containing 2 mmol/L glucose. Islets were introduced into a temperature-controlled microfluidic device and mounted on an inverted epifluorescence microscope (Leica DMI 4000B). Islets were then perifused by a continuous flow of KRB at 37°C (pH 7.4) for 10 min. KRB containing 14 mmol/L glucose or other stimulators was administered to the islets using a peristaltic pump or a syringe.
Islets were observed with 10× objectives. Dual-wavelength Fura-2 was excited ratiometrically at 340 and 380 nm, and changes in [Ca2+] were expressed as F340/F380 (% changes). Rh123 was excited using a 495-nm filter (Chroma Technology) mounted in a Lambda DG-4 wavelength switcher. Emission of Fura-2/AM and Rh123 fluorescence was filtered using a Fura-2/FITC polychroic beam splitter and double-band emission filter (Chroma Technology; Part number: 73.100 bs). The images were captured using Retiga-SRV, Fast 1394 (QImaging). Simple PCI software (Hamamatsu Corp.) was used for analysis. Aliquots were collected from the device by a fraction collector to measure insulin secretion.
Immunoelectron Microscopy
Immunoelectron microscopy was performed as described previously (35). Briefly, islets were cultured overnight in the absence of glucose and then stimulated with 14 mmol/L glucose for 5 min and fixed in 4% paraformaldehyde and 0.5% glutaraldehyde in 0.1 mol/L phosphate buffer at room temperature. The fixed islets were dehydrated in graded ethyl alcohol and placed in a mixture of acrylic resin (LR White: 70% alcohol, 1:2) for 1 h and then in pure acrylic resin for 48 h at room temperature. Ultrathin 70-nm sections were mounted on 200-mesh nickel grids and stained using protein A–gold technique (36–38). Sections were stained with a guinea pig anti-mouse insulin antibody (1:5,000) diluted in PBS, 1% bovine albumin, and 1% glycine, at pH 7.4, followed by washing and immersion for 1 h in a solution of 15-nm diameter colloidal gold particles (CGP) covered with protein A (Janssen Pharmaceutica, Olen, Belgium) at a dilution of 1:50 (pH 8.2) in a moist chamber at 37°C. The grids were subjected to immunoelectron microscopy (Zeiss CEM 902).
Statistical Analysis
Data are expressed as means ± SE. Significance was tested by unpaired or paired two-way Student t tests using Excel software or ANOVA test using SPSS 17 software. A P < 0.05 was considered significant.
Results
IG20/MADD Can Be Deleted in Pancreatic β-Cells Upon Tamoxifen Injection in KMA1ko Mice
Exons 3 and 4 of the IG20/MADD gene were flanked by Flox sites (Fig. 1A). The homozygous floxed mice were crossed with transgenic (Ins2-cre/Esr1)1Dam/J mice to generate KMA1het (KMA1flox+/−: Cre+) mice, and the KMA1het was intercrossed to generate WT (KMA1flox−/−: Cre−), KMA1het, and KMA1ko (KMA1flox+/+: Cre+) mice (Fig. 1B). WT, KMA1het, and KMA1ko mice were treated with tamoxifen for 5 days (27). Because an earlier report had shown that rat insulin promoter activity may also be seen in the hypothalamus (39), total RNA isolated from pancreatic islets and the hypothalamus was subjected to RT-PCR. The floxed allele was deleted only in pancreatic islets and not in the hypothalamus of KMA1ko mice or in tissue in KMA1het or WT mice (Fig. 1C).
KMA1ko Mice Show Hyperglycemia With Reduced Fasting Insulin Levels
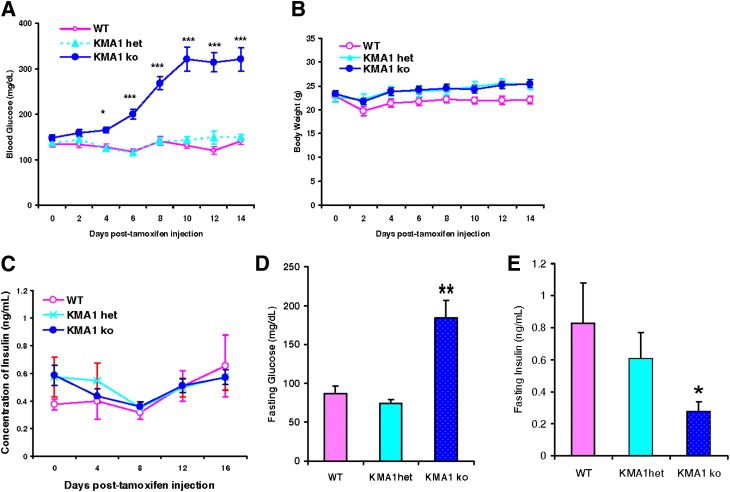
Mice were treated with tamoxifen, and the nonfasting blood glucose level was monitored. Starting on day 6, relative to WT and KMA1het mice, KMA1ko mice showed elevated blood glucose levels, which reached peak levels (321.08 ± 25.96 mg/dL in KMA1ko mice vs. 140.42 ± 6.94 mg/dL in WT mice) by day 10 (Fig. 2A). The body weights were similar among different groups of mice (Fig. 2B). Interestingly, nonfasting blood insulin levels in the KMA1ko mice remained unaltered and were comparable to the levels found in the other groups of control mice (Fig. 2C). The fasting plasma glucose levels were also elevated in the KMA1ko mice compared with the WT mice (184.55 ± 22.55 mg/dL in KMA1ko vs. 87.1 ± 9.5 mg/dL in WT mice; P < 0.01) (Fig. 2D). However, in contrast to nonfasting insulin levels, the fasting blood insulin levels in the KMA1ko mice were significantly lower than those in the WT mice (0.12 ± 0.02 ng/mL in KMA1ko vs. 0.26 ± 0.05 ng/mL in WT; P < 0.05) (Fig. 2E).
Figure 2.
Conditional knockout IG20/MADD gene in β-cells leads to hyperglycemia in KMA1ko mice. Mice were injected with 40 mg/kg/day tamoxifen in 100 μL peanut oil for 5 consecutive days and then tested for body weight and levels of blood glucose and insulin. Results are shown for nonfasting blood glucose levels (A), body weight (B), nonfasting blood insulin levels (C), fasting (16 h) blood glucose levels (D), and fasting blood insulin levels (E). The x-axis shows days after the first injection of tamoxifen. Data are expressed as means ± SE. One-way ANOVA analysis was performed. *P < 0.05, **P < 0.01, ***P < 0.01 with KMA1ko mice vs. WT; n = 10–12 mice per genotype.
KMA1ko Mice Show Delayed Blood Glucose Clearance and Glucose-Stimulated Insulin Release
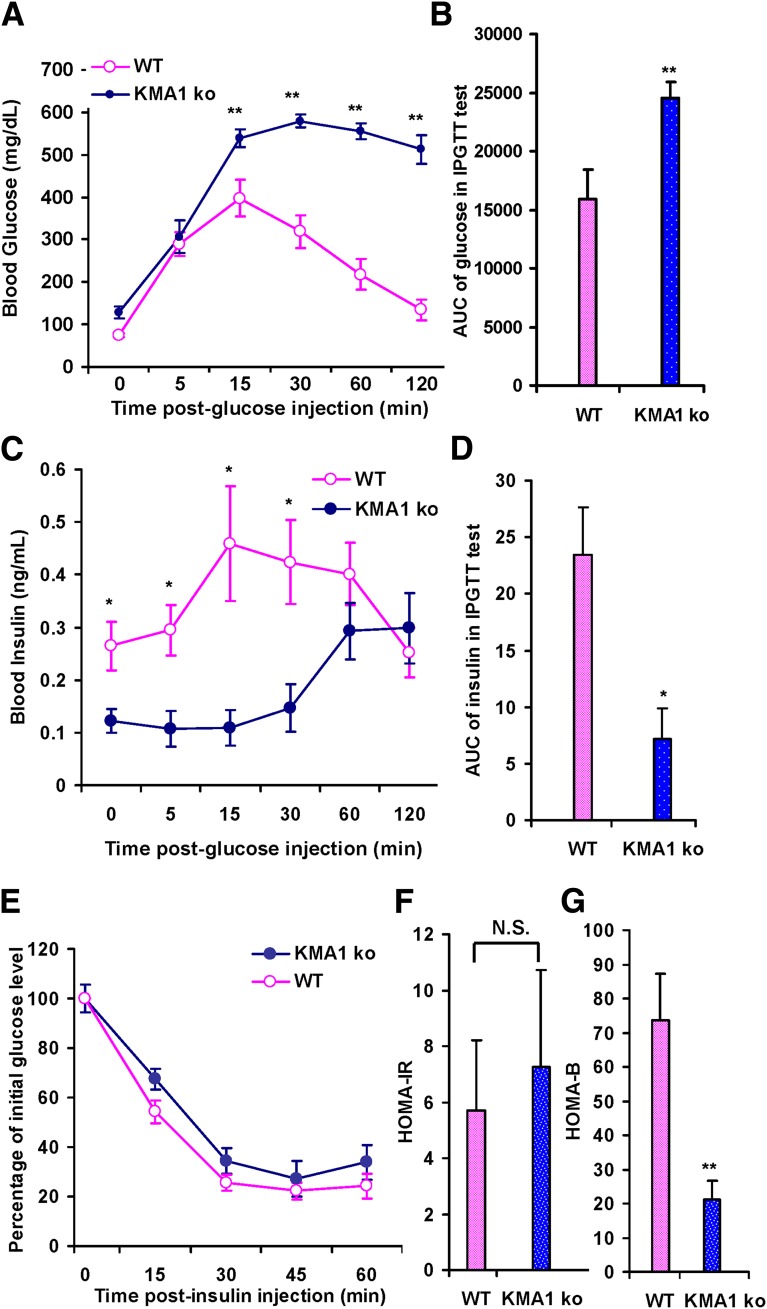
To determine the underlying cause of hyperglycemia and fasting hypoinsulinemia in KMA1ko mice, these mice underwent an intraperitoneal glucose tolerance test. At 15 min after a glucose injection (2 g/kg) into the peritoneal cavity of mice that were fasted for 16 h, the blood glucose levels were dramatically increased in KMA1ko mice and remained at ∼500 mg/dL even after 2 h (Fig. 3A). The area under the curve (AUC) of glucose in KMA1ko mice was significantly larger compared with the WT mice (24,550.56 ± 1,302.99 min ⋅ ng/mL for KMA1ko vs. 15,927.78 ± 2,453.12 min ⋅ ng/mL for WT; P < 0.01; Fig. 3B). Moreover, the rise in blood insulin levels in KMA1ko mice was significantly delayed compared with WT mice. Interestingly, the highest levels of insulin in KMA1ko did not reach the peak levels found in WT mice even after 120 min (Fig. 3C). Although the serum insulin levels were comparable in WT and KO mice (Fig. 3C) at 2 hours after the glucose challenge, they were inappropriately lower in KMA1ko mice given that the serum glucose concentration at that time point was much higher in those mice (Fig. 3A). This is reflected in the AUC for insulin being significantly less for KMA1ko mice than for the WT mice (7.16 ± 2.71 vs. 23.44 ± 4.17 min ⋅ ng/mL; P < 0.05; Fig. 3D) and suggested that IG20/MADD deletion in β-cells can cause hyperglycemia secondary to a delayed insulin release and inappropriately lower level of circulating insulin in response to a glucose challenge.
Figure 3.
Glucose and insulin tolerance test. Mice were injected with 40 mg/kg/day tamoxifen in 100 μL peanut oil for 5 consecutive days, and underwent glucose and insulin tolerance tests 14–22 days later. Results are shown for blood glucose levels at different times after intraperitoneal glucose injection (A), the AUC for blood glucose during intraperitoneal glucose tolerance test (IPGTT) (B), insulin levels at different times after intraperitoneal glucose injection (C), and the AUC for insulin levels (D). A–D: Mice were fasted overnight. E: Blood glucose levels at different times after insulin injection into KMA1ko and WT mice. Overnight fasting blood glucose and plasma insulin were used to assess HOMA-IR (F) or HOMA-B (G). Male and female mice 6 to 8 weeks of age were used. Data are expressed as means ± SE. *P < 0.05 and **P < 0.01 with KMA1ko mice vs. WT; N.S., not significant; n = 7–10 mice per group.
KMA1ko Mice Do Not Exhibit Peripheral Insulin Resistance but Show Defective β-Cell Function
Next, these mice underwent an insulin tolerance test and HOMA test to detect β-cell function and insulin resistance. KMA1ko and WT mice both showed similar degrees of insulin sensitivity (Fig. 3E) and a comparable HOMA-IR score (7.27 ± 3.43 vs. 5.68 ± 2.54; P = N.S.; Fig. 3F). However, the HOMA-B score was significantly lower in the KMA1ko mice than in the WT mice (21.05 ± 5.59 vs. 73.62 ± 13.75; P < 0.01; Fig. 3G). These results indicated that peripheral insulin sensitivity was preserved but that the KMA1ko mice may have defective β-cell function.
KMA1ko Mice Show No Apparent Change in β-Cell Morphology or Insulin Processing
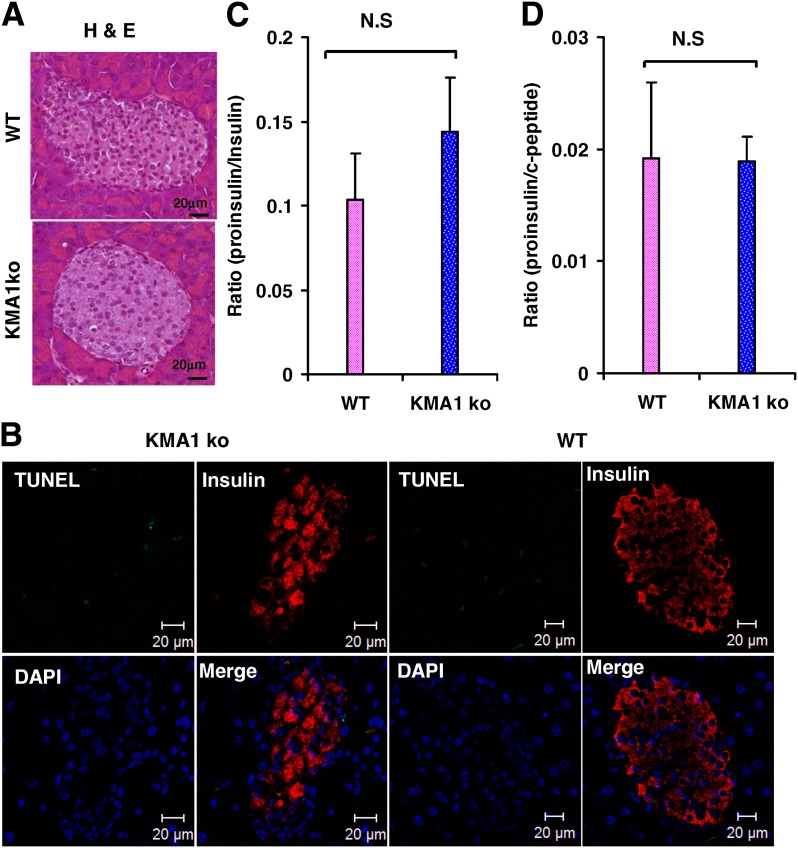
Because IG20/MADD knockdown has been shown to cause spontaneous apoptosis of various cancer cells (6,7,14,15,17), we wondered if this accounted for the suboptimal function of β-cells in the KMA1ko mice. Our results showed that pancreatic islets from the KMA1ko mice or the WT mice had no evidence of significant changes in morphology (Fig. 4A) or apoptosis (Fig. 4B).
Figure 4.
Islet morphology and apoptosis and insulin processing. Mice were injected with 40 mg/kg/day tamoxifen in 100 μL peanut oil for 5 consecutive days, and their pancreata were examined 30 days later. A: Hematoxylin and eosin (H & E) stained sections of islets from KMA1ko and WT mice show normal morphology. B: TUNEL staining shows no difference in the frequency of apoptotic cells in the islets of KMA1ko and WT mice (n = 3 mice per group). At 14 to 22 days after the last tamoxifen injection, blood was collected to determine the fasting levels of plasma proinsulin, insulin, and C-peptide by ELISA. C: Ratio of proinsulin to insulin. D: Ratio of proinsulin to C-peptide. N.S., not significant (n = 10 mice per group).
To test if posttranslational processing of insulin in the KMA1ko mice was affected, plasma samples from KMA1ko or WT mice, fasted overnight, were tested for proinsulin, insulin, and C-peptide levels. There was no significant difference in the proinsulin-to-insulin (Fig. 4C) and proinsulin-to-C-peptide (Fig. 4D) ratios between KMA1ko and WT mice, indicating that IG20/MADD knockout in pancreatic β-cells did not affect posttranslational processing of insulin.
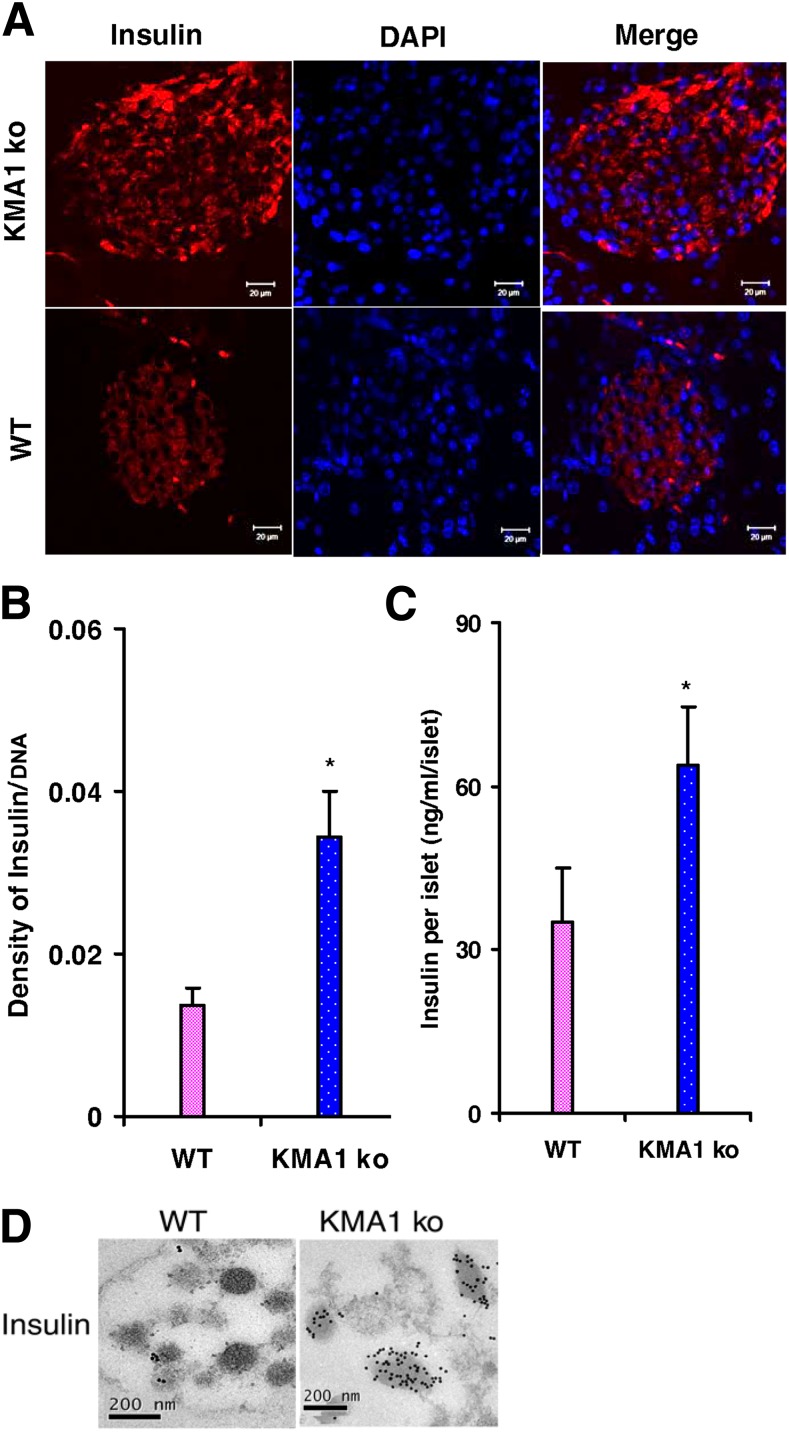
β-Cells From KMA1ko Mice Show Insulin Accumulation
To test if IG20/MADD knockout affected β-cell insulin content, we stained pancreas sections with an anti-insulin antibody and analyzed them using immunofluorescence microscopy. There was increased accumulation of insulin in KMA1ko β-cells relative to β-cells from WT mice (Fig. 5A). The ratio of insulin intensity to DAPI intensity in the β-cells was increased in KMA1ko mice compared with WT mice β-cells (Fig. 5B). Moreover, the total insulin content was significantly higher in KMA1ko islets relative to WT islets (Fig. 5C). Electron microscopy of islet cells revealed much denser insulin granules in the β-cells from KMA1ko mice compared with those from WT mice (Fig. 5D). Similarly, insulin content was increased in the pancreas of KMA1ko mice relative to the levels found in the pancreas of KMA1het or WT mice (Supplementary Fig. 6). These results indicated that insulin accumulation in β-cells of KMA1ko mice was not due to increased transcription of the insulin gene (Fig. 1C) but was likely due to a defect in glucose-induced insulin release.
Figure 5.
Insulin content in pancreatic β-cells. Mice were injected with 40 mg/kg/day tamoxifen in 100 μL peanut oil for 5 consecutive days, and the animals were killed 28 to 31 days later. A: Pancreatic sections were stained with anti-insulin antibody and further stained using a TRITC-conjugated anti-guinea pig antibody; DAPI was used to stain the nucleus. B: Average intensity of insulin staining relative to nuclear DAPI staining. C: Insulin content in the freshly isolated islets from KMA1ko and WT mice. D: Electron micrograph shows insulin-protein A–gold staining of insulin granules within the selected area of β-cells. *P < 0.05 (n = 3 to 4 mice per group).
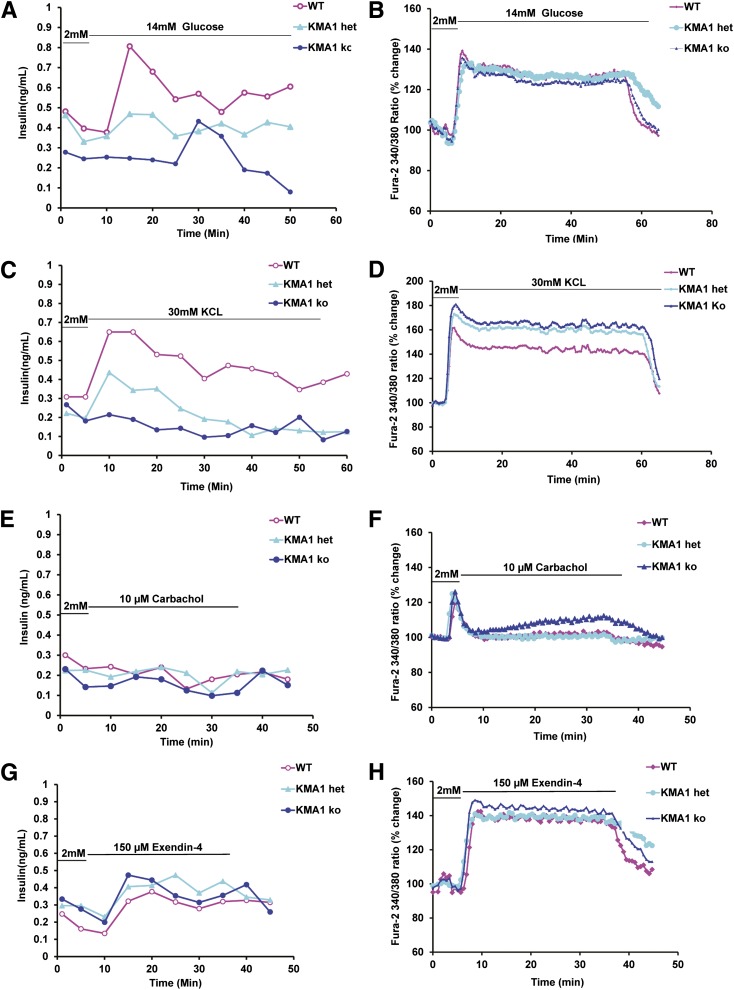
KMA1ko Islet Shows Defect in Glucose-Induced Insulin Secretion but Normal Glucose Metabolism
The islets from WT or KMA1ko mice were subjected to a microperfusion assay and stimulated with 14 mmol/L of glucose for 45 min (Fig. 6A), 30 mmol/L KCL for 60 min (Fig. 6C), 10 µmol/L carbachol (Fig. 6E), or 150 µmol/L exendin-4 (Fig. 6G) for 45 min. The perfusates were collected every 5 min and analyzed for insulin levels. Although the islets from WT mice showed a robust and expected pattern of insulin secretion, the islets from KMA1ko or KMA1het mice failed to show a similar timely increase in insulin secretion in response to the glucose challenge (Fig. 6A). Upon treatment with 30 mmol/L KCL, islets from WT mice showed a pattern of insulin release analogous to that found upon stimulation with 14 mmol/L glucose, whereas KMA1het and KMA1ko islets failed to show a similar timely increase in insulin release (Fig. 6C). In contrast, no significant increase in insulin secretion was noted upon stimulation with 10 µmol/L carbachol alone in the absence of glucose (Fig. 6E). However, islets from all three groups of mice showed a similar pattern of insulin release upon stimulation with 150 μmol/L exendin-4 (Fig. 6G).
Figure 6.
KMA1ko in pancreatic β-cells results in defective glucose-induced insulin secretion but normal calcium influx. Mice were injected with 40 mg/kg/day tamoxifen in 100 μL peanut oil for 5 consecutive days. Pancreatic islets isolated on day 7 from WT, KMA1het, and KMA1ko mice were cultured overnight and then preincubated with 2 mmol/L glucose for 5 min and stimulated with 14 mmol/L glucose (A and B), 30 mmol/L KCl for 60 min (C and D), 10 μmol/L carbachol (E and F), or 150 μmol/L exendin-4 (G and H) for 45 min. The insulin secretion (A, C, E, and G) and calcium influx (B, D, F, and H) were measured. Percentage changes in the mean value from the initial levels are expressed. A representative figure from 3 mice is shown.
We repeated stimulation of the islets from WT and KMA1ko mice by treating them alternatively with 14 mmol/L of glucose for 20 min and 2 mmol/L of glucose for 10 min. The perfusate was collected every 5 min and analyzed for insulin levels. The islets from WT mice showed a robust and expected pattern of insulin secretion (Supplementary Fig. 7A, pink line); however, the islets from KMA1ko mice failed to increase insulin secretion in response to glucose challenge (Supplementary Fig. 7A, blue line). The AUCs of the first peak (Supplementary Fig. 7B) and the second peak (Supplementary Fig. 7C) were significantly lower in islets from KMA1ko mice compared with those from WT mice. These results indicated that targeted deletion of IG20/MADD in pancreatic β-cells resulted in defective glucose-stimulated insulin secretion by those cells.
Mitochondrial energetics and intracellular calcium levels play critical roles in insulin secretion in response to glucose (40) and can serve as reliable indicators of glucose metabolism by β-cells. We tested to determine if the hyperglycemia noted in KMA1ko mice was due to a defect in β-cell glucose metabolism. Islet microperfusion assays showed no significant difference between the groups in their response to stimulation with 14 mmol/L glucose, 30 mmol/L KCl, 10 μmol/L carbachol, or 150 μmol/L exendin-4 (Fig. 6B, D, F, and H). These data showed that calcium signaling in islets from KMA1ko mice is normal. Collectively, our results indicated that IG20/MADD KO causes defective glucose-stimulated insulin secretion but does not affect glucose metabolism in pancreatic β-cells.
Discussion
The current study clearly showed that targeted deletion of IG20/MADD in mouse β-cells can result in type 2 diabetes phenotype. The nonfasting insulin levels in KMA1ko mice were comparable to those found in WT mice, but the fasting insulin levels were significantly lower. This suggested that KMA1ko mice suffered from a severe defect in insulin release in response to normal homeostatic triggers. This was substantiated by the very slow rise and lower peak in insulin levels in the KMA1ko mice after a prolonged glucose challenge and further affirmed by a lack of optimal and timely insulin secretion by β-cells from KMA1ko mice upon repeated islet perifusion with glucose (Fig. 6 and Supplementary Fig. 7). Because the total β-cell insulin content was higher and the peripheral proinsulin-to-insulin and proinsulin-to-C-peptide ratios in KMA1ko mice were similar to those found in WT mice, it was unlikely that the defect was in insulin production or processing. Collectively, these results demonstrated that the hyperglycemia seen in KMA1ko mice was a consequence of slower and suboptimal glucose-induced insulin release from β-cells.
Interestingly, KMA1ko islets failed to show normal insulin release upon stimulation with 14 mmol/L glucose or 30 µmol/L KCL. However, they did respond in a manner similar to that of KMA1het and WT islets in response to stimulation with 10 µmol/L carbachol (Fig. 6E) or 150 µmol/L of exendin-4 (Fig. 6G). These differential responses to stimulation by different secretagogues could be because glucose and KCL induce insulin release by primarily facilitating the influx of extracellular Ca2+, whereas exendin-4 and carbachol cause Ca2+ release from intracellular storage by upregulating EPAC2 and PKC, respectively (41). Although these results do not definitively show the underlying mechanism of action of MADD, they strongly suggest that the mode 1, and not the mode 2, exocytosis was impaired in the KMA1ko islets (42–44).
Although how loss of IG20/MADD function can affect insulin release is not yet known, from earlier studies we do know that IG20/MADD can act as a guanosine triphosphate exchange protein for Rab3A (45) and Rab27A (46), which play critical roles in glucose-stimulated insulin release from β-cells (47,48). It is therefore possible that the pancreatic β-cells of KMA1ko mice likely had reduced levels of “active” Rab3A and Rab27A, which resulted in inefficient glucose-induced insulin release. Nevertheless, our findings, for the first time, show the critical importance of IG20/MADD in regulating glucose-induced insulin release.
A recent report by the Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) analyzed the results from 21 different genome-wide association studies with information on fasting glucose and insulin, indices of β-cell function (HOMA-B), and IR (HOMA-IR) and identified 16 loci, including the MADD rs7944584 SNP, associated with fasting glucose and HOMA-B (25). Subsequent analyses found a highly significant association between IG20/MADD rs7944584 and insulin processing that resulted in higher proinsulin levels with no effect on insulin secretion (49,50). Interestingly, the KMA1ko mice showed no apparent reduction in β-cell mass or defect in insulin processing, suggesting that the insulin-processing defect might be associated with genetic variation(s) other than the IG20/MADD rs7944584 SNP or that the rs7944584 SNP might be affecting the function of an as-yet-unidentified gene. However, IG20/MADD rs7944584 did not increase the risk for developing type 2 diabetes in European subjects (49–51). A notable exception is a study involving a large population of Shanghai Han Chinese, which showed a clear association of IG20/MADD rs7944584 SNP with type 2 diabetes (26). Several explanations for the observed differences were suggested but failed to provide a clear insight into how the IG20/MADD rs7944584 SNP might affect glucose homeostasis in humans (52).
Localization of IG20/MADD rs7944584 SNP to an intron suggests that it might affect the transcription or alternative splicing of IG20/MADD or act on the expression of other genes. In addition, individuals who harbor this SNP also harbor other mutations, and thus, attributing the observed association with a given phenotype to this particular genetic variation is nearly impossible. This putative interplay may partly explain the association of the IG20/MADD rs7944584 SNP with related but different phenotypes in different subpopulations discussed above. Therefore, functional studies have to be done to establish a causal relationship between a particular gene and the observed phenotype (53). Our current findings clearly show that the loss of Ig20/Madd function in β-cells alone is sufficient to cause hyperglycemia and diabetes. Our findings may have clinical relevance, in that the GLP-1 analog may be used to treat IG20/MADD rs7944584 SNP-associated hyperglycemia. Further studies are required to fully elucidate the role of IG20/MADD in clinical diabetes and its potential to serve as a therapeutic target for type 2 diabetes.
Article Information
Acknowledgments. The authors thank Dr. Jianzhong Qin for his help with some animal studies.
Funding. This work was supported in part by National Institutes of Health grant R01-DK-091526 to J.O.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. L.-c.L. designed the experiment, researched data, and drafted the manuscript. Y.W., R.C., C.S.H., Z.L., L.Q., and Q.W. researched data. A.V.M. contributed to discussion. J.O. contributed to discussion and reviewed and edited the manuscript. B.S.P. conceived the project, designed the experiments, reviewed the results, and wrote the manuscript. B.S.P. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db13-0707/-/DC1.
References
- 1.Goto Y, De Silva MG, Toscani A, Prabhakar BS, Notkins AL, Lan MS. A novel human insulinoma-associated cDNA, IA-1, encodes a protein with “zinc-finger” DNA-binding motifs. J Biol Chem 1992;267:15252–15257 [PubMed] [Google Scholar]
- 2.Cunningham SJ. Cloning and Characterization of a Novel cDNA Isolated from Human Beta Cells Galveston, TX, University of Texas Medical Branch, Graduate School of Biomedical Sciences, 1996 [Google Scholar]
- 3.Al-Zoubi AM, Efimova EV, Kaithamana S, et al. Contrasting effects of IG20 and its splice isoforms, MADD and DENN-SV, on tumor necrosis factor alpha-induced apoptosis and activation of caspase-8 and -3. J Biol Chem 2001;276:47202–47211 [DOI] [PubMed] [Google Scholar]
- 4.Chow VT, Lim KM, Lim D. The human DENN gene: genomic organization, alternative splicing, and localization to chromosome 11p11.21-p11.22. Genome 1998;41:543–552 [DOI] [PubMed] [Google Scholar]
- 5.Schievella AR, Chen JH, Graham JR, Lin LL. MADD, a novel death domain protein that interacts with the type 1 tumor necrosis factor receptor and activates mitogen-activated protein kinase. J Biol Chem 1997;272:12069–12075 [DOI] [PubMed] [Google Scholar]
- 6.Mulherkar N, Ramaswamy M, Mordi DC, Prabhakar BS. MADD/DENN splice variant of the IG20 gene is necessary and sufficient for cancer cell survival. Oncogene 2006;25:6252–6261 [DOI] [PubMed] [Google Scholar]
- 7.Li LC, Sheng JR, Mulherkar N, Prabhakar BS, Meriggioli MN. Regulation of apoptosis and caspase-8 expression in neuroblastoma cells by isoforms of the IG20 gene. Cancer Res 2008;68:7352–7361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurada BR, Li LC, Mulherkar N, Subramanian M, Prasad KV, Prabhakar BS. MADD, a splice variant of IG20, is indispensable for MAPK activation and protection against apoptosis upon tumor necrosis factor-alpha treatment. J Biol Chem 2009;284:13533–13541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Efimova E, Martinez O, Lokshin A, Arima T, Prabhakar BS. IG20, a MADD splice variant, increases cell susceptibility to gamma-irradiation and induces soluble mediators that suppress tumor cell growth. Cancer Res 2003;63:8768–8776 [PubMed] [Google Scholar]
- 10.Efimova EV, Al-Zoubi AM, Martinez O, et al. IG20, in contrast to DENN-SV, (MADD splice variants) suppresses tumor cell survival, and enhances their susceptibility to apoptosis and cancer drugs. Oncogene 2004;23:1076–1087 [DOI] [PubMed] [Google Scholar]
- 11.Murakami-Mori K, Mori S, Bonavida B, Nakamura S. Implication of TNF receptor-I-mediated extracellular signal-regulated kinases 1 and 2 (ERK1/2) activation in growth of AIDS-associated Kaposi’s sarcoma cells: a possible role of a novel death domain protein MADD in TNF-alpha-induced ERK1/2 activation in Kaposi’s sarcoma cells. J Immunol 1999;162:3672–3679 [PubMed] [Google Scholar]
- 12.Brown TL, Howe PH. MADD is highly homologous to a Rab3 guanine-nucleotide exchange protein (Rab3-GEP). Curr Biol 1998;8:R191. [PubMed] [Google Scholar]
- 13.Levivier E, Goud B, Souchet M, Calmels TP, Mornon JP, Callebaut I. uDENN, DENN, and dDENN: indissociable domains in Rab and MAP kinases signaling pathways. Biochem Biophys Res Commun 2001;287:688–695 [DOI] [PubMed] [Google Scholar]
- 14.Li LC, Jayaram S, Ganesh L, et al. Knockdown of MADD and c-FLIP overcomes resistance to TRAIL-induced apoptosis in ovarian cancer cells. Am J Obstet Gynecol 2011;205:362 e312–e325 [DOI] [PMC free article] [PubMed]
- 15.Subramanian M, Pilli T, Bhattacharya P, et al. Knockdown of IG20 gene expression renders thyroid cancer cells susceptible to apoptosis. J Clin Endocrinol Metab 2009;94:1467–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prabhakar BS, Mulherkar N, Prasad KV. Role of IG20 splice variants in TRAIL resistance. Clin Cancer Res 2008;14:347–351 [DOI] [PubMed] [Google Scholar]
- 17.Li L, Jayarama S, Pilli T, Qian L, Pacini F, Prabhakar BSP. Down-modulation of expression, or dephosphorylation, of IG20/MADD in tumor necrosis factor-related apoptosis-inducing ligand-resistant thyroid cancer cells makes them susceptible to treatment with this ligand. Thyroid 2013;23:70–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lim KM, Yeo WS, Chow VT. Antisense abrogation of DENN expression induces apoptosis of leukemia cells in vitro, causes tumor regression in vivo and alters the transcription of genes involved in apoptosis and the cell cycle. Int J Cancer 2004;109:24–37 [DOI] [PubMed] [Google Scholar]
- 19.Li P, Jayarama S, Ganesh L, et al. Akt-phosphorylated mitogen-activated kinase-activating death domain protein (MADD) inhibits TRAIL-induced apoptosis by blocking Fas-associated death domain (FADD) association with death receptor 4. J Biol Chem 2010;285:22713–22722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyoshi J, Takai Y. Dual role of DENN/MADD (Rab3GEP) in neurotransmission and neuroprotection. Trends Mol Med 2004;10:476–480 [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Zhou L, Miller CA. A splicing variant of a death domain protein that is regulated by a mitogen-activated kinase is a substrate for c-Jun N-terminal kinase in the human central nervous system. Proc Natl Acad Sci U S A 1998;95:2586–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Del Villar K, Miller CA. Down-regulation of DENN/MADD, a TNF receptor binding protein, correlates with neuronal cell death in Alzheimer’s disease brain and hippocampal neurons. Proc Natl Acad Sci U S A 2004;101:4210–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka M, Miyoshi J, Ishizaki H, et al. Role of Rab3 GDP/GTP exchange protein in synaptic vesicle trafficking at the mouse neuromuscular junction. Mol Biol Cell 2001;12:1421–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlüter OM, Schmitz F, Jahn R, Rosenmund C, Südhof TC. A complete genetic analysis of neuronal Rab3 function. J Neurosci 2004;24:6629–6637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dupuis J, Langenberg C, Prokopenko I, et al. DIAGRAM Consortium. GIANT Consortium. Global BPgen Consortium. Anders Hamsten on behalf of Procardis Consortium. MAGIC Investigators New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 2010;42:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu C, Zhang R, Wang C, et al. Variants from GIPR, TCF7L2, DGKB, MADD, CRY2, GLIS3, PROX1, SLC30A8 and IGF1 are associated with glucose metabolism in the Chinese. PLoS ONE 2010;5:e15542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersson KB, Winer LH, Mørk HK, Molkentin JD, Jaisser F. Tamoxifen administration routes and dosage for inducible Cre-mediated gene disruption in mouse hearts. Transgenic Res 2010;19:715–725 [DOI] [PubMed] [Google Scholar]
- 28.Song Y, Manson JE, Tinker L, et al. Insulin sensitivity and insulin secretion determined by homeostasis model assessment and risk of diabetes in a multiethnic cohort of women: the Women’s Health Initiative Observational Study. Diabetes Care 2007;30:1747–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallace TM, Levy JC, Matthews DR. Use and abuse of HOMA modeling. Diabetes Care 2004;27:1487–1495 [DOI] [PubMed] [Google Scholar]
- 30.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985;28:412–419 [DOI] [PubMed] [Google Scholar]
- 31.Adewola AF, Lee D, Harvat T, et al. Microfluidic perifusion and imaging device for multi-parametric islet function assessment. Biomed Microdevices 2010;12:409–417 [DOI] [PubMed] [Google Scholar]
- 32.Szot GL, Koudria P, Bluestone JA. Murine pancreatic islet isolation. J Vis Exp 2007:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kenny AJ. Extractable glucagon of the human pancreas. J Clin Endocrinol Metab 1955;15:1089–1105 [DOI] [PubMed] [Google Scholar]
- 34.Matsui K, Oda T, Nishizawa E, et al. Pancreatic function of spontaneously diabetic torii rats in pre-diabetic stage. Exp Anim 2009;58:363–374 [DOI] [PubMed] [Google Scholar]
- 35.Saccomanno K, Taccagni G, Bosi E, Preti P, Dozio N, Cantaboni A. Immunoelectron microscopy: a new method for detection of insulin antibodies. J Histochem Cytochem 1993;41:1233–1239 [DOI] [PubMed] [Google Scholar]
- 36.Bendayan M, Garzon S. Protein G-gold complex: comparative evaluation with protein A-gold for high-resolution immunocytochemistry. J Histochem Cytochem 1988;36:597–607 [DOI] [PubMed] [Google Scholar]
- 37.Roth J. Subcellular organization of glycosylation in mammalian cells. Biochim Biophys Acta 1987;906:405–436 [DOI] [PubMed] [Google Scholar]
- 38.Roth J. Post-embedding cytochemistry with gold-labelled reagents: a review. J Microsc 1986;143:125–137 [DOI] [PubMed] [Google Scholar]
- 39.Wicksteed B, Brissova M, Yan W, et al. Conditional gene targeting in mouse pancreatic β-cells: analysis of ectopic Cre transgene expression in the brain. Diabetes 2010;59:3090–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacDonald PE, Joseph JW, Rorsman P. Glucose-sensing mechanisms in pancreatic beta-cells. Philos Trans R Soc Lond B Biol Sci 2005;360:2211–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 2006;444:840–846 [DOI] [PubMed] [Google Scholar]
- 42.Chepurny OG, Kelley GG, Dzhura I, et al. PKA-dependent potentiation of glucose-stimulated insulin secretion by Epac activator 8-pCPT-2′-O-Me-cAMP-AM in human islets of Langerhans. Am J Physiol Endocrinol Metab 2010;298:E622–E633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kasai H, Suzuki T, Liu TT, Kishimoto T, Takahashi N. Fast and cAMP-sensitive mode of Ca(2+)-dependent exocytosis in pancreatic beta-cells. Diabetes 2002;51(Suppl. 1):S19–S24 [DOI] [PubMed] [Google Scholar]
- 44.Tian Y, Laychock SG. Protein kinase C and calcium regulation of adenylyl cyclase in isolated rat pancreatic islets. Diabetes 2001;50:2505–2513 [DOI] [PubMed] [Google Scholar]
- 45.Coppola T, Perret-Menoud V, Gattesco S, et al. The death domain of Rab3 guanine nucleotide exchange protein in GDP/GTP exchange activity in living cells. Biochem J 2002;362:273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshimura S, Gerondopoulos A, Linford A, Rigden DJ, Barr FA. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J Cell Biol 2010;191:367–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kasai K, Ohara-Imaizumi M, Takahashi N, et al. Rab27a mediates the tight docking of insulin granules onto the plasma membrane during glucose stimulation. J Clin Invest 2005;115:388–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yaekura K, Julyan R, Wicksteed BL, et al. Insulin secretory deficiency and glucose intolerance in Rab3A null mice. J Biol Chem 2003;278:9715–9721 [DOI] [PubMed] [Google Scholar]
- 49.Strawbridge RJ, Dupuis J, Prokopenko I, et al. DIAGRAM Consortium. GIANT Consortium. MuTHER Consortium. CARDIoGRAM Consortium. C4D Consortium Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes 2011;60:2624–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner R, Dudziak K, Herzberg-Schäfer SA, et al. Glucose-raising genetic variants in MADD and ADCY5 impair conversion of proinsulin to insulin. PLoS ONE 2011;6:e23639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boesgaard TW, Grarup N, Jørgensen T, Borch-Johnsen K, Hansen T, Pedersen O, Meta-Analysis of Glucose and Insulin-Related Trait Consortium (MAGIC) Variants at DGKB/TMEM195, ADRA2A, GLIS3 and C2CD4B loci are associated with reduced glucose-stimulated beta cell function in middle-aged Danish people. Diabetologia 2010;53:1647–1655 [DOI] [PubMed] [Google Scholar]
- 52.Ingelsson E, Langenberg C, Hivert MF, et al. MAGIC investigators Detailed physiologic characterization reveals diverse mechanisms for novel genetic Loci regulating glucose and insulin metabolism in humans. Diabetes 2010;59:1266–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Billings LK, Florez JC. The genetics of type 2 diabetes: what have we learned from GWAS? Ann N Y Acad Sci 2010;1212:59–77 [DOI] [PMC free article] [PubMed] [Google Scholar]