Abstract
Molecular genetic testing informs diagnosis, prognosis, and risk assessment for patients and their family members. Recent advances in low-cost, high-throughput DNA sequencing and computing technologies have enabled the rapid expansion of genetic test content, resulting in dramatically increased numbers of DNA variants identified per test. To address this challenge, our laboratory has developed a systematic approach to thorough and efficient assessments of variants for pathogenicity determination. We first search for existing data in publications and databases including internal, collaborative and public resources. We then perform full evidence-based assessments through statistical analyses of observations in the general population and disease cohorts, evaluation of experimental data from in vivo or in vitro studies, and computational predictions of potential impacts of each variant. Finally, we weigh all evidence to reach an overall conclusion on the potential for each variant to be disease-causing. In this report, we highlight the principles of variant assessment, address the caveats and pitfalls, and provide examples to illustrate the process. By sharing our experience and providing a framework for variant assessment, including access to a freely available customizable tool, we hope to help move towards standardized and consistent approaches to variant assessment.
Keywords: Clinical Interpretation, Gain of Function (GOF), Genetic Variant, Loss of Function (LOF), Next-Generation Sequencing (NGS), Sequence Analysis, Variant Assessment, Variant of Uncertain Significance (VUS)
Introduction
Molecular genetic testing informs medical decision-making in the diagnosis of symptomatic individuals, in the prediction of disease risk, in reproductive genetic counseling, and in determining pharmacogenetic profiles for treatment guidance. Until recently, the majority of clinically available molecular genetic tests have either analyzed known DNA variants, such as cystic fibrosis carrier screening panels (1), or sequenced the coding regions and splicing boundaries of a limited set of well-known disease-associated genes. Recent technological advances in low-cost, high-throughput sequencing and computing have enabled testing for targeted panels of >100 disease-area genes, as well as exomes and genomes. While these next-generation sequencing (NGS) technologies have increased diagnostic sensitivity (2, 3), the number of genetic variants with uncertain clinical significance (VUS) per test has also increased. For example, expanding testing for dilated cardiomyopathy from 5 to 46 genes in our laboratory resulted in a 3-fold increase in clinical sensitivity and an even more dramatic increase in inconclusive cases, many with multiple VUSs. In addition, exome and genome sequencing tests add a new layer of complexity, as the genes interrogated may not have been carefully assessed for their role in disease until variants are identified.
Although molecular genetic testing has a unique place in the diagnosis, management, and prevention of genetic disorders, the field is compromised by the absence of a standard, comprehensive, and efficient variant assessment protocol approved and shared by the community. However, guidelines for variant interpretation are available and being updated as variant-level knowledge expands, including those from the American College of Medical Genetics and Genomics (ACMG) (4–7). To supplement these guidelines and capture the evolving state of the field, we developed a variant assessment tool (VAT) that systematically evaluates multiple parameters for each variant and facilitates the capture of new knowledge in the literature and databases (Supplementary Information).
The clinical significance of a variant in relation to a disease or phenotype can be determined by answering three core questions. 1) Does the variant alter the function of the gene (i.e. loss-of-function (LOF) or gain-of-function (GOF))? 2) Can the functional change result in disease or another phenotype? 3) Is the associated disease or phenotype relevant to the specific clinical condition present in the tested individual? In some cases variant assessment in a clinical laboratory may only be focused on the first two questions; however, for maximal benefit to the patient, a careful assessment of the third dimension can be highly informative, particularly for VUSs. Here, we share our decade-long experience with variant assessment, highlighting key points and challenges of clinical interpretation. We have evaluated 245 genes associated with 53 diseases while testing greater than 22,000 cases. We have iteratively developed a framework through clinical assessments of over 17,000 variants, including >8,000 that have been validated and reported in patients. Using our semi-automated tool, it takes on average 40 minutes to perform a thorough evidence-based clinical variant assessment for variants being returned after disease-targeted testing. When assessments with literature are excluded, the average time decreases to 22 minutes. An overview of the process is presented in Figure 1 and useful online resources are presented in Table 1. In addition, the approaches described below refer to the provided VAT available for download through the Supplemental Information.
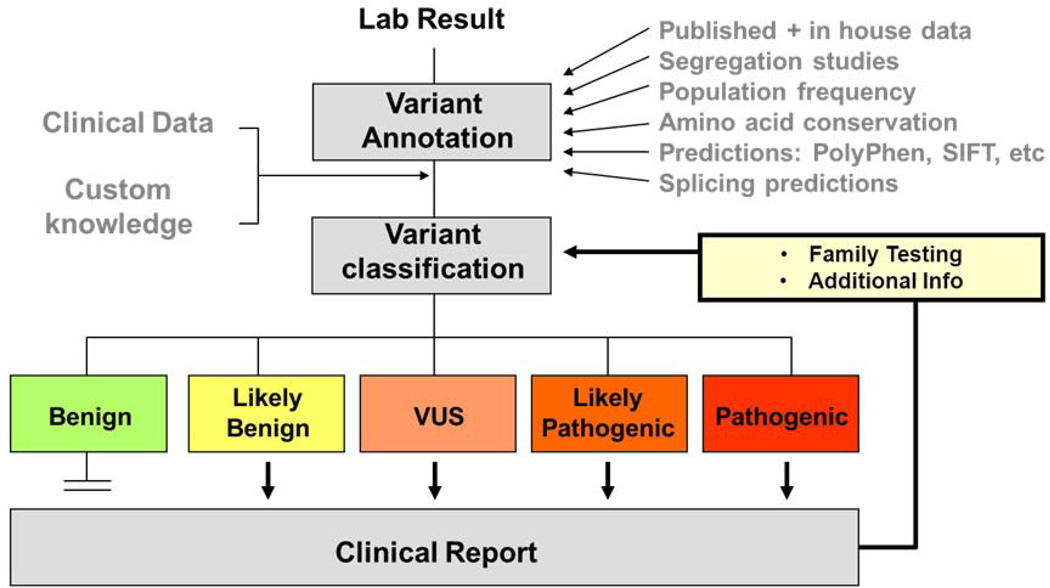
Figure 1.
Variant assessment workflow. Genetic variants identified by laboratory testing are annotated with information from various sources including publications, computational prediction algorithms, and public, collaborative and internal databases. After evaluation of all pertinent information in conjunction with patient specific clinical and family information, a professionally trained individual will classify the variant into one of the five clinical categories and combine all variants for a clinical report.
Table 1.
Useful online resources for variant assessment
Linking genes to disease
As a first step in variant assessment, it is necessary to determine which disease phenotypes are associated with a gene and which types of variation may result in clinically relevant consequences. This includes the types of variants that are known to cause disease in the gene (truncating/LOF, non-truncating, etc.), the inheritance patterns observed for variants in the gene, the protein domains that are implicated in disease, and any genotype-phenotype correlations described.
To characterize disease phenotypes, it is important to review the literature for common clinical features as well as phenotypic variation among affected individuals. Large cohort studies may provide expressivity, age-of-onset, penetrance, and prevalence information, while detailed reports of families with multiple affected individuals help determine the mode of inheritance and strength of association. Comparison of the variant spectrum in affected individuals against that in the general population may be useful in identifying the types of mutations that are disease-causing. For instance, heterozygous LOF variants in MYBPC3 have been reported in 14% (311/2302) of patients with hypertrophic cardiomyopathy (HCM) tested in our laboratory, but in <0.1% (6/6500) of the general population per the NHLBI Exome Sequencing Project (ESP), supporting that LOF MYBPC3 variants are a common mechanism in HCM (8). However, external information must be carefully vetted. An apparent frameshift variant in MYBPC3 NM_000256:c.2854_2858del reported to occur in 7% of the general population in ESP is likely a technical artifact, as we have never observed it sequencing the region by NGS and/or Sanger in over 2,000 cases.
Different types of variants in the same gene may be associated with distinct phenotypes or inheritance patterns. For example, missense GOF variants in PTPN11 cause RASopathies, such as Noonan syndrome, whereas LOF variants lead to an entirely different phenotype, a cartilage tumor syndrome (metachondromatosis) characterized by enchondromas and exostoses (9). Certain missense variants in TECTA lead to autosomal dominant hearing loss (10), whereas LOF variants result in autosomal recessive hearing loss (11). Similarly, variants in different regions or domains of a gene may cause different phenotypes (10, 12). Important questions to consider when analyzing gene-disease associations and specific variants within a gene can be found in Table 2.
Table 2.
Variant assessment checklist
Gene-level information
|
Variant validation
|
Genetic data
|
Functional data
|
Computational data
|
Validating variants to ensure accuracy
As test complexity has increased, so has the need to ensure variants identified and included on a clinical report are technically accurate. This is especially important for sequencing tests where the variants are not part of a pre-defined list. It is essential to review raw assay results (e.g. chromatographs of Sanger sequencing traces or NGS reads) to verify the variants and their nomenclature. Prior to variant assessment, the laboratory should predefine the genome build, gene name, and reference transcript that will be used in interpretation and reporting, along with a method linking genomic coordinates to cDNA and amino acid level annotations. Laboratories should also be aware of homologous and repetitive regions particularly from pseudogenes and segmental duplications, which may result in lack of coverage, alignment difficulties, and incorrect variant calls. These steps will enable validation of the correct variant call, zygosity and nomenclature according to the Human Genome Variation Society (HGVS) guidelines (13). Validation information is captured in the “Variant” tab of the VAT.
Because standards for variant nomenclature have only recently been widely adopted and still do not address all modifications, variants may have differing names in publications and databases. The amino acid position may not be numbered according to the start codon to be consistent with current recommendations. For example, TTR variants were originally numbered according to the position within the mature protein lacking the 20 amino acid signal peptide (14). Partial cloning of a gene may have led to inconsistent nomenclature in early publications (15, 16). Nucleotide gene numbering may have been determined using the transcription start site instead of the translation start site, which was particularly challenging given transcriptional start site variability. Furthermore, for many small insertions and deletions, it is not possible to determine the exact location of the inserted or deleted base(s). This can lead to multiple potential names for the same variant, highlighting the importance for following standard HGVS nomenclature rules such as attributing alternations within a repetitive stretch to the most 3’ possible position. Legacy terms and alternative aliases are useful to maintain association with the correctly named variant both to facilitate searching the literature and databases, as well as communicating with ordering physicians and other laboratories.
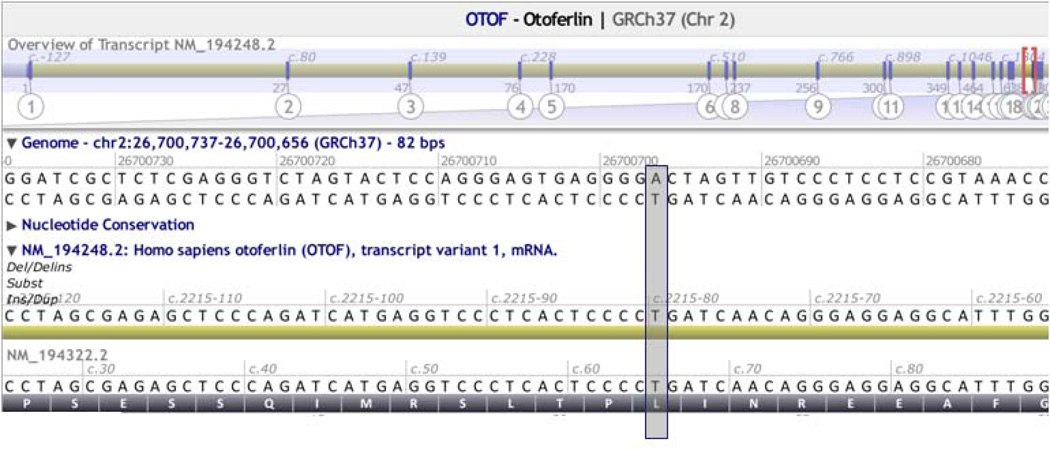
Genes may have multiple transcripts, some of which are tissue-specific and associated with distinct phenotypes. For example, the shorter USH1C transcript (NM_005709) is expressed in both the retina and inner ear, whereas the longer transcript (NM_153676) is expressed exclusively in the inner ear (17). Accordingly, variants in exons common to both transcripts lead to Usher syndrome type 1C, characterized by profound deafness, retinitis pigmentosa, and vestibular dysfunction, whereas variants in exons unique to NM_153676 lead to non-syndromic hearing loss (18). Variants should be reported according to a single primary transcript. The reported reference is typically the major transcript unless a more severe impact is predicted on an alternative transcript, in which case the variant should be defined according to the alternative transcript, noting an alias to the primary transcript (Figure 2A).
Figure 2.
A. Reference transcript selection. Two transcripts for OTOF are shown: NM_194248, the longest transcript selected as the primary transcript, and NM_194322, a shorter alternate transcript. Position g.26799794 (grey box) is non-coding in NM_194248 (c.2250–80) but coding in NM_194322 (c.66); therefore NM_194322 should be selected while evaluating this variant.
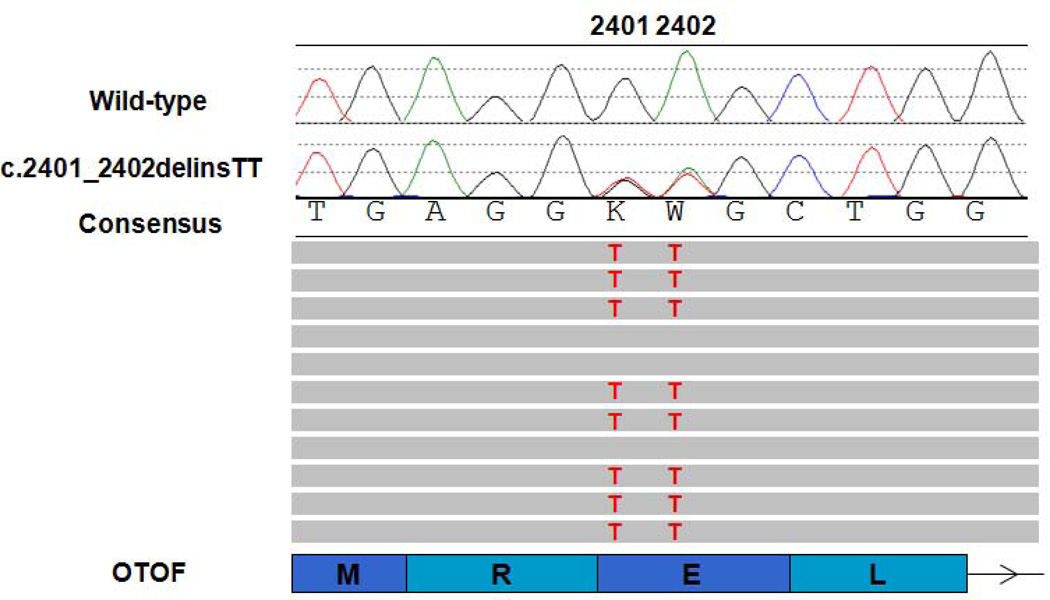
Figure 2B. Phasing multiple variants. Two variants are present at positions c.2401 and c.2402 in OTOF (NM_194248). The top traces show the chromatographs from Sanger sequencing with the consensus reference sequence shown underneath. Representative aligned NGS reads are shown below. Grey bars represent reference sequence with variants highlighted in red. The bottom schematic shows associated OTOF coding exons (rectangles) and the reference amino acid sequence. The arrow indicates the 5’ to 3’ direction. The c.2401G>T (p.Glu801*) is listed in dbSNP (rs75624587) and ESP as a nonsense variant but with an allele frequency of 10% in African Americans. However, NGS reads reveal that the variants are in cis and should therefore be named c.2401_2402delinsTT (p.Glu891Leu).
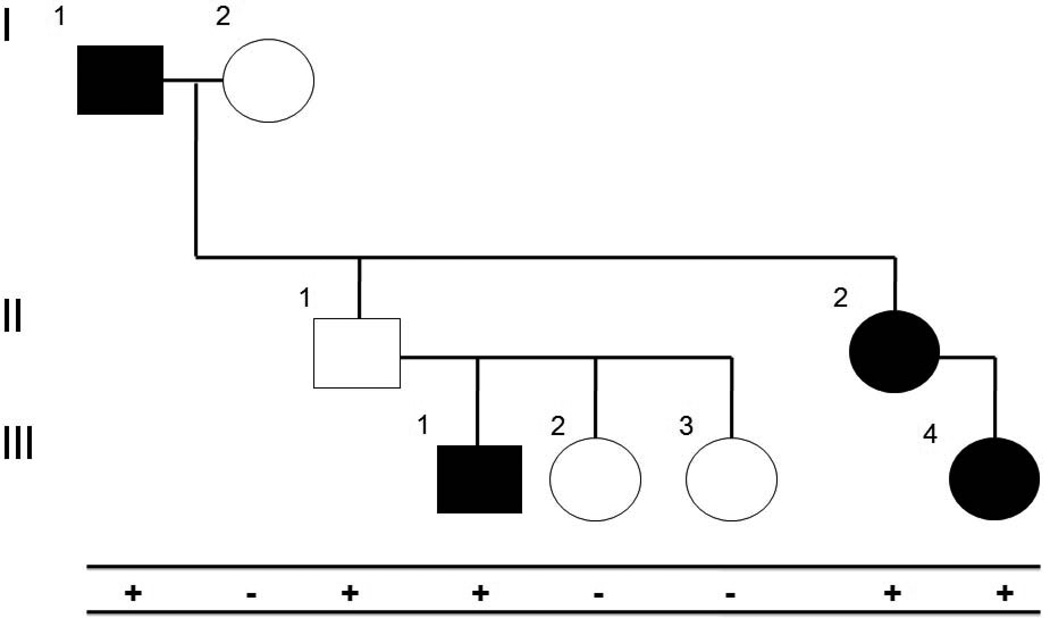
Figure 2C. Segregation analysis with incomplete penetrance. A family with hypertrophic cardiomyopathy is shown. Affected individuals are indicated by filled squares (males) or circles (females). Mutation-positive individuals are indicated by a “+”, while mutation-negative individuals are indicated by a “−“. All mutation-positive individuals are affected, with the exception of individual II-1. Because HCM can display reduced penetrance, individual II-1 would not be considered a non-segregation.
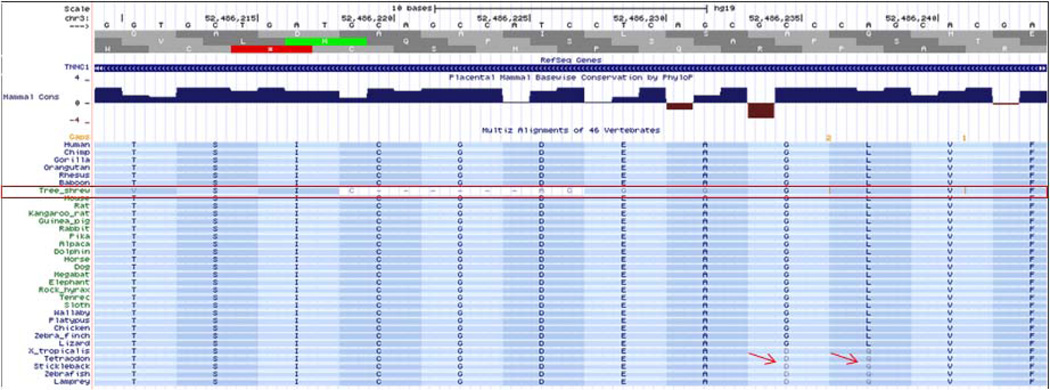
Figure 2D. Conservation based on multiple species alignment. An example of alignment of TNNC1 in UCSC Genome Browser is shown. Tree shrew sequence shows poor alignment (red box). Arrows point to non-conserved residues.
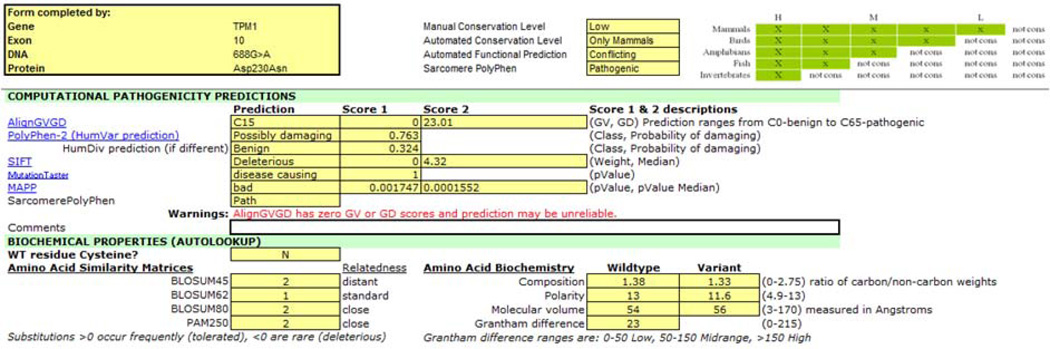
Figure 2E. Conflicting computational predictions of a missense variant. The results of multiple computational tools are captured in the VAT. They provide conflicting predictions for the NM_000366:c.688G>A (p.Asp230Asn) variant in TPM1, suggesting that at least some tools are not reliable.
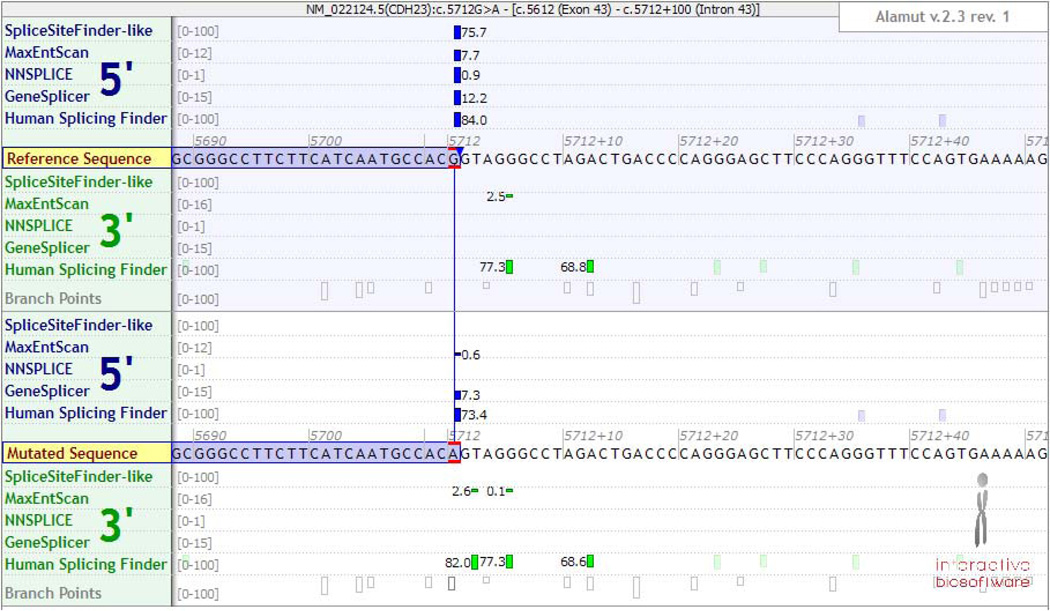
Figure 2F. Predicted splicing effect of a coding variant. Splicing prediction tools indicate that the NM_022124:c.5712G>A variant in CDH23, which affects the last base in exon 43, may impact splicing. However, not all programs agree in the potential effect on splicing, and they cannot predict whether it would lead to exon skipping, intron retention or use of cryptic splice sites.
When multiple variants in the same gene are identified, the phase of the variants (i.e. on the same chromosome – in cis – or on homologous chromosomes – in trans) may influence the interpretation, especially for autosomal recessive traits. If variants are within the same NGS fragment, the phase may be determined without parental samples (Figure 2B).
Collecting evidence to determine the likelihood of pathogenicity
Once the variant call is validated, literature, variant databases, and population control studies should be evaluated. This information is used to determine whether and under what context the variant has been previously observed. The population, literature and internal case data are captured in the “Control_Freq”, “DB” and “Publ+Internal_data” tabs of the VAT.
Recent large-scale population studies such as the NHLBI Exome Sequencing Project (9), the 1000 Genomes Project (19), the ClinSeq Project (20) and others found in dbSNP (21) have catalogued large amounts of sequence variation (Table 1). Because these populations may include presymptomatic individuals with late onset diseases, asymptomatic individuals with low penetrance diseases or younger than typical age-of-onset, and heterozygous carriers of recessive traits, variants should not be assumed benign simply because of their presence in large population studies. Information on affected individuals with the variant can be obtained from internal and public variant databases (e.g., ClinVar, HGMD (22) or locus-specific databases), as well as from the literature. Public variant databases are of varying quality and may be outdated or contain contradictory data. Recent studies have demonstrated a large number of false-positive variants incorrectly identified as clinically relevant in these databases (23–27). Therefore, databases available today should be used to identify relevant primary literature rather than directly reference a variant classification.
For Mendelian disorders, the pathogenicity of a variant can be ruled out if its frequency in the general population exceeds what can be accounted for by inheritance pattern, age-of-onset, prevalence, penetrance, and heterogeneity. Large sample sizes without selection bias towards individuals with disease phenotypes are required to achieve confidence in estimating the population allele frequency. Moreover, disease prevalence is not always known, accurate, or applicable across all populations. Because one affected allele is sufficient to cause an autosomal dominant trait, a pathogenic allele must present at a frequency lower than the disease prevalence in the general population. HCM is primarily an autosomal dominant condition occurring in 1 in 500 individuals (1/1000 chromosomes or 0.1% allele frequency) (28). We consider a variant likely benign if the allele frequency is >0.3% which is a conservative 1.5 times above the highest frequency expected even if penetrance was only 50% and the disease was due to one pathogenic variant. In contrast, both paternal and maternal alleles need to be affected to cause an autosomal recessive disorder. The heterozygous carrier frequency of any pathogenic allele must be less than twice the square root of the disease prevalence (which is the hypothetical allele frequency if only one disease allele accounts for all cases). For example, the prevalence of congenital hearing loss with a genetic etiology is roughly 1 in 1,000 and half of these cases are due to GJB2 variants. Therefore, the estimated prevalence of GJB2-related hearing loss is 1 in 2,000. Accordingly, pathogenic variants in GJB2 are expected to occur no more than 4% in the general population. It is not surprising that the carrier frequency for the c.35delG variant in GJB2 could be as high as 2% (29). Population data pertaining to a specific ethnic composition are particularly useful. The 1000 Genomes Project has revealed many variants common in certain ethnic groups, but rare in general (30). If a subpopulation does not have an increased occurrence of the associated disease and affected individuals are not under-diagnosed, variant classification based on the allele frequency in the subpopulation can be applied more broadly.
While a high allele frequency in the general population may rule out pathogenicity of a variant for a rare disorder, absence or a very low frequency of a variant in the broad population cannot be used to assume pathogenicity. While coding variants below 1% allele frequency in the seven populations examined by the 1000 Genomes Project are enriched for functional variants (31), lack of a variant from population datasets cannot be used to assume absence from the population unless it is determined that the study technically interrogated the position sufficiently to rule out a potential false-negative result. A variant is statistically more likely pathogenic if it occurs in affected individuals more than expected by chance. The likelihood of random occurrence can be calculated as the probability of co-incidence of rare events, as the logarithm of odds (LOD) score through linkage analysis or as p-values through case-control studies using a Fisher’s exact or chi-square test. Low probabilities of co-incidence statistically demonstrate non-random occurrences of the variant in affected individuals.
The presence of de novo variants may support disease association due to their rarity. The de novo point mutation rate is ~1 per exome (32), consistent with an average rate of 1.2×10−8 per nucleotide per generation in human genome (33). Therefore, confirmed de novo status of a variant in a disease-associated gene strongly increases the likelihood of pathogenicity in rare conditions when the patient’s disease is de novo and matches the associated phenotypes. Testing of biological parents and excluding the possibilities of non-paternity and sample swap (e.g. genotyping with microsatellite markers) are necessary for confirmation of de novo variants. Similarly, for rare recessive disorders, if a rare variant is confirmed in trans with another pathogenic variant in the same disease gene, it is more likely pathogenic.
Significant co-segregation of a variant with disease provides strong genetic linkage evidence to support pathogenicity. Linkage analysis programs can be used to calculate the LOD scores, but a simple count of informative segregations can provide an estimate. As a rule of thumb, 10 informative segregations would achieve a LOD score > 3.0, necessary to establish linkage between a genetic locus and a disease. For established disease genes, given the a priori probability of disease association, fewer informative segregations may be acceptable in combination with other supporting evidence. Because genotype-phenotype correlation may be masked by incomplete penetrance, variable expressivity, and late age-of-onset in genotype-positive individuals, unaffected family members should not contribute segregation information under these circumstances (34) (Figure 2C). Additional evidence may still be required to establish pathogenicity, as any variant in linkage disequilibrium with the causative variant will segregate with the disease. For example, the Ile148Thr variant in CFTR was removed from the original cystic fibrosis carrier-testing panel because it was later determined its association with the disease was due to tight linkage with another pathogenic variant (35, 36).
Functional evidence that links the variant to disease phenotypes is important to establish causality. However, this information is typically unavailable for individual variants in routine diagnostic testing. When studies regarding a specific variant have been published, it is important to determine the type of assay used and whether the results and conclusions drawn are applicable to the mechanism and presentation of the disease. In general, direct assays on patient tissues provide the strongest functional evidence because they reveal true biological consequences of a variant within a human individual. In vivo studies in mammals may add more evidence at the system level. In vitro studies can be useful, especially in cases where the in vitro assay directly tests an established molecular mechanism of disease (e.g. structural proteins or ion channels (37)), but may not accurately represent the biological environment or directly prove causation of disease.
In summary, population, statistical and functional evidence need to be carefully evaluated to determine the clinical significance of a variant. Table 2 lists some important considerations when collecting this data.
Predicting disease association using bioinformatics tools
If the evidence for disease association from existing data is not strong or the mechanism of gene function is unclear, a number of bioinformatics tools may be used to predict the possible impact of the variant on the gene or protein. Computational predictions are generally based on the type of change, the domain structure, sequence conservation, and biochemical properties of the affected amino acid residues. Computational information is captured in the “Conserv_Biochem” and “Splicing” tabs of the VAT.
At both the nucleotide and amino acid level, sequence conservation may indicate regions and positions of functional importance, as negative selection removes changes that are deleterious to proper biological function, leading to high evolutionary conservation (38). Computationally derived alignments can indicate when a specific sequence is important to the underlying gene or protein function (Figure 2D). Conversely, presence of the variant amino acid in other species, particularly primates and other mammals, may indicate a tolerance to that change.
Many algorithms are available to classify missense substitutions and potential splicing alterations (Figure 2E). Use of multiple prediction algorithms is recommended. Because most of the programs use similar underlying datasets and assumptions, they should not be regarded as independent evidence, though some may include additional features. The datasets used for training the algorithms are mostly from non-clinical grade databases that may not be accurate or comprehensive. Disease specific algorithms can be applied to a specific set of genes with significantly enhanced performance (39, 40), though these are limited in availability. Predicting the effect of variants occurring near the splice region can be particularly challenging as it is often unclear what kind of abnormal transcript may be produced (Figure 2E).
In summary, although computational predictions are useful in guiding classification, they are not able to determine or rule out pathogenicity. Table 2 addresses specific questions for consideration when examining computational data.
Combining multiple lines of evidence to reach an overall interpretation
Final interpretation of the clinical significance of a variant requires examination of all the available evidence. While some data can be strong enough to determine or rule out pathogenicity, most information only moderately influences final conclusions and is valuable in combination. Table 3 lists some of the possible types of evidence available and the general weight we assign to their ability to indicate a pathogenic or benign assertion.
Table 3.
Strength of evidence for variant assessment*
| Evidence leading to a patdogenic assertion | Strengtd of evidence |
| Significant segregation in affected family members (LOD >2) | +++ |
| Confirmed de novo inheritance in relevant disease-associated gene | +++ |
| In vivo data from mammalian model organisms suggest an impact on function | ++ |
| Case-control studies significantly associate the variant to disease | ++ |
| Nucleotide and amino acid strongly conserved in distantly related species | ++ |
| In vitro data from recombinant DNA constructs or proteins suggest an impact on function | + |
| Variant is present in trans with an established pathogenic variant in recessive disease | + |
| Variant is rare or absent in large population studies | + |
| Computational tools predict an impact on function and/or splicing | + |
| Evidence leading to a benign assertion | Strength of evidence |
| Frequency of variant in general population is too high to cause disease, accounting for penetrance and prevalence of disease | +++ |
| Variant is present in unaffected adults in fully penetrant early onset dominant disease | ++ |
| Variant is present in a homozygous state in unaffected adults in fully penetrant early onset recessive disease | ++ |
| Variant is absent in affected family members (non-segregation) after ruling out potential phenocopy | ++ |
| Variant amino acid is present at this position in multiple mammalian species | ++ |
| Adequately powered case-control studies show no association to disease | + |
| In vitro/in vivo assay for variant does not implicate effect on function or disease | + |
| Variant type is not part of known disease mechanism | + |
| Computational tools predict no impact on splicing or function | + |
Strength of evidence is based on the correlation of the type of evidence with the accuracy of variant classification. Direct evidence with sufficient statistical power or from proper biological experiments is deemed “strong”, while supportive studies or in silico predictions that cannot prove true biological consequences are considered moderate or weak.
For instance, a synonymous variant in exon 16 of TECTA, p.Leu1777Leu, may not be expected to be pathogenic because it does not alter the amino acid. However, it is predicted to lead to loss of an exonic splice enhancer binding site, has not been reported in large population studies, and has been reported to segregate with disease in 10 affected family members with autosomal dominant hearing loss (41). In addition, examination of mRNA from patient lymphocytes revealed skipping of exon 16, leading to an in-frame deletion in the amino acid sequence. Protein impairment, but not total LOF, is associated with TECTA-related autosomal dominant hearing loss, consistent with this prediction. This example demonstrates the importance of evaluating clinical data as well as functional evidence to make a definitive classification.
Conclusions and future perspectives
Variant assessment has become the bottleneck of large scale sequencing tests. Using the VAT described here has served to decrease the average time of variant assessment in our laboratory to 22 minutes by utilizing hyperlinks to perform database and literature searches and providing a platform to compile, analyze, and interpret variant data. However, it may take longer than 2 hours if a large collection of literature needs to be reviewed. Large gene panels may produce >10 variants that need review, and even after filtration strategies, exome and genome sequencing may produce 100s of variants. Further automation to retrieve relevant variant information directly from the literature and databases will speed the process. Clinically validated prediction algorithms trained on variants with well-established pathogenic or benign classifications (39) will improve the accuracy of computational prediction. Routine and standardized functional assays will provide necessary evidence to classify VUSs, but it is challenging to establish and support these assays in clinical diagnostics laboratories.
Information sharing and collaboration amongst laboratories will reduce the number of unique assessments performed. ClinVar, a recent NCBI initiative aiming to share clinical-grade variant information, is expected to support the molecular diagnostics community through genotype-phenotype associations aided by actual patient data. This may in turn inspire and accelerate the development of automated diagnostic prediction algorithms. Software is currently being developed to support the aggregation of internal and external variant information to enable sharing of clinical-grade variant data between different laboratories without jeopardizing patient identity (Table 1). Collectively, these approaches will greatly facilitate variant classification.
Conventionally, each clinical laboratory has had the liberty to develop, validate and perform diagnostic tests following recommendations by national or international agencies such as ACMG, CAP, CLIA, CLSI, EMNQ and WHO. Although proficiency testing has addressed the consistency in raw test output between different laboratories, there is still lack of agreement in variant assessment procedures and parameters, as well as final classification criteria. ACMG has provided guidelines for variant assessment (4, 5), but a consensus structured framework ensuring evidence-based classifications that can be easily adopted by individual laboratories is currently missing. While working groups have been formed to address this issue and we are optimistic that current variant classification guidelines will evolve into a consensus variant grading system based on the feedback recently provided by individual laboratories (ACMG 2013 Interpreting Sequencing Variants Open Forum), we hope that the framework above provides some rules and examples to partially fill the existing gap.
Supplementary Material
Acknowledgments
We thank Jordan Lerner-Ellis, Sami Amr, and Mark Bowser for their help in developing and maintaining the NVA form. We also thank all of our colleagues past and present at the LMM for their contributions to our variant assessment process over the past decade. This work was supported in part by National Institutes of Health grants HG006834 and HG006500.
Footnotes
Conflict of interest statement
HD, JS, HM, MAK, TJP, BHF, HLR and MSL are employed by fee-for-service laboratories performing clinical sequencing services. Several individuals serve on advisory boards or in other capacities for companies providing sequencing or other genetic services (HLR – BioBase, Clinical Future, Complete Genomics, GenomeQuest, Illumina, Ingenuity, Knome, Omicia; BF – InVitae; JS – LabCorp).
Supporting information
Variant Assessment Tool
Variant Assessment Static Data
Variant Assessment SOP
REFERENCES
- 1.Richards CS, Bradley LA, Amos J, Allitto B, Grody WW, Maddalena A, McGinnis MJ, Prior TW, Popovich BW, Watson MS, et al. Standards and guidelines for CFTR mutation testing. Genet Med. 2002;4:379–391. doi: 10.1097/00125817-200209000-00010. [DOI] [PubMed] [Google Scholar]
- 2.Huang T. Next generation sequencing to characterize mitochondrial genomic DNA heteroplasmy. Curr Protoc Hum Genet. 2011;Chapter 19(Unit19):Unit18. doi: 10.1002/0471142905.hg1908s71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valencia CA, Ankala A, Rhodenizer D, Bhide S, Littlejohn MR, Keong LM, Rutkowski A, Sparks S, Bonnemann C, Hegde M. Comprehensive mutation analysis for congenital muscular dystrophy: a clinical PCR-based enrichment and next-generation sequencing panel. PLoS One. 2013;8:e53083. doi: 10.1371/journal.pone.0053083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, Lyon E, Ward BE. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 5.Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST Working Group of the American College of Medical Genetics Laboratory Quality Assurance, C. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med. 2011;13:680–685. doi: 10.1097/GIM.0b013e3182217a3a. [DOI] [PubMed] [Google Scholar]
- 6.Plon SE, Eccles DM, Easton D, Foulkes WD, Genuardi M, Greenblatt MS, Hogervorst FB, Hoogerbrugge N, Spurdle AB, Tavtigian SV, et al. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat. 2008;29:1282–1291. doi: 10.1002/humu.20880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rehm HL, Bale SJ, Bayrak-Toydemir P, Berg JS, Brown KK, Deignan JL, Friez MJ, Funke BH, Hegde MR, Lyon E. ACMG clinical laboratory standards for next-generation sequencing. Genet Med. 2013 doi: 10.1038/gim.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niimura H, Bachinski LL, Sangwatanaroj S, Watkins H, Chudley AE, McKenna W, Kristinsson A, Roberts R, Sole M, Maron BJ, et al. Mutations in the gene for cardiac myosin-binding protein C and late-onset familial hypertrophic cardiomyopathy. N Engl J Med. 1998;338:1248–1257. doi: 10.1056/NEJM199804303381802. [DOI] [PubMed] [Google Scholar]
- 9.Sobreira NL, Cirulli ET, Avramopoulos D, Wohler E, Oswald GL, Stevens EL, Ge D, Shianna KV, Smith JP, Maia JM, et al. Whole-genome sequencing of a single proband together with linkage analysis identifies a Mendelian disease gene. PLoS Genet. 2010;6:e1000991. doi: 10.1371/journal.pgen.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verhoeven K, Van Laer L, Kirschhofer K, Legan PK, Hughes DC, Schatteman I, Verstreken M, Van Hauwe P, Coucke P, Chen A, et al. Mutations in the human alpha-tectorin gene cause autosomal dominant non-syndromic hearing impairment. Nat Genet. 1998;19:60–62. doi: 10.1038/ng0598-60. [DOI] [PubMed] [Google Scholar]
- 11.Mustapha M, Weil D, Chardenoux S, Elias S, El-Zir E, Beckmann JS, Loiselet J, Petit C. An alpha-tectorin gene defect causes a newly identified autosomal recessive form of sensorineural pre-lingual non-syndromic deafness, DFNB21. Hum Mol Genet. 1999;8:409–412. doi: 10.1093/hmg/8.3.409. [DOI] [PubMed] [Google Scholar]
- 12.Balciuniene J, Dahl N, Jalonen P, Verhoeven K, Van Camp G, Borg E, Pettersson U, Jazin EE. Alpha-tectorin involvement in hearing disabilities: one gene--two phenotypes. Hum Genet. 1999;105:211–216. doi: 10.1007/s004390051091. [DOI] [PubMed] [Google Scholar]
- 13.Taschner PE, den Dunnen JT. Describing structural changes by extending HGVS sequence variation nomenclature. Hum Mutat. 2011;32:507–511. doi: 10.1002/humu.21427. [DOI] [PubMed] [Google Scholar]
- 14.Mita S, Maeda S, Shimada K, Araki S. Cloning and sequence analysis of cDNA for human prealbumin. Biochem Biophys Res Commun. 1984;124:558–564. doi: 10.1016/0006-291x(84)91590-0. [DOI] [PubMed] [Google Scholar]
- 15.Joensuu T, Hamalainen R, Yuan B, Johnson C, Tegelberg S, Gasparini P, Zelante L, Pirvola U, Pakarinen L, Lehesjoki AE, et al. Mutations in a novel gene with transmembrane domains underlie Usher syndrome type 3. Am J Hum Genet. 2001;69:673–684. doi: 10.1086/323610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fields RR, Zhou G, Huang D, Davis JR, Moller C, Jacobson SG, Kimberling WJ, Sumegi J. Usher syndrome type III: revised genomic structure of the USH3 gene and identification of novel mutations. Am J Hum Genet. 2002;71:607–617. doi: 10.1086/342098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verpy E, Leibovici M, Zwaenepoel I, Liu XZ, Gal A, Salem N, Mansour A, Blanchard S, Kobayashi I, Keats BJ, et al. A defect in harmonin, a PDZ domain-containing protein expressed in the inner ear sensory hair cells, underlies Usher syndrome type 1C. Nat Genet. 2000;26:51–55. doi: 10.1038/79171. [DOI] [PubMed] [Google Scholar]
- 18.Ouyang XM, Xia XJ, Verpy E, Du LL, Pandya A, Petit C, Balkany T, Nance WE, Liu XZ. Mutations in the alternatively spliced exons of USH1C cause non-syndromic recessive deafness. Hum Genet. 2002;111:26–30. doi: 10.1007/s00439-002-0736-0. [DOI] [PubMed] [Google Scholar]
- 19.Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biesecker LG, Mullikin JC, Facio FM, Turner C, Cherukuri PF, Blakesley RW, Bouffard GG, Chines PS, Cruz P, Hansen NF, et al. The ClinSeq Project: piloting large-scale genome sequencing for research in genomic medicine. Genome Res. 2009;19:1665–1674. doi: 10.1101/gr.092841.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2012;40:D13–D25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stenson PD, Ball EV, Mort M, Phillips AD, Shaw K, Cooper DN. The Human Gene Mutation Database (HGMD) and its exploitation in the fields of personalized genomics and molecular evolution. Curr Protoc Bioinformatics. 2012;Chapter 1(Unit1 13) doi: 10.1002/0471250953.bi0113s39. [DOI] [PubMed] [Google Scholar]
- 23.Andreasen C, Nielsen JB, Refsgaard L, Holst AG, Christensen AH, Andreasen L, Sajadieh A, Haunso S, Svendsen JH, Olesen MS. New population-based exome data are questioning the pathogenicity of previously cardiomyopathy-associated genetic variants. Eur J Hum Genet. 2013 doi: 10.1038/ejhg.2012.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bell CJ, Dinwiddie DL, Miller NA, Hateley SL, Ganusova EE, Mudge J, Langley RJ, Zhang L, Lee CC, Schilkey FD, et al. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci Transl Med. 2011;3:65ra64. doi: 10.1126/scitranslmed.3001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue Y, Chen Y, Ayub Q, Huang N, Ball EV, Mort M, Phillips AD, Shaw K, Stenson PD, Cooper DN, et al. Deleterious- and disease-allele prevalence in healthy individuals: insights from current predictions, mutation databases, and population-scale resequencing. Am J Hum Genet. 2012;91:1022–1032. doi: 10.1016/j.ajhg.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunt KA, Smyth DJ, Balschun T, Ban M, Mistry V, Ahmad T, Anand V, Barrett JC, Bhaw-Rosun L, Bockett NA, et al. Rare and functional SIAE variants are not associated with autoimmune disease risk in up to 66,924 individuals of European ancestry. Nat Genet. 2012;44:3–5. doi: 10.1038/ng.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kenna KP, McLaughlin RL, Hardiman O, Bradley DG. Using reference databases of genetic variation to evaluate the potential pathogenicity of candidate disease variants. Hum Mutat. 2013;34:836–841. doi: 10.1002/humu.22303. [DOI] [PubMed] [Google Scholar]
- 28.Maron BJ. Hypertrophic cardiomyopathy: a systematic review. JAMA. 2002;287:1308–1320. doi: 10.1001/jama.287.10.1308. [DOI] [PubMed] [Google Scholar]
- 29.Gasparini P, Rabionet R, Barbujani G, Melchionda S, Petersen M, Brondum-Nielsen K, Metspalu A, Oitmaa E, Pisano M, Fortina P, et al. High carrier frequency of the 35delG deafness mutation in European populations. Genetic Analysis Consortium of GJB2 35delG. Eur J Hum Genet. 2000;8:19–23. doi: 10.1038/sj.ejhg.5200406. [DOI] [PubMed] [Google Scholar]
- 30.1000 Genomes Project Consortium. Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marth GT, Yu F, Indap AR, Garimella K, Gravel S, Leong WF, Tyler-Smith C, Bainbridge M, Blackwell T, Zheng-Bradley X, et al. The functional spectrum of low-frequency coding variation. Genome Biol. 2011;12:R84. doi: 10.1186/gb-2011-12-9-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, Karakoc E, Mackenzie AP, Ng SB, Baker C, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, Gudjonsson SA, Sigurdsson A, Jonasdottir A, Jonasdottir A, et al. Rate of de novo mutations and the importance of father's age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caleshu C, Day S, Rehm HL, Baxter S. Use and interpretation of genetic tests in cardiovascular genetics. Heart. 2010;96:1669–1675. doi: 10.1136/hrt.2009.190090. [DOI] [PubMed] [Google Scholar]
- 35.Rohlfs EM, Zhou Z, Sugarman EA, Heim RA, Pace RG, Knowles MR, Silverman LM, Allitto BA. The I148T CFTR allele occurs on multiple haplotypes: a complex allele is associated with cystic fibrosis. Genet Med. 2002;4:319–323. doi: 10.1097/00125817-200209000-00001. [DOI] [PubMed] [Google Scholar]
- 36.Buyse IM, McCarthy SE, Lurix P, Pace RP, Vo D, Bartlett GA, Schmitt ES, Ward PA, Oermann C, Eng CM, et al. Use of MALDI-TOF mass spectrometry in a 51-mutation test for cystic fibrosis: evidence that 3199del6 is a disease-causing mutation. Genet Med. 2004;6:426–430. doi: 10.1097/01.gim.0000139508.61701.bd. [DOI] [PubMed] [Google Scholar]
- 37.Mann SA, Castro ML, Ohanian M, Guo G, Zodgekar P, Sheu A, Stockhammer K, Thompson T, Playford D, Subbiah R, et al. R222Q SCN5A mutation is associated with reversible ventricular ectopy and dilated cardiomyopathy. J Am Coll Cardiol. 2012;60:1566–1573. doi: 10.1016/j.jacc.2012.05.050. [DOI] [PubMed] [Google Scholar]
- 38.Reed FA, Akey JM, Aquadro CF. Fitting background-selection predictions to levels of nucleotide variation and divergence along the human autosomes. Genome Res. 2005;15:1211–1221. doi: 10.1101/gr.3413205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jordan DM, Kiezun A, Baxter SM, Agarwala V, Green RC, Murray MF, Pugh T, Lebo MS, Rehm HL, Funke BH, et al. Development and validation of a computational method for assessment of missense variants in hypertrophic cardiomyopathy. Am J Hum Genet. 2011;88:183–192. doi: 10.1016/j.ajhg.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crockett DK, Lyon E, Williams MS, Narus SP, Facelli JC, Mitchell JA. Utility of gene-specific algorithms for predicting pathogenicity of uncertain gene variants. J Am Med Inform Assoc. 2012;19:207–211. doi: 10.1136/amiajnl-2011-000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Collin RW, de Heer AM, Oostrik J, Pauw RJ, Plantinga RF, Huygen PL, Admiraal R, de Brouwer AP, Strom TM, Cremers CW, et al. Mid-frequency DFNA8/12 hearing loss caused by a synonymous TECTA mutation that affects an exonic splice enhancer. Eur J Hum Genet. 2008;16:1430–1436. doi: 10.1038/ejhg.2008.110. [DOI] [PubMed] [Google Scholar]
- 42.Tavtigian SV, Deffenbaugh AM, Yin L, Judkins T, Scholl T, Samollow PB, de Silva D, Zharkikh A, Thomas A. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet. 2006;43:295–305. doi: 10.1136/jmg.2005.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sunyaev S, Lathe W, 3rd, Bork P. Integration of genome data and protein structures: prediction of protein folds, protein interactions and "molecular phenotypes" of single nucleotide polymorphisms. Curr Opin Struct Biol. 2001;11:125–130. doi: 10.1016/s0959-440x(00)00175-5. [DOI] [PubMed] [Google Scholar]
- 44.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.