Abstract
Recently the use of engineered viral scaffolds in biotechnology and medical applications has been increasing dramatically. T4 phage capsid derived nanoparticles (NPs) have potential advantages as sensors and in biotechnology. These applications require that the physical properties and cellular uptake of these NPs be understood. In this study we used a T4 deletion mutant to investigate the effects of removing both the Hoc and Soc proteins from the capsid surface on T4 tailless NPs. The surface charge, zeta potential, size, and cellular uptake efficiencies for both the T4 NP and T4ΔHocΔSoc NP mutant were measured and compared using dynamic light scattering and flow cytometry and significant differences were detected.
Keywords: T4 NPs, Hoc, Soc, Zeta potentials, Cellular uptake
1. Introduction
Viruses are widely used scaffolds for biotechnology applications due to their diverse sizes, shapes, and versatile surface functional groups allowing chemical modification. Such chemically modified viral based nanoparticles (VNPs) are biocompatible, have low cytotoxicity, and are suitable for various biotechnology and sensor applications [1–3]. However, variability among VNPs resulting from their intrinsic chemical and physical properties define the limitations of their bio-applications, and therefore it is necessary to understand the role of these variables on the cellular uptake effects in order to fine-tune the function of VNPs for various bio-applications.
Among VNPs, phages are preferred due to their robustness and easy accessibility as materials. Based on their shapes, phages can be divided into icosahedral and filamentous types. Filamentous phages, such as M13 and fd, which can be micrometers in length, and icosahedral phage heads often less than 100 nm diameter, are extensively used as scaffolds for chemical modifications for cellular imaging [3,4] and drug delivery [5,6]. In particular, the icosahedral phage based NPs are very similar in many respects to other icosahedral viruses, such as Cowpea Mosaic virus (CPMV) and Cowpea chlorotic mottle virus (CCMV), that are used more often in many biotechnology applications. However by taking advantage of the power of phage genetics and the ability to obtain large quantities of phages from bacterial hosts, icosahedral phages are now being utilized more frequently for imaging and bio-medical applications.
The Myoviridae bacteriophage T4 particle consists of an icosahedral capsid containing a 170 kb dsDNA genome and a ~100 nm long contractile tail. T4 has one of the largest head surface areas among the bacteriophage capsids~75 nm in width and 100 nm in length), which accommodates more than 1 × 105 functional groups suitable for chemical modification [3]. Moreover, it has a flexible genetic display system allowing peptide display [7,8]. More remarkably, the head can also package more than 100 copies of foreign protein inside the capsid with the DNA [9]. Therefore T4 is an ideal scaffold with broad bio-applications. While the T4 phage head has no bacterial infectivity, the capsid may determine its biological properties related to binding to mammalian cells. Thus mutation of the Lambda (λ) phage capsid protein resulted in escape from entrapment by the mammalian reticuloendothelium [10], suggesting the head protein controls the interaction with the eukaryotic cells by an unknown mechanism. In addition, one T4 head surface protein is known to be highly immunogenic [11]. T4 capsids consist of two major essential capsid proteins, gp23 at the hexamer and gp24 at the pentamer positions, and two non-essential capsid surface proteins that bind to the mature capsid lattice, Hoc (highly immunogenic outer capsid protein) and Soc (small outer capsid protein) [12,13]. It is known that the deletion of Hoc and Soc does not affect the overall structure and assembly of the T4 capsid [14], but the alteration of the physical properties and the effect on cell interaction is unknown. To investigate the effects of the protein content on the physical properties and interaction with mammalian cells, a Hoc-Soc mutant (T4ΔHocΔSoc) NP was compared to T4 NP. Dynamic light scattering was used to investigate the size and zeta-potential of the T4 derived NPs and flow cytometry was used to measure their uptake efficiency into tumor cells.
2. Materials and methods
2.1. Preparation of T4 derived NPs
T4 mutantsK10 (38– 51–denA– denB–) and K10ΔHocΔSoc (38– 51–denA– denB–Hoc–Soc–), were grown in the non-suppressor Escherichia coli Rosetta, to make tailless T4 NPs according to our previously established procedure [2,15]. In brief, Rosetta were grown in M9S supplemented with 1/3 volume of Luria Broth, then the cells were infected with phage, followed by centrifugation and cell lysis. Potassium phosphate buffer (50 KP/10 MgCl2) containing 50 mM potassium phosphate (pH 7.5) supplemented with 10 mM MgCl2 and 2 mM CaCl2, CHCl3 (1/20 of total volume), DNase I (40 μg/mL), RNase I (50 μg/mL), and 0.4 μg/mL of PMSF (phenylmethanesulfonylfluoride or phenylmethylsulfonyl fluoride) was used for resuspending the resulting T4 NPs. The cell debris was removed by spinning and the cell lysate containing the head scaffolds was concentrated through a Microcon YM-100 membrane according to the manufacturer's procedure (Millipore Corp., MA). The T4 NPs were then purified by gel filtration with Superose 6 (GE Healthcare Biosciences, NJ). The flow-through and eluted fractions were then assessed using a 1.2% Tris–acetate agarose gel stained with either ethidium bromide or Coomassie blue.
2.2. Dye conjugation
Purified tailless T4 NP solutions in 50 KP/10 MgCl2were reacted with Alexa 488 or Alexa 546 (Invitrogen, Carlsbad, CA) in the same buffer with the addition of 10% DMSO (Sigma–Aldrich, St. Louis, MO) with the ratio of 5 dyes to each amine group and incubated at 22 °C overnight [3]. The dye-labeled T4 NPs were then loaded separately into a Superose™ 6 prep grade (GE Healthcare Biosciences, Pittsburgh, PA) column and the first 1.5 mL fractions were collected and analyzed by gel electrophoresis, and UV–Vis spectroscopy.
2.3. Flow cytometry
A549 cells were treated with dye-conjugated T4 based NPs at 50000 T4/cell according to our previously published protocol [3]. The stained cells were then run on an Accuri C6 flow cytometer using standard lasers and filters for PE (FL-2) and APC (FL-4). For each sample 2 × 104 events were collected in a gate corresponding to the live cell population based on forward vs. side scatter. The percentage of cells positive for the dye-T4 NPs was calculated using the histogram subtraction tool on FSC Express V3.
2.4. Size analysis of suspended particles and aggregates
The size of suspended T4 based NPs was investigated by dynamic light scattering spectroscopy (DLS) using a Malvern Zetasizernano-ZS (Westborough, MA) equipped with MPT-2 titrator at 25 °C. The details of DLS techniques can be found elsewhere [16]. The effect of pH on the sizes of suspended T4 based NPs was determined by the stepwise-addition of 0.1 N NaOH or HCl with the help of MPT-2 titrator and subsequent DLS analysis. After each NaOH or HCl addition, the mixed aqueous solution was stirred for 8 min. After mixing the solution, the pH was determined and the mixed aqueous solution was introduced to the Zetasizer sample cell with the circulation system integrated into the MPT-2 titrator for the DLS size analysis.
2.5. Zeta potential analysis of suspended particles and colloids
The analysis of zeta (z)-potential as a function of pH was conducted by laser Doppler velocimetry (LDV) using a Malvern Zetasizernano-ZS equipped with a MPT-2 titrator at 25 °C according to Furukawa et al. [17]. Each suspension sample, with an appropriate adjustment to pH with either 0.1 N NaOH or HCl, was loaded into a capillary cell with embedded electrodes at either of the two ends using the MPT-2 titrator. Suspended particles move towards the electrode of the opposite charge when the potential is applied, and their velocity is measured and expressed in unit field strength as a function of their mobility. By knowing the physical properties of the suspension medium, the velocity can be converted to the z-potential using the Smolchowski equation [16].
3. Results
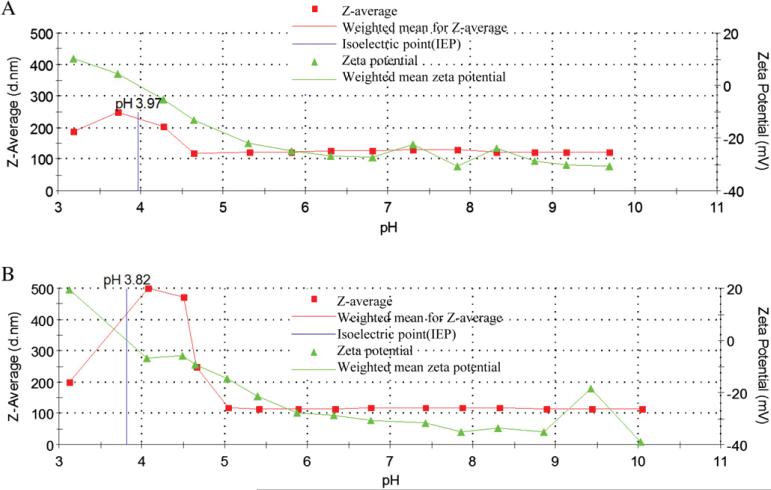
Tailless T4 mutants, K10 (T4 NP), and K10 lacking Hoc and Soc capsid proteins (T4ΔHocΔSoc NP) were used to make tailless T4 based NPs for our experiments. There is no Hoc or Soc gene expression in the T4ΔHocΔSoc mutant examined by PCR and Western blotting (data not shown). The T4ΔHocΔSoc NPs were able to withstand the purification procedures and chemical reaction with Alexa 546 (A546) as shown in Fig. 1 without producing lower molecular weight bands. The deletion of Hoc and Soc does not affect the head assembly pathway and does not change the interior head dimension, therefore this mutant will package the same amount of DNA as wild type (wt) T4 head using head-full packaging mechanism [8]. Our results indicated that T4ΔHocΔSoc NPs appeared to be more negative relative to T4 wt NPs and migrated faster on the agarose gel due to the alteration of surface protein content (Lane 2 in Fig. 1). For unknown reasons, we also observed that each T4 based NP ran as doublet bands, although they appeared to be homogenous in size in atomic force microscopy images [2]. Using a dynamic light scattering instrument, the average size for tailless T4 NPs and T4ΔHocΔSoc NPs between pH 5–9 were found to be 125 nm and 116 nm respectively (Fig. 2). Large aggregates appear to form around the capsid isoelectric point at pH 4 (Fig. 2A and B) and could be reversibly dispersed into separated NPs at pH 5 or greater. The average zeta potential for T4 NPs is ~–27 mV and T4ΔHocΔSoc NPs is ~–33 mV at between pH 7–8. This is consistent with the result from electrophoretic mobility measurement in agarose (Figs. 1 and 2).
Fig. 1.
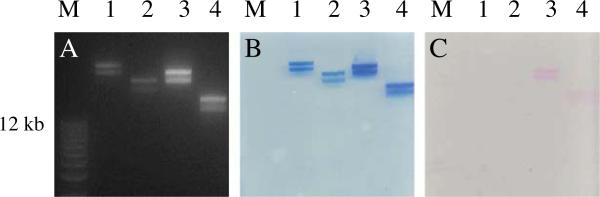
Electrophoretic mobility on agarose gel for phage T4 derived NPs. Panel A, B and C were derived from the same gel. A is the image of the gel subjected to ethidium bromide staining to visualize DNA bands. B is the image of the gel stained with coomassie blue to visualize the protein bands. C is the image of the gel without any staining. The pink bands represent the T4 NPs cross-linked with Alexa 546. Lanes M, 1, 2, 3 and 4 represent DNA size marker, T4 NPs, T4ΔHocΔSoc NPs, Alexa 546-T4 NPs, and Alexa 546-T4ΔHocΔSoc NPs.
Fig. 2.
Measurement of zeta potential and size for phage T4 derived NPs. A. Size and charges for wt T4 NPs: titrated from pH 10 to pH 3 using 0.1 N HCl. The zeta potential at pH 7 is ~–25 mV. The isoelectric point is pH 3.97. The colloid size is ~125 nm. However, near or below the isoelectric point (IEP), the colloids aggregate. B. Size and charges for T4ΔHocΔSoc NPs. Titrated from pH 3 to pH 10 using 0.1 N NaOH. The zeta potential at pH 7 is ~–31 mV. The colloids are initially aggregated below the isoelectric point (pH 3.82) but as the pH increases to above IEP; they disaggregate to individual colloid particles of ~116 nm.
The Alexa 546 conjugated T4 NPs appeared to be more negative, due to the contribution of one negative charge from Alexa 546 (Fig. 1C). Although there is no measurement of the zeta potential and size for dye-T4 NPs, we can estimate that the zeta potential for Alexa 546-T4 NPs is ~–30 mV (between –27 and –33 mV) and the zeta potential for Alexa 546-T4ΔHocΔSoc NPs is ~–39 mV, judging from migration distance on agarose gel (Fig. 1).
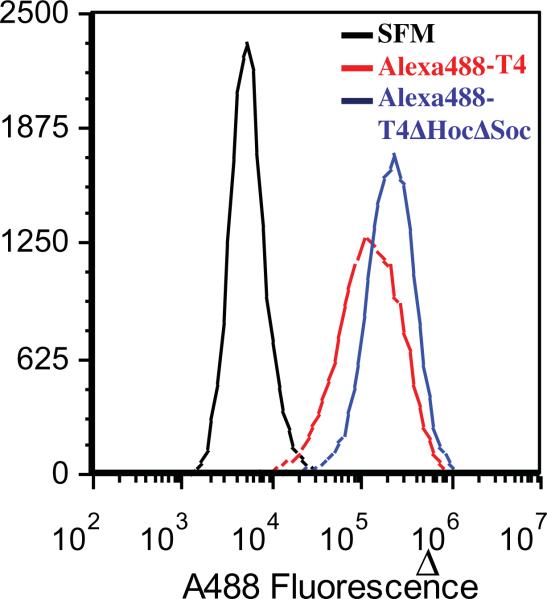
Flow cytometry was used to quantify the uptake of T4 and T4ΔHocΔSoc NPs, labeled with Alexa 488 dye, into A549 cells (lung cancer epithelial cells). The cells were incubated with 50,000 T4 or T4ΔHocSoc NPs per cell for 6hrs and cells containing the NPs were able to be distinguished from control cells with no NPs in serum free media (SFM). The plots for the two Alexa 488-T4 NPs and no T4 NPs (SFM) are shown in Fig. 3 and the percentage of dye-T4 NPs uptake was quantified by comparing the fluorescence to the cells without dye-T4 NPs. The number of the dyes/virus (D/V) for both Alexa 488-T4 and Alexa 488-T4ΔHocΔSoc NPs were similar (~5000 D/V) so the intensities should be comparable. Our results indicated that the percentage of cells that internalized Alexa 488-T4ΔHocΔSoc NPs was approximately 95% which is approximately 5% higher than the percentage of cells that internalized Alexa 488-T4 NPs (significantly different as judged by the t-test, p < 0.05). In addition, the median fluorescence intensity for the cell population treated with A488-T4ΔHocΔSoc NPs is approximately three times higher than those cells treated with A488-T4 NPs (statistically significantly by t-test, p < 0.05) (Table 1).
Fig. 3.
Quantification of phage T4 derived NPs using Flow cytometry. Both wt T4 and T4ΔHocΔSoc NPs were cross-linked with Alexa 488 (~5000 D/V). A549 cells were treated with T4 NPs for 6hrs with a ratio of 50,000 to one cell. The black line is the serum free media (SFM) representing the cell population without uptake of the labeled T4 NPs. The red line is the cell population uptake of Alexa 488-T4 NPs and the blue line is the cell population uptake of the Alexa 488-T4ΔHocΔSoc NPs.
Table 1.
Companion of T4 NPs uptake by A549 cells.
| Average % positive cells | Median fluorescent intensity | |||
|---|---|---|---|---|
| Sample | % positive (n = 3)3 | St.Dev | Intensity4 | St.Dev |
| Alexa 488-T41 | 93.13 | 3.41 | 78681.00 | 39339.32 |
| Alexa 488-T4ΔHocΔSoc2 | 98.63 | 0.21 | 210930.33 | 52448.82 |
5285 dyes/virus for T4.
5063 dyes/virus for T4ΔHocΔSoc.
t-test, p < 0.05.
t-test, p < 0.05.
4. Discussion
The properties of VNPs have a tremendous effect on their use for biotechnology applications. Therefore, it is important that the surface properties and uptake mechanisms of these nanoparticles are explored. We measured zeta potential, size, and cellular uptake efficiency for the wt T4 NP and a Hoc-Soc capsid protein deletion mutant.
Zeta potential (Zp) is an electrokinetic property of the electrical double layer (EDL) surrounding the particle. It is a potential at the slip plane that divides the diffuse layer into two regions: the inner diffuse layer where ions move with the particle movement, and the outer diffuse layer where ions are still influenced by the particle due to long range forces but are not part of the coherent unit that moves with the particle. Even though zp is defined as such and is strictly different from the surface potential, it is often used as a proxy for the surface potential as: (i) it represents the average electrokinetic behavior of the particles; and (ii) it can be determined experimentally unlike surface potential. It influences the interaction between two molecules and two different phases [16]. Prior to this study, there is no information about how the change of zp in VNPs affects the interaction with the eukaryotic cells.
Hoc and Soc are two non-essential T4 capsid surface proteins which do not affect the phage infectivity and viability. Hoc protein can attach to the center of the capsid hexamer in vivo or in vitro [18] and contain an immunoglobulin-like domain, which can widely interact with other surface proteins on mammalian cells and bacteria [13,19]. This interaction may help facilitate the cellular uptake of the wt T4 NP shown in Table 1. The function of Soc is to stabilize the head structure; however, our results indicate that the Soc deleted T4 heads remain stable enough to endure the chemical conjugation reaction in the presence of 10% DMSO.
For the first time, we successfully demonstrated that the alteration of the surface protein content in T4 derived NPs changes their biological activity during cellular uptake. Although it was proposed that Hoc can enhance the binding to bacterial host [13], our results indicate that the Hoc deletion mutant entered eukaryotic cells more efficiently than the wt. The deletion of both Hoc and Soc from the T4 head scaffolds results in the decrease of the electrical charge and a reduction in size of the NP. Although the smaller size of T4ΔHocΔSoc NPs may help to increase the uptake [20], the increase of the surface negative charges of T4ΔHocΔSoc heads, which repel the interaction of negatively charged membrane, can cancel the positive effect on the cellular uptake. Therefore, we believe that there is another mechanism to govern the cellular uptake of tailless T4 NPs. It was also hypothesized that there is a β-integrin receptor binding site (KGD) residing in the gp24, an essential capsid surface protein, and the loss of Hoc facilitates the exposure of gp24 binding site to the cellular receptor, thus increasing the uptake [21]. Our results are consistent with this hypothesis and more studies are on the way to investigate the detail of T4 NP uptake mechanism.
Acknowledgments
We thank Drs. Patricia Legler and Stella North for their comments on the manuscript. This work is supported by NRL 6.2 base program. The opinions expressed here are those of authors and do not represent those of the US Navy, the US Department of Defense, or the US government.
References
- 1.Soto CM, Ratna BR. Virus hybrids as nanomaterials for biotechnology. Curr. Opin. Biotechnol. 2010;21:426–438. doi: 10.1016/j.copbio.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Archer MJ, Liu JL. Bacteriophage T4 nanoparticles as materials in sensor applications: variables that influence their organization and assembly on surfaces. Sensors. 2009;9:6298–6311. doi: 10.3390/s90806298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robertson KL, Soto CM, Archer MJ, Odoemene O, Liu JL. Engineered T4 viral nanoparticles for cellular imaging and flow cytometry. Bioconjug. Chem. 2011;22:595–604. doi: 10.1021/bc100365j. [DOI] [PubMed] [Google Scholar]
- 4.Li K, Chen Y, Li S, Nguyen HG, Niu Z, You S, Mello CM, Lu X, Wang Q. Chemical modification of M13 bacteriophage and its application in cancer cell imaging. Bioconjug. Chem. 2010;21:1369–1377. doi: 10.1021/bc900405q. [DOI] [PubMed] [Google Scholar]
- 5.Paillard F. Bacteriophage: tools toward a cell-targeted delivery. Hum. Gene Ther. 1998;9:2307–2308. doi: 10.1089/hum.1998.9.16-2307. [DOI] [PubMed] [Google Scholar]
- 6.Ashley CE, Carnes EC, Phillips GK, Durfee PN, Buley MD, Lino CA, Padilla DP, Phillips B, Carter MB, Willman CL, Brinker CJ, Caldeira Jdo C, Chackerian B, Wharton W, Peabody DS. Cell-specific delivery of diverse cargos by bacteriophage MS2 virus-like particles. ACS Nano. 2011;5:5729–5745. doi: 10.1021/nn201397z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malys N, Chang DY, Baumann RG, Xie D, Black LW. A bipartite bacteriophage T4 SOC and HOC randomized peptide display library: detection and analysis of phage T4 terminase (gp17) and late sigma factor (gp55) interaction. J. Mol. Biol. 2002;319:289–304. doi: 10.1016/S0022-2836(02)00298-X. [DOI] [PubMed] [Google Scholar]
- 8.Rao VB, Black LW. Structure and assembly of bacteriophage T4 head. Virol. J. 2010;7:356. doi: 10.1186/1743-422X-7-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mullaney JM, Black LW. Activity of foreign proteins targeted within the bacteriophage T4 head and prohead: implications for packaged DNA structure. J. Mol. Biol. 1998;283:913–929. doi: 10.1006/jmbi.1998.2126. [DOI] [PubMed] [Google Scholar]
- 10.Merril CR, Biswas B, Carlton R, Jensen NC, Creed GJ, Zullo S, Adhya S. Long-circulating bacteriophage as antibacterial agents. Proc. Natl. Acad. Sci. USA. 1996;93:3188–3192. doi: 10.1073/pnas.93.8.3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamaguchi Y, Yanagida M. Head shell protein hoc alters the surface charge of bacteriophage T4. Composite slab gel electrophoresis of phage T4 and related particles. J Mol Biol. 1980;141:175–193. doi: 10.1016/0022-2836(80)90384-8. [DOI] [PubMed] [Google Scholar]
- 12.Fokine A, Battisti AJ, Bowman VD, Efimov AV, Kurochkina LP, Chipman PR, Mesyanzhinov VV, Rossmann MG. Cryo-EM study of the Pseudomonas bacteriophage phiKZ. Structure. 2007;15:1099–1104. doi: 10.1016/j.str.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Fokine A, Islam MZ, Zhang Z, Bowman VD, Rao VB, Rossmann MG. Structure of the three N-terminal immunoglobulin domains of the highly immunogenic outer capsid protein from a T4-like bacteriophage. J. Virol. 2011;85:8141–8148. doi: 10.1128/JVI.00847-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwasaki K, Trus BL, Wingfield PT, Cheng N, Campusano G, Rao VB, Steven AC. Molecular architecture of bacteriophage T4 capsid: vertex structure and bimodal binding of the stabilizing accessory protein. Soc. Virol. 2000;271:321–333. doi: 10.1006/viro.2000.0321. [DOI] [PubMed] [Google Scholar]
- 15.Lin H, Rao VB, Black LW. Analysis of capsid portal protein and terminase functional domains: interaction sites required for DNA packaging in bacteriophage T4. J. Mol. Biol. 1999;289:249–260. doi: 10.1006/jmbi.1999.2781. [DOI] [PubMed] [Google Scholar]
- 16.Berne BJ, Pecora R. Dynamic Ligth Scattering, Robert E. Krieger Publishing Company; Malabar, FL: 1990. [Google Scholar]
- 17.Furukawa Y, Watkins JL, Kim J, Curry KJ, Bennett RH. Aggregation of montmorillonite and organic matter in aqueous media containing artificial seawater. Geochem. Trans. 2009;10:2–13. doi: 10.1186/1467-4866-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fokine A, Chipman PR, Leiman PG, Mesyanzhinov VV, Rao VB, Rossmann MG. Molecular architecture of the prolate head of bacteriophage T4. Proc. Natl. Acad. Sci. USA. 2004;101:6003–6008. doi: 10.1073/pnas.0400444101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bateman A, Eddy SR, Mesyanzhinov VV. A member of the immunoglobulin superfamily in bacteriophage T4. Virus Genes. 1997;14:163–165. doi: 10.1023/a:1007977503658. [DOI] [PubMed] [Google Scholar]
- 20.Chan WCW, Chithrani BD, Ghazani AA. Determining the size and shape dependence of gold nanoparticle uptake into mammalian cells. Nano Lett. 2006;6:662–668. doi: 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- 21.Gorski KDA, Switala-Jele K, Nowaczyk M, Weber-Dabrowska B, Boratynski J, Wietrzyk J, Opolski A. New insights into the possible role of bacteriophages in host defense and disease. Med. Immunol. 2003;2:2–5. doi: 10.1186/1476-9433-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]