Abstract
Many plant species in temperate climate regions require vernalization over winter to initiate flowering. Flowering Locus C (FLC) and FLC-like genes are key regulators of vernalization requirement and growth habit in winter-annual and perennial Brassicaceae. In the biennial crop species Beta vulgaris ssp. vulgaris in the evolutionarily distant Caryophyllales clade of core eudicots growth habit and bolting time are controlled by the vernalization and photoperiod response gene BTC1 and the downstream BvFT1-BvFT2 module. B. vulgaris also contains a vernalization-responsive FLC homolog (BvFL1). Here, to further elucidate the regulation of vernalization response and growth habit in beet, we functionally characterized BvFL1 by RNAi and over-expression in transgenic plants. BvFL1 RNAi neither eliminated the requirement for vernalization of biennial beets nor had a major effect on bolting time after vernalization. Over-expression of BvFL1 resulted in a moderate late-bolting phenotype, with bolting after vernalization being delayed by approximately 1 week. By contrast, RNAi-induced down-regulation of the BvFT1-BvFT2 module led to a strong delay in bolting after vernalization by several weeks. The data demonstrate for the first time that an FLC homolog does not play a major role in the control of vernalization response in a dicot species outside the Brassicaceae.
Keywords: Beta vulgaris, bolting, Flowering Locus C (FLC), photoperiod, vernalization
Introduction
Vernalization is the process by which the exposure of a plant to a prolonged period of cold temperatures over winter promotes the initiation of flowering. In temperate climate regions vernalization is an integral part of life cycle strategies as an evolutionary adaptation to changing seasons. In the annual dicotyledonous species Arabidopsis thaliana and its perennial relative Arabis alpina, the vernalization response is regulated by the MADS-box gene Flowering Locus C (FLC) and its ortholog Perpetual Flowering 1 (PEP1), respectively (Michaels and Amasino, 1999; Sheldon et al., 1999; Wang et al., 2009; Zografos and Sung, 2012). By contrast, the vernalization response in monocotyledonous species like barley or wheat requires the Vernalization 1-3 (VRN1-3) genes, with VRN1 being the only MADS-box gene of these three (Yan et al., 2003, 2004, 2006). Recent studies in Beta vulgaris, which on an evolutionary scale is similarly distantly related to Arabidopsis (~120 million years of evolution) and the monocots (~140 million years; Chaw et al., 2004; Davies et al., 2004), revealed a new mode of life cycle control in dicotyledonous species. In B. vulgaris, the pseudo-response regulator (PRR) gene Bolting Time Control 1 (BTC1) determines whether floral transition occurs in the first year of growth, as in annual accessions, or in the second year, as in biennials (Pin et al., 2012). BTC1 mediates bolting and flowering by regulation of an antagonistic pair of Flowering Locus T (FT) homologs first described by Pin et al. (2010). Bienniality in beet derives from a recessive BTC1 allele (btc1) with a reduced responsiveness to the floral inductive stimulus of long days and/or reduced activity of the BTC1 protein compared to annual beets. The perception of prolonged cold over winter after the first growing season restores the competence to bolt and flower in biennial beets. This process was suggested to involve up-regulation of BTC1, leading to suppression of the flowering repressor BvFT1 and expression of the flowering activator BvFT2. Life cycle control by BTC1 thus involves the integration of both photoperiod and vernalization signals. By contrast, the PRR genes in monocots such as PPD1 in Hordeum vulgare or SbPRR37 in Sorghum bicolor are only known to mediate photoperiod response, while a role in vernalization response or life cycle control has not been described (Turner et al., 2005; Murphy et al., 2011).
In Arabidopsis, FLC represses flowering by binding to cis-regulatory sequences in the floral integrator genes FT and Suppressor of Overexpression of Constans 1 (SOC1) (Helliwell et al., 2006; Searle et al., 2006). During vernalization, FLC is down-regulated and the repressed state is epigenetically maintained after vernalization. The repression of FLC allows activation of FT under long-day conditions through the photoperiod pathway and its central regulator Constans (CO). FT protein expressed in the phloem companion cells of the leaves moves to the shoot apical meristem as part of the “florigen” signal and initiates flowering (Andrés and Coupland, 2012). Besides their highly conserved function as day length-induced floral activators, FT and FT-like genes also control other processes like stomatal opening in Arabidopsis or tuberization in potato (Pin and Nilsson, 2012).
A dose-dependent positive correlation between FLC expression and circadian period length was shown by using genotypes with different functional and non-functional allele compositions at FLC and the FLC-regulatory locus FRIGIDA (FRI) as well as a 35S::FLC over-expressor line (Salathia et al., 2006). Furthermore, El-Assal et al. (2003) showed that FLC negatively and dose-dependently regulates expression of the photoreceptor gene Cryptochrome 2 (CRY2). CRY2 co-regulates circadian period length together with CRY1 and tends to act as a negative regulator of period length (Devlin and Kay, 2000; Gould et al., 2013), suggesting that the FLC-induced increase in circadian period length may be mediated through cryptochromes. Vernalization resulted in a significant decrease in circadian period length, which was suggested to reduce the day length threshold required for photoperiodic induction of flowering and thus to accelerate flowering in spring (Yanovsky and Kay, 2002; Salathia et al., 2006). Finally, mutations in photoperiod pathway genes affected expression of FLC, providing further indication for the crosstalk between vernalization and photoperiod pathways (Rouse et al., 2002).
FLC and FLC-like genes belong to a major MADS-box gene clade that was recently shown to also include monocot genes (Ruelens et al., 2013). In dicots, FLC-like genes have been identified in two species outside the Brassicaceae, i.e., B. vulgaris in the Caryophyllales clade of core eudicots (Reeves et al., 2007) and the asterid species Cichorium intybus (Périlleux et al., 2013), which includes the biennial crop root chicory. Complementation analyses of B. vulgaris FLC-LIKE 1 (BvFL1) and C. intybus FLC-LIKE (CiFL1) in Arabidopsis and down-regulation of BvFL1 and CiFL1 by vernalization in beet or chicory, respectively, suggested a conserved floral repressor function of these genes. However, instead of being epigenetically maintained in a transcriptionally silent state after vernalization, the expression of BvFL1 and CiFL1 after vernalization reverted to pre-vernalization levels (Reeves et al., 2007; Périlleux et al., 2013). Interestingly, the FLC ortholog PEP1 in A. alpina also reverts to pre-vernalization expression levels after return to warm temperatures, which correlates with unstable histone modifications at the PEP1 locus (Wang et al., 2009). Unstable repression of PEP1 after vernalization was suggested to correlate with perennial life cycle strategies (Wang et al., 2009).
A more complex pattern of BvFL1 regulation in beet emerged from a study of BvFL1 expression in the shoot apical meristem (Trap-Gentil et al., 2011). According to this study, “bolting sensitive” biennial beet genotypes, which only require relatively short periods of vernalization for bolting to occur, are first down-regulated during vernalization, but up-regulated during a later stage of vernalization. The authors suggested that the early transient decrease in BvFL1 expression during vernalization may account for the relatively high susceptibility to bolting in these genotypes. By contrast, “bolting resistant” biennial genotypes that require relatively long periods of vernalization exhibited a gradual increase in expression during vernalization. Furthermore, RNA methylation of BvFL1 mRNA was detected in the shoot apical meristem of a bolting-resistant genotype after vernalization and was proposed to contribute to the control of vernalization response in sugar beet (Hébrard et al., 2013). However, a clear picture of the functional role of BvFL1 in beet has not yet emerged, and a characterization of this gene's function through transgenic or mutational analyses in beet is still lacking.
Here, we further dissect the vernalization response in beet by over-expression and RNAi-mediated down-regulation of BvFL1 and down-regulation of the FT homologs BvFT1 and BvFT2 in transgenic plants. Phenotypic analysis revealed a delay in bolting after vernalization by 1 week in transformants over-expressing BvFL1, while BvFL1 RNAi neither had a major effect on bolting time after vernalization nor did it lead to bolting without vernalization. RNAi-induced concomitant down-regulation of the floral repressor BvFT1 and the floral activator BvFT2 resulted in a bolting delay by up to 7 weeks and a high percentage of non-bolting plants in a subset of transformation events. Taken together, our data support a dominant role of the BvFT1-BvFT2 module in the control of vernalization response and show that, by contrast, BvFL1 is not a major regulator of vernalization response in beet.
Materials and methods
Vector construction and plant transformation
For the BvFL1 over-expression construct a 616 bp cDNA fragment covering the whole coding sequence of the splice variant BvFL1_v3 (Reeves et al., 2007) was inserted downstream of a Cauliflower Mosaic Virus (CaMV) 35S promoter and the Tobacco Mosaic Virus (TMV) 5′UTR and upstream of an Agrobacterium tumefaciens nos 3′ terminator. In Arabidopsis, over-expression of BvFL1_v3 caused the strongest delay in flowering among BvFL1 splice variants (Reeves et al., 2007). RNAi vectors were constructed by insertion of a 332 bp fragment of the BvFL1 3′UTR or a 361 bp cDNA fragment spanning most of the phosphatidylethanolamine-binding protein (PEBP) domain of BvFT1 (Pin et al., 2010), respectively, as inverted repeats between the regulatory elements described above. A 91 bp sugar beet intron sequence was used as spacer between the sense and antisense repeat units. The phosphinothricin acetyl transferase (PAT) gene was inserted downstream of the RNAi cassettes for selection of transgenic plants with glufosinate. The constructs were introduced into the biennial sugar beet genotype SES01 (SESVanderHave, Tienen, Belgium) by polyethylene glycol-mediated DNA transfer as described previously (Hall et al., 1996; Pin et al., 2012). Transgenic protoplasts, calli and regenerating plantlets were selected using glufosinate and transgene integration was confirmed by PCR. Low copy number (1–3 transgene copies) transformants were selected by quantitative PCR using TaqMan® assays (Life Technologies, Carlsbad, California, USA) and DNA gel blot analysis for the effector transgene and the PAT gene (Table 1). For DNA gel blot analysis, genomic DNA was digested with two different restriction enzymes, EcoRI and NcoI, separated by gel electrophoresis and transferred to Hybond™-N membranes (GE Healthcare, Little Chalfont, UK). Construct-specific probes were amplified from the corresponding plasmid DNA using primers 5′-CTATTTACAATTACACC ATGGCAGGCG and 5′-TGAACGATCGGGGAAATTCGAGCTCGG for analysis of BvFL1 over-expression transformants, 5′-GGTTTTATATGTACTACTGTTGTAGCTG and 5′-TGAACGATCGGGGAAATTCGAGCTCGG for BvFL1 RNAi transformants, and 5′-GGTTTTATATGTACTACTGTTGTAGCTG and 5′-TGAACGATCGGGGAAATTCGAGCTCGG for BvFT1-BvFT2 RNAi transformants. A PAT gene-specific probe was amplified using primers 5′-AGATTAGGCCAGCTACAGCAGCTGATA and 5′-GCCTTGGAGGAGCTGGCAACTCAAAAT. Probes were radioactively labeled by random primer labeling (Feinberg and Vogelstein, 1983) using α-32P-dCTP and the large (Klenow) fragment of DNA polymerase I (Life Technologies, Carlsbad, California, USA). Copy number was determined as the number of discrete bands after hybridization. In cases where the number of detectable bands for a given transformant differed between the two enzymes, the detected range of copy numbers is given in Table 1. Transgenic and non-transgenic control plants were clonally multiplied in vitro and transferred to soil according to standard procedures (Hall et al., 1996).
Table 1.
Bolting time after vernalization in independent sugar beet transformants carrying BvFL1 or BvFT1-BvFT2 RNAi or over-expression constructs.
| Effector construct type | Transgenic event number | Total number of vern. plantsa | Number of bolting plants | DTBb (mean ± SDc | Unpaired t-test value for DTB (probabilityd) | Number of non-bolting plants | Total number of non-vern. plantse | PAT gene copy numberf | Effector transgene copy numberf | Target gene expression level |
|---|---|---|---|---|---|---|---|---|---|---|
| BvFL1 | ||||||||||
| RNAi | 019-07G | 15 | 15 | 35.60 ± 3.20 | 6.85 (p < 0.0001) | 0 | n.a. | 2 | 2 | unchanged |
| RNAi | 021-11G | 15 | 15 | 37.00 ± 2.71 | 5.45 (p < 0.0001) | 0 | 9 | 2 | 1 | down |
| RNAi | 021-12A | 15 | 15 | 39.87 ± 4.84 | 1.16 (p = 0.2500) | 0 | n.a. | 2 | 2 | down |
| RNAi | 021-12H | 15 | 15 | 37.67 ± 4.13 | 3.72 (p = 0.0005) | 0 | n.a. | 2 | 2 | down |
| RNAi | 022-10F | 12 | 12 | 38.75 ± 3.24 | 2.68 (p = 0.0100) | 0 | n.a. | 2 | 3 | unchanged |
| RNAi | 024-11E | 20 | 20 | 36.55 ± 2.22 | 7.15 (p < 0.0001) | 0 | n.a. | 1 | 1 | down |
| RNAi | 024-12E | 20 | 20 | 44.15 ± 3.47 | −4.06 (p = 0.0002) | 0 | 6 | 1 | 1 | down |
| Over-expression | 016-05C | 20 | 19 | 49.21 ± 6.31 | −6.96 (p < 0.0001) | 1 | 5 | 1 | 1 | up |
| Over-expression | 016-10A | 15 | 14 | 41.07 ± 3.81 | −0.05 (p = 0.9600) | 1 | 8 | 2 | 3 | up |
| Over-expression | 017-06C | 20 | 20 | 48.55 ± 5.62 | −7.05 (p < 0.0001) | 0 | n.a. | 2–3 | 2–3 | up |
| Over-expression | 017-07C | 15 | 15 | 39.93 ± 4.19 | 1.20 (p = 0.2400) | 0 | n.a. | 1 | 2 | up |
| BvFT1-BvFT2 | ||||||||||
| RNAi | 014-02G | 20 | 18 | 48.83 ± 9.21 | −4.79 (p < 0.0001) | 2 | n.a. | 1 | 1 | down |
| RNAi | 014-07F | 15 | 13 | 73.46 ± 18.37 | −10.3 (p < 0.0001) | 2 | 14 | 1–2 | 1 | down |
| RNAi | 014-08B | 20 | 20 | 43.80 ± 5.78 | −2.54 (p = 0.0140) | 0 | n.a. | 3 | 2 | unchanged |
| RNAi | 018-06E | 15 | 4 | 68.75 ± 5.80 | −18.5 (p < 0.0001) | 11 | n.a. | 1 | 2 | downg |
| RNAi | 018-09A | 15 | 15 | 52.40 ± 4.99 | −11.2 (p < 0.0001) | 0 | 10 | 2 | 1 | down |
| RNAi | 019-01E | 16 | 9 | 93.67 ± 36.73 | −8.46 (p < 0.0001) | 7 | n.a. | 3–4 | 3 | downg |
| RNAi | 020-01E | 15 | 13 | 63.00 ± 8.49 | −14.1 (p < 0.0001) | 2 | 6 | 2 | 2 | down |
| RNAi | 018-12H | 15 | 15 | 57.73 ± 7.64 | −11.9 (p < 0.0001) | 0 | 9 | 2 | 1 | down |
| RNAi | 020-01C | 15 | 14 | 63.93 ± 25.27 | −5.35 (p < 0.0001) | 1 | 8 | 2 | 1 | down |
| RNAi | 020-05G | 15 | 12 | 60.00 ± 15.04 | −7.28 (p < 0.0001) | 3 | n.a. | 2–3 | 2 | down |
| Non-transgenic control | ||||||||||
| n.a. | n.a. | 37 | 37 | 41.03 ± 2.21 | n.a. | 0 | n.a. | n.a. | n.a. | n.a. |
Plants were generated by in-vitro multiplication (cloning) of primary transformants.
Days to bolting after vernalization.
Standard deviation of the mean.
Probability that the DTB value is significantly different from that obtained using the non-transgenic control.
For a subset of transformants, the number of plants indicated in this column was grown without vernalization in the greenhouse over spring and summer under optimal conditions for more than 6 months, but all of these plants failed to bolt.
Copy numbers for the PAT gene and effector transgenes were determined by DNA gel blot analysis.
High transgene expression.
n.a., not applicable.
Growth conditions and phenotyping
Transgenic plants and non-transgenic SES01 control plants were grown side-by-side in the greenhouse in early spring with supplementary lighting under 16 h light/8 h dark cycles. Vernalization and phenotyping was as described previously for Bvbtc1 RNAi transformants (Pin et al., 2012). In brief, vernalization was carried out in a climate chamber at 4°C and 16 h light for 3 months. To avoid devernalization several acclimation steps were performed over a period of 6 weeks during which the temperature was raised from 4 to 25°C during the light cycle and from 4 to 15°C during the dark cycle. Plants were phenotyped for the occurrence and time of bolting three times per week until 6 months after vernalization. Bolting time was defined as the number of days after the end of vernalization for a plant to reach a stem height of 5 cm. For each independent transgenic event 12–20 plants were phenotyped for bolting time. The non-transgenic control comprised 37 clones of the host genotype that was used for transformation. The Student's t-test was used for statistical analysis of phenotypic data. A subset of plants was not vernalized but instead continued to be grown in the greenhouse over spring and summer for more than 6 months under natural daylight conditions with supplementary lighting (16 h).
Gene expression analysis
For each transgenic event and the non-transgenic control genotype, leaf samples of three clones each were harvested before vernalization 2 months after transfer to soil and again at the end of a 12 week vernalization period at Zeitgeber time (ZT) 6-8. For diurnal expression analysis, leaf samples of three individual clones of the BvFL1 over-expressing transformant 016-05C were taken every 2 h over a period of 24 h 4 weeks after the end of vernalization. RNA extraction and cDNA synthesis were done for each of the three biological replicates (clones) separately and exactly as described for Bvbtc1 RNAi transformants (Pin et al., 2012). Primer annealing temperatures and elongation times are given in Table S1. Three technical replicates were performed for each RT-qPCR reaction. RT-qPCR was performed on a CFX96 Real-Time PCR detection system (Bio-Rad Laboratories, Hercules, California, USA) as described in Pin et al. (2012). To determine RT-qPCR efficiencies and serve as positive controls, the endogenous target transcript regions analyzed by RT-qPCR were cloned into the pGEM-T vector (Promega Corporation, Madison, Wisconsin, USA). All plant samples were assayed for expression of the respective RNAi or over-expression target gene and the housekeeping gene BvGAPDH, which was used as a reference gene for normalization. The comparative CT(2−ΔΔCT) method was applied for analysis (Schmittgen and Livak, 2008).
Results
RNAi and over-expression of vernalization response genes in biennial beets
BvFL1 and the FT-like gene pair BvFT1-BvFT2 were analyzed by RNAi or over-expression in a biennial genetic background. Four to ten independent transformants were pre-selected for the presence of transgene inserts and low transgene copy numbers by PCR, TaqMan® assays and DNA gel blot analysis as described in Materials and Methods. Transformants with low copy numbers (1–3) were multiplied by clonal propagation and analyzed for changes in target gene expression and bolting time (Table 1; Table S1).
BvFL1
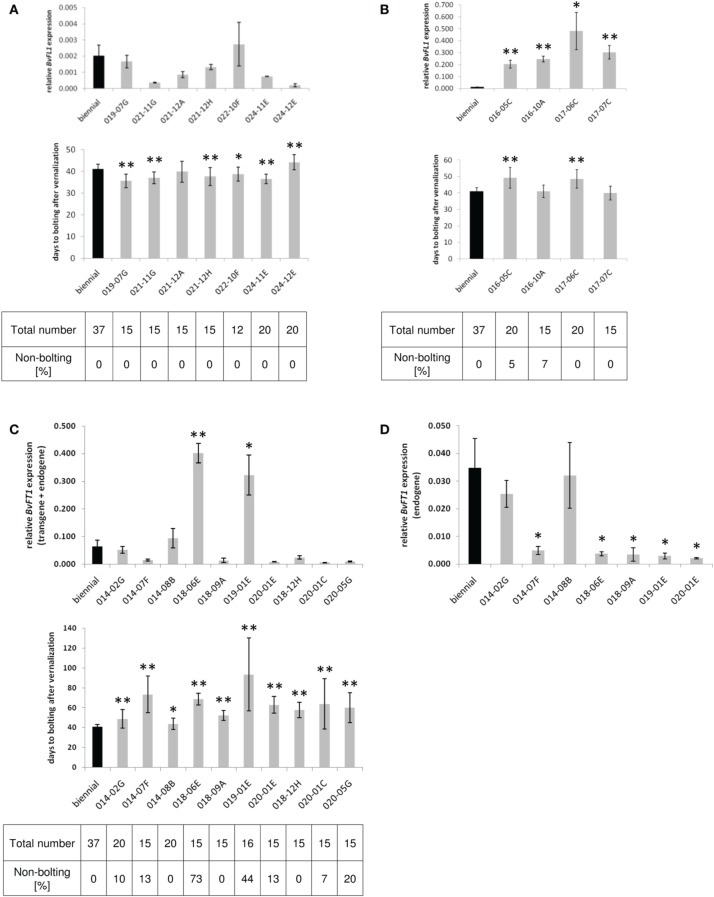
Down-regulation of BvFL1 by RNAi to less than half of the expression level in the non-transgenic control plants was successful in four out of seven transgenic events (Figure 1A). Two of these events (021-11G, 024-12E) showed a reduction to less than 20% of the expression level in the control plants. Following vernalization, all BvFL1 RNAi transformants bolted (Figure 1A; Table 1). The mean days to bolting after the end of vernalization varied from 35.60 to 44.15 days, whereas bolting occurred on average 41.03 days after vernalization in the non-transgenic control plants. In one BvFL1 RNAi event, the mean days to bolting did not deviate significantly from the control plants. Five events bolted 2.28–5.43 days earlier and one event bolted 3.12 days later than the control plants. Of the four BvFL1 RNAi events in which BvFL1 was down-regulated most, two (021-11G, 021-12H) bolted 3–4 days earlier than the control, one (024-12E) bolted 3 days later, and one (021-12A) did not deviate significantly from the control. Together, the data suggest a certain level of experimental noise but did not reveal a clear and consistent phenotypic effect of reduced BvFL1 expression.
Figure 1.
Gene expression and bolting time phenotypes in BvFL1 RNAi (A), BvFL1 over-expression (B), and BvFT1-BvFT2 RNAi transformants (C,D). Leaf samples of non-vernalized plants derived from independent sugar beet transformation events and the non-transgenic biennial control genotype were taken under long-day conditions at Zeitgeber time (ZT) 6-8. For each transgenic event, three clones were analyzed as biological replicates, and each RT-qPCR reaction was run in triplicate. Gene expression was normalized using the house-keeping gene BvGAPDH and the 2−ΔΔCT method (Schmittgen and Livak, 2008). Error bars represent mean ± SE of the mean. Expression of BvFT1 in BvFT1-BvFT2 RNAi plants was determined with primers which co-amplify endogenous and transgenic BvFT1 transcripts (C) and with primers which specifically amplify the endogenous BvFT1 transcript (D). Bolting time was measured in days to bolting after the end of vernalization. The mean of days to bolting and the SE of the mean are shown for plants which bolted within 6 months after the end of vernalization. Significant differences between expression levels in the transformants and the control plants and between bolting time are indicated by asterisks (*α = 0.05, **α = 0.01 according to Student's t-test). The total number of plants per transgenic event and the percentage of plants which failed to bolt within this period are given in the tables below the bar graphs.
Of the four events derived from transformation with a BvFL1 over-expression construct, all showed strong up-regulation of BvFL1 expression (Figure 1B). Bolting time after vernalization varied from 39.93 to 49.21 days. In two events (016-05C, 017-06C), bolting was delayed by approximately 8 days, whereas in the two other events (016-10A, 017-07C) bolting time did not deviate significantly from the control (Table 1). Two events (016-05C, 016-10A) included one plant each which failed to bolt until the end of the experiment 6 months after the end of vernalization.
Of the two events with down-regulation of BvFL1 to less than 20% of the control (021-11G, 024-12E) and two BvFL1 over-expression events (016-05C, 016-10A), an additional 5–9 plants each were grown in parallel for more than 6 months over spring and summer under long-day conditions and without vernalization in the greenhouse, but none of these plants initiated bolting (Table 1).
BvFT1-BvFT2
Out of ten transgenic events derived from transformation with an RNAi construct carrying part of the BvFT1 cDNA, seven exhibited down-regulation of the BvFT1 endogene (Figure 1C). A further analysis of several BvFT1 RNAi transformants revealed that not only BvFT1 but also BvFT2 was down-regulated in these plants, presumably due to RNAi off-target effects (see further below). Therefore, the RNAi transformants expose the effects of co-silencing of both constituents of the BvFT1-BvFT2 module and will be referred to as BvFT1-BvFT2 RNAi events.
Bolting was delayed in all seven events and occurred approximately 8–32 days later than in the control plants (Figure 1C; Table 1). Besides a delay in bolting time, five of the seven BvFT1-BvFT2 RNAi events also included one to three non-bolting plants each among the 15–20 plants that were phenotyped for each of the BvFT1-BvFT2 RNAi events. An additional 47 plants (6–14 plants each of events 014-07F, 018-09A, 020-01E, 018-12H, 020-01C) were grown for more than 6 months without vernalization in the greenhouse and side-by-side with the BvFL1 events mentioned above, but like these did not initiate bolting (Table 1).
Two of the transgenic events (018-06E and 019-01E) stood out in that they appeared to show over-expression of BvFT1 rather than down-regulation (Figure 1C). The primer binding sites of the RT-qPCR assay for BvFT1 expression were located within the segment of the cDNA that was used for construction of the inverted repeat cassette in the RNAi construct. Thus, both the endogenous BvFT1 transcript and the transgene-derived transcript can be co-amplified, suggesting that transcription from the transgene may contribute to the observed high levels of transcript accumulation. To test this possibility, BvFT1 expression was re-analyzed by RT-qPCR using endogene-specific primers (with binding sites outside the cDNA fragment used for RNAi transgene construction) in the two events in question as well as five of the events in which BvFT1 expression was either down-regulated or largely unchanged. For the latter five events, this analysis confirmed the previous expression data. However, for the events in question the endogene-specific RT-qPCR now revealed clear down-regulation of the endogene (Figure 1D). Transcript accumulation in these two events was similarly low as in other transformants in which BvFT1 was down-regulated (<20% of transcript accumulation in the control). Interestingly, the same two events also contained exceptionally high percentages of non-bolting plants (73% in 018-06E and 44% in 019-01E; Figure 1C; Table 1).
Pre- and post-vernalization expression of floral regulators
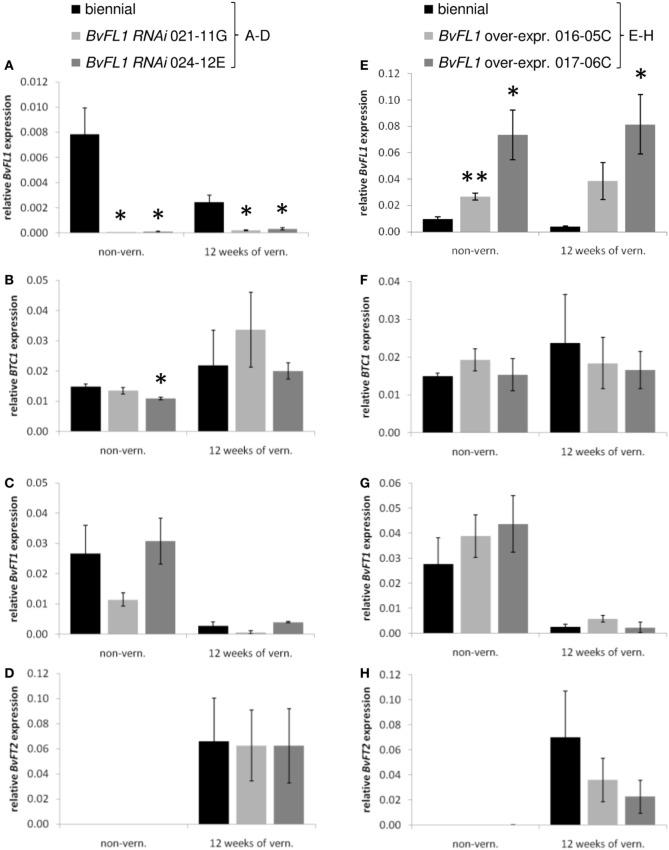
Two independent transgenic events each which showed either clear down- or up-regulation of the gene of interest were analyzed further. These events were assayed for target gene expression before vernalization and at the end of a 12-week vernalization period. Expression of BvFL1 in the non-transgenic control plants was lower at the end of vernalization than before vernalization (Figure 2A). The strong down-regulation of BvFL1 by RNAi in the transgenic events 021-11G and 024-12E when compared to the control plants was evident both before and at the end of vernalization. To test for possible regulatory effects on the three central flowering time control genes thus far identified in beet, the BvFL1 RNAi plants were analyzed for expression of BTC1, BvFT1, and BvFT2 (Figures 2B–D). In the non-transgenic control plants, expression differences between samples harvested before and at the end of vernalization were largely consistent with previous reports (Pin et al., 2010, 2012), i.e., BTC1 and BvFT2 expression levels were higher at the end of vernalization than before vernalization, whereas BvFT1 expression was strongly reduced at the end of vernalization. Down-regulation of BvFL1 by RNAi did not result in consistent changes in expression of any of the central floral regulators.
Figure 2.
Expression of floral regulators in BvFL1 RNAi (A–D) and BvFL1 over-expression (E–H) transformants. Expression of BvFL1 (A,E) and the floral regulators BTC1 (B,F), BvFT1 (C,G), and BvFT2 (D,H) was measured in leaves of non-vernalized plants and at the end of a 12-week vernalization period at ZT 6-8 under long-day conditions. Expression analysis, normalization, and statistical analysis was performed as described for Figure 1. Significant differences between expression levels in the transformants and the control plants are indicated by asterisks (*α = 0.05, **α = 0.01 according to Student's t-test).
In the two 35S::BvFL1 events which were further analyzed (016-05C and 017-06C) BvFL1 was stably over-expressed both before and at the end of vernalization (Figure 2E). The difference in expression levels between the two events was in approximate accordance with the respective transgene copy numbers (1 in 016-05C and 2–3 in 017-06C; Table 1). BTC1 expression did not appear to be majorly affected by BvFL1 over-expression (Figure 2F). BvFT1 expression before vernalization was slightly higher in the 35S::BvFL1 transformants than in the control plants but BvFL1 over-expression did not prevent down-regulation of BvFT1 by vernalization (Figure 2G). BvFT2 expression was not detectable before vernalization in either the control or over-expression plants. BvFT2 was expressed at the end of vernalization and was lower in the BvFL1 transformants than in the controls (Figure 2H). None of the expression levels in the transformants deviated significantly from the control plants.
Expression analysis of BvFT1-BvFT2 RNAi plants before and at the end of vernalization showed down-regulation of both of the FT genes (Figure 3). Because BvFT2 is only expressed after vernalization, down-regulation of this gene was only detectable in the post-vernalization samples (Figure 3C). As described above, BvFT1-BvFT2 RNAi transformants showed low accumulation of both endogene- and transgene-derived transcripts (including 014-07F and 020-01E; Figure 3) except for two events (018-06E, 019-01E) in which the transgene-derived transcripts accumulated to higher levels. The distinction between these two types of events was evident both before and at the end of vernalization.
Figure 3.
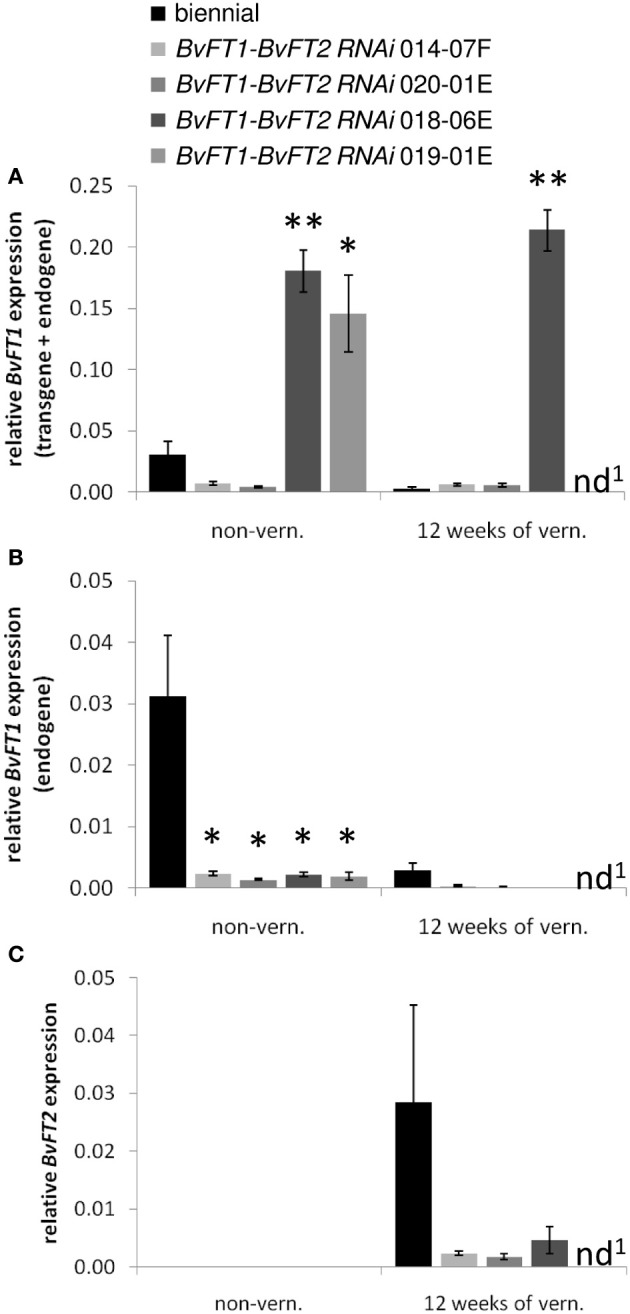
Expression of BvFT1 (A,B) and BvFT2 (C) in BvFT1-BvFT2 RNAi transformants. Expression was measured in leaves of non- vernalized plants and at the end of a 12-week vernalization period at ZT 6-8 under long-day conditions. BvFT1 expression was determined either with primers which co-amplify endogenous and transgenic BvFT1 transcripts (A) or with primers which specifically amplify the endogenous BvFT1 transcript (B). Expression analysis, normalization, and statistical analysis was performed as described for Figure 1. nd, not determined. Significant differences between expression levels in the transformants and the control plants are indicated by asterisks (*α = 0.05, **α = 0.01 according to Student's t-test).
Diurnal expression profiles of floral regulators in BvFL1 over-expression plants
Previous reports for Arabidopsis indicated a regulation of the circadian clock by FLC (Swarup et al., 1999; El-Assal et al., 2003; Salathia et al., 2006). Therefore, the late-bolting BvFL1 over-expression event 016-05C, which carries a single copy of the transgene, was assayed for changes in the diurnal expression profiles of the beet homolog of the circadian clock gene GIGANTEA (GI) (Pin et al., 2012) and the photoperiod response gene BTC1. BvFT1, BvFT2 and BvLHP1, a homolog of the vernalization pathway gene LIKE HETEROCHROMATIN 1 in Arabidopsis (GenBank accession number KJ636469), were also included in the analysis. Diurnal expression was analyzed under long-day conditions (16 h light, 8 h darkness) 4 weeks after the end of vernalization.
In the non-transgenic control plants, BvFL1 had two broad peaks of expression at mid-day to mid-afternoon and in the second half of the night until early morning (Figure 4A), indicating that BvFL1 itself is diurnally regulated. Over-expression of BvFL1 resulted in strongly increased transcript accumulation during the entire course of the day (Figure 4B). Transcript accumulation was not constant but peaked at ZT 12. Diurnal fluctuations of similar amplitude in expression from a CaMV 35S promoter in transgenic plants were observed before (Millar et al., 1992; Lu et al., 2011). BvLHP1 transcript accumulation exhibited two peaks in the early afternoon (ZT 10) and in the middle of the night (ZT 20; Figure 4C). Over-expression of BvFL1 correlated with a phase shift by approximately 2 h in BvLHP1 expression during the light cycle compared to the control plants, resulting in a peak of expression at ZT 12. BvLHP1 expression in the dark was in phase with the control. Expression of BvGI (Figure 4D) and BTC1 (Figure 4E) was similar as reported previously (Pin et al., 2012). While expression of both genes reached its maximum around mid-afternoon, the peak of expression was broader for BTC1. Overall, the expression profiles of both BvGI and BTC1 were similar in the BvFL1 over-expressing plants and the control plants.
Figure 4.
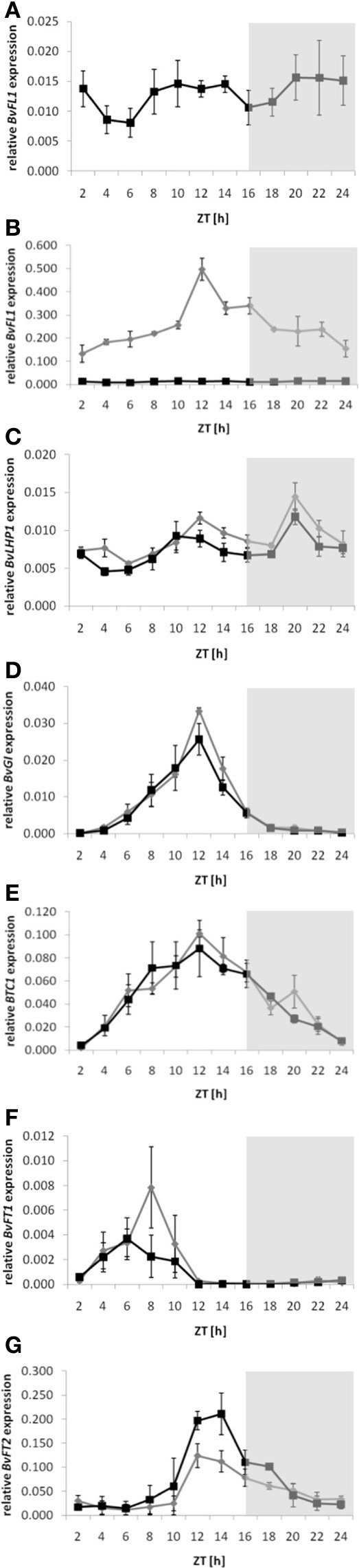
Diurnal expression profiles of floral regulator genes or candidate genes in BvFL1 over-expressing plants. Expression in the BvFL1 over-expressing transgenic event 016-05C (gray line and diamonds) and the biennial control genotype (black line and squares) was determined 4 weeks after the end of vernalization under long-day conditions. (A,B) BvFL1, (C) BvLHP1, (D) BvGI, (E) BTC1, (F) BvFT1, and (G) BvFT2. Expression analysis and normalization was performed as described for Figure 1.
As reported before (Pin et al., 2010, 2012), the floral repressor BvFT1 is only relatively weakly expressed after vernalization but exhibits detectable transcript accumulation in the morning hours, whereas expression of the floral activator BvFT2 peaks around mid-afternoon. Expression of these genes in the control plants of the current study were in accordance with the previous reports (Figures 4F,G). However, the BvFL1 over-expressing plants revealed the following deviations from the regular expression patterns in the control plants: (1) BvFT1 expression showed a sharp peak of increased expression around ZT 8, and (2) BvFT2 showed a reduction in gene expression in the afternoon and evening when compared to the control plants.
Discussion
Recent studies have revealed a central role of the B locus gene BTC1 and its downstream target gene BvFT1 in vernalization response and bolting control in beet (Pin et al., 2010, 2012). Furthermore, in contrast to Arabidopsis, where vernalization requirement and growth habit is governed by natural variation at FLC or its upstream activator FRI, life cycle control in beet is determined by allelic variants of BTC1. Despite the apparent differences in the genetic make-up of the core regulatory modules in Arabidopsis and beet, an FLC homolog has been identified in beet (Reeves et al., 2007). Complementation analysis in an early-flowering flc mutant in Arabidopsis showed that the FLC-like gene BvFL1 was able to rescue the wild-type phenotype, but the function of BvFL1 has not yet been analyzed in beet. The current study aimed to address the functional role of BvFL1 and possible regulatory interactions with BTC1 and/or the BvFT1-BvFT2 module by transgenic analyses in beet. The main findings are that (1) down-regulation of BvFL1 neither affects bolting time majorly after vernalization nor enables bolting without vernalization, (2) over-expression of BvFL1 is not sufficient to prevent bolting after vernalization but can result in a moderate delay of bolting, and (3) co-silencing of the BvFT1-BvFT2 module in BvFT1 RNAi transformants leads to a stronger bolting delay than BvFL1 over-expression and high percentages of non-bolting plants in some events.
The observed lack of a floral inductive effect in BvFL1 RNAi transformants stands in contrast to observations in Arabidopsis, where mutation or antisense-mediated down-regulation of FLC strongly accelerates flowering (Michaels and Amasino, 1999; Sheldon et al., 1999, 2000) and can eliminate the very late-flowering phenotype found in winter-annual (vernalization-responsive) accessions (Michaels and Amasino, 2001). This observation corroborates the notion from the work on BTC1 and BvFT1 that in beet a different regulatory switch has evolved for the control of growth habit, and shows for the first time that in beet altered regulation of the FLC-like gene is not sufficient to promote an early-bolting (annual) growth habit. The fact that BvFL1 RNAi transformants are responsive to vernalization further suggests that vernalization can promote bolting through a BvFL1-independent pathway. In Arabidopsis, despite the regulatory role of FLC in the vernalization pathway, flc null mutants are also vernalization-responsive, which suggested the presence of an FLC-independent vernalization response pathway also in this species (Michaels and Amasino, 2001). Later work implicated other MADS-box genes in FLC-independent regulation of vernalization response (Alexandre and Hennig, 2008). In beet, a BvFL1-independent vernalization response pathway is likely to involve at least in part the actions of BTC1 and BvFT1.
While BvFL1 may not have a key role in the regulation of vernalization requirement and response in beet, the moderate delay in bolting that was observed in transformants over-expressing BvFL1 suggests that the gene has retained a functional role in the control of floral transition, and is consistent with the earlier complementation studies in Arabidopsis (Reeves et al., 2007). However, phenotypic effects of over-expression are not a definite proof of a gene's function in an endogenous biological process. For example, ectopic expression of the A. thaliana gene FLC in rice also delayed flowering despite the absence of FLC-like genes in rice (Tadege et al., 2003). With regard to growth habit, the biennial, vernalization-responsive sugar beet accession used in the current study is similar to winter-annual Arabidopsis accessions. Over-expression of FLC in winter-annual Arabidopsis accessions frequently resulted in transformants which completely failed to flower (Michaels and Amasino, 1999; Sheldon et al., 1999). In beet, however, complete suppression of bolting by BvFL1 over-expression was not observed. The notion that BvFL1 expression is not sufficient to prevent floral transition in beet also appears consistent with the previous finding that the temporary down-regulation of BvFL1 during vernalization is reversed upon transfer to warmer temperatures (Reeves et al., 2007). The moderate phenotypic effects of altered BvFL1 expression may suggest that the strong floral inhibitory effect of BvFT1 (Pin et al., 2010) masks or overrides a possible contributory role of BvFL1 in the repression of bolting.
In Arabidopsis, allelic variation at FLC was suggested to affect circadian period length (Swarup et al., 1999) and over-expression of FLC lengthened the circadian period by approximately 1 h (Salathia et al., 2006). Salathia et al. further argued that repression of FLC in response to vernalization and the resultant shorter circadian periods may reduce the critical daylength required for the photoperiod pathway to promote flowering, thus accelerating flowering in spring. In our study, expression of BvFL1 showed diurnal oscillations both in the biennial control plants and the BvFL1 over-expressing plants. While strong diurnal oscillations of FLC have not been reported in Arabidopsis (e.g., Fujiwara et al., 2010), a similar expression profile to that observed here for BvFL1 was found by Lu et al. (2011), with peaks of FLC expression in the afternoon and at the end of the night. Among the putative clock-regulated genes analyzed in BvFL1 over-expression plants, neither BvGI nor BTC1 were majorly affected in their diurnal expression profiles. Expression of both of these genes and of BvLHP1 was slightly elevated in the afternoon hours in the BvFL1 over-expressing plants, but the differences were too subtle to be conclusive.
FLC inhibits floral transition at least in part by repression of FT in leaves, which involves a direct interaction of FLC protein with FT chromatin (Helliwell et al., 2006; Searle et al., 2006). Our data for beet tentatively suggest that BvFL1 over-expression leads to a reduction of BvFT2 expression, which is apparent both at the end of vernalization (Figure 2H) and in the diurnal expression profile 4 weeks after vernalization (Figure 4G). BvFT2 expression in BvFL1 over-expressing transformants rises more slowly in the mid-day hours and is reduced compared to the control plants during the afternoon and evening hours. This suggests that the observed bolting delay may also be mediated by negative regulation of BvFT2 by BvFL1 in beet. However, although BvFT2 down-regulation is consistent in all samples and is apparent at multiple consecutive time points in the diurnal expression profile, the differences are not statistically significant in pairwise comparisons with the respective controls. Similarly, the analysis of BvFT1 in BvFL1 over-expression plants showed an increase in BvFT1 expression which however was not statistically significant. Thus, it remains speculative whether changes in BvFT1 and/or BvFT2 expression mediate the observed bolting delay in BvFL1 over-expression plants.
The current study also revealed phenotypic effects of co-silencing of BvFT1 and BvFT2. BvFT1 and BvFT2 share 80% sequence identity at the nucleotide sequence level within the 361 bp region of the coding sequence that was used for RNAi vector construction, including a 23 bp tract of perfect sequence identity, suggesting that down-regulation of BvFT2 is due to off-target effects. Down-regulation of BvFT1 by RNAi had not been achieved previously (cf. Pin et al., 2010). Although down-regulation of BvFT1 in the RNAi transformants investigated here was accompanied by down-regulation of BvFT2, the data provide new evidence for the critical role in bolting control of the BvFT1-BvFT2 module in beet, and show that the concomitant down-regulation of both activities inhibits rather than promotes bolting. The data also suggest that BvFT1 expression before vernalization as it is typical for biennial beets is not necessary for pre-vernalization development and that the main function of BvFT1 is its role in the control of vernalization response. This notion is consistent with the apparent lack of BvFT1 expression in annual beets throughout development (under long-day conditions; Pin et al., 2010). All BvFL1 RNAi, BvFL1 over-expression and BvFT1-BvFT2 RNAi transformants investigated here were grown and analyzed side-by-side with each other as well as with the btc1 RNAi transformants described by Pin et al. (2012), thus facilitating a comparative view. The strong phenotypic effect of altered regulation of the BvFT1-BvFT2 module in BvFT1 RNAi transformants when compared to BvFL1 RNAi or over-expression point at the predominant role of the FT genes in bolting control in beet. Finally, it is also interesting to note that among all the BvFL1, BvFT1-BvFT2, and btc1 RNAi transformants, it was clearly the btc1 RNAi transformants which showed the strongest suppression of bolting, with multiple transgenic events in which bolting was completely suppressed until the end of the experiment 6 months after the end of vernalization (Pin et al., 2012).
Perhaps noteworthily, the strongest inhibitory effect on bolting was found in two transgenic events (018-06E and 019-01E) in which the BvFT1 transgene was highly expressed despite a strong reduction in accumulation of the endogenous BvFT1 transcript. A large number of plants derived from these events failed to bolt after vernalization [11 out of 15 plants (73%) and 7 out of 16 plants (44%), respectively], while the remaining plants of these events bolted very late and showed a stunted phenotype similar to btc1 RNAi transformants (Pin et al., 2012). The concomitant accumulation of the transgene transcript and silencing of the endogenous transcript may suggest that in these transformants transgenic and endogenous transcripts are not co-suppressed, but that the transgene transcript may trigger RNAi of the endogenous transcript without itself being a target of (efficient) RNAi-mediated transcript degradation. Because the two transformants carry multiple copies of the transgene it is conceivable that at least one of these copies carries the complete BvFT1 inverted repeat cassette and effects RNAi, whereas another copy may have integrated only partially and escaped silencing. The cDNA fragment used for RNAi transgene construction spans ~67% of the full-length coding sequence and covers 88% of the central PEBP domain, including the functionally important amino acids in the fourth exon (Pin et al., 2010). The putative translation product, starting with the first in-frame ATG codon downstream of the 35S promoter, is predicted to contain 92 amino acids (~51%) of the full-length protein and ~67% of the PEBP domain. Thus, it is conceivable that expression of a partial BvFT1 protein at least contributes to the particularly strong inhibition of bolting observed in these transformants. In this scenario, the protein sequence outside the 92 amino acid region would appear to be dispensable for repression of bolting by BvFT1.
In conclusion, our data show that BvFL1 is not a major regulator of vernalization response in beet. A comparison with phenotypic data from BvFT1-BvFT2 RNAi plants and our previously described btc1 RNAi transformants further suggests that in beet the BvFT1-BvFT2 module and its upstream regulator BTC1 have evolved a more dominant role in the control of vernalization reponse and bolting time. Future comparative studies between both species may help to uncouple the contributions of FLC and FLC-like genes to floral regulation through direct effects on FT genes or upstream interactions between vernalization- and photoperiod responsive flowering time control mechanisms. From an evolutionary perspective, knowledge of conservation and divergence of floral control mechanisms between model species and the phylogenetically distant dicot species B. vulgaris is casting an increasingly interesting light on one of the best studied developmental processes in plants.
Author contributions
Sebastian H. Vogt designed and performed experiments and wrote the manuscript. Guy Weyens and Bettina Bork designed and performed experiments. Marc Lefèbvre and Axel Schechert designed and supervised experiments. Andreas E. Müller designed and supervised the project and wrote the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Bettina Rohardt, Murielle Lommel, Sigrid Vanstraelen, Marlies Athmer and Markus Wolf for technical assistance, support and discussions, Gretel Schulze-Buxloh for BvLHP1 sequence information, and Christian Jung for continuous support throughout the project. The Institute of Clinical Molecular Biology, University of Kiel, Germany, is acknowledged for performing the Sanger-based sequence analyses of this study. Andreas E. Müller was funded through a grant by the Deutsche Forschungsgemeinschaft (DFG JU 205/14-1). The project was funded by the German Federal Ministry of Education and Research (grant number 0315058A/B).
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00146/abstract
References
- Alexandre C. M., Hennig L. (2008). FLC or not FLC: the other side of vernalization. J. Exp. Bot. 59, 1127–1135 10.1093/jxb/ern070 [DOI] [PubMed] [Google Scholar]
- Andrés F., Coupland G. (2012). The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639 10.1038/nrg3291 [DOI] [PubMed] [Google Scholar]
- Chaw S. M., Chang C. C., Chen H. L., Li W. H. (2004). Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J. Mol. Evol. 58, 424–441 10.1007/s00239-003-2564-9 [DOI] [PubMed] [Google Scholar]
- Davies T. J., Barraclough T. G., Chase M. W., Soltis P. S., Soltis D. E., Savolainen V. (2004). Darwin's abominable mystery: insights from a supertree of the angiosperms. Proc. Natl. Acad. Sci. U.S.A. 101, 1904–1909 10.1073/pnas.0308127100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin P. F., Kay S. A. (2000). Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell 12, 2499–2509 10.2307/3871244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Assal S. E. D., Alonso-Blanco C., Peeters A. J. M., Wagemaker C., Weller J. L., Koornneef M. (2003). The role of Cryptochrome 2 in flowering in arabidopsis. Plant Physiol. 133, 1504–1516 10.1104/pp.103.029819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. (1983). A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132, 6–13 10.1016/0003-2697(83)90418-9 [DOI] [PubMed] [Google Scholar]
- Fujiwara S., Nakagawa M., Oda A., Kato K., Mizoguchi T. (2010). Photoperiod pathway regulates expression of MAF5 and FLC that encode MADS-box transcription factors of the FLC family in Arabidopsis. Plant Biotechnol. 27, 447–454 10.5511/plantbiotechnology.10.0823b [DOI] [Google Scholar]
- Gould P. D., Ugarte N., Domijan M., Costa M., Foreman J., Macgregor D., et al. (2013). Network balance via CRY signalling controls the Arabidopsis circadian clock over ambient temperatures. Mol. Syst. Biol. 9, 650 10.1038/msb.2013.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R. D., Riksen-Bruinsma T., Weyens G. J., Rosquin I. J., Denys P. N., Evans I. J., et al. (1996). A high efficiency technique for the generation of transgenic sugar beets from stomatal guard cells. Nat. Biotechnol. 14, 1133–1138 10.1038/nbt0996-1133 [DOI] [PubMed] [Google Scholar]
- Hébrard C., Trap-Gentil M.-V., Lafon-Placette C., Delaunay A., Joseph C., Lefèbvre M., et al. (2013). Identification of differentially methylated regions during vernalization revealed a role for RNA methyltransferases in bolting. J. Exp. Bot. 64, 651–656 10.1093/jxb/ers363 [DOI] [PubMed] [Google Scholar]
- Helliwell C. A., Wood C. C., Robertson M., Peacock W. J., Dennis E. S. (2006). The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecularweight protein complex. Plant J. 46, 183–192 10.1111/j.1365-313X.2006.02686.x [DOI] [PubMed] [Google Scholar]
- Lu S. X., Knowles S. M., Webb C. J., Celaya R. B., Cha C., Siu J. P., et al. (2011). The jumonji C domain-containing protein JMJ30 regulates period length in the arabidopsis circadian clock. Plant Physiol. 155, 906–915 10.1104/pp.110.167015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S. D., Amasino R. M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956 10.2307/3870827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S. D., Amasino R. M. (2001). Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13, 935–941 10.2307/3871350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A. J., Short S. R., Chua N. H., Kay S. A. (1992). A novel circadian phenotype based on firefly luciferase expression in transgenic plants. Plant Cell 4, 1075–1087 10.2307/3869476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R. L., Klein R. R., Morishige D. T., Brady J. A., Rooney W. L., Miller F. R., et al. (2011). Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc. Natl. Acad. Sci. U.S.A. 108, 16469–16474 10.1073/pnas.1106212108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Périlleux C., Pieltain A., Jacquemin G., Bouché F., Detry N., D'Aloia M., et al. (2013). A root chicory MADS box sequence and the Arabidopsis flowering repressor FLC share common features that suggest conserved function in vernalization and de-vernalization responses. Plant J. 75, 390–402 10.1111/tpj.12208 [DOI] [PubMed] [Google Scholar]
- Pin P. A., Benlloch R., Bonnet D., Wremerth-Weich E., Kraft T., Gielen J. J. L., et al. (2010). An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330, 1397–1400 10.1126/science.1197004 [DOI] [PubMed] [Google Scholar]
- Pin P. A., Nilsson O. (2012). The multifaceted roles of FLOWERING LOCUS T in plant development. Plant Cell Environ. 35, 1742–1755 10.1111/j.1365-3040.2012.02558.x [DOI] [PubMed] [Google Scholar]
- Pin P. A., Zhang W., Vogt S. H., Dally N., Büttner B., Schulze-Buxloh G., et al. (2012). The role of a pseudoresponse regulator gene in life cycle adaptation and domestication of beet. Curr. Biol. 22, 1095–1101 10.1016/j.cub.2012.04.007 [DOI] [PubMed] [Google Scholar]
- Reeves P. A., He Y., Schmitz R. J., Amasino R. M., Panella L. W., Richards C. M. (2007). Evolutionary conservation of the FLOWERING LOCUS C mediated vernalization response: evidence from the sugar beet (Beta vulgaris). Genetics 176, 295–307 10.1534/genetics.106.069336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse D. T., Sheldon C. C., Bagnall D. J., Peacock W. J., Dennis E. S. (2002). FLC, a repressor of flowering, is regulated by genes in different inductive pathways. Plant J. 29, 183–191 10.1046/j.0960-7412.2001.01210.x [DOI] [PubMed] [Google Scholar]
- Ruelens P., de Maagd R. A., Proost S., Theissen G., Geuten K., Kaufmann K. (2013). FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nat. Commun. 4, 2280 10.1038/ncomms3280 [DOI] [PubMed] [Google Scholar]
- Salathia N., Davis S. J., Lynn J. R., Michaels S. D., Amasino R. M., Millar A. J. (2006). FLOWERING LOCUS C-dependent and -independent regulation of the circadian clock by the autonomous and vernalization pathways. BMC Plant Biol. 6:10 10.1186/1471-2229-6-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Searle I., He Y., Turck F., Vincent C., Fornara F., Kröber S., et al. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20, 898–912 10.1101/gad.373506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon C. C., Burn J. E., Perez P. P., Metzger J., Edwards J. A., Peacock W. J., et al. (1999). The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458 10.2307/3870872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon C. C., Rouse D. T., Finnegan E. J., Peacock W. J., Dennis E. S. (2000). The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. U.S.A. 97, 3753–3758 10.1073/pnas.97.7.3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K., Alonso-Blanco C., Lynn J. R., Michaels S. D., Amasino R. M., Koornneef M., et al. (1999). Natural allelic variation identifies new genes in the Arabidopsis circadian system. Plant J. 20, 67–77 10.1046/j.1365-313X.1999.00577.x [DOI] [PubMed] [Google Scholar]
- Tadege M., Sheldon C. C., Helliwell C. A., Upadhyaya N. M., Dennis E. S., Peacock W. J. (2003). Reciprocal control of flowering time by OsSOC1 in transgenic Arabidopsis and by FLC in transgenic rice. Plant Biotechnol. J. 1, 361–369 10.1046/j.1467-7652.2003.00034.x [DOI] [PubMed] [Google Scholar]
- Trap-Gentil M. V., Hébrard C., Lafon-Placette C., Delaunay A., Hagège D., Joseph C., et al. (2011). Time course and amplitude of DNA methylation in the shoot apical meristem are critical points for bolting induction in sugar beet and bolting tolerance between genotypes. J. Exp. Bot. 62, 2585–2597 10.1093/jxb/erq433 [DOI] [PubMed] [Google Scholar]
- Turner A., Beales J., Faure S., Dunford R. P., Laurie D. A. (2005). The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310, 1031–1034 10.1126/science.1117619 [DOI] [PubMed] [Google Scholar]
- Wang R., Farrona S., Vincent C., Joecker A., Schoof H., Turck F., et al. (2009). PEP1 regulates perennial flowering in Arabis alpina. Nature 459, 423–427 10.1038/nature07988 [DOI] [PubMed] [Google Scholar]
- Yan L., Fu D., Li C., Blechl A., Tranquilli G., Bonafede M., et al. (2006). The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc. Natl. Acad. Sci. U.S.A. 103, 19581–19586 10.1073/pnas.0607142103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Loukoianov A., Blechl A., Tranquilli G., Ramakrishna W., SanMiguel P., et al. (2004). The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303, 1640–1644 10.1126/science.1094305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Loukoianov A., Tranquilli G., Helguera M., Fahima T., Dubcovsky J. (2003). Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. U.S.A. 100, 6263–6268 10.1073/pnas.0937399100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky M. J., Kay S. A. (2002). Molecular basis of seasonal time measurement in Arabidopsis. Nature 419, 308–312 10.1038/nature00996 [DOI] [PubMed] [Google Scholar]
- Zografos B. R., Sung S. (2012). Vernalization-mediated chromatin changes. J. Exp. Bot. 63, 4343–4348 10.1093/jxb/ers157 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.