Abstract
That each of us is truly biologically unique, extending to even monozygotic, “identical” twins, is not fully appreciated. Now that it is possible to perform a comprehensive “omic” assessment of an individual, including one's DNA and RNA sequence, and at least some characterization of one's proteome, metabolome, microbiome, autoantibodies, and epigenome, it has become abundantly clear that each of us has truly one-of-a-kind biological content. Well beyond the allure of the matchless fingerprint or snowflake concept, these singular, individual data and information set up a remarkable and unprecedented opportunity to improve medical treatment and develop preventive strategies to preserve health.
From Digital to Biological to Individualized Medicine
In 2010, Eric Schmidt of Google said “The power of individual targeting—the technology will be so good it will be very hard for people to watch or consume something that has not in some sense been tailored for them (Jenkins, 2010).” Although referring to the capability of digital technology, we have now reached a time of convergence of the digital and biologic domains. It has been well established that 0,1 are interchangeable with A, C, T and G in books and Shakespeare sonnets, and that DNA may represent the ultimate data storage system (Church, 2012, Goldman et al., 2013b). Biological transistors, also known as genetic logic gates, have now been developed that make a computer from a living cell (Bonnet et al., 2013). The convergence of biology and technology was further captured by one of the protagonists of the digital era, Steve Jobs, who said “I think the biggest innovations of the 21st century will be at the intersection of biology and technology. A new era is beginning (Issacson, 2011).”
With whole genome DNA sequencing and a variety of omic technologies to define aspects of each individual's biology at many different levels, we have indeed embarked on a new era of medicine. The term “personalized medicine” has been used for many years, but has engendered considerable confusion. A recent survey indicated that only 4% of the public understand what the term is intended to mean (Stanton, 2013) and the hackneyed, commercial use of “personalized” makes many people think this refers to a concierge service of medical care. While “person” refers to a human being, “personalized” can mean anything from having monogrammed stationary or luggage to ascribing personal qualities. Therefore, It was not surprising that a committee representing the National Academy of Sciences proposed using the term “precision medicine” as defined by “tailoring of medical treatment to the individual characteristics of each patient (Disease, 2011).” Although the term “precision” denotes the objective of exactness, ironically, it, too, can be viewed as ambiguous in this context because it does not capture the sense that the information is derived from the individual. For example, many laboratory tests could be made more precise by assay methodology, and treatments could be made more precise by avoiding side effects-- without having anything to do with a specific individual. Other terms that have been suggested include genomic, digital and stratified medicine, but all of these have a similar problem or appear to be too narrowly focused.
The definition of individual is a single human being, derived from the latin word individu, or indivisible. I propose individualized medicine as the preferred term, since it has a useful double entendre. It relates not only to medicine that is particularized to a human being, but also the future impact of digital technology on individual's driving their health care. There will increasingly be the flow of one's biologic data and relevant medical information directly to the individual. Be it a genome sequence on a tablet, or the results of a biosensor for blood pressure or another physiologic metric displayed on a smartphone, the digital convergence with biology will definitively anchor the individual as a source of salient data, the conduit of information flow, and a, if not the, principal driver of medicine in the future.
The Human GIS
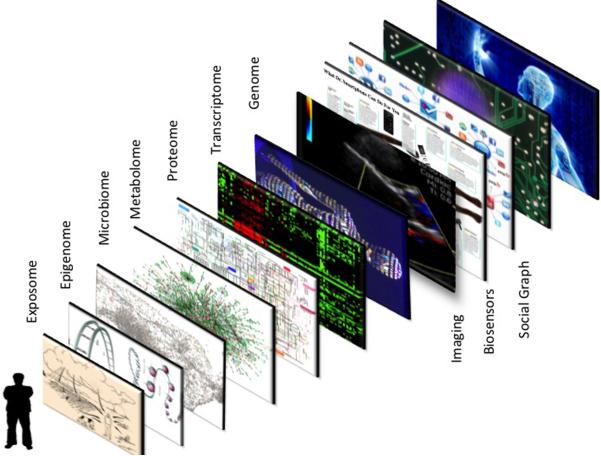
Perhaps the most commonly used geographic information systems (GIS) are Google maps, which provide a layered approach to data visualization, such as viewing a location via satellite overlaid with street names, landmarks, and real-time traffic data. This GIS exemplifies the concept of gathering and transforming large bodies of data to provide exquisite temporal and location information. With the multiple virtual views, it gives one the sense of physically being on site. While Google has digitized and thus created a GIS for the Earth, it is now possible to digitize a human being. As shown in Figure 1, there are multiple layers of data that can now be obtained for any individual. This includes data from biosensors, scanners, electronic medical records, social media, and the various omics that include DNA sequence, transcriptome, proteome, metabolome, epigenome, microbiome, and exposome. Going forward, I will use the term “panoromic” to denote the multiple biologic omic technologies. This term closely resembles and is adopted from panoramic, which refers to a wide-angle view or comprehensive representation across multiple applications and repositories. Or more simply, according to the Merriam-Webster definition of panoramic, it “includes a lot of information and covers many topics.” Thus the term panoromic may be well suited for portraying the concept of big biological data.
Figure 1. Geographic information system (GIS) of a human being.
The ability to digitize the medical essence of a human being is predicated on the integration of multi-scale data, akin to a Google map, which consists of superimposed layers of data such as street, traffic and satellite views. For a human being, these layers include demographics and the social graph, biosensors to capture the individual's physiome, imaging to depict the anatomy (often along with physiologic data), and the biology from the various omics (genome-DNA sequence, transcriptome, proteome, metabolome, microbiome, and epigenome). In addition to all these layers, there is one's important environmental exposure data, known as the “exposome.”
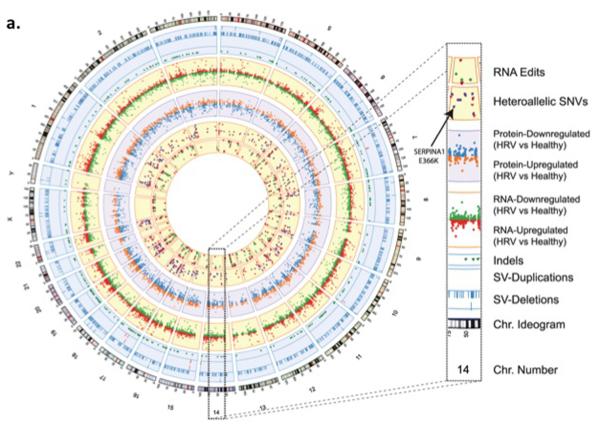
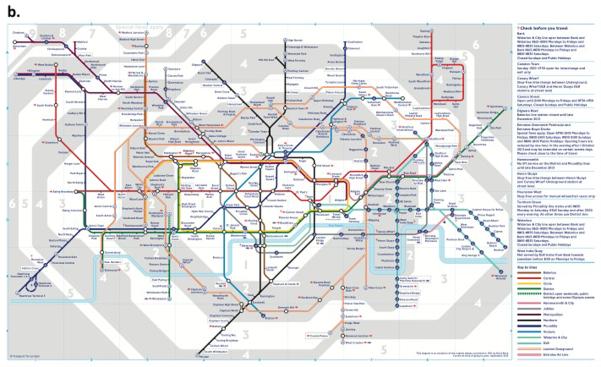
The first individual who had a human GIS-like construct was Michael Snyder. Not only was his whole genome sequenced, he also collected serial gene expression, autoantibody, proteomic and metabolomic (Chen et al., 2012) samples. A portion of the data deluge that was generated is represented in the Circos plot of Figure 2 or an adoption of the London Tube map (Shendure and Lieberman Aiden, 2012). The integrated personal omics profiling (iPOP) or “Snyderome,” as it became known, proved to be useful for connecting viral infections to markedly elevated glucose levels. With this integrated analysis in hand Michael Snyder changed his lifestyle, eventually restoring normal glucose homeostasis. Since that report in 2012, Snyder and his team have proceeded to obtain further omic data, including whole genome DNA methylation data at multiple time points, serial microbiome (gut, urine, nasal, skin and tongue) sampling and the use of biosensors for activity tracking and heart rhythm. Snyder also discovered several extended family members had smoldering, unrecognized glucose intolerance, thereby changing medical care for multiple individuals.
Figure 2. Plots of panoromic information.
a) Circos plot from the Snyderome
Circos plot summarizing the Snyder genome. From outer to inner rings: chromosome ideogram; genomic data (pale blue ring), structural variants >50 bp (deletions [blue tiles], duplications [red tiles]), indels (green triangles); transcriptomic data (yellow ring), expression ratio of viral infection to healthy states; proteomic data (light purple ring), ratio of protein levels during human rhinovirus (HRV) infection to healthy states; transcriptomic data (yellow ring), differential heteroallelic expression ratio of alternative allele to reference allele for missense and synonymous variants (purple dots) and candidate RNA missense and synonymous edits (red triangles, purple dots, orange triangles and green dots, respectively). From From (Chen et al., 2012) with permission.
(b) Adopted London Tube Model of Integrated Omics from Shendure and Aiken
Integration of the many applications of next-generation DNA sequencing which include: sites of DNA methylation (Methyl-Seq), Protein-DNA interactions (CHIP-Seq), 3-dimensional genome structure (Hi-C), genetically targeted purification of polysomal mRNAs (TRAP), the B-cell and T-cell repertoires (Immuno-Seq), and functional consequences of genetic variation (Synthetic saturation mutagenesis) with a small set of core techniques, represented as open circles of “stations.” Like subway lines, individual sequencing experiments move from station to station, until they ultimately arrive at a common terminal: DNA sequencing.
From (Shendure and Lieberman Aiden, 2012) with permission.
Of note, to obtain the data and process this first panoromic study, it required an armada of 40 experienced co-authors and countless hours of bioinformatics and analytical work. To give context to the digital data burden, it took 1 TB for DNA sequence, 2 TB for the epigenomic data, 0.7 TB for the transcriptome, and 3 TB for the microbiome. Accordingly, this first human GIS can be considered a remarkable academic feat and yielded key diagnostic medical information for the individual. But, it can hardly be considered practical or scalable at this juncture. With the cost of storing information continuing to drop substantially, the bottleneck for scalability will likely be automating the analysis. On the other hand, each omic technology can readily be undertaken now and has the potential of providing meaningful medical information for an individual.
The Omic Tools
Whole Genome and Exome Sequencing
Perhaps the greatest technologic achievement in the biomedical domain has been the extraordinary progress in our ability to sequence a human genome over the past decade. Far exceeding the pace of Moore's Law for the relentless improvement in transistor capacity, there has been a >4-log order (or 0.00007th) reduction in cost of sequencing (Butte, 2013), with a cost in 2004 of approximately $28.8 million compared with the cost as low as $1000 in 2014 (Hayden, 2014). However, despite this incomparable progress, there are still major limitations to how rapid, accurate, and complete sequencing can be accomplished. High-throughput sequencing involves chopping the DNA into small fragments, which are then amplified by PCR. Currently it takes 3–4 days in our lab to do the sample preparation and sequencing at 30 to 40X coverage of a human genome. The read length of the fragments is now ~250 base pairs for the most cost-effective sequencing methods, but this is still suboptimal in determining maternal vs. paternal alleles, or what is known as phasing. Since so much of understanding diseases involves compound heterozygote mutations, cis-acting sequence variant combinations, and allele-specific effects, phasing the diploid genome, or what we have called “diplomics” (Tewhey et al., 2011) is quite important. Recently, Moleculo introduced a method for synthetically stitching together DNA sequencing reads yielding fragments as long as 10,000 basepairs. These synthetic long reads are well suited for phasing. Unfortunately, the term “whole genome sequencing” is far from complete since about 900 genes, or 3–4% of the genome, are not accessible (Marx, 2013). These regions are typically in centromeres or telomeres. Other technical issues that detract from accuracy include long sequences of repeated bases (“homopolymers”), and regions rich in guanine and cytosine. Furthermore, the accuracy for medical grade sequencing still needs to be improved. A missed call rate of 1 in 10,000, which may not seem high, translates into a substantial number of errors when considering the 6 billion bases in a diploid genome. These errors obfuscate rare, but potentially functional variants. Beyond this issue, the accurate determination of insertions, deletions and structural variants is impaired, in part due to the relatively short reads that are typically obtained. The Clinical Sequencing Exploratory Research (CSER) program at the National Institutes of Health is aimed at improving the accuracy of sequencing for medical applications (Institute, 2013).
Despite these shortcomings, the ability to identify rare or low frequency variants that are pathogenic has been a major outgrowth of high throughput sequencing. Well beyond the genome scans and genome-wide association studies that identified common variants associated with most complex, polygenic diseases and human traits, sequencing leads to high definition of the uncommon variants that typically have much higher penetrance. For example, rare Mendelian conditions have seen a remarkable surge of definition of their genomic underpinnings (Boycott et al., 2013). In the first half of 2010, the basis for 4 rare diseases was published, but in the first half of 2012 that number jumped to 68 (Boycott et al., 2013). With the power of sequencing, it is anticipated that the molecular basis for most of the 7000 known Mendelian diseases will be unraveled in the next few years.
The 1.5% versus 98.5% Genome Sequencing Dilemma
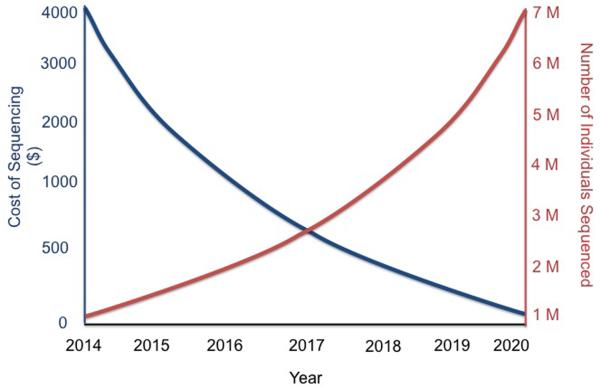
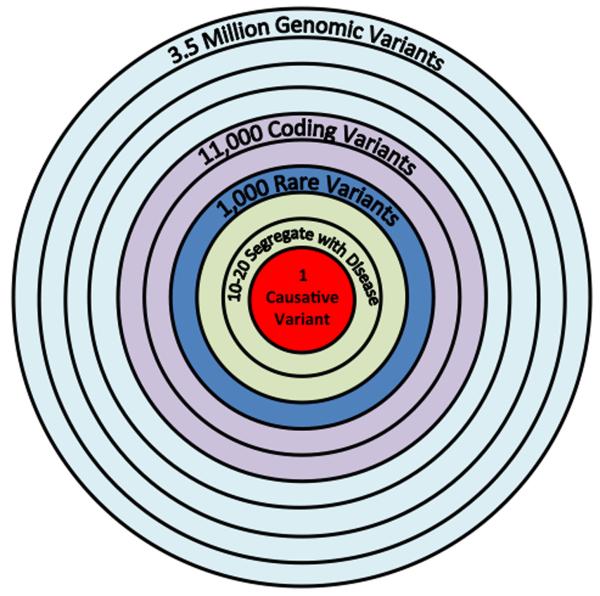
The exome consists of only ~40 Mb, or 1.5% of the human genome. There is continued debate over the use of whole exome sequencing compared with whole genome sequencing, given the lower cost of sequencing an exome, that can be readily captured via kits from a few different companies (Agilent SureSelect, Illumina TruSeq, Roche NimbleGen). Exome sequencing is typically performed at much deeper coverage, >100× (as compared with 30–40× for whole genome), which enhances accuracy, and the interpretation of variants that affect coding elements is far more advanced compared with the rest of the genome. However, the collective output from genome-wide association studies of complex traits have indicated that about 80% of the incriminated loci are in non-coding regions, outside the confines of genes (Koboldt et al., 2013). It is fair to say that we have long under-estimated the importance of the rest of the genome, but its high density of key regulatory features provides intricate and extraordinarily tight control over how genes operate. Recent whole genome sequencing studies have identified many critical variants in non-coding portions of the genome (Khurana et al., 2013). A typical whole human genome sequence will contain about 3.5 million variants compared with the reference genome, predominantly comprised of single nucleotide polymorphisms but also including insertion-deletions, copy number variants, and other types of structural variants (Frazer et al., 2009). Today, analysis of most of the 3.5 million variants is left with the “variant of unknown significant” (VUS) diagnosis. As more people get sequenced, with the full range of disease phenotypes, the proportion of VUS will drop and each sequence will become more informative. Figure 3 provides a theoretical plot of how further reduction of the cost of whole genome sequencing will also be accompanied by large numbers of individuals undergoing sequencing. In 2014, still well under 100,000 people have had whole genome sequencing with only a very limited number of phenotypes addressed. At some point in the future, sequence data gets progressively more informative at a lower price point, thus establishing particular value of whole genome sequencing. It's not just about getting a large number of people with diverse medical conditions and diverse ancestries sequenced. The drive to informativeness will clearly be enhanced by incorporating family genomic assessment, especially for determining the whether rare variants are meaningful. Here too much focus on the individual can result in a loss of context, back to our analogy of the Google map of maximal zoom obscuring understanding. By anchoring the genomics of family members, such as was done with the important discovery of PCSK9 rare variants (Hall, 2013) in cholesterol metabolism, progress in genomic medicine will be catalyzed.
Figure 3. Hypothetical plot of Cost of Sequencing and Number of Individuals Sequenced over the next 6 years.
As of early 2014, <100,000 individuals have had whole genome sequencing with leaving the information difficult to fully interpret (of limited informativeness or value). When millions of people undergo sequencing, with the full gamut of diverse phenotypes and ancestries, and the cost for sequencing continues to drop, a virtuous cycle of informativeness is established. With the new capability in 2014 to have whole genome sequencing at a cost of $1000 along with extremely high throughput, it is likely that millions of individuals will be sequenced in the next 3–4 years. The cost of sequencing will continue to drop throughout this time, as the increasing numbers of individuals undergo sequencing. Projections suggest that at least 20,000 individuals with each phenotype may be necessary to reliably identify rare, functional genomic variants. Accordingly, once millions of individuals across all main phenotypes and ancestries are sequenced, there is a new set point, or threshold, of informativeness.
At this juncture, however, it appears that exome and whole genome sequencing provide complementary information. As the cost of whole genome sequencing is further reduced, along with the availability of enhanced analytical tools for the non-gene 98.5% content interpretation, exome sequencing may ultimately become obsolete.
Single Cell Sequencing
The ability to perform sequencing of individual cells has provided remarkable new insights about human biology and disease (Shapiro et al., 2013, Battich et al., 2013, Owens, 2012). The unexpected heterogeneity in DNA sequence from one cell to another, such as has been well documented in tumor tissue, and even somatic cells in healthy individuals, has enlightened us about intra-individual genomic variation. The concept of “mosaicism” has gained rapid acceptance, with multiple mechanisms, ranging from gamete formation, embryonic development to somatic mutation in cells in adulthood, that account for why each of us has cells with different DNA sequences (Lupski, 2013, Wang et al., 2012, Macosko and McCarroll, 2012, Poduri et al., 2013). It remains unclear whether mosaicism has functional significance beyond being tied to certain congenital conditions and cancers, but this is an active area of research that is capitalizing on single cell sequencing technology. This is especially the case in neuroscience in order to explain the observed frequent finding of transposons, which appear to involve between 80 and 300 unique insertions for each neuron, and are potentially associated with neurologic diseases (Poduri et al., 2013).
Sperm, which tend to swim solo, are particularly well suited for single cell genomics. This work has quantified recombination rates of approximately 25 events per sperm, identified the hotspots where these events are most likely to occur, and determination of genomic instability as reflected by the rate of de novo mutations (Wang et al., 2012, Poduri et al., 2013). Such de novo mutations, which increase in sperm with paternal age, are associated with autism, schizophrenia and intellectual disability (Poduri et al., 2013, Kong et al., 2012, Veltman and Brunner, 2012, de Ligt et al., 2012).
Intriguing, and possibly revolutionary, single cell methods using in situ sequencing protocols are set to offer precise spatial information in addition to linear sequence data. In situ sequencing holds the potential to resolve the spatial distribution of: copy number variants, circular DNA, tumor heterogeneity, and RNA localization. A number of methods have been published in the last year and progress is likely to accelerate in the near future.
Transcriptomics, Proteomics and Metabolomics
As opposed to the DNA sequence, which is relatively static, RNA reflects the dynamic state of the cell. Gene expression of a particular tissue of the whole genome has been available via microarrays for several years, but RNA-seq is a relatively new tool that transcends simple expression by capturing data on gene fusions, alternative spliced transcripts and post-transcriptional changes, along with the whole gamut of RNAs (including microRNA [e.g. miRNAseq], small RNA, lincRNA, ribosomal RNA, and transfer RNA).
A particularly valuable metric related to RNA is the expression quantitative trait locus (eQTL). By having both genome-wide association study (GWAS) data and whole genome gene expression, at baseline, with or without particular stimuli, functional genomic assessment has been enabled. For example, Westra and colleagues used eQTLs and loci derived from GWAS to provide functional genomic, mechanistic insights for multiple complex traits including lupus and type 1 diabetes (Westra et al., 2013).
The proteome, metabolome, and autoantibody landscape can be assessed for an individual approaching the whole genome level via recent advances in mass spectrometry and protein arrays. Using these techniques, post-translational modifications of proteins, protein-protein interactions, or the small molecule metabolites produced by these proteins can be revealed. Emerging technologies such as RASL-seq, barcoded shRNA libraries, and combinatorial antibody libraries provide inexpensive and efficient views of biology. Longer read sequencing provides the opportunity to sequence antibodies, which typically have variable and constant regions combined of ~2000 nucleotides.
Microbiome
Perhaps no area of biology has received more attention in recent years than the microbiome. Just the gut microbiome has orders of magnitude more DNA content than germline human DNA, and markedly heightened diversity. Our commensal bacterial flora have been shown to play an important role in various medical conditions (Cho and Blaser, 2012). From fecal samples using a 16S ribosomal amplicon sequencing method, the gut microbiome has been the subject of intensive prospective clinical assessment. It was determined that there were 3 major enterotypes of the intestinal microbiome based on the predominant bacterial species, such as Bacteroides, Ruminococcus or Prevotella (Arumugam et al., 2011). The resident species appear to be quite stable over an extended period of time and initially transmitted via the mother at childbirth (Faith et al., 2013). As the interface between genomics and the host's environment, the microbiome clearly plays a pivotal role in defining each individual. The influence of the diet on the gut microbiome, such as the content of fiber, along with the underpinning of malnutrition has been documented (Gordon et al., 2012, Ridaura et al., 2013). For example, even an individual's response to medications, such as digoxin (Haiser et al., 2013), or multiple drugs used for cancer, has been shown to be linked to the bacterial flora of the gut microbiome (Viaud et al., 2013, Iida et al., 2013).
Epigenome
There has been extraordinary progress in our ability to map the human epigenome, from DNA methylation to histone modifications and chromatin structure (Ziller et al., 2013, Rivera and Ren, 2013). The prolific ENCODE project has provided troves of data detailing the role of regulatory elements such as enhancers and insulators, and how they are tied to DNA methylation and histone changes (Dawson and Kouzarides, 2012). Like gene expression, epigenomic findings are highly cell type specific, with over 200 different cell types in the human body. For methylation, whole genome bisulphite sequencing has recently been performed for 30 diverse human cell types (Rivera and Ren, 2013). Epigenomic reprogramming has a clearcut role in cancer, be it via transcription factors or chromatin regulators (Dawson and Kouzarides, 2012, Suva et al., 2013). Although access to tissue to define epigenomic signatures is a limiting factor outside of the cancer space, it is apparent that many other diseases are affected by epigenomic dynamics, such as complications of diabetes, rheumatoid arthritis or hypertension (Pirola et al., 2010, Liu et al., 2013, Fratkin et al., 2012). Furthermore, epigenomic changes affect susceptibility to diseases, as has been shown with open chromatin related to the TCF7L2 gene (Groop, 2010) and parental origin of sequence variants for Type 2 diabetes mellitus, breast and prostate cancer (Kong et al., 2009). This parent-of-origin issue may be tied to trans-generational epigenomic instability, as has been well documented in plants, and is certainly a key element of human biology and heritability (Schmitz et al., 2011).
Physiome and Exposome
Understanding and quantifying an individual's physiology and environmental interactions are crucial to digitizing a human being. Through wearable biosensors and smartphones, this has become eminently practical. Continuous tracking is now obtainable for most key physiologic metrics including blood pressure, heart rhythm, glucose, blood oxygen saturation, brain waves, intraocular eye pressure, and lung function indices. Similarly, there are environmental sensors that connect with smartphones to quantify such indices as air pollution, pollen count, radiation, water quality, ambient humidity, electromagnetic fields, and the presence of pesticides in food.
Bioinformatics
Fundamental to individualized medicine is the ability to analyze the immense data sets and extract all of the useful, salient information. This is exemplified by the task of sifting through a trio of whole genome sequences to find a causative mutation in a proband with an undiagnosed disease. Typically this translates to finding 1 critical nucleotide variant out of well over 1 to 2 million single base variants, and simplifying the analysis by only considering variants that change amino-acid sequence or lead to obvious splicing defects (Maher, 2011). Identifying the signal from the noise, with the vast majority of variants categorized as “unknown significance” (VUS), is the crux of the challenge. Moreover, the tools to assess structural variants and indels are not as extensively developed and validated. So there are considerably more data that come from the sequencer for an individual than can be fully and accurately mined. Beyond this, there is the need for better integration of the multiple GIS layers, such as panoromic and biosensor data, and the ability to provide an integrative multi-scale approach to an individual's data set. While not the comprehensive multi-layer as depicted in Figure 2, Zhang et al recently used an integrated systems approach including omics of both human and mice brains to discover genetic networks in Alzheimer's disease (Zhang et al., 2013). Although in the past there were generally insufficient efforts to understand epistasis, gene-gene interactions have been upstaged by the complexity of a higher order bioinformatics challenge.
How the Omic Tools Reboot Medicine
A Pre-Womb to Tomb Assessment
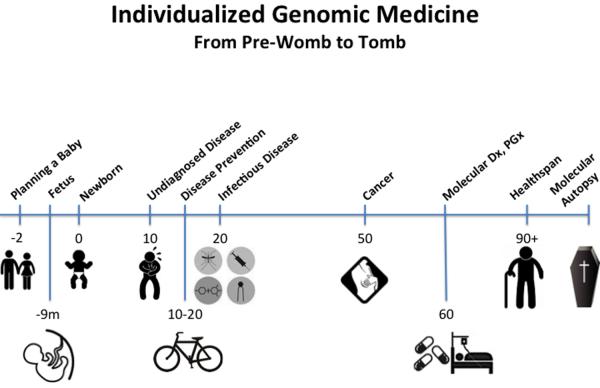
At many points in the span of a lifetime, the unique biology of the individual will play an increasing role. As depicted in the timeline of Figure 4, I will go through each topic sequentially.
Figure 4. Timeline of Sequencing Applications in Medicine from Pre-Womb to Tomb.
The medical application of genomics is relevant to many points during an individual's lifespan. Prior to conception, a couple can have genomic screening for important recessive alleles. An expectant mother, at 8–12 weeks of pregnancy, can now have single tube of blood used to accurately assess chromosomal abnormalities of the fetus, determine gender, and even have whole genome sequencing of the fetus performed. At birth, sequencing the genome of the newborn can be used to rapidly diagnosis many critical conditions for which a time delay, which frequently can occur with the present heel stick screening methods, might lead to irrevocable damage. The molecular basis for serious, undiagnosed conditions can often be established by sequencing the individual with parents of siblings. Ultimately, omic information at a young age will be useful by providing susceptibility to various medical conditions that have actionable prevention strategies. Sequencing can be done to define a pathogen for more rapid and accurate approaches to infectious diseases. The driver mutations and key biologic underpinning pathways of an individual's cancer can frequently be pinpointed by omics. The root causes of common polygenic conditions such as diabetes or coronary heart disease may ultimately be defined at the individual level. Specific sequence variants of germline DNA or the gut microbiome have relevance for response to prescription medication (both efficacy and safety). Defining the genomics of healthspan, rather than the traditional focus on diseases, may prove to be especially worthwhile to understand protective alleles and modifier genes. For an individual with sudden death, a molecular autopsy via sequencing can be performed, along with family survivors, to determine the cause of death and potentially prevent untimely or avoidable deaths of members of the family and subsequent generations.
Pre-Conception
The ability to determine carrier mutations for each prospective parent has been greatly enhanced through multiple direct-to-consumer sources, including 23andMe, Counsyl, GenePeeks and Good Start Genetics. This can be considered the ultimate form of prevention for major recessive conditions and has only received modest attention to date. Counsyl screens for over 100 recessive Mendelian traits; 23andMe screens 50 carrier conditions. The carrier rates for many serious conditions are higher than most people would suspect, such as 1 in 35 for spinal muscular atrophy, 1 in 40 for cystic fibrosis, and 1 in 125 individuals for Fragile X syndrome (Test, 2013). GenePeeks uses carrier data from ~100,000 DNA sequence variants for each prospective parent to perform a computer simulation of 10,000 “digital babies,” determining the probabilistic odds of significant Mendelian disorders (Couzin-Frankel, 2012). They are already using their analytic methodology to screen sperm from the Manhattan cryobank; until now sperm banks have been completely unregulated and without genomic assessment (Almeling, 2013, Rincon, 2013). The concept of higher DNA resolution pre-conception screening is attractive, given that there are many more pathogenic variants in the genes that are implicated in disease, such as cystic fibrosis (~2000 variants), than are conventionally assessed (Sosnay et al., 2013).
Fetal Sequencing
While the diagnosis of chromosomal aberrations such as trisomy-21, 18, 13 required amniocentesis or chorionic villi sampling, there are now 4 different maternal blood sampling assays to accomplish the same assessment with extremely high (>99%) accuracy (Morain et al., 2013). Relying on the plasma fetal DNA present in adequate quantity from a maternal blood sample at 8–10 weeks of pregnancy, such testing has been transformative, preempting the need for amniocentesis in all but the rare exception when results are ambiguous. Here is a great example of using plasma free DNA sequencing to avoid an invasive test that carries a small but important risk of miscarriage. However, with over 4 million births in the United States each year, only a tiny fraction (<2%) of prenatal maternal blood sampling has yet been performed in clinical practice. Multiple groups have demonstrated the ability to do a fetal whole exome sequence from a maternal blood sample (Fan et al., 2012), or whole genome sequencing from both parents DNA along with the maternal plasma free DNA (Kitzman et al., 2012, Lo et al., 2010), but this takes a rather extensive computing and bioinformatics effort that is not presently scalable. Undoubtedly, that will be resolved over time, but will engender the serious bioethical issues of what constitutes the appropriate reasons for termination of pregnancy. But at the same time it will afford the opportunity to make the molecular diagnoses of conditions in utero and facilitate treatment then or at the earliest time after birth.
Neonatal Sequencing
Monogenic diseases, many of which present in the first month of life, are a major cause of neonatal fatality and morbidity (Saunders et al., 2012). Despite routine heel sticks for blood sent out for analysis, with attendant delays of several days to weeks in obtaining results, there has not been any improvement in reducing neonatal mortality related to genetic disorders in the past 20 years (Kaiser, 2013, Sosnay et al., 2013). Now it has been shown that whole genome sequencing of newborns can be accomplished in <48 hours and lead to highly actionable information for managing a neonate's condition, such as in the classic example of phenylketonuria or galactosemia whereby irrevocable damage might otherwise occur (Saunders et al., 2012, Kaiser, 2013, Sosnay et al., 2013).
Undiagnosed, Idiopathic and Rare Diseases
The diagnosis of an XIAP mutation in a child with fulminant pan-colitis, with successful, curative treatment, is often cited as the first case of sequencing to save an individual's life (Worthey et al., 2011). Since that report, there have been several other cases that used whole genome or exome sequencing, along with other omic tools, for making the molecular diagnosis of idiopathic conditions (Jacob et al., 2013). For example, the National Institutes of Health Undiagnosed Disease Program uses exome sequencing to facilitate the diagnosis (Gahl and Tifft, 2011). Recently, the group at Baylor College of Medicine published a series of 250 individuals, of whom 80% were pediatric and largely affected by neurologic conditions, who underwent whole exome sequencing. In that cohort there were many affected patients with a known Mendelian trait but without a specific root cause established. A molecular diagnosis was made in 25% of the cohort (Yang et al., 2013). At Scripps, we have screened more than 100 individuals for the potential of having an idiopathic disease. This requires review by a multi-disciplinary physician panel to assure that a comprehensive evaluation of the patient has been performed before turning to DNA sequencing. The first of 15 individuals who we enrolled into our protocol was 16 years old and had an incapacitating neurologic condition. She and her parents were sequenced and in Figure 5 the bioinformatics challenge of interpreting the 3 whole genome sequences is presented, along with the molecular diagnosis of an ADCY5 mutation that had not been previously described. In our 15 probands at Scripps, we have had a successful molecular diagnosis (using criteria as described by the Baylor group) in 8 individuals. However, establishing the diagnosis represents only the first step of the desired strategy, as providing an effective treatment is the fundamental goal. Unfortunately, the number of individuals for whom that has been achieved is quite limited to date, but strategies using repurposing of existing drugs, drugs that were partially developed but not commercialized, or acceleration of the development of genomically-guided therapies are all actively being pursued. With an estimate of at least 1 million individuals in the United States with a serious medical condition but left without a diagnosis, such progress is encouraging (Jacob et al., 2013).
Figure 5. Legend From Whole Genome Sequencing to Identification of a Causative Variant.
Following sequencing, alignment and annotation, the 3.5 million variants, all genetic variants in the family (unaffected and affected individuals) are analyzed to identify known disease variants. Then inheritance-based and population-based filters are applied. Phenotype-informed ranking and functional filters are used to then determine the root-cause variant. The timeline for this involves (1) sample preparation of 2 days; (2) sequencing of 2 days in fast output mode; and the (3) preliminary analysis in ~24 hours (6 hrs for variant calling, 1 hr annotation, 1 hr for each candidate variant) and (4) literature review to exclude and include genes hit by potential candidate variants, an additional ~5 days. The cost for analysis is predominantly personnel and compute time on the cloud, estimated to be ~$300.
Disease Prevention
At some point in the future it is hoped that having DNA sequence information will pave the way for prevention of an individual's predisposed conditions. To date, however, that concept has not been actualized for a few principal reasons. First, most of the complex, polygenic traits have not had much more than 10% of their heritability explained by the common variants assessed by GWAS. The “missing” or unsolved heritability detracts from the ability to assign an individual any certainty or risk, or protection from a particular condition. There are some notable exceptions, such as age-related macular degeneration or type 1 diabetes mellitus, where combinations of common and rare variants can provide a well-characterized, quantified risk profile. Second, there is the appropriate question of whether the knowledge of a risk allele is actionable. Prototypic here is the apoε4 allele, which carries an unequivocal high risk for Alzheimer's disease, yet there is no proven strategy to prevent the disease. So even armed with known risk there is a lack of knowledge for how to mitigate it. Third, the way data for genomic susceptibility are analyzed via a population approach, makes it difficult to extrapolate such average findings to a particular individual. For example, someone may have low frequency modifier genes for a condition at risk, or unusual environmental interactions, that markedly affect susceptibility. Notwithstanding these issues, as millions of individuals with diverse phenotypes undergo whole genome sequencing (Figure 3), the ability to provide meaningful risk data will increase. Having the full GIS of each individual will further enrich the probabilistic approach of providing vulnerability data early in one's life. For example, if one's sequence data indicates a risk for hypertension, that risk may be further modulated by knowledge of his/her proteins, metabolites, microbiome and epigenomics. The use of a biosensor watch to passively collect continuous blood pressure measurements could make diagnosis at the earliest possible time, avoiding any end-organ damage to the heart or kidney. Specific treatments could be used that are biologically based from one's GIS. Similarly, for asthma the panoromic information, coupled with biosensors that track air quality, pollution, forced expiratory volume, and other relevant physiologic metrics could prove useful to prevent an attack. A futuristic way in which genomics and biosensors will ultimately converge is through injectable nanosensors that put the blood in continuous surveillance mode (Ferguson et al., 2013). Such sensors have the ability to detect a DNA, RNA, auto-antibody, or protein signal and wirelessly transmit the signal to the individual's smartphone. This sets up the potential for detecting endothelial sloughing from an artery before a heart attack (Damani et al., 2012), plasma tumor DNA in a patient being treated for or in remission from cancer, or a child with known genomic risk of autoimmune diabetes that is developing auto-antibodies to pancreatic β-islet cells long before there has been destruction. The blood under continuous surveillance concept highlights the potential ability to temporally detect a risk signal for a major clinical event, and implement true preventive therapy. That could be intensive anti-platelet medication to prevent a heart attack, genomic-guided treatment of cancer recurrence at the earliest possible juncture, or immunomodulation therapy for autoimmune diabetes.
Infectious Diseases
Whole genome sequencing has proven to be particularly useful for tracking pathogen outbreaks, such as for tuberculosis (Gardy et al., 2011), methicillin-resistant Staphylococcus aureus, antibiotic resistant Klebsiella pneumonia, and Clostridium difficile (Eyre et al., 2013, Harris et al., 2013, Snitkin et al., 2012). Beyond identifying the particular bacterial or viral strain that accounts for an outbreak's origin and spread, sequencing is likely to prove to be quite useful for rapid, early characterization of the cause of infection and specific, effective antibiotic therapy. For sepsis, the current standard of care is to take blood or other body fluids for culture, which typically takes 2 days to grow out. Additionally, there is at least another day required to determine sensitivities to a range of antibiotics. In the future, with lab-on-a-chip sequencing platforms that attach to or are integrated with a smartphone (Biomeme and QuantuMD), it may be feasible to do rapid sequencing of the pathogen and determination of the optimal treatment. Such a strategy would preempt the need for broad-spectrum antibiotic use, and the rapid diagnosis and targeted treatment would likely have a favorable impact on prognosis in these very high-risk, critically ill patients.
Cancer
With cancer's basis in genomics, there have been extensive efforts to characterize the principal driver mutations and biologic pathways, especially through The Cancer Genome Atlas (TCGA) (Kandoth et al., 2013, Alexandrov et al., 2013). Our understanding of the biology of cancer has expanded exponentially, and with it so has the appreciation for its extreme complexity. Perhaps the two classical “Hallmarks of Cancer” reviews in Cell, one in 2000 and the sequel in 2011, best exemplify this (Hanahan and Weinberg, 2000, Hanahan and Weinberg, 2011). The diagram to explain the principal mechanisms of cancer was already exceptionally complex in 2000, and became at least a log order more intricate a decade later. In a more recent review of the cancer genome landscape, the Johns Hopkins group provided perspective for the 84 known oncogenes, and 54 tumor suppressor genes, that have been fully validated (Vogelstein et al., 2013). There will unquestionably be more, but estimates of the total number of genes involved in pivotal mutations may wind up being ~200. Beyond this the principal pathways, involving cell survival, cell fate, and DNA damage repair, are recognized. Certain cancers have a relative low burden of mutations per megabase of the tumor genome, such as acute myelogenous leukemia (<1), while other are quite high, like lung adenocarcinoma or squamous cell carcinoma (~50) (Kandoth et al., 2013, Vogelstein et al., 2013). Mutations of certain genes, such as the P53 tumor suppressor, are found in some patients with any of 12 common forms of cancer (Kandoth et al., 2013). Our old taxonomy of cancer based upon the organ of origin may be considered inapt, for knowledge of the driver mutation(s) and pathway could be more useful for individualizing treatment. “N of 1” case reports with whole genome sequencing have been particularly illuminating for the clonal origin of an individual's cancer (Brannon and Sawyers, 2013, Haffner et al., 2013). In the past 2 years, the Food and Drug Administration has approved almost 20 new drugs that target a specific mutation for cancer. So with these leaps in understanding biology, and introduction of new therapies, why has there been relatively little impact in the clinic to date? One major barrier is that we do not have drugs that can target tumor suppressor genes, making up ~40% of mix of principal, driver mutations. Sometimes there are workarounds for this issue, such as the tumor suppressor gene PTEN, which results in PI3-kinase activation, but more often this is not the case. Even for oncogenes, less than 40% have a specific drug antagonist, as most are part of protein complexes (Vogelstein et al., 2013). A second critical issue is that there is marked heterogeneity in tumors, both within an individual's primary tumor, and certainly inter-metastatic. This appears to be a foundation for the common occurrence of relapse after an initial marked response, reflecting success directed to an oncogene, but that other undetected mutations become capable of propagating the tumor. The BRAF mutations, which are drivers in a variety of tumors, notably melanoma, thyroid and colon, can be treated with a specific BRAF inhibitor. In the first 2 weeks of oral therapy, there is usually a marked response but at 9–12 months a relapse is quite typical (Sosman et al., 2012). Interestingly, when targeting BRAF for colon cancer there appears to be primary resistance to these inhibitors (Prahallad et al., 2012), related to EGFR expression, and emphasizing that the stroma, micro-environment of the tumor can still exert an important role. The issues of heterogeneity and resistance lend credence to the use of combinations of targeted drugs in the future, but that has yet to be explored at scale in prospective trials. A third largely unaddressed issue in the clinic is the involvement of the epigenome in tumorigenesis. At least 40 epigenome regulator genes are known that have highly recurrent somatic mutations in tumors, across a variety of cancers, affecting multiple target genes simultaneously (Kandoth et al., 2013, Vogelstein et al., 2013, Garraway and Lander, 2013, Shen and Laird, 2013). These are not screened for clinically, nor are there drugs available to modulate their effect.
In the clinic today, the bare bones of mutation screening are typically used, such as HER2 for breast cancer of KRAS for colon cancer. Recently Foundation Medicine commercialized a targeted gene panel of 287 genes that have an established role in cancer (Frampton et al., 2013). Using predominantly fixed-formalin, paraffin-embedded samples, mutation cell concordance was established compared with mass spectrometric methodology (Sequenom) and the typical driver mutations were identified in a cohort of over 2000 individuals, such as TP53, KRAS, CDKN2A, and PIK3CA (Frampton et al., 2013). However, there was a long tail of uncommon mutations that was identified reflecting the profound diversity of cancers.
This panel represents a step forward compared with a very limited gene mutation screen for commonly occurring drivers, which might even miss other pathogenic mutations within incriminated genes. The 287 genes assessed represent only <15% of genes, and only the coding elements. This is in contrast to research studies of whole genome and whole exome sequencing, with paired germline DNA for each individual, to more precisely determine driver mutations (Kandoth et al., 2013). Further, multiple recent studies have highlighted the role of non-coding elements of the genome to play a prominent role, such as TERT promoters in melanoma (Huang et al., 2013), a long, non-coding RNA SChLAP1 for aggressive prostate cancer (Prensner et al., 2013), and identification of ~100 non-coding driver variants for cancer using a new bioinformatics tool known as FunSeq (Khurana et al., 2013). Clearly, even a comprehensive exome would only represent a limited swath of sampling for root causes of cancer in an individual.
Cancer genomic medicine of the future will likely involve a GIS of the tumor, with assessment of DNA sequence, gene expression, RNA-seq, microRNAs, proteins, copy number variations, and DNA methylation, cross-referenced with the individual's germline DNA. But the issue of addressing heterogeneity still looms (Vogelstein et al., 2013, Bedard et al., 2013), and for that there are a few possible steps, including deep sequencing of the tumor at multiple locations, single cell sequencing, or the use of the “liquid biopsy” of cancer (Schwarzenbach et al., 2011, Leary et al., 2010, Forshew et al., 2012).
Cell free tumor DNA in plasma, which is present in the vast majority of patients with cancer, has been shown to be a useful biomarker for following patients (Schwarzenbach et al., 2011, Leary et al., 2010, Forshew et al., 2012) and appears to have independent prognostic significance (Dawson et al., 2013). It may be that plasma tumor DNA is the best representative of the cancer for targeted treatment, since avoidance of metastasis is of utmost concern. The ease of isolating and sequencing cell free tumor DNA is likely to make this very attractive and routine in the future. Especially appealing is the ability to sample on a more frequent, and even continuous basis, using biosensors. Whether the “liquid biopsy” will help override the challenges related to intra-tumor heterogeneity, and recurrence of cancer, awaits prospective evaluation. Also of particular interest, at some point, will be screening healthy people for cell free tumor DNA to determine whether we are constantly facing microscopic tumor burden but are able to effectively keep the disease in check by a variety of homeostatic mechanisms.
Molecular Diagnosis
When a patient receives a diagnosis of a chronic illness today, it is non-specific, based on clinical and not molecular features. Take for example diabetes mellitus, type 2, which could reflect anything from insulin resistance, failure of β-islet cells, or a variety of subtypes including α-adrenergic receptor (ADRA2A) diabetes (Gribble, 2010) or a zinc transporter subtype (SLC30A8) (Sladek et al., 2007). Common genomic variants have been identified in pathways involving signal transduction, cell proliferation, glucose sensing, and circadian rhythm (Dupuis et al., 2010). Some individuals with a high fasting glucose have a G6PC2 variant that is associated with protection from diabetes (Bouatia-Naji et al., 2008). A genotype score, amalgamating the number of risk variants, has been shown helpful for identifying high susceptibility (Meigs et al., 2008). Moreover, there are 13 classes of drugs to treat diabetes and the treatment could be made considerably more rational with knowledge of the individual's underlying mechanism(s).
This brief summary of the diabetes example reflects the need for a new molecular taxonomy across all diseases. When an individual is diagnosed (or at some point when risk can be defined), the molecular basis will be assessed and, ideally, when possible, the root cause will be established. Clearly for many common diseases there are multiple pathways implicated and this may prove to be difficult. But there has yet to be a systematic attempt of providing such a molecular diagnosis in clinical care. Despite multiple reports of molecular subtypes of asthma (Wenzel, 2012), multiple sclerosis (Ottoboni et al., 2012), colon (Sadanandam et al., 2013) and uterine cancer (Cancer Genome Atlas Research et al., 2013), which appear to be linked with therapy and prognosis, this has yet to be made part of medical practice.
Pharmacogenomics
Just as molecular subtyping of chronic disease is not part of medical practice, pharmacogenomics screening for either assurance of efficacy or avoidance of major side effects, is predominantly ignored. With the use of GWAS, there has been an avalanche of discovery of alleles that are pivotal to individual drug response. Unlike polygenic disease, for which the penetrance for a common sequence variant is quite low (approximate odds ratio of 1.15), the typical genotype odds ratio for prescription drugs can be as high as 80, and for many the range is 3–40 fold (reviewed in depth, (Harper and Topol, 2012)). The likely explanation for this pronounced impact of common variants on individual drug response is based on selection: as compared with diseases, the human genome has had very limited time to adapt to medication exposure. Despite there being over 100 drugs that carry a genomic “label” by the Food and Drug Administration, meaning that there is a recommendation for genotype assessment before the drug is used, there is rarely any pharmacogenomic assessment in clinical practice. This needs to improve, and perhaps the availability of point-of-care testing will help, along with reduced cost, to eventually promote routine use. Beyond this barrier, there needs to be more genomic sequencing for commonly used drugs, with associated phenotypic determination of efficacy and side effects, along with systematic omic assessment for drugs in development. From the marked success of discovering genomic-drug interactions found to date, there is certainly the sense that the more you look, the more that will be found. The potential here is to reduce the waste of pharmaceuticals, not just by avoiding drugs that will not provide efficacy for particular individuals, but also avoiding serious toxicity that can be either fatal or life-threatening.
Healthspan
The human reference genome is based upon multiple young individuals who had no phenotypic characterization. Accordingly, we know nothing about the reference human's natural history of disease, and one can consider this as a flawed standard for comparison. Ideally, we should have a reference genome that has had rigorous phenotyping. This is especially the case in an era of using sequencing in medical practice, but with an inadequate comparator. Perhaps the optimal phenotype would be healthspan. At Scripps, we have defined healthy elderly as age >80 years, with no history of chronic illness or use of medications. The cohort (known as “Wellderly”) that we have assembled over the past 7 years of 1400 individuals has an average age of 88, and we have completed whole genome sequencing for 500 of these individuals. The intent is to provide a more useful reference genome with a clearly defined, uniform, relevant phenotype.
Moreover, there is another important application of healthspan genomics. Multiple studies have established that a research investment in understanding healthy aging would be more prudent than in any specific disease category (Goldman et al., 2013a). Since the cohort that we have enrolled carries a similar burden of common risk variants for chronic diseases compared with the general population, there are most likely a substantial number of modifier genes and protective alleles that may be ultimately identified. One example is from APP, a gene that has a variant for both high risk of Alzheimer's and another rare variant with marked protection from cognitive impairment of Alzheimer's (De Strooper and Voet, 2012). Unquestionably, there are many more such variants left to be discovered, and therein lies the potential for drug discovery efforts that can follow such findings from Nature of particular genes and pathways that prevent diseases.
Molecular Autopsies
While physical autopsies have lost favor and become exceptionally rare, there is an opportunity to use sequencing to determine the cause of death, particularly when this occurs suddenly. Targeted or whole genome sequencing for heritable heart disorders implicated in sudden death, including ion channel mutations and hypertrophic cardiomyopathy, can be performed in the deceased individual and family members. This approach is now actively being pursued in New York City for all sudden cardiac deaths (Erdmann, 2013) and may prove helpful in preventing this condition in family members.
Future Directions
This pre-womb to tomb review has emphasized that there is a disproportionate relationship between knowledge and implementation into clinical practice. For individualized medicine to take hold, it will require intensive, rigorous validation that these new approaches improve patient outcomes and are demonstrated to be cost-effective. This proof will be essential for the medical community to embrace the opportunities, but also will require educational programs that squarely address the knowledge chasm that currently exists for practicing physicians. A second theme is that our efforts have been largely sequence-centric and have not adequately taken into account or integrated the data from other omics, no less biosensors and imaging. Related to this deficiency, there is a profound shortage of data scientists in biomedicine, with unparalleled opportunities to process enormous, high yield data sets. While we will increasingly rely on algorithms, artificial intelligence, and machine learning, the rate-limiting step necessitates talented biocomputing and bioinformatic expertise.
One of the most attractive outgrowths of defining each individual's unique biology in an era with unprecedented digital infrastructure, is to be able to share the data. By taking the de-identified data from each individual, including panoromic, biosensors, social graph, treatment and outcomes, an extraordinary resource can now be developed. Such a massive open on-line medical information (MOOM) repository could provide matching capability to approximate a newly diagnosed individual's data as compared with all of those previously amalgamated. For a patient with cancer, for example, this could provide closest matches to the tumor GIF, demographics, treatment and outcomes to select an optimal strategy; this would potentially take Bayesian principles to a new, enriched potential. Such a MOOM resource does not need to be confined to cancer, but the first to be announced was with the Leukemia & Lymphoma Society and Oregon Health Sciences University for 900 patients with liquid tumors (Winslow, 2013). Hopefully this will be one of many data sharing initiatives in medicine to go forward, now that such rich unique information can be captured at the individual level, and our computing infrastructure is so well suited to perform such functions. While we are still at the nascent stages of individualized medicine, there has never been more promise and opportunity to reboot the way health care can be rendered. Only with systematic validation of these approaches at the intersection of biology and digital technology, can we actualize this more precise, futuristic version of medicine.
Acknowledgements
I want to express my gratitude to my colleagues Ali Torkamani, PhD and Erick Scott, MD who reviewed the manuscript and offered editorial input, and to Katrina Schreiber, who helped prepare the typescript and graphics. This work was supported by NIH/NCATS 1 UL1 TR001114.
Funding by NIH/NCATS 1 UL1 TR001114
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- ALEXANDROV LB, NIK-ZAINAL S, WEDGE DC, APARICIO SA, BEHJATI S, BIANKIN AV, BIGNELL GR, BOLLI N, BORG A, BORRESEN-DALE AL, BOYAULT S, BURKHARDT B, BUTLER AP, CALDAS C, DAVIES HR, DESMEDT C, EILS R, EYFJORD JE, FOEKENS JA, GREAVES M, HOSODA F, HUTTER B, ILICIC T, IMBEAUD S, IMIELINSKI M, JAGER N, JONES DT, JONES D, KNAPPSKOG S, KOOL M, LAKHANI SR, LOPEZ-OTIN C, MARTIN S, MUNSHI NC, NAKAMURA H, NORTHCOTT PA, PAJIC M, PAPAEMMANUIL E, PARADISO A, PEARSON JV, PUENTE XS, RAINE K, RAMAKRISHNA M, RICHARDSON AL, RICHTER J, ROSENSTIEL P, SCHLESNER M, SCHUMACHER TN, SPAN PN, TEAGUE JW, TOTOKI Y, TUTT AN, VALDES-MAS R, VAN BUUREN MM, VAN 'T VEER L, VINCENT-SALOMON A, WADDELL N, YATES LR, AUSTRALIAN PANCREATIC CANCER GENOME, I. CONSORTIUM IBC, CONSORTIUM IM-S, PEDBRAIN I, ZUCMAN-ROSSI J, FUTREAL PA, MCDERMOTT U, LICHTER P, MEYERSON M, GRIMMOND SM, SIEBERT R, CAMPO E, SHIBATA T, PFISTER SM, CAMPBELL PJ, STRATTON MR. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALMELING R. The Unregulated Sperm Industry. Times; New York: 2013. [Accessed 12/01/2013]. Online. Available: http://ow.ly/rsnVh. [Google Scholar]
- ARUMUGAM M, RAES J, PELLETIER E, LE PASLIER D, YAMADA T, MENDE DR, FERNANDES GR, TAP J, BRULS T, BATTO JM, BERTALAN M, BORRUEL N, CASELLAS F, FERNANDEZ L, GAUTIER L, HANSEN T, HATTORI M, HAYASHI T, KLEEREBEZEM M, KUROKAWA K, LECLERC M, LEVENEZ F, MANICHANH C, NIELSEN HB, NIELSEN T, PONS N, POULAIN J, QIN J, SICHERITZ-PONTEN T, TIMS S, TORRENTS D, UGARTE E, ZOETENDAL EG, WANG J, GUARNER F, PEDERSEN O, DE VOS WM, BRUNAK S, DORE J, META HITC, ANTOLIN M, ARTIGUENAVE F, BLOTTIERE HM, ALMEIDA M, BRECHOT C, CARA C, CHERVAUX C, CULTRONE A, DELORME C, DENARIAZ G, DERVYN R, FOERSTNER KU, FRISS C, VAN DE GUCHTE M, GUEDON E, HAIMET F, HUBER W, VAN HYLCKAMA-VLIEG J, JAMET A, JUSTE C, KACI G, KNOL J, LAKHDARI O, LAYEC S, LE ROUX K, MAGUIN E, MERIEUX A, MELO MINARDI R, M'RINI C, MULLER J, OOZEER R, PARKHILL J, RENAULT P, RESCIGNO M, SANCHEZ N, SUNAGAWA S, TORREJON A, TURNER K, VANDEMEULEBROUCK G, VARELA E, WINOGRADSKY Y, ZELLER G, WEISSENBACH J, EHRLICH SD, BORK P. Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BATTICH N, STOEGER T, PELKMANS L. Image-based transcriptomics in thousands of single human cells at single-molecule resolution. Nat Methods. 2013;10:1127–33. doi: 10.1038/nmeth.2657. [DOI] [PubMed] [Google Scholar]
- BEDARD PL, HANSEN AR, RATAIN MJ, SIU LL. Tumour heterogeneity in the clinic. Nature. 2013;501:355–64. doi: 10.1038/nature12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BONNET J, YIN P, ORTIZ ME, SUBSOONTORN P, ENDY D. Amplifying genetic logic gates. Science. 2013;340:599–603. doi: 10.1126/science.1232758. [DOI] [PubMed] [Google Scholar]
- BOUATIA-NAJI N, ROCHELEAU G, VAN LOMMEL L, LEMAIRE K, SCHUIT F, CAVALCANTI-PROENCA C, MARCHAND M, HARTIKAINEN AL, SOVIO U, DE GRAEVE F, RUNG J, VAXILLAIRE M, TICHET J, MARRE M, BALKAU B, WEILL J, ELLIOTT P, JARVELIN MR, MEYRE D, POLYCHRONAKOS C, DINA C, SLADEK R, FROGUEL P. A polymorphism within the G6PC2 gene is associated with fasting plasma glucose levels. Science. 2008;320:1085–8. doi: 10.1126/science.1156849. [DOI] [PubMed] [Google Scholar]
- BOYCOTT KM, VANSTONE MR, BULMAN DE, MACKENZIE AE. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat Rev Genet. 2013;14:681–91. doi: 10.1038/nrg3555. [DOI] [PubMed] [Google Scholar]
- BRANNON AR, SAWYERS CL. “N of 1” case reports in the era of whole-genome sequencing. J Clin Invest. 2013;123:4568–70. doi: 10.1172/JCI70935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUTTE AJ. [Accessed 02/15/2013];Should Healthy People Have Their Genomes Sequenced At This Time? 2013 [Online]. Available: http://ow.ly/qnEu2.
- CANCER GENOME ATLAS RESEARCH, N. KANDOTH C, SCHULTZ N, CHERNIACK AD, AKBANI R, LIU Y, SHEN H, ROBERTSON AG, PASHTAN I, SHEN R, BENZ CC, YAU C, LAIRD PW, DING L, ZHANG W, MILLS GB, KUCHERLAPATI R, MARDIS ER, LEVINE DA. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN R, MIAS GI, LI-POOK-THAN J, JIANG L, LAM HY, CHEN R, MIRIAMI E, KARCZEWSKI KJ, HARIHARAN M, DEWEY FE, CHENG Y, CLARK MJ, IM H, HABEGGER L, BALASUBRAMANIAN S, O'HUALLACHAIN M, DUDLEY JT, HILLENMEYER S, HARAKSINGH R, SHARON D, EUSKIRCHEN G, LACROUTE P, BETTINGER K, BOYLE AP, KASOWSKI M, GRUBERT F, SEKI S, GARCIA M, WHIRL-CARRILLO M, GALLARDO M, BLASCO MA, GREENBERG PL, SNYDER P, KLEIN TE, ALTMAN RB, BUTTE AJ, ASHLEY EA, GERSTEIN M, NADEAU KC, TANG H, SNYDER M. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHO I, BLASER MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–70. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHURCH GM, GAO Y, KOSURI S. Science. 2012;337:1628. doi: 10.1126/science.1226355. [DOI] [PubMed] [Google Scholar]
- COUZIN-FRANKEL J. Genetics. New company pushes the envelope on pre-conception testing. Science. 2012;338:315–6. doi: 10.1126/science.338.6105.315. [DOI] [PubMed] [Google Scholar]
- DAMANI S, BACCONI A, LIBIGER O, CHOURASIA AH, SERRY R, GOLLAPUDI R, GOLDBERG R, RAPEPORT K, HAASER S, TOPOL S, KNOWLTON S, BETHEL K, KUHN P, WOOD M, CARRAGHER B, SCHORK NJ, JIANG J, RAO C, CONNELLY M, FOWLER VM, TOPOL EJ. Characterization of circulating endothelial cells in acute myocardial infarction. Sci Transl Med. 2012;4:126ra33. doi: 10.1126/scitranslmed.3003451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAWSON MA, KOUZARIDES T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- DAWSON SJ, TSUI DW, MURTAZA M, BIGGS H, RUEDA OM, CHIN SF, DUNNING MJ, GALE D, FORSHEW T, MAHLER-ARAUJO B, RAJAN S, HUMPHRAY S, BECQ J, HALSALL D, WALLIS M, BENTLEY D, CALDAS C, ROSENFELD N. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368:1199–209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- DE LIGT J, WILLEMSEN MH, VAN BON BW, KLEEFSTRA T, YNTEMA HG, KROES T, VULTO-VAN SILFHOUT AT, KOOLEN DA, DE VRIES P, GILISSEN C, DEL ROSARIO M, HOISCHEN A, SCHEFFER H, DE VRIES BB, BRUNNER HG, VELTMAN JA, VISSERS LE. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–9. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- DE STROOPER B, VOET T. Alzheimer's disease: A protective mutation. Nature. 2012;488:38–9. doi: 10.1038/488038a. [DOI] [PubMed] [Google Scholar]
- DISEASE NRCUCOAFFDANTO. Toward Precision Medicine. The National Academies Press; Washington, D.C.: 2011. [Google Scholar]
- DUPUIS J, LANGENBERG C, PROKOPENKO I, SAXENA R, SORANZO N, JACKSON AU, WHEELER E, GLAZER NL, BOUATIA-NAJI N, GLOYN AL, LINDGREN CM, MAGI R, MORRIS AP, RANDALL J, JOHNSON T, ELLIOTT P, RYBIN D, THORLEIFSSON G, STEINTHORSDOTTIR V, HENNEMAN P, GRALLERT H, DEHGHAN A, HOTTENGA JJ, FRANKLIN CS, NAVARRO P, SONG K, GOEL A, PERRY JR, EGAN JM, LAJUNEN T, GRARUP N, SPARSO T, DONEY A, VOIGHT BF, STRINGHAM HM, LI M, KANONI S, SHRADER P, CAVALCANTI-PROENCA C, KUMARI M, QI L, TIMPSON NJ, GIEGER C, ZABENA C, ROCHELEAU G, INGELSSON E, AN P, O'CONNELL J, LUAN J, ELLIOTT A, MCCARROLL SA, PAYNE F, ROCCASECCA RM, PATTOU F, SETHUPATHY P, ARDLIE K, ARIYUREK Y, BALKAU B, BARTER P, BEILBY JP, BEN-SHLOMO Y, BENEDIKTSSON R, BENNETT AJ, BERGMANN S, BOCHUD M, BOERWINKLE E, BONNEFOND A, BONNYCASTLE LL, BORCH-JOHNSEN K, BOTTCHER Y, BRUNNER E, BUMPSTEAD SJ, CHARPENTIER G, CHEN YD, CHINES P, CLARKE R, COIN LJ, COOPER MN, CORNELIS M, CRAWFORD G, CRISPONI L, DAY IN, DE GEUS EJ, DELPLANQUE J, DINA C, ERDOS MR, FEDSON AC, FISCHER-ROSINSKY A, FOROUHI NG, FOX CS, FRANTS R, FRANZOSI MG, GALAN P, GOODARZI MO, GRAESSLER J, GROVES CJ, GRUNDY S, GWILLIAM R, GYLLENSTEN U, HADJADJ S, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–16. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ERDMANN J. Telltale hearts. Nat Med. 2013;19:1361–4. doi: 10.1038/nm1113-1361. [DOI] [PubMed] [Google Scholar]
- EYRE DW, CULE ML, WILSON DJ, GRIFFITHS D, VAUGHAN A, O'CONNOR L, IP CL, GOLUBCHIK T, BATTY EM, FINNEY JM, WYLLIE DH, DIDELOT X, PIAZZA P, BOWDEN R, DINGLE KE, HARDING RM, CROOK DW, WILCOX MH, PETO TE, WALKER AS. Diverse sources of C. difficile infection identified on whole-genome sequencing. N Engl J Med. 2013;369:1195–205. doi: 10.1056/NEJMoa1216064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAITH JJ, GURUGE JL, CHARBONNEAU M, SUBRAMANIAN S, SEEDORF H, GOODMAN AL, CLEMENTE JC, KNIGHT R, HEATH AC, LEIBEL RL, ROSENBAUM M, GORDON JI. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAN HC, GU W, WANG J, BLUMENFELD YJ, EL-SAYED YY, QUAKE SR. Non-invasive prenatal measurement of the fetal genome. Nature. 2012;487:320–4. doi: 10.1038/nature11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FERGUSON BS, HOGGARTH DA, MALINIAK D, PLOENSE K, WHITE RJ, WOODWARD N, HSIEH K, BONHAM AJ, EISENSTEIN M, KIPPIN TE, PLAXCO KW, SOH HT. Real-time, aptamer-based tracking of circulating therapeutic agents in living animals. Sci Transl Med. 2013;5:213ra165. doi: 10.1126/scitranslmed.3007095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FORSHEW T, MURTAZA M, PARKINSON C, GALE D, TSUI DW, KAPER F, DAWSON SJ, PISKORZ AM, JIMENEZ-LINAN M, BENTLEY D, HADFIELD J, MAY AP, CALDAS C, BRENTON JD, ROSENFELD N. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4:136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- FRAMPTON GM, FICHTENHOLTZ A, OTTO GA, WANG K, DOWNING SR, HE J, SCHNALL-LEVIN M, WHITE J, SANFORD EM, AN P, SUN J, JUHN F, BRENNAN K, IWANIK K, MAILLET A, BUELL J, WHITE E, ZHAO M, BALASUBRAMANIAN S, TERZIC S, RICHARDS T, BANNING V, GARCIA L, MAHONEY K, ZWIRKO Z, DONAHUE A, BELTRAN H, MOSQUERA JM, RUBIN MA, DOGAN S, HEDVAT CV, BERGER MF, PUSZTAI L, LECHNER M, BOSHOFF C, JAROSZ M, VIETZ C, PARKER A, MILLER VA, ROSS JS, CURRAN J, CRONIN MT, STEPHENS PJ, LIPSON D, YELENSKY R. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–31. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRATKIN E, BERCOVICI S, STEPHAN DA. The implications of ENCODE for diagnostics. Nat Biotechnol. 2012;30:1064–5. doi: 10.1038/nbt.2418. [DOI] [PubMed] [Google Scholar]
- FRAZER KA, MURRAY SS, SCHORK NJ, TOPOL EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10:241–51. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- GAHL WA, TIFFT CJ. The NIH Undiagnosed Diseases Program: lessons learned. JAMA. 2011;305:1904–5. doi: 10.1001/jama.2011.613. [DOI] [PubMed] [Google Scholar]
- GARDY JL, JOHNSTON JC, HO SUI SJ, COOK VJ, SHAH L, BRODKIN E, REMPEL S, MOORE R, ZHAO Y, HOLT R, VARHOL R, BIROL I, LEM M, SHARMA MK, ELWOOD K, JONES SJ, BRINKMAN FS, BRUNHAM RC, TANG P. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med. 2011;364:730–9. doi: 10.1056/NEJMoa1003176. [DOI] [PubMed] [Google Scholar]
- GARRAWAY LA, LANDER ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- GOLDMAN DP, CUTLER D, ROWE JW, MICHAUD PC, SULLIVAN J, PENEVA D, OLSHANSKY SJ. Substantial health and economic returns from delayed aging may warrant a new focus for medical research. Health Aff (Millwood) 2013a;32:1698–705. doi: 10.1377/hlthaff.2013.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GOLDMAN N, BERTONE P, CHEN S, DESSIMOZ C, LEPROUST EM, SIPOS B, BIRNEY E. Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature. 2013b;494:77–80. doi: 10.1038/nature11875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GORDON JI, DEWEY KG, MILLS DA, MEDZHITOV RM. The human gut microbiota and undernutrition. Sci Transl Med. 2012;4:137ps12. doi: 10.1126/scitranslmed.3004347. [DOI] [PubMed] [Google Scholar]
- GRIBBLE FM. Alpha2A-adrenergic receptors and type 2 diabetes. N Engl J Med. 2010;362:361–2. doi: 10.1056/NEJMcibr0911499. [DOI] [PubMed] [Google Scholar]
- GROOP L. Open chromatin and diabetes risk. Nat Genet. 2010;42:190–2. doi: 10.1038/ng0310-190. [DOI] [PubMed] [Google Scholar]
- HAFFNER MC, MOSBRUGER T, ESOPI DM, FEDOR H, HEAPHY CM, WALKER DA, ADEJOLA N, GUREL M, HICKS J, MEEKER AK, HALUSHKA MK, SIMONS JW, ISAACS WB, DE MARZO AM, NELSON WG, YEGNASUBRAMANIAN S. Tracking the clonal origin of lethal prostate cancer. J Clin Invest. 2013;123:4918–22. doi: 10.1172/JCI70354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAISER HJ, GOOTENBERG DB, CHATMAN K, SIRASANI G, BALSKUS EP, TURNBAUGH PJ. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science. 2013;341:295–8. doi: 10.1126/science.1235872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HALL SS. Genetics: a gene of rare effect. Nature. 2013;496:152–5. doi: 10.1038/496152a. [DOI] [PubMed] [Google Scholar]
- HANAHAN D, WEINBERG RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- HANAHAN D, WEINBERG RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- HARPER AR, TOPOL EJ. Pharmacogenomics in clinical practice and drug development. Nat Biotechnol. 2012;30:1117–24. doi: 10.1038/nbt.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARRIS SR, CARTWRIGHT EJ, TOROK ME, HOLDEN MT, BROWN NM, OGILVY-STUART AL, ELLINGTON MJ, QUAIL MA, BENTLEY SD, PARKHILL J, PEACOCK SJ. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect Dis. 2013;13:130–6. doi: 10.1016/S1473-3099(12)70268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAYDEN EC. [Accessed 01/15/2014];Is the $1,000 genome for real? 2014 [Online]. Available: http://www.nature.com/news/is-the-1-000-genome-for-real-1.14530.
- HUANG FW, HODIS E, XU MJ, KRYUKOV GV, CHIN L, GARRAWAY LA. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–9. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IIDA N, DZUTSEV A, STEWART CA, SMITH L, BOULADOUX N, WEINGARTEN RA, MOLINA DA, SALCEDO R, BACK T, CRAMER S, DAI RM, KIU H, CARDONE M, NAIK S, PATRI AK, WANG E, MARINCOLA FM, FRANK KM, BELKAID Y, TRINCHIERI G, GOLDSZMID RS. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–70. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- INSTITUTE NHGR. [Accessed 01/23/2014];Clinical Sequencing Exploratory Research. 2013 [Online]. Available: http://www.genome.gov/27546194.
- ISSACSON W. Steve Jobs. Simon & Schuster; New York: 2011. [Google Scholar]
- JACOB HJ, ABRAMS K, BICK DP, BRODIE K, DIMMOCK DP, FARRELL M, GEURTS J, HARRIS J, HELBLING D, JOERS BJ, KLIEGMAN R, KOWALSKI G, LAZAR J, MARGOLIS DA, NORTH P, NORTHUP J, ROQUEMORE-GOINS A, SCHARER G, SHIMOYAMA M, STRONG K, TAYLOR B, TSAIH SW, TSCHANNEN MR, VEITH RL, WENDTANDRAE J, WILK B, WORTHEY EA. Genomics in clinical practice: lessons from the front lines. Sci Transl Med. 2013;5:194–5. doi: 10.1126/scitranslmed.3006468. [DOI] [PubMed] [Google Scholar]
- JENKINS HW. Google and the Search for the Future [Online] [Accessed 08/14/2010 2010];Wall Street Journal. 2010 Available: http://ow.ly/qglOu.
- KAISER J. Genomics. Researchers to explore promise, risks of sequencing newborns' DNA. Science. 2013;341:1163. doi: 10.1126/science.341.6151.1163. [DOI] [PubMed] [Google Scholar]
- KANDOTH C, MCLELLAN MD, VANDIN F, YE K, NIU B, LU C, XIE M, ZHANG Q, MCMICHAEL JF, WYCZALKOWSKI MA, LEISERSON MD, MILLER CA, WELCH JS, WALTER MJ, WENDL MC, LEY TJ, WILSON RK, RAPHAEL BJ, DING L. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–9. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KHURANA E, FU Y, COLONNA V, MU XJ, KANG HM, LAPPALAINEN T, SBONER A, LOCHOVSKY L, CHEN J, HARMANCI A, DAS J, ABYZOV A, BALASUBRAMANIAN S, BEAL K, CHAKRAVARTY D, CHALLIS D, CHEN Y, CLARKE D, CLARKE L, CUNNINGHAM F, EVANI US, FLICEK P, FRAGOZA R, GARRISON E, GIBBS R, GUMUS ZH, HERRERO J, KITABAYASHI N, KONG Y, LAGE K, LILUASHVILI V, LIPKIN SM, MACARTHUR DG, MARTH G, MUZNY D, PERS TH, RITCHIE GR, ROSENFELD JA, SISU C, WEI X, WILSON M, XUE Y, YU F, GENOMES PROJECT C, DERMITZAKIS ET, YU H, RUBIN MA, TYLER-SMITH C, GERSTEIN M. Integrative annotation of variants from 1092 humans: application to cancer genomics. Science. 2013;342:1235587. doi: 10.1126/science.1235587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KITZMAN JO, SNYDER MW, VENTURA M, LEWIS AP, QIU R, SIMMONS LE, GAMMILL HS, RUBENS CE, SANTILLAN DA, MURRAY JC, TABOR HK, BAMSHAD MJ, EICHLER EE, SHENDURE J. Noninvasive whole-genome sequencing of a human fetus. Sci Transl Med. 2012;4:137–76. doi: 10.1126/scitranslmed.3004323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOBOLDT DC, STEINBERG KM, LARSON DE, WILSON RK, MARDIS ER. The next-generation sequencing revolution and its impact on genomics. Cell. 2013;155:27–38. doi: 10.1016/j.cell.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KONG A, FRIGGE ML, MASSON G, BESENBACHER S, SULEM P, MAGNUSSON G, GUDJONSSON SA, SIGURDSSON A, JONASDOTTIR A, JONASDOTTIR A, WONG WS, SIGURDSSON G, WALTERS GB, STEINBERG S, HELGASON H, THORLEIFSSON G, GUDBJARTSSON DF, HELGASON A, MAGNUSSON OT, THORSTEINSDOTTIR U, STEFANSSON K. Rate of de novo mutations and the importance of father's age to disease risk. Nature. 2012;488:471–5. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KONG A, STEINTHORSDOTTIR V, MASSON G, THORLEIFSSON G, SULEM P, BESENBACHER S, JONASDOTTIR A, SIGURDSSON A, KRISTINSSON KT, JONASDOTTIR A, FRIGGE ML, GYLFASON A, OLASON PI, GUDJONSSON SA, SVERRISSON S, STACEY SN, SIGURGEIRSSON B, BENEDIKTSDOTTIR KR, SIGURDSSON H, JONSSON T, BENEDIKTSSON R, OLAFSSON JH, JOHANNSSON OT, HREIDARSSON AB, SIGURDSSON G, CONSORTIUM D, FERGUSON-SMITH AC, GUDBJARTSSON DF, THORSTEINSDOTTIR U, STEFANSSON K. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462:868–74. doi: 10.1038/nature08625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEARY RJ, KINDE I, DIEHL F, SCHMIDT K, CLOUSER C, DUNCAN C, ANTIPOVA A, LEE C, MCKERNAN K, DE LA VEGA FM, KINZLER KW, VOGELSTEIN B, DIAZ LA, JR, VELCULESCU VE. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2:20–14. doi: 10.1126/scitranslmed.3000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU Y, ARYEE MJ, PADYUKOV L, FALLIN MD, HESSELBERG E, RUNARSSON A, REINIUS L, ACEVEDO N, TAUB M, RONNINGER M, SHCHETYNSKY K, SCHEYNIUS A, KERE J, ALFREDSSON L, KLARESKOG L, EKSTROM TJ, FEINBERG AP. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol. 2013;31:142–7. doi: 10.1038/nbt.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LO YM, CHAN KC, SUN H, CHEN EZ, JIANG P, LUN FM, ZHENG YW, LEUNG TY, LAU TK, CANTOR CR, CHIU RW. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med. 2010;2:61–91. doi: 10.1126/scitranslmed.3001720. [DOI] [PubMed] [Google Scholar]
- LUPSKI JR. Genetics. Genome mosaicism--one human, multiple genomes. Science. 2013;341:358–9. doi: 10.1126/science.1239503. [DOI] [PubMed] [Google Scholar]
- MACOSKO EZ, MCCARROLL SA. Exploring the variation within. Nat Genet. 2012;44:614–6. doi: 10.1038/ng.2311. [DOI] [PubMed] [Google Scholar]
- MAHER B. Human genetics: Genomes on prescription. Nature. 2011;478:22–4. doi: 10.1038/478022a. [DOI] [PubMed] [Google Scholar]
- MARX V. Next-generation sequencing: The genome jigsaw. Nature. 2013;501:263–8. doi: 10.1038/501261a. [DOI] [PubMed] [Google Scholar]
- MEIGS JB, SHRADER P, SULLIVAN LM, MCATEER JB, FOX CS, DUPUIS J, MANNING AK, FLOREZ JC, WILSON PW, D'AGOSTINO RB, SR, CUPPLES LA. Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med. 2008;359:2208–19. doi: 10.1056/NEJMoa0804742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORAIN S, GREENE MF, MELLO MM. A new era in noninvasive prenatal testing. N Engl J Med. 2013;369:499–501. doi: 10.1056/NEJMp1304843. [DOI] [PubMed] [Google Scholar]
- OTTOBONI L, KEENAN BT, TAMAYO P, KUCHROO M, MESIROV JP, BUCKLE GJ, KHOURY SJ, HAFLER DA, WEINER HL, DE JAGER PL. An RNA profile identifies two subsets of multiple sclerosis patients differing in disease activity. Sci Transl Med. 2012;4:153–131. doi: 10.1126/scitranslmed.3004186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OWENS B. Genomics: The single life. Nature. 2012;491:27–9. doi: 10.1038/491027a. [DOI] [PubMed] [Google Scholar]
- PIROLA L, BALCERCZYK A, OKABE J, EL-OSTA A. Epigenetic phenomena linked to diabetic complications. Nat Rev Endocrinol. 2010;6:665–75. doi: 10.1038/nrendo.2010.188. [DOI] [PubMed] [Google Scholar]
- PODURI A, EVRONY GD, CAI X, WALSH CA. Somatic mutation, genomic variation, and neurological disease. Science. 2013;341:1237758. doi: 10.1126/science.1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PRAHALLAD A, SUN C, HUANG S, DI NICOLANTONIO F, SALAZAR R, ZECCHIN D, BEIJERSBERGEN RL, BARDELLI A, BERNARDS R. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–3. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- PRENSNER JR, IYER MK, SAHU A, ASANGANI IA, CAO Q, PATEL L, VERGARA IA, DAVICIONI E, ERHO N, GHADESSI M, JENKINS RB, TRICHE TJ, MALIK R, BEDENIS R, MCGREGOR N, MA T, CHEN W, HAN S, JING X, CAO X, WANG X, CHANDLER B, YAN W, SIDDIQUI J, KUNJU LP, DHANASEKARAN SM, PIENTA KJ, FENG FY, CHINNAIYAN AM. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45:1392–8. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RIDAURA VK, FAITH JJ, REY FE, CHENG J, DUNCAN AE, KAU AL, GRIFFIN NW, LOMBARD V, HENRISSAT B, BAIN JR, MUEHLBAUER MJ, ILKAYEVA O, SEMENKOVICH CF, FUNAI K, HAYASHI DK, LYLE BJ, MARTINI MC, URSELL LK, CLEMENTE JC, VAN TREUREN W, WALTERS WA, KNIGHT R, NEWGARD CB, HEATH AC, GORDON JI. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RINCON P. Genepeeks firm to offer 'digital baby' screen for sperm donors [Online] [Accessed 10/04/2013];BBC News. 2013 Available: http://ow.ly/qi9wm.
- RIVERA CM, REN B. Mapping human epigenomes. Cell. 2013;155:39–55. doi: 10.1016/j.cell.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SADANANDAM A, LYSSIOTIS CA, HOMICSKO K, COLLISSON EA, GIBB WJ, WULLSCHLEGER S, OSTOS LC, LANNON WA, GROTZINGER C, DEL RIO M, LHERMITTE B, OLSHEN AB, WIEDENMANN B, CANTLEY LC, GRAY JW, HANAHAN D. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19:619–25. doi: 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAUNDERS CJ, MILLER NA, SODEN SE, DINWIDDIE DL, NOLL A, ALNADI NA, ANDRAWS N, PATTERSON ML, KRIVOHLAVEK LA, FELLIS J, HUMPHRAY S, SAFFREY P, KINGSBURY Z, WEIR JC, BETLEY J, GROCOCK RJ, MARGULIES EH, FARROW EG, ARTMAN M, SAFINA NP, PETRIKIN JE, HALL KP, KINGSMORE SF. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med. 2012;4:154–135. doi: 10.1126/scitranslmed.3004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHMITZ RJ, SCHULTZ MD, LEWSEY MG, O'MALLEY RC, URICH MA, LIBIGER O, SCHORK NJ, ECKER JR. Transgenerational epigenetic instability is a source of novel methylation variants. Science. 2011;334:369–73. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHWARZENBACH H, HOON DS, PANTEL K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–37. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- SHAPIRO E, BIEZUNER T, LINNARSSON S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet. 2013;14:618–30. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- SHEN H, LAIRD PW. Interplay between the cancer genome and epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHENDURE J, LIEBERMAN AIDEN E. The expanding scope of DNA sequencing. Nat Biotechnol. 2012;30:1084–94. doi: 10.1038/nbt.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SLADEK R, ROCHELEAU G, RUNG J, DINA C, SHEN L, SERRE D, BOUTIN P, VINCENT D, BELISLE A, HADJADJ S, BALKAU B, HEUDE B, CHARPENTIER G, HUDSON TJ, MONTPETIT A, PSHEZHETSKY AV, PRENTKI M, POSNER BI, BALDING DJ, MEYRE D, POLYCHRONAKOS C, FROGUEL P. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–5. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- SNITKIN ES, ZELAZNY AM, THOMAS PJ, STOCK F, GROUP NCSP, HENDERSON DK, PALMORE TN, SEGRE JA. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci Transl Med. 2012;4:148–116. doi: 10.1126/scitranslmed.3004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SOSMAN JA, KIM KB, SCHUCHTER L, GONZALEZ R, PAVLICK AC, WEBER JS, MCARTHUR GA, HUTSON TE, MOSCHOS SJ, FLAHERTY KT, HERSEY P, KEFFORD R, LAWRENCE D, PUZANOV I, LEWIS KD, AMARAVADI RK, CHMIELOWSKI B, LAWRENCE HJ, SHYR Y, YE F, LI J, NOLOP KB, LEE RJ, JOE AK, RIBAS A. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–14. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SOSNAY PR, SIKLOSI KR, VAN GOOR F, KANIECKI K, YU H, SHARMA N, RAMALHO AS, AMARAL MD, DORFMAN R, ZIELENSKI J, MASICA DL, KARCHIN R, MILLEN L, THOMAS PJ, PATRINOS GP, COREY M, LEWIS MH, ROMMENS JM, CASTELLANI C, PENLAND CM, CUTTING GR. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nat Genet. 2013;45:1160–7. doi: 10.1038/ng.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STANTON D. [Accessed 08/22/2013];GFK Survey [Online] 2013 Available: http://ow.ly/qgnw4.
- SUVA ML, RIGGI N, BERNSTEIN BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–70. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TEST TC. [Accessed 10/29/2013];The Counsyl Test [Online] 2013 Available: https://http://www.counsyl.com.
- TEWHEY R, BANSAL V, TORKAMANI A, TOPOL EJ, SCHORK NJ. The importance of phase information for human genomics. Nat Rev Genet. 2011;12:215–23. doi: 10.1038/nrg2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VELTMAN JA, BRUNNER HG. De novo mutations in human genetic disease. Nat Rev Genet. 2012;13:565–75. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- VIAUD S, SACCHERI F, MIGNOT G, YAMAZAKI T, DAILLERE R, HANNANI D, ENOT DP, PFIRSCHKE C, ENGBLOM C, PITTET MJ, SCHLITZER A, GINHOUX F, APETOH L, CHACHATY E, WOERTHER PL, EBERL G, BERARD M, ECOBICHON C, CLERMONT D, BIZET C, GABORIAU-ROUTHIAU V, CERFBENSUSSAN N, OPOLON P, YESSAAD N, VIVIER E, RYFFEL B, ELSON CO, DORE J, KROEMER G, LEPAGE P, BONECA IG, GHIRINGHELLI F, ZITVOGEL L. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–6. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGELSTEIN B, PAPADOPOULOS N, VELCULESCU VE, ZHOU S, DIAZ LA, JR, KINZLER KW. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG J, FAN HC, BEHR B, QUAKE SR. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell. 2012;150:402–12. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WENZEL SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. 2012;18:716–25. doi: 10.1038/nm.2678. [DOI] [PubMed] [Google Scholar]
- WESTRA HJ, PETERS MJ, ESKO T, YAGHOOTKAR H, SCHURMANN C, KETTUNEN J, CHRISTIANSEN MW, FAIRFAX BP, SCHRAMM K, POWELL JE, ZHERNAKOVA A, ZHERNAKOVA DV, VELDINK JH, VAN DEN BERG LH, KARJALAINEN J, WITHOFF S, UITTERLINDEN AG, HOFMAN A, RIVADENEIRA F, T HOEN PA, REINMAA E, FISCHER K, NELIS M, MILANI L, MELZER D, FERRUCCI L, SINGLETON AB, HERNANDEZ DG, NALLS MA, HOMUTH G, NAUCK M, RADKE D, VOLKER U, PEROLA M, SALOMAA V, BRODY J, SUCHY-DICEY A, GHARIB SA, ENQUOBAHRIE DA, LUMLEY T, MONTGOMERY GW, MAKINO S, PROKISCH H, HERDER C, RODEN M, GRALLERT H, MEITINGER T, STRAUCH K, LI Y, JANSEN RC, VISSCHER PM, KNIGHT JC, PSATY BM, RIPATTI S, TEUMER A, FRAYLING TM, METSPALU A, VAN MEURS JB, FRANKE L. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–43. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WINSLOW R. Patients Share DNA for Cures [Online] [Accessed 09/16/2013];Wall Street Journal. 2013 Available: http://ow.ly/rsrLA.
- WORTHEY EA, MAYER AN, SYVERSON GD, HELBLING D, BONACCI BB, DECKER B, SERPE JM, DASU T, TSCHANNEN MR, VEITH RL, BASEHORE MJ, BROECKEL U, TOMITA-MITCHELL A, ARCA MJ, CASPER JT, MARGOLIS DA, BICK DP, HESSNER MJ, ROUTES JM, VERBSKY JW, JACOB HJ, DIMMOCK DP. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2011;13:255–62. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- YANG Y, MUZNY DM, REID JG, BAINBRIDGE MN, WILLIS A, WARD PA, BRAXTON A, BEUTEN J, XIA F, NIU Z, HARDISON M, PERSON R, BEKHEIRNIA MR, LEDUC MS, KIRBY A, PHAM P, SCULL J, WANG M, DING Y, PLON SE, LUPSKI JR, BEAUDET AL, GIBBS RA, ENG CM. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369:1502–11. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG B, GAITERI C, BODEA LG, WANG Z, MCELWEE J, PODTELEZHNIKOV AA, ZHANG C, XIE T, TRAN L, DOBRIN R, FLUDER E, CLURMAN B, MELQUIST S, NARAYANAN M, SUVER C, SHAH H, MAHAJAN M, GILLIS T, MYSORE J, MACDONALD ME, LAMB JR, BENNETT DA, MOLONY C, STONE DJ, GUDNASON V, MYERS AJ, SCHADT EE, NEUMANN H, ZHU J, EMILSSON V. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell. 2013;153:707–20. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZILLER MJ, GU H, MULLER F, DONAGHEY J, TSAI LT, KOHLBACHER O, DE JAGER PL, ROSEN ED, BENNETT DA, BERNSTEIN BE, GNIRKE A, MEISSNER A. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–81. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]