Abstract
Genes that alter disease risk only in combination with certain environmental exposures may not be detected in genetic association analysis. By using methods accounting for gene-environment (G × E) interaction, we aimed to identify novel genetic loci associated with breast cancer risk. Up to 34,475 cases and 34,786 controls of European ancestry from up to 23 studies in the Breast Cancer Association Consortium were included. Overall, 71,527 single nucleotide polymorphisms (SNPs), enriched for association with breast cancer, were tested for interaction with 10 environmental risk factors using three recently proposed hybrid methods and a joint test of association and interaction. Analyses were adjusted for age, study, population stratification, and confounding factors as applicable. Three SNPs in two independent loci showed statistically significant association: SNPs rs10483028 and rs2242714 in perfect linkage disequilibrium on chromosome 21 and rs12197388 in ARID1B on chromosome 6. While rs12197388 was identified using the joint test with parity and with age at menarche (P-values = 3 × 10−07), the variants on chromosome 21 q22.12, which showed interaction with adult body mass index (BMI) in 8,891 postmenopausal women, were identified by all methods applied. SNP rs10483028 was associated with breast cancer in women with a BMI below 25 kg/m2 (OR = 1.26, 95% CI 1.15–1.38) but not in women with a BMI of 30 kg/m2 or higher (OR = 0.89, 95% CI 0.72–1.11, P for interaction = 3.2 × 10−05). Our findings confirm comparable power of the recent methods for detecting G × E interaction and the utility of using G × E interaction analyses to identify new susceptibility loci.
Keywords: breast cancer risk, gene-environment interaction, polymorphisms, body mass index, case-control study
Introduction
The risk of breast cancer, the most common malignant disease in women, is known to be influenced by multiple genetic and nongenetic (environmental3) factors. Among the most important environmental risk factors are reproductive factors, such as parity (the number of births) and age at menarche, but exogenous hormone use, anthropometric factors, such as body height and body mass index (BMI), and several other lifestyle factors are also associated with breast cancer risk [Bakken et al., 2011; Bergström et al., 2001; Clavel-Chapelon, 2002; Collaborative Group on Hormonal Factors in Breast Cancer, 1996; Ewertz et al., 1990; Key et al., 2006; Ursin et al., 1995; van den Brandt et al., 2000]. Nevertheless, one of the strongest risk factors for breast cancer is having a family member with a diagnosis of breast cancer [Pharoah et al., 1997]. Several high-penetrance genes, such as BRCA1 and BRCA2, as well as moderate penetrance genetic risk variants have been identified. Disease-causing mutations in BRCA1 and BRCA2 increase breast cancer risk up to 20-fold [Mavaddat et al., 2010; Stratton and Rahman, 2008]. However, due to the low frequency of the high-risk and moderate risk variants, they account for only about 20% of familial breast cancer. Genetic association analyses have additionally identified a number of common genetic susceptibility variants. Recently, the large-scale Collaborative Oncological Gene-environment Study (COGS) validated 23 of 27 previously established breast cancer loci and identified 41 new loci associated with overall breast cancer risk, 4 additional loci for estrogen receptor negative breast cancer, and 2 loci for BRCA1 and BRCA2 mutation carriers [Couch et al., 2013; Garcia-Closas et al., 2013; Gaudet et al., 2013; Michailidou et al., 2013]. All the common genetic loci, taken together, have been estimated to explain about 30% of familial risk [Michailidou et al., 2013]. Gene-gene and gene-environment (G × E) interactions may explain a further part of the remaining familial breast cancer risk [Mavaddat et al., 2010]. Testing for interactions with previously identified common susceptibility variants for breast cancer has led to very few consistent results [Campa et al., 2011; Marian et al., 2011; Milne et al., 2010; Nickels et al., 2013; Prentice et al., 2010, 2009; Rebbeck et al., 2009; Travis et al., 2010].
An agnostic approach to identify G × E interactions using existing genome-wide association data has been considered a largely untapped potential means to detect new genetic variants associated with disease risk [Thomas et al., 2012]. As the standard case-control approach is known to have low power for detecting multiplicative G × E interactions, alternative methods with greater power have been developed for testing for G × E interactions in large-scale association studies [Mukherjee et al., 2012]. For large-scale scans, two-step procedures attempt to gain power through enrichment of possible G × E interaction after a first screening step for marginal genetic association and/or G × E correlation [Gauderman et al., 2013; Hsu et al., 2012; Murcray et al., 2011]. Testing jointly for marginal genetic association and G × E interaction in a two degree of freedom (df) test has been shown to achieve good power in gene discovery [Dai et al., 2012; Kraft et al., 2007].
We, therefore, aimed to identify new breast cancer susceptibility loci using about 71,500 single nucleotide polymorphisms (SNPs) enriched for association with breast cancer, by employing different recently proposed methods that account for G × E interaction in a large pooled dataset from studies participating in the Breast Cancer Association Consortium (BCAC).
Methods
Study Participants
We analyzed primary data from 21 case-control and 2 cohort studies in European populations participating in BCAC (Supplementary Table S1). These studies fulfilled the criteria of comprising individuals of European descent and having at least 200 cases and 200 controls with information on age and at least one of the environmental risk factors of interest. All studies were approved by the relevant ethics committees and informed consent was obtained from all participants. All studies collected data with standardized questionnaires. To reconcile differences in study questionnaires, a multistep harmonization procedure was applied to data submitted by all studies according to a common data dictionary. All time-dependent variables were assessed at reference age, which was defined as the age at diagnosis for cases and the age at enrolment for controls in cohort studies, and age at diagnosis for cases and age at interview for controls in case-control studies [Nickels et al., 2013]. Menopausal status was defined based on reference age: women aged ≤54 years were considered as being premenopausal and women aged >54 years as being postmenopausal [Nickels et al., 2013]. To calculate adult BMI, we used the variable “usual weight.” For this variable, women were asked for their usual weight in adulthood or their weight a year ago. Participants were excluded from analysis if they were male, were prevalent cases at recruitment in Melbourne Collaborative Cohort Study (MCCS), were not of European descent, or had a missing value for reference age, the specific environmental variable of interest, or the related adjustment variables. The number of women included in analyses therefore varied according to the environmental factor being studied (Table 1).
Table 1.
Description of the environmental risk factors by case-control status from 23 studies in the BCAC
| Risk factor | Category | Cases | Controls | All |
|---|---|---|---|---|
| Reference age | N | 34,475 | 34,786 | 69,261 |
| N premenopausal | 13,954 | 14,532 | 28,486 | |
| N postmenopausal | 20,521 | 20,254 | 40,775 | |
| Mean (SD)a | 56.2 (11.2) | 55.5 (11.5) | 55.9 (11.4) | |
| Number of births (parity) | N b | 27,174 | 28,508 | 55,682 |
| Mean (SD)a | 1.9 (1.3) | 2.0 (1.3) | 1.9 (1.3) | |
| Age at menarche (menarche) | N b | 21,942 | 23,109 | 45,051 |
| Mean (SD)a | 13.1 (1.6) | 13.1 (1.6) | 13.1 (1.6) | |
| Adult body height (cm, height) | N b | 24,016 | 20,178 | 44,194 |
| Mean (SD)a | 164 (6.6) | 165 (6.6) | 164 (6.6) | |
| BMI (kg/m2, postmenopausal women, BMI post) | N b | 4,423 | 4,468 | 8,891 |
| Mean (SD)a | 25.2 (4.5) | 24.8 (4.2) | 25.0 (4.4) | |
| BMI (kg/m2, premenopausal women, BMI pre) | N b | 1,759 | 1,446 | 3,205 |
| Mean (SD)a | 24.7 (5.1) | 25.5 (5.6) | 25.0 (5.4) | |
| Use of oral contraceptives (years, oral contraceptive duration) | N b | 11,017 | 11,911 | 22,928 |
| Mean (SD)a | 5.3 (7.0) | 5.9 (7.1) | 5.6 (7.1) | |
| Estrogen-progesterone therapy (years, postm. women,a estrogen-progesterone therapy duration) | N b | 3,790 | 4,057 | 7,847 |
| Mean (SD)a | 1.7 (4.3) | 1.2 (3.7) | 1.4 (4.0) | |
| Estrogen therapy (years, postm. women,a estrogen therapy duration) | N b | 3,876 | 4,085 | 7,961 |
| Mean (SD)a | 1.3 (4.1) | 1.0 (3.5) | 1.1 (3.8) | |
| Alcohol consumption (grams per day, alcohol) | N b | 3,812 | 4,055 | 7,867 |
| Mean (SD)a | 7.3 (16.0) | 6.8 (11.3) | 7.1 (13.8) | |
| Family history of breast cancer (famhist) | N b | 20,108 | 18,522 | 38,630 |
| Yes (%) | 4,213 (21%) | 1,606 (9%) | 5,819 (15%) |
Women who stopped hormone therapy before diagnosis/interview were assigned 0 years of therapy.
N is the final sample size for analysis without individuals with unknown values in the variable or any of the adjustment variables.
Genetic Information
Genotyping was carried out in BCAC with the collaboration of three other consortia as part of the COGS. Details of initial SNP selection, genotyping, and quality-control criteria are available in the supplementary material of a recent publication [Michailidou et al., 2013]. Briefly, genotyping of 211,155 SNPs proposed by the four consortia was carried out using an Illumina iSelect genotyping array (iCOGS). Of the 70,862 SNPs proposed by BCAC, 61,240 SNPs had originally been selected from a meta-analysis of nine genome-wide association studies of breast cancer risk, which has led to the discovery of 41 new susceptibility loci for breast cancer [Michailidou et al., 2013]. The remaining SNPs were (i) for fine mapping of known susceptibility loci, (ii) in selected candidate genes or pathways (iii) potentially related to prognosis, or (iii) associated with cancer-related quantitative traits or other cancers. After genotyping, standard quality-control measures were applied to all SNPs and all samples genotyped. SNPs were excluded from the database if their genotypes were discrepant in more than 2% of the duplicate samples across all consortia using this array. SNPs were also excluded if their call rates were below 95% or if their distribution in controls strongly deviated from Hardy–Weinberg Equilibrium (P < 10−6). Study participants were excluded from all analyses if the overall call rate was below 95% or if heterozygosity deviated significantly from that expected in the general population (either lower or higher, P < 10−6). We used genotype data of 87,658 SNPs nominated by BCAC as well as SNPs of common interest, for example, because of possible association with breast cancer related traits or other cancer sites, which remained after application of quality-control criteria. The present analysis aimed to identify new breast cancer susceptibility loci by considering G × E interaction, therefore fine mapping SNPs for the previously identified regions were excluded from analysis, leading to a final number of 71,527 SNPs. Genotype intensity cluster plots were checked manually for SNPs in each new region yielding a statistically significant G × E interaction using any one of the methods employed and SNPs were eliminated if the clustering was judged to be poor. SNP annotations were checked using HaploReg v2 [Ward and Kellis, 2012], and the UCSC Genome Browser [Meyer et al., 2013]. Information on linkage disequilibrium (LD) structure around identified SNPs was obtained using SNP Annotation and Proxy Search (SNAP) [Johnson et al., 2008].
Statistical Analysis
Ten established environmental risk factors for breast cancer were considered. The specific risk variables were selected based on the marginal effects for these risk factors derived from meta-analyses of the nine population-based studies and included number of full-term pregnancies, age at menarche, adult body height, adult BMI (separately for postmenopausal and premenopausal women), duration of oral contraceptive use, duration of menopausal hormone therapy in current users (separately for estrogen-progesterone therapy and estrogen-only therapy), average daily alcohol intake, and family history of breast cancer in first-degree relatives. All 10 environmental variables were evaluated as continuous variables with the exception of family history of breast cancer.
SNPs were assessed using a log-additive model, in which the SNPs are coded according to the number of minor alleles (0–1–2) and analyzed as continuous variables. All analyses were adjusted for reference age, study, and six principal components (PCs) to account for population stratification, with an additional PC for the study Leuven Multidisciplinary Breast Centre (LMBC). The PCs had initially been derived by analyzing 37,000 uncorrelated SNPs that had been genotyped on the same array for other consortia [Michailidou et al., 2013]. Further adjustment variables or restrictions were applied according to the environmental variables assessed (Supplementary Table S2).
Four recently proposed methods that exploit G × E interaction to detect new disease-associated SNPs were applied. Three methods were designed to test for G × E interaction: (i) the hybrid two-step (H2) approach, (ii) a cocktail method (Cocktail), and (iii) a joint screening (EDG × E) approach [Gauderman et al., 2013; Hsu et al., 2012; Murcray et al., 2011]. The fourth method was designed to test jointly for genetic main effect and G × E interaction: the 2df test [Dai et al., 2012]. The H2, Cocktail, and EDG × E approaches are two-step approaches, which combine a testing step with a screening step and a multiple testing correction module. All three methods use the same two tests in the screening step. The first test is a marginal test for genetic association, where the association of the SNPs with the disease of interest is tested without inclusion of the environmental factor. The second test in the screening step tests for correlation between the environmental factor and the SNP, where one is used as an explanatory variable for the other. This test is performed in combined cases and controls, and takes advantage of the oversampling of cases as compared with the general population.
The H2 approach sets certain P-value thresholds for SNPs to pass the marginal and the correlation screening step [Murcray et al., 2011]. Only those SNPs that pass at least one of the screening steps are further tested for G × E interaction. For the screening step for H2, the proposed thresholds of 10−5 for the marginal screening step and 10−3 for the correlation screening were used. G × E interaction is tested using the likelihood ratio test to compare the logistic regression models with and without an interaction term, as in standard case-control analysis. The P-value thresholds for the testing step are calculated by dividing the desired P-value level by the number of SNPs that passed the respective screening step. As two screening steps are performed, a weighting factor of 0.5 is applied to both (giving them equal weight) in order to maintain the overall significance level. SNPs that pass both screening steps are assigned the higher P-value.
In the Cocktail approach, the common screening P-value is assigned the P-value of the correlation screening if this P-value is below a predefined threshold (in our case 10−3) [Hsu et al., 2012]. Otherwise, it is assigned the P-value from the marginal screening test. For the testing step, either standard case-control analysis or a case-only analysis is applied depending on the P-values in the screening tests [Hsu et al., 2012]. If the screening P-value corresponds to the P-value from the marginal screening, SNPs are tested with case-only analysis and case-control analysis otherwise. Subsequently, all SNPs are sorted in ascending order by the screening P-value. According to the weighted hypothesis testing, j groups of increasing size are formed by the equation: sizej = 5 × 2(j−1). All SNPs of j groups are assigned identical alpha thresholds by the formula: aj = 0.05/[5 × 2(2j−1)], which ensures that the overall desired alpha level of 0.05 is maintained [Ionita-Laza et al., 2007]. A SNP is considered significant in the Cocktail approach if the P-value from the testing step is below the alpha threshold for the respective group determined in the screening step.
The EDG × E approach combines the chi-square values from both screening tests into one value and compares it with the chi-square distribution at 2 df [Gauderman et al., 2013]. Resulting P-values are sorted in ascending order and alpha thresholds for j groups are calculated according to the weighted hypothesis testing approach. In the testing step, the EDG × E approach uses case-control analysis and the resulting P-values are compared to the thresholds calculated based on the screening step.
The 2df test jointly tests marginal association and G × E interaction [Kraft et al., 2007]. We employed the newly proposed procedure to combine the two independent tests for the marginal genetic association and for the G × E interaction, exploiting the independence between the two tests [Dai et al., 2012]. This is a chi-squared test applied to the sum of the two squared z scores or log P-values. To correct for multiple testing, Bonferroni correction was applied leading to a P-value threshold of about 7 × 10−7 in the present analysis. Dai et al. offered three different options to test for G × E interaction, of which the standard case-control logistic regression was chosen to avoid biased results due to violation of the G × E independence assumption in the population [Dai et al., 2012].
For comparison, standard case-control logistic regression (CC) for G × E interaction with the Bonferroni-corrected P-value threshold of 7 × 10−7 was also applied. To assess study heterogeneity, we estimated odds ratios (OR) for the per-allele genetic main effect and G × E interaction for each individual study, adjusting for age, and assessed P-values for heterogeneity using a Q-test. Subjects with missing data for a particular SNP or environmental factor were excluded from the respective analysis. We also calculated stratum-specific per-allele ORs for each SNP tested statistically significant using any one of the methods employed. Data preparation and statistical analyses were performed with SAS software (release 9.2) and the R Language and Environment for Statistical Computing, version 2.15.0.
Results
The mean age at recruitment of the study participants was 56 years (Table 1). The sample size and number of studies included for the analyses of the 10 different environmental variables varied between 3,205 women from 4 studies for BMI among premenopausal women and 55,682 women from 22 studies for the number of full-term pregnancies. The exact numbers by study are shown in Supplementary Table S3.
Overall, three SNPs showing a statistically significant association were detected in the analysis of G × E interaction between 10 environmental variables and 71,527 SNPs. Not all were detected by all four methods applied and none was detected using the standard CC approach. All three SNPs were found with the 2df test (Table 2). The latter two of these SNPs were also found to show statistically significant interaction using the other three approaches (Supplementary Tables S4a–c). One SNP is located on chromosome 6 and the other two SNPs are located on chromosome 21q22.12. The latter two SNPs lie in a distance of about 4,000 base pairs, which makes recombination unlikely [Li and Freudenberg, 2009], and are in perfect LD (r2 = 1.0) [Hapmap, 2013]. The SNPs, which were all found to be statistically significantly associated using the 2df test, have not been identified previously as being associated with breast cancer risk and are not in LD with known susceptibility loci.
Table 2.
Significantly associated SNPs with P < 7 × 10−7 in the analysis of multiplicative interaction between 71,527 SNPs and 10 environmental risk factors for breast cancer, using the 2df test
| SNP | Chromosome | Region | Position build 36 | Environmental variable | P-value marginal | OR marginal | P-value interaction | OR interaction | P-value 2df test |
|---|---|---|---|---|---|---|---|---|---|
| rs12197388 | 6 | ARID1B | 161630341 | Parity | 5.69 × 10−8 | 1.09 | 0.37 | 1.01 | 2.68 x 10−7 |
| Menarche | 5.24 × 10−8 | 1.09 | 0.52 | 0.99 | 2.99 × 10−7 | ||||
| rs10483028a | 21 | 21q22.12 | 35595443 | BMI post | 1.70 × 10−5 | 1.17 | 3.19 × 10−5 | 0.84 | 1.68 × 10−8 |
| rs2242714a | 21 | 21q22.12 | 35599557 | BMI post | 2.47 × 10−5 | 1.16 | 4.12 × 10−5 | 0.84 | 3.07 × 10−8 |
These two SNPs were also identified to show statistically significant multiplicative interaction with BMI using the H2, Cocktail, and EDG × E approaches (see Supplementary Tables S4a–c).
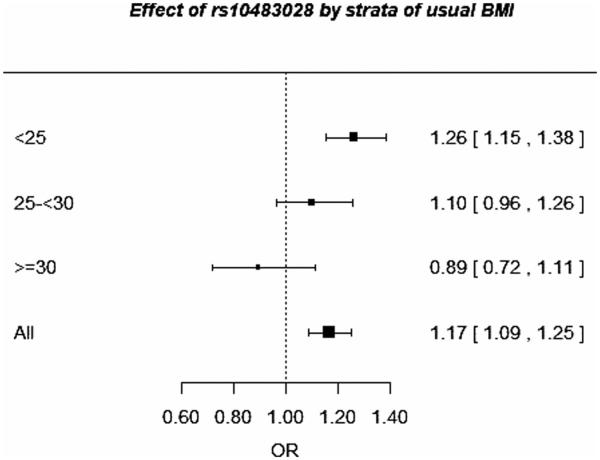
The two associated SNPs on chromosome 21q22.12 (rs10483028 and rs2242714) were identified by analyzing interaction with adult BMI in a sample of 8,891 postmenopausal women from seven studies. Considering the G × E interaction effect (OR = 0.84) was essential for the identification of the two SNPs. The SNP rs10483028 on chromosome 21 showed a decreased effect with increasing BMI, the per-allele ORs being 1.26 (95% CI 1.15–1.38) in women with BMI <25 kg/m2, 1.10 (95% CI 0.96–1.26) in women with BMI between 25 kg/m2 and 30 kg/m2, and 0.89 (95% CI 0.72–1.11) in women with BMI >30 kg/m2 (Fig. 1).
Figure 1.
Effect of rs10483028 on breast cancer risk by strata of adult BMI in 8,891 postmenopausal women from BCAC.
SNP rs12197388 on chromosome 6 was identified in interaction analyses with age at menarche and with parity. This SNP did not show a clear G × E interaction (OR = 1.09) with either risk factor but passed the threshold of the 2df test (7 × 10−7) due to its highly significant marginal association (P < 6 × 10−8).
There was little or no evidence for heterogeneity by study in the G × E interaction ORs for the three identified SNPs. This was also true for the marginal associations of the SNPs with breast cancer risk (Supplementary Table S5 and Supplementary Figure S1 [panel A1–A4]). None of the three identified SNPs had been selected for COGS Illumina iSelect genotyping array (iCOGS) with respect to the environmental factors studied and none was found to be substantially correlated with parity/age at menarche and adult BMI in postmenopausal women, respectively (Supplementary Table S6).
Discussion
To our knowledge, this is the first large-scale agnostic search for G × E interaction to identify new susceptibility loci for breast cancer. To gain power, three recently developed two-step approaches for testing for G × E interaction as well as a joint test for marginal association and G × E interaction were used. We identified three SNPs representing two genetic loci associated with breast cancer risk.
The two SNPs rs10483028 and rs2242714 on chromosome 21q22.12 showing strong G × E interaction effects are located outside known genes. Nevertheless, as shown for the region 8q24, these regions might contain enhancer elements, which may affect the expression of genes in the vicinity [Ahmadiyeh et al., 2010]. There are several SNPs in strong LD (r2 > 0.8) with rs10483028 and rs2242714 (Supplementary Figure S3). However, none of them is located in a known regulatory element (Supplementary Figure S4). The RUNX1 gene is located approximately 300 kb upstream of the two SNPs and has a tumor suppressor role reflected by many somatic mutations in breast tumors. The tumor suppressor activity of RUNX1 is considered to be mediated in part by antagonism of estrogen signaling [Chimge and Frenkel, 2013]. Recurrent mutations in the CBFB transcription factor gene and deletions of its partner RUNX1 also indicated inactivation of this transcription factor complex in breast cancer [Banerji et al., 2012].
The identified SNP rs12197388 is located on chromosome 6 in an intronic region of ARID1B, which belongs to the SWI/SNF chromatin remodeling complex family. SWI/SNF complexes have the ability to enhance or suppress gene transcription by mobilizing nucleosomes [Weissman and Knudsen, 2009]. ARID1B has recently been implicated in breast cancer development through the identification of driver mutations, which confer clonal selective advantage on cancer cells [Stephens et al., 2012]. This gene has been shown to act as a tumor suppressor in pancreatic cancer cell lines [Khursheed et al., 2013]. Mutations in the SWI/SNF complex have also been associated with certain types of syndromes, among those the ARID1B-related intellectual disability syndrome [Kosho et al., 2013] as well as with early treatment failure and decreased survival in children with neuroblastoma [Sausen et al., 2013]. Whether rs12197388 potentially influences ARID1B function is unclear, as it does not seem to be associated with regulatory elements, and there are no further SNPs in at least moderate LD (r2 > 0.6) with rs12197388 based on data from the 1000 Genomes Project (Supplementary Figure S2).
Our results indicate that accounting for G × E interaction using two-step/hybrid methods can lead to the identification of new susceptibility loci. All three methods that test only for G × E interaction identified the two SNPs on chromosome 21 because of a strong interaction between the SNPs and BMI in postmenopausal women but not the SNP rs12197388 on chromosome 6 because of the absence of G × E interaction. This suggests comparable power of these methods based on empirical evidence, which was also demonstrated by simulation studies [Mukherjee et al., 2012]. The consistency of the results between methods provides some support for the robustness of the finding. The SNP rs12197388, on the other hand, was identified through its marginal effect on breast cancer risk. The association between rs12197388 and breast cancer risk was weaker if all subjects of European descent from BCAC were included, irrespective of the availability of information on the respective environmental risk factors (OR = 1.05, P = 7.2×10−5). Because the genetic association was not genome-wide statistically significant, rs12197388 was not identified as susceptibility locus for breast cancer in the recent publication [Michailidou et al., 2013]. Restricted to studies that collected information on epidemiologic risks, our finding could be due to chance or through introducing a selection bias that we are currently not able to explain. However, both the marginal association with breast cancer risk of rs12197388 and the estimates for G × E with number of births and age at menarche were not heterogeneous between studies in the current analysis (Supplementary Figure S1). For the other two SNPs, rs10483028 and rs2242714, which showed statistical interaction with adult BMI in postmenopausal women, the association with breast cancer risk was weaker but still apparent when analyzed in all postmenopausal subjects of European descent in BCAC (OR = 1.06, P-value = 0.001).
Large sample sizes, comprising more than 20,000 cases and controls, were available for the present interaction analyses with number of births, age at menarche, and adult body height. However, sample size was moderate for analyses with most of the other risk factors, such as BMI and menopausal hormone therapy. Multiplicative interactions identified to date between environmental risk factors and common breast cancer susceptibility alleles have been weak or at most moderate [Nickels et al., 2013]. An at least fourfold larger sample size has been shown to be necessary for the identification of G × E effects of the same order of magnitude as compared to marginal effects [Smith and Day, 1984]. Therefore, statistical power to detect an interaction with the other risk factors was still limited [Hein et al., 2008].
It is likely that further susceptibility loci for breast cancer that predominantly act through G × E interactions can be identified in the human genome. Of the set of SNPs in the present analysis, approximately 61,240 were selected based on evidence of association with breast cancer or specifically estrogen receptor negative disease [Garcia-Closas et al., 2013; Michailidou et al., 2013]. A detectable genetic effect, however, is not a prerequisite for the identification of G × E interaction effects. Thus, further SNPs with G × E interaction markers could be identified when expanding the set of genetic considered.
As the present analyses were based on preselected SNPs, the parameters used for the methods, which are designed for genome-wide G × E detection, might not have been optimal in this setting. The H2 and the Cocktail approach require thresholds for the screening step P-values, which can be arbitrary. For the H2 approach, we used the thresholds for the screening steps, which were proposed by the authors and found to be optimal in most of their simulation configurations [Murcray et al., 2011]. Similarly, for the Cocktail approach we used the threshold that had been originally proposed for the Cocktail I approach [Hsu et al., 2012].
All methods employed correct inherently for multiple comparisons introduced by testing large numbers of SNPs, but the number of environmental variables tested was not taken into account. It could be argued that all thresholds should be reduced by one decimal power to correct for multiple testing of environmental factors. However, all 10 environmental variables in our analysis are known breast cancer risk factors. Both SNPs on chromosome 21 would remain significant at the 5% level even if the P-value threshold was reduced by one decimal power to 7 × 10−8. This would not be the case for rs12197388. Although four different methods were used, correction of multiple testing due to the use of different methods did not seem appropriate because all the methods for assessing G × E interaction are highly correlated.
Several studies that contributed to the present analyses were nonpopulation-based. However, selection bias is not expected to influence estimates of G × E interactions in most circumstances [Morimoto et al., 2003]. We did not observe pronounced differences between results from population-based and nonpopulation-based studies in G × E interaction analyses (Supplementary Figure S1). In a previous publication on G × E interactions with known breast cancer SNPs, we also did not observe between-study heterogeneity in interaction ORs. In sensitivity analyses, G × E estimates were not found to change substantially after restriction to population-based studies only [Nickels et al., 2013]. Differential misclassification would rather have led to an underestimation of interaction effects [Garcia-Closas et al., 1998]. In BCAC, risk factor information is harmonized thoroughly in a standardized fashion. For cases in case-control and cohort studies, the reference time was always time at diagnosis. For controls, reference time was time at interview and therefore at baseline recruitment for cohort studies. Misclassification of the menopausal status by using an age surrogate was therefore unproblematic. But specifically for risk factors that are likely to change over time (e.g., smoking behavior and menopausal hormone therapy use), different referent times for assessment could lead to heterogeneity of results derived from cohort vs. case-control studies. As shown in Supplementary Figure S1 (panel A1, A2, B1, B2), we did not observe heterogeneity between case-control and cohort studies. Therefore, the differing reference times did not bias our results to a great extent.
The present analyses were restricted to subjects of European ancestry and adjusted for study to reduce bias due to population stratification. The present results were consistent with previous results on marginal SNP associations from BCAC [Michailidou et al., 2013]. Most of the previously identified breast cancer susceptibility alleles were again detected by application of the 2df test, which also considers the marginal genetic association.
To conclude, the identification of the new breast cancer associated loci supports the hypothesis that new risk loci can be identified by methods that account for G × E interaction in the association analysis. In addition to GWAS for genetic main effects, this approach may facilitate identifying a proportion of the susceptibility loci contributing to polygenic susceptibility to breast cancer, where association differs according to the presence or absence of a particular environmental factor, or is restricted to those with the environmental factor. Replication of the susceptibility loci identified through G × E interaction will however require large sample sizes with environmental risk factor data to achieve adequate power, which might not be trivial to recruit.
Supplementary Material
Acknowledgments
Funding for the iCOGS infrastructure came from the European Community’s Seventh Framework Programme under grant agreement 223175 (HEALTH-F2-2009-223175, COGS), Cancer Research UK (C1287/A10118, C1287/A 10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692), the National Institutes of Health (CA128978), and Post-Cancer GWAS initiative (1U19 CA148537, 1U19 CA148065, and 1U19 CA148112—the GAME-ON initiative), the Department of Defence (W81XWH-10-1-0341), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer, Komen Foundation for the Cure, the Breast Cancer Research Foundation, and the Ovarian Cancer Research Fund.
This study would not have been possible without the contributions of the following: Per Hall (COGS); Douglas F. Easton, Paul Pharoah, Kyriaki Michailidou, Manjeet K. Bolla, Qin Wang (BCAC); Andrew Berchuck (OCAC); Rosalind A. Eeles, Douglas F. Easton, Ali Amin Al Olama, Zsofia Kote-Jarai, Sara Benlloch (PRACTICAL); Georgia Chenevix-Trench, Antonis Antoniou, Lesley McGuffog, Fergus Couch and Ken Offit (CIMBA); Joe Dennis, Alison M. Dunning, Andrew Lee, and Ed Dicks, Craig Luccarini, and the staff of the Centre for Genetic Epidemiology Laboratory; Javier Benitez, Anna Gonzalez-Neira, and the staff of the CNIO genotyping unit; Jacques Simard and Daniel C. Tessier, Francois Bacot, Daniel Vincent, Sylvie LaBoissière, and Frederic Robidoux and the staff of the McGill University; and Génome Québec Innovation Centre, Stig E. Bojesen, Sune F. Nielsen, Borge G. Nordestgaard, and the staff of the Copenhagen DNA laboratory; and Julie M. Cunningham, Sharon A. Windebank, Christopher A. Hilker, Jeffrey Meyer, and the staff of Mayo Clinic Genotyping Core Facility.
The ABCFS work was supported by the United States National Cancer Institute, National Institutes of Health (NIH) under RFA-CA-06–503 and through cooperative agreements with members of the Breast Cancer Family Registry (BCFR) and Principal Investigators, including Cancer Care Ontario (U01 CA69467), Cancer Prevention Institute of California (U01 CA69417), University of Melbourne (U01 CA69638). The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the BCFR, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government or the BCFR. The ABCFS was also supported by the National Health and Medical Research Council of Australia, the New South Wales Cancer Council, the Victorian Health Promotion Foundation (Australia), and the Victorian Breast Cancer Research Consortium. J.L.H. is a National Health and Medical Research Council (NHMRC) Australia fellow and a Victorian Breast Cancer Research Consortium group leader. M.C.S. is an NHMRC senior research fellow and a Victorian Breast Cancer Research Consortium group leader. The ABCS was funded by the Dutch Cancer Society Grant NKI2007–3839 and BBMRI-NL complementation project 11; BBMRI-NL is a research infrastructure financed by the Dutch government (NWO 184.021.007); MKS was funded by Dutch Cancer Society Grant NKI2009–4363. The work of the BBCC was partly funded by ELAN-Fond of the University Hospital of Erlangen. The BBCS is funded by Cancer Research U.K. and Breakthrough Breast Cancer and acknowledges NHS funding to the NIHR Biomedical Research Centre, and the National Cancer Research Network (NCRN). The CECILE study was funded by the Fondation de France, the French National Institute of Cancer (INCa), the National League against Cancer, the National Agency for Environmental and Occupational Health and Food Safety (ANSES), the National Agency for Research (ANR), and the Association for Research against Cancer (ARC). The CGPS was supported by the Chief Physician Johan Boserup and Lise Boserup Fund, the Danish Medical Research Council, and Herlev Hospital. The CNIO-BCS was supported by the Genome Spain Foundation, the Red Temática de Investigación Cooperativa en Cáncer, and grants from the Asociación Española Contra el Cáncer and the Fondo de Investigación Sanitario (PI11/00923 and PI081120). The Human Genotyping-CEGEN Unit, CNIO is supported by the Instituto de Salud Carlos III. ESTHER was supported in part by the Baden-Württemberg State Ministry of Science, Research, and Arts, and by the German Federal Ministry of Education and Research. Additional cases were recruited in the context of the VERDI study, which was supported by a grant from the German Cancer Aid (Deutsche Krebshilfe). The GENICA was funded by the Federal Ministry of Education and Research (BMBF) Germany grants 01KW9975/5, 01KW9976/8, 01KW9977/0, 01KW0114, 01KH0401, 01KH0410, and 01KH0411, the Robert Bosch Foundation, Stuttgart, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, Institute for Prevention and Occupational Medicine of the German Social Accident Insurance (IPA), Bochum, as well as the Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany. KBCP was funded by the special government funding (EVO) of Kuopio University Hospital grants, Cancer Fund of North Savo, the Finnish Cancer Organizations, the Academy of Finland, and by the strategic funding of the University of Eastern Finland. kConFab is supported by grants from the National Breast Cancer Foundation, the National Health and Medical Research Council (NHMRC) and by the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, the Cancer Foundation of Western Australia. AOCS is funded by the U.S. Army Medical Research and Materiel Command (DAMD 170110729 and W81XWH0610220 [AUS]). LMBC is supported by the “Stichting tegen Kanker” (232–2008 and 196–2010). MARIE was supported by the Deutsche Krebshilfe e.V. (70–2892-BR I), the Hamburg Cancer Society, the German Cancer Research Center, and the genotype work in part by the Federal Ministry of Education and Research (BMBF) Germany (01KH0402). MCBCS is funded by the NCI specialized program of research excellence (SPORE) in breast cancer (P50 CA116201), NIH R01 CA128978, and the Breast Cancer Research Foundation. MCCS cohort recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further supported by Australian NHMRC grants 209057, 251553, and 504711 and by infrastructure provided by Cancer Council Victoria. MTLGEBCS: The Canadian Breast Cancer Research Initiative supported the initial case-control study. M.G. gratefully acknowledges receipt of an Investigator Award from the CIHR and a Health Scholar Award from the Fonds de la recherche en santé du Québec. The work of the OFBCR was supported by grant UM1 CA164920 from the National Cancer Institute. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the BCFR. The PBCS was funded by Intramural Research Funds of the National Cancer Institute, Department of Health and Human Services, United States. pKARMA is a combination of the KARMA and LIBRO-1 studies. KARMA was supported by Märit and Hans Rausings Initiative Against Breast Cancer. KARMA and LIBRO-1 were supported the Cancer Risk Prediction Center (CRisP; www.crispcenter.org), a Linnaeus Centre (Contract ID 70867902) financed by the Swedish Research Council. SASBAC was supported by funding from the Agency for Science, Technology and Research of Singapore (A*STAR), the U.S. National Institute of Health (NIH), and the Susan G. Komen Breast Cancer Foundation. KC was financed by the Swedish Cancer Society (5128-B07–01PAF). The SBCS was supported by Yorkshire Cancer Research S305PA. SEARCH was funded by Cancer Research UK:C490/A10119, C490/A10124, C8197/A10123, C1287/A12014, C1287/A10118. UKBGS was funded by Breakthrough Breast Cancer and Institute of Cancer Research.
ABCFS: The authors thank Maggie Angelakos, Judi Maskiell, and Gillian Dite. ABCS wishes to thank Sten Cornelissen, Richard van Hien, Frans Hogervorst, Senno Verhoef, Laura van‘t Veer, Emiel Rutgers, and Ellen van der Schoot. BBCC: The authors thank Sonja Oeser and Silke Landrith. BBCS: We thank Lorna Gibson, Eileen Williams, Elaine Ryder-Mills, and Kara Sargus. CGPS: The authors thank the Danish Breast Cancer Group. CNIO-BCS: The authors thank Charo Alonso, Tais Moreno, Guillermo Pita, Primitiva Menendez, and Anna González-Neira. ESTHER: We thank all the individuals who took part in this study and all the researchers, clinicians, technicians and administrative staff who have enabled this work to be carried out. GENICA: The authors thank the GENICA network—Dr. Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart and University of Tuebingen, Germany (Hiltrud Brauch, Wing-Yee Lo, Christina Justenhoven [current address: Bioscientia Center for Human Genetics, Ingelheim]); Molecular Genetics of Breast Cancer, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, Germany (Ute Hamann); Institute for Prevention and Occupational Medicine of the German Social Accident Insurance; Institute of the Ruhr University Bochum (IPA), Bochum, Germany (Thomas Brüning, Beate Pesch, Sylvia Rabstein, Anne Lotz); Institute for Occupational Medicine and Maritime Medicine, University Medical Center Hamburg-Eppendorf, Germany (Volker Harth); Institute of Pathology, Medical Faculty of the University of Bonn, Bonn, Germany (Hans-Peter Fischer), Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany (Yon-Dschun Ko, Christian Baisch). KBCP: We thank Eija Myöhänen and Helena Kemiläinen for technical assistance. kConFab/AOCS: We wish to thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow Up Study (funded 2001–2009 by NHMRC and currently by the National Breast Cancer Foundation and Cancer Australia 628333) for their contributions to this resource, and the many families who contribute to kConFab. The Australian group gratefully acknowledges the members of the Australian Ovarian Cancer Study Group. LMBC: The authors thank Gilian Peuteman, Dominiek Smeets, Thomas Van Brussel, and Kathleen Corthouts. MARIE: We thank Muhabbet Celik for technical assistance. MCBCS: The authors thank the Mayo Clinic Breast Cancer Patient Registry, David F. and Margaret T. Grohne Family Foundation and Ting Tsung and Wei Fong Chao Foundation. MTLGEBCS: The authors gratefully acknowledge the assistance of Lesley Richardson and Marie-Claire Goulet in conducting the study. OFBCR: The authors thank Teresa Selander, Nayana Weerasooriya, and the OFBCR staff and participants. PBCS: The authors thank Louise Brinton, Neonila Szeszenia-Dabrowska, Beata Peplonska, Witold Zatonski, Pei Chao, and Michael Stagner. SBCS: The authors thank Sue Higham, Simon S. Cross, and Malcolm W.R. Reed. UKBGS: The authors thank Breakthrough Breast Cancer and the Institute of Cancer Research for support and funding of the Breakthrough Generations Study, and the study participants, study staff, and the doctors, nurses and other health care providers and health information sources who have contributed to the study. ICR acknowledges NHS funding to the NIHR Biomedical Research Center.
Footnotes
Supporting Information is available in the online issue at wileyonlinelibrary.com.
Environmental factors include all factors that are not directly measurable from genomic DNA but could nevertheless be partly genetically determined.
The authors declare no conflict of interest.
References
- Ahmadiyeh N, Pomerantz MM, Grisanzio C, Herman P, Jia L, Almendro V, He HH, Brown M, Liu XS, Davis M. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proc Natl Acad Sci USA. 2010;107(21):9742–9746. doi: 10.1073/pnas.0910668107. and others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakken K, Fournier A, Lund E, Waaseth M, Dumeaux V, Clavel-Chapelon F, Fabre A, Hemon B, Rinaldi S, Chajes V. Menopausal hormone therapy and breast cancer risk: impact of different treatments. The European Prospective Investigation into Cancer and Nutrition. Int J Cancer. 2011;128(1):144–156. doi: 10.1002/ijc.25314. and others. [DOI] [PubMed] [Google Scholar]
- Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, Lawrence MS, Sivachenko AY, Sougnez C, Zou L. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486(7403):405–409. doi: 10.1038/nature11154. and others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergström A, Pisani P, Tenet V, Wolk A, Adami HO. Overweight as an avoidable cause of cancer in Europe. Int J Cancer. 2001;91(3):421–430. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1053>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Campa D, Kaaks R, Le ML, Haiman CA, Travis RC, Berg CD, Buring JE, Chanock SJ, Diver WR, Dostal L. Interactions between genetic variants and breast cancer risk factors in the breast and prostate cancer cohort consortium. J Natl Cancer Inst. 2011;103(16):1252–1263. doi: 10.1093/jnci/djr265. and others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimge NO, Frenkel B. The RUNX family in breast cancer: relationships with estrogen signaling. Oncogene. 2013;32(17):2121–1230. doi: 10.1038/onc.2012.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavel-Chapelon F. Differential effects of reproductive factors on the risk of preand postmenopausal breast cancer. Results from a large cohort of French women. Br J Cancer. 2002;86(5):723–727. doi: 10.1038/sj.bjc.6600124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Group on Hormonal Factors in Breast Cancer Breast cancer and hormonal contraceptives: collaborative reanalysis of individual data on 53 297 women with breast cancer and 100 239 women without breast cancer from 54 epidemiological studies. Lancet. 1996;347(9017):1713–1727. doi: 10.1016/s0140-6736(96)90806-5. [DOI] [PubMed] [Google Scholar]
- Couch FJ, Wang X, McGuffog L, Lee A, Olswold C, Kuchenbaecker KB, Soucy P, Fredericksen Z, Barrowdale D, Dennis J. Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genet. 2013;9(3):e1003212. doi: 10.1371/journal.pgen.1003212. and others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai JY, Logsdon BA, Huang Y, Hsu L, Reiner AP, Prentice RL, Kooperberg C. Simultaneously testing for marginal genetic association and gene-environment interaction. Am J Epidemiol. 2012;176(2):164–173. doi: 10.1093/aje/kwr521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewertz M, Duffy SW, Adami HO, Kvale G, Lund E, Meirik O, Mellemgaard A, Soini I, Tulinius H. Age at first birth, parity and risk of breast cancer: a meta-analysis of 8 studies from the Nordic countries. Int J Cancer. 1990;46(4):597–603. doi: 10.1002/ijc.2910460408. [DOI] [PubMed] [Google Scholar]
- Garcia-Closas M, Thompson WD, Robins JM. Differential misclassification and the assessment of gene-environment interactions in case-control studies. Am J Epidemiol. 1998;147(5):426–433. doi: 10.1093/oxfordjournals.aje.a009467. [DOI] [PubMed] [Google Scholar]
- Garcia-Closas M, Couch FJ, Lindstrom S, Michailidou K, Schmidt MK, Brook MN, Orr N, Rhie SK, Riboli E, Feigelson HS. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat Genet. 2013;45(4):392–398. doi: 10.1038/ng.2561. and others. 398e1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauderman WJ, Zhang P, Morrison JL, Lewinger JP. Finding novel genes by testing G × E interactions in a genome-wide association study. Genet Epidemiol. 2013;37(6):603–613. doi: 10.1002/gepi.21748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet MM, Kuchenbaecker KB, Vijai J, Klein RJ, Kirchhoff T, McGuffog L, Barrowdale D, Dunning AM, Lee A, Dennis J. Identification of a BRCA2-specific modifier locus at 6p24 related to breast cancer risk. PLoS Genet. 2013;9(3):e1003173. doi: 10.1371/journal.pgen.1003173. and others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HapMap International HapMap Project; 2006. 2013 Available at: http://hapmap.ncbi. nlm.nih.gov/cgi-perl/gbrowse/hapmap27_B36/. Accessed on June 12, 2013.
- Hein R, Beckmann L, Chang-Claude J. Sample size requirements for indirect association studies of gene-environment interactions (G × E) Genet Epidemiol. 2008;32(3):235–245. doi: 10.1002/gepi.20298. [DOI] [PubMed] [Google Scholar]
- Hsu L, Jiao S, Dai JY, Hutter C, Peters U, Kooperberg C. Powerful cocktail methods for detecting genome-wide gene-environment interaction. Genet Epidemiol. 2012;36(3):183–194. doi: 10.1002/gepi.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionita-Laza I, McQueen MB, Laird NM, Lange C. Genome-wide weighted hypothesis testing in family-based association studies, with an application to a 100K scan. Am J Hum Genet. 2007;81(3):607–614. doi: 10.1086/519748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24(24):2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Key J, Hodgson S, Omar RZ, Jensen TK, Thompson SG, Boobis AR, Davies DS, Elliott P. Meta-analysis of studies of alcohol and breast cancer with consideration of the methodological issues. Cancer Causes Control. 2006;17(6):759–770. doi: 10.1007/s10552-006-0011-0. [DOI] [PubMed] [Google Scholar]
- Khursheed M, Kolla JN, Kotapalli V, Gupta N, Gowrishankar S, Uppin SG, Sastry RA, Koganti S, Sundaram C, Pollack JR. ARID1B, a member of the human SWI/SNF chromatin remodeling complex, exhibits tumour-suppressor activities in pancreatic cancer cell lines. Br J Cancer. 2013;108(10):2056–2062. doi: 10.1038/bjc.2013.200. and others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosho T, Okamoto N, Ohashi H, Tsurusaki Y, Imai Y, Hibi-Ko Y, Kawame H, Homma T, Tanabe S, Kato M. Clinical correlations of mutations affecting six components of the SWI/SNF complex: detailed description of 21 patients and a review of the literature. Am J Med Genet A. 2013;161(6):1221–1237. doi: 10.1002/ajmg.a.35933. and others. [DOI] [PubMed] [Google Scholar]
- Kraft P, Yen YC, Stram DO, Morrison J, Gauderman WJ. Exploiting gene-environment interaction to detect genetic associations. Hum Hered. 2007;63(2):111–119. doi: 10.1159/000099183. [DOI] [PubMed] [Google Scholar]
- Li W, Freudenberg J. Two-parameter characterization of chromosome-scale recombination rate. Genome Res. 2009;19(12):2300–2307. doi: 10.1101/gr.092676.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marian C, Ochs-Balcom HM, Nie J, Kallakury BV, Ambrosone CB, Trevisan M, Edge S, Shields PG, Freudenheim JL. FGFR2 intronic SNPs and breast cancer risk: associations with tumor characteristics and interactions with exogenous exposures and other known breast cancer risk factors. Int J Cancer. 2011;129(3):702–712. doi: 10.1002/ijc.25686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavaddat N, Antoniou AC, Easton DF, Garcia-Closas M. Genetic susceptibility to breast cancer. Mol Oncol. 2010;4(3):174–191. doi: 10.1016/j.molonc.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, Sloan CA, Rosen-bloom KR, Roe G, Rhead B. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41:D64–D69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, Schmidt MK, Chang-Claude J, Bojesen SE, Bolla MK. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genet. 2013;45(4):353–361. doi: 10.1038/ng.2563. and others. 361e1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne RL, Gaudet MM, Spurdle AB, Fasching PA, Couch FJ, Benitez J, Arias Perez JI, Zamora MP, Malats N, dos SSI. Assessing interactions between the associations of common genetic susceptibility variants, reproductive history and body mass index with breast cancer risk in the breast cancer association consortium: a combined case-control study. Breast Cancer Res. 2010;12(6):R110. doi: 10.1186/bcr2797. and others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto LM, White E, Newcomb PA. Selection bias in the assessment of gene-environment interaction in case-control studies. Am J Epidemiol. 2003;158(3):259–263. doi: 10.1093/aje/kwg147. [DOI] [PubMed] [Google Scholar]
- Mukherjee B, Ahn J, Gruber SB, Chatterjee N. Testing gene-environment interaction in large-scale case-control association studies: possible choices and comparisons. Am J Epidemiol. 2012;175(3):177–190. doi: 10.1093/aje/kwr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcray CE, Lewinger JP, Conti DV, Thomas DC, Gauderman WJ. Sample size requirements to detect gene-environment interactions in genome-wide association studies. Genet Epidemiol. 2011;35(3):201–210. doi: 10.1002/gepi.20569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickels S, Truong T, Hein R, Stevens K, Buck K, Behrens S, Eilber U. Evidence of gene-environment interactions between common breast cancer susceptibility loci and established environmental risk factors. PloS Genet. 2013;9(3):e1003284. doi: 10.1371/journal.pgen.1003284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharoah PD, Day NE, Duffy S, Easton DF, Ponder BA. Family history and the risk of breast cancer: a systematic review and meta-analysis. Int J Cancer. 1997;71(5):800–809. doi: 10.1002/(sici)1097-0215(19970529)71:5<800::aid-ijc18>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Prentice RL, Huang Y, Hinds DA, Peters U, Pettinger M, Cox DR, Beilharz E, Chlebowski RT, Rossouw JE, Caan B. Variation in the FGFR2 gene and the effects of postmenopausal hormone therapy on invasive breast cancer. Cancer Epidemiol Biomarkers Prev. 2009;18(11):3079–3085. doi: 10.1158/1055-9965.EPI-09-0611. and others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice RL, Huang Y, Hinds DA, Peters U, Cox DR, Beilharz E, Chlebowski RT, Rossouw JE, Caan B, Ballinger DG. Variation in the FGFR2 gene and the effect of a low-fat dietary pattern on invasive breast cancer. Cancer Epidemiol Biomarkers Prev. 2010;19(1):74–79. doi: 10.1158/1055-9965.EPI-09-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbeck TR, DeMichele A, Tran TV, Panossian S, Bunin GR, Troxel AB, Strom BL. Hormone-dependent effects of FGFR2 and MAP3K1 in breast cancer susceptibility in a population-based sample of post-menopausal African-American and European-American women. Carcinogenesis. 2009;30(2):269–274. doi: 10.1093/carcin/bgn247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sausen M, Leary RJ, Jones S, Wu J, Reynolds CP, Liu X, Blackford A, Parmigiani G, Diaz LA, Jr., Papadopoulos N. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet. 2013;45(1):12–17. doi: 10.1038/ng.2493. and others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith PG, Day NE. The design of case-control studies: the influence of confounding and interaction effects. Int J Epidemiol. 1984;13(3):356–365. doi: 10.1093/ije/13.3.356. [DOI] [PubMed] [Google Scholar]
- Stephens PJ, Tarpey PS, Davies H, van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486(7403):400–404. doi: 10.1038/nature11017. and others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, Rahman N. The emerging landscape of breast cancer susceptibility. Nat Genet. 2008;40(1):17–22. doi: 10.1038/ng.2007.53. [DOI] [PubMed] [Google Scholar]
- Thomas DC, Lewinger JP, Murcray CE, Gauderman WJ. Invited commentary: GE-Whiz! Ratcheting gene-environment studies up to the whole genome and the whole exposome. Am J Epidemiol. 2012;175(3):203–207. doi: 10.1093/aje/kwr365. discussion 208-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis RC, Reeves GK, Green J, Bull D, Tipper SJ, Baker K, Beral V, Peto R, Bell J, Zelenika D. Gene-environment interactions in 7610 women with breast cancer: prospective evidence from the Million Women Study. Lancet. 2010;375(9732):2143–2151. doi: 10.1016/S0140-6736(10)60636-8. and others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursin G, Longnecker MP, Haile RW, Greenland S. A meta-analysis of body mass index and risk of premenopausal breast cancer. Epidemiology. 1995;6(2):137–141. doi: 10.1097/00001648-199503000-00009. [DOI] [PubMed] [Google Scholar]
- van den Brandt PA, Spiegelman D, Yaun SS, Adami HO, Beeson L, Folsom AR, Fraser G, Goldbohm RA, Graham S, Kushi L. Pooled analysis of prospective cohort studies on height, weight, and breast cancer risk. Am J Epidemiol. 2000;152(6):514–527. doi: 10.1093/aje/152.6.514. and others. [DOI] [PubMed] [Google Scholar]
- Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(Database issue):D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman B, Knudsen KE. Hijacking the chromatin remodeling machinery: impact of SWI/SNF perturbations in cancer. Cancer Res. 2009;69(21):8223–8230. doi: 10.1158/0008-5472.CAN-09-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.