Abstract
Aims
Regulated intramembrane proteolysis has been shown to be an important mechanism for oncogenic activation of epithelial cell adhesion molecule (EpCAM) through nuclear translocation of the intracellular domain EpICD. Recent studies have identified new membrane-bound EpCAM variants. To evaluate the prevalence of two membranous EpCAM variants in human tumours, we performed a large-scale expression analysis using specific antibodies against the extracellular domain EpEX (MOC-31 clone) and the intracellular domain EpICD (9-2 clone) of the EpCAM antigen by immunohistochemistry.
Material and methods
Two multi-tissue microarrays (TMA) series containing 1564 tissue samples each of 53 different histological tumour types were stained and compared. One TMA was stained for EpEX and one for EpICD. Membranous full-length EpCAM (EpCAMMF) expression in tissues was defined by the expression of EpEX and EpICD, while the truncated variant of EpCAM (EpCAMMT) was characterised by a significant loss of membranous EpICD expression compared with EpEX expression.
Results
We defined tumours with high EpCAMMT expression (ie, cancers of the endometrium and bladder), tumours with intermediate (ie, gastric, pancreatic, colorectal and oesophageal cancer) and tumours with low rates of expression of the EpCAMMT variant (ie, lung, ovarian, gallbladder, breast and prostate cancer).
Conclusions
Our results indicate that loss of membranous EpICD expression is a common event in human epithelial carcinomas, arguing for the expression of different degrees of EpCAMMF and EpCAMMT variants across the most important tumour entities. Future studies evaluating the prognostic and predictive role of these variants in human malignancies, especially in patients treated with EpCAM-specific antibodies, are clearly warranted.
Keywords: CANCER, CELL ADHESION MOLECULES, IMMUNOHISTOCHEMISTRY
Introduction
Epithelial cell adhesion molecule (EpCAM) is a type I transmembrane glycoprotein which is expressed on the basolateral membrane in most normal epithelial tissues and is overexpressed in many human carcinomas.1 The corresponding EpCAM gene encodes a 40 kDa protein consisting of a 289 amino acid long extracellular domain called EpEX and a short intracellular domain (EpICD) of 26 amino acids.2 Several biological functions of the carcinoma-associated antigen EpCAM have been described including cell adhesion as well as mitogenic signalling.2 3 Recently, EpCAM has also been identified as a marker for cancer-initiating stem cells, making it an interesting target for cancer therapy.4 5
In 2009, the trifunctional anti-EpCAM-specific antibody catumaxomab has been approved for the treatment of malignant ascites in cancer patients with EpCAM positive tumours.6 However, despite advances in the understanding of the biology of EpCAM, results from clinical trials using other EpCAM-specific targeting agents have been disappointing.7 Recent studies have demonstrated that activation of EpCAM follows regulated intramembrane proteolysis.3 Cleavage within the transmembrane domain of EpCAM results in shedding of the extracellular domain EpEX and accumulation of the intracellular domain EpICD to the nucleus. Hence, the cleaved intracellular domain EpICD has been proposed as a novel marker for treatment response in EpCAM positive tumours8 and there is now growing evidence that subcellular compartmental accumulation of EpICD may be involved in development of epithelial carcinomas.9 Very recently, different variants of the EpCAM protein have been described in cell lines with different biological functions.10 A better characterisation of the expression of different EpCAM variants that may predict response to EpCAM-targeted therapies is crucial to optimise the selection of patients for these treatments. Routinely, EpCAM expression is determined by immunohistochemistry through evaluating its membranous staining intensity. However, diagnostic as well therapeutic antibodies that target EpCAM are usually directed against its ectodomain EpEX and these antibodies alone are not sufficient to discriminate between different EpCAM variants. By combining EpEX antibody with EpICD antibody staining on two separate tissue slides of the same tumour specimen, two EpCAM variants can be differentiated: the membrane-bound full-length protein (EpCAMMembranous full-length; EpCAMMF) and its truncated variant (EpCAMMembranous truncated; EpCAMMT) which have lost the intracellular domain but still have a remnant transmembranous and integral extracellular domain.
Defining EpCAM variants by this double-staining procedure, the aim of this study was to elucidate the expression of EpCAMMF and EpCAMMT in human cancers. By the use of two series of multi-tissue microarrays (TMA), we were able to investigate simultaneously the presence of the extracellular (EpEX) as well as the intracellular domain (EpICD) of EpCAM applying specific monoclonal antibodies and comparing the staining results under optimised conditions. Our results show for the first time that loss of membranous EpICD expression is a frequent event in human cancers across a large panel of tumour samples reflecting the different degree of expression of the membranous variants EpCAMMT and EpCAMMF.
Material and methods
Ethics statement
This study was approved by the ethic committee of Basel, Switzerland, and oral and written consent was obtained from all patients. Retrieval of tissue and clinical data was performed according to the regulations of the local institutional review board and data safety laws.
Material and microarray construction
Formalin-fixed and paraffin-embedded tissue probes were sampled from the archives of the Institute for Pathology of the University Hospital Basel (Switzerland). A total of 1564 samples were arranged into a TMA format as described elsewhere.11 Briefly, tissue cylinders with a diameter of 0.6 mm were punched from representative tumour areas of each ‘donor’ tissue block and brought into four different recipient paraffin blocks. Multiple 4 µm sections of the resulting TMA blocks were cut and mounted to an adhesive-coated slide system. In addition, whole tissue sections of tumours from the colon (n=5), stomach (n=5), endometrium (n=10), pancreas (n=5) and ovary (n=5) were analysed.
Calibration of EpICD and EpEX immunohistochemical staining
The development and specificity of the 9-2 antibody clone for the EpICD domain (distributed by HVD Life Science GmbH, Vienna, Austria; for more information: http://www.oncotyrol.at/business/produkte/anti-epicd-epcam-c-term/) were described previously.12 The staining intensity of this antibody was calibrated to be equivalent to the expression observed with the commercially available anti-EpEX antibody MOC-31 (clone: MOC-31, 1:50, Dako).13 Calibration relied on increase of EpICD antibody dilution from 1:25 to 1:50, 1:100, 1:250, 1:500 and 1:1000 on whole tissue sections from normal colon mucosa and colorectal cancer patients (figure 1A) to obtain similar staining pattern to that observed with the EpEX antibody. An optimised concentration of the EpICD antibody of 1:100 was then used to stain one TMA series and the second TMA series was stained with the EpEX antibody MOC-31 as described below.
Figure 1.
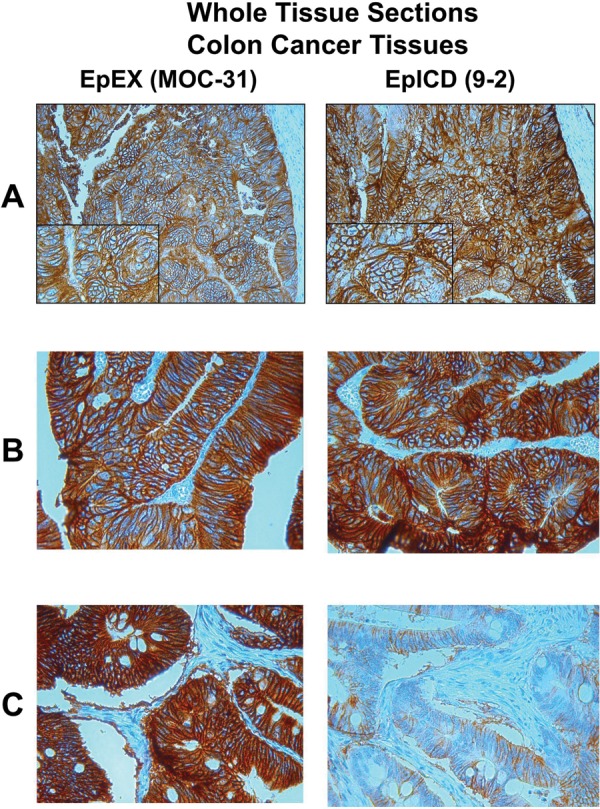
Two tissue sections of the same tumour sample stained with the EpEX antibody MOC-31 and the EpICD antibody 9-2. (A) Calibration of the staining intensity of both antibodies using a colorectal cancer sample. (B) Colon cancer sample with the predominant expression of the full-length variant EpCAMMF. (C) Colon cancer sample with the predominant expression of the truncated variant EpCAMMT.
Immunohistochemistry
Immunohistochemistry of tumour samples with the antibody clone MOC-31 directed against the extracellular domain (EpEX) of EpCAM was performed according to manufacturer's instructions. This staining protocol is used routinely in pathology laboratories for evaluation of pathological specimens.13
For detection of the C-terminus of the intracellular domain (EpICD) of EpCAM, the mouse monoclonal antibody clone 9-2 (distributed by HVD Life Science GmbH; for more information: http://www.oncotyrol.at/business/produkte/anti-epicd-epcam-c-term/) was designed in our laboratory and generated by Biogenes GmbH as described previously.12 Briefly, 4 µm thick sections from formalin-fixed paraffin-embedded tissue specimens were stained using the automated immunostainer Leica BOND-MAX™ (Leica Microsystems, Wetzlar, Germany). Leica provides all the solutions used for specific staining, with the exception of the primary antibody clone 9-2 directed against EpICD. The following steps were programmed on the staining machine: dewaxing at 72°C; antigen retrieval with ER2 (epitope retrieval, pH 9) 20 min at 100°C; wash solution; peroxide blocking solution for 5 min at room temperature; wash solution; treatment with the primary antibody 9-2 at a concentration of 1:100 for staining of microarrays; wash solution; postprimary antibody treatment over 8 min at room temperature; wash solution; Leica BOND-MAX Polymer treatment over 8 min at room temperature; wash solution; mixed DAB treatment over 10 min at room temperature; washing with distillate water; counterstaining with HTX over 8 min at room temperature; washing with distillate water; dehydratation in alcohol and xilene and mounting on glass slides with entelan.
Scoring
EpEX and EpICD antigen expression was evaluated by two independent observers (GM and LT) using light microscopy in a blinded fashion. Discordant cases were re-evaluated on a double-headed microscope to achieve a consensus. Antigen expression was defined as the presence of specific staining on surface membranes of tumour cells as described previously.14 15 Briefly, the expression of EpEX and EpICD was evaluated by calculating a Total Immunostaining Score (TIS) as the product of a Proportion Score (PS) and an Intensity Score (IS). The PS describes the estimated fraction of positive stained tumour cells (0, none; 1, <10%; 2, 10%–50%; 3, 51%–80%; 4, >80%). The IS represents the estimated staining intensity as compared with control cell lines (0, no staining; 1, weak; 2, moderate; 3, strong). The TIS (TIS=PS×IS) ranges from 0 to 12 with only nine possible values (ie, 0, 1, 2, 3, 4, 6, 8, 9 and 12). Using the total immunoreactive score we defined four groups (TIS Group; TISG): no expression (TISG=0, TIS=0); weak expression (TISG=1, TIS 1–4); moderate expression (TISG=2, TIS score=6 and 8); and intense expression (TISG=3, TIS score=9 and 12). A predominant expression of the EpCAMMT variant in a tumour sample was defined if membranous staining was present for EpEX and the corresponding tissue showed a reduction in membranous EpICD staining intensity by at least two TISG scoring points (EpEX+/EpICD− phenotype). Accordingly, the predominant expression of the EpCAMMF variant in a tumour sample was defined by the presence of positive membranous staining for both epitopes without a significant reduction of EpICD expression (ie, loss of <2 TISG scores; EpEX+/EpICD+ phenotype). Samples with weak EpEX expression (TISG=1) and no EpICD expression (TISG=0) were classified as EpCAMMF. Figure 1 and Figure 2 show representative tumour samples with predominant expression of EpCAMMF and samples with the predominant expression of EpCAMMT variants. Differences between histological subtypes of a single tumour entity, if available, were statistically analysed using the χ2 test.
Figure 2.
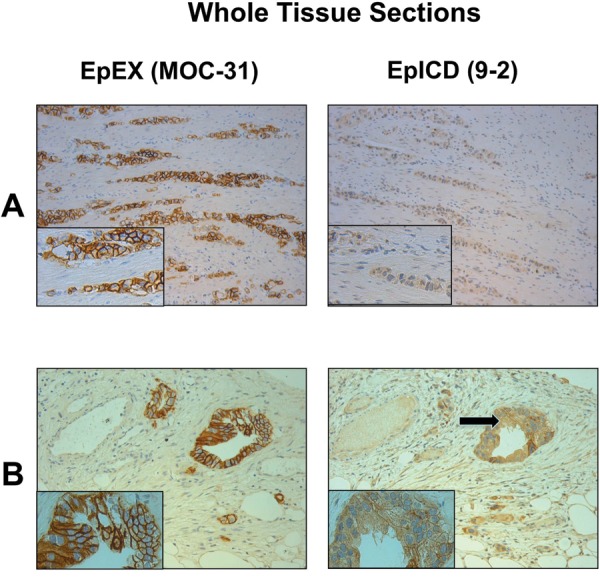
(A) Two tissue sections of the same gastric cancer sample stained with the EpEX antibody MOC-31 and the EpICD antibody 9-2 showing the predominant expression of the truncated variant EpCAMMT. (B) Two tissue sections of a pancreatic cancer sample showing the predominant expression of the truncated variant EpCAMMT. Note the EpICD expression in the cytoplasm (arrow).
Results
Whole tissue sections
To test the suitability and specificity of the 9-2 antibody on formalin-fixed material, a total of 30 whole tissue sections from patients with tumours of the colon, stomach, endometrium, pancreas and ovary were analysed. The known expression status of EpEX staining with the monoclonal antibody MOC-31 of normal colonic epithelium and colon carcinomas served as control. In addition, to validate the results obtained by immunostaining, western blot analysis was performed for EpICD antibody specificity as described previously.12 In concordance with previous reports,11 15 a strong homogenous membranous expression of the extracellular domain EpEX of the EpCAM antigen was detected in the majority of the tumour samples. However, some tumours concurrently revealed differences in the expression pattern of the intracellular domain EpICD. Based on these observations, we defined two membrane-bound EpCAM variants: the full-length variant EpCAMMF defined by a similar expression of both EpEX and EpICD and the truncated variant EpCAMMT defined by a significant loss of EpICD expression compared with EpEX expression (figures 1 and 2). No nuclear expression of EpICD was observed in these tumour samples.
TMA analysis
To further evaluate the prevalence of these two EpCAM variants in human solid tumours, expression of EpEX and EpICD was investigated in two TMA series composed of 1564 independent cases. A total of 1564 human tumour and normal tissue specimens were successfully analysed. For most tumour types, representative number of cases (n>30) was available within the array. EpEX/EpICD staining was predominantly membranous, but weak cytoplasmic staining could also be observed in some tumour types. No nuclear expression of EpICD was observed in these tumour samples. For all analyses, only membranous staining was considered (table 1). As described previously, mesenchymal tissue and tumours as well as tumours of neural origin were EpCAM-negative. As expected, colon cancer demonstrated the most frequent (>90%) expression of EpEX followed by tumours of the female genital tract (table 1). To further evaluate the frequency of loss of EpICD in EpEX positive tissues, a detailed analysis for the subgroup of EpEX-positive tissue samples (n=713) was performed (table 2).
Table 1.
EpEX and EpICD expression defined by MOC-31 and 9-2 antibodies in human malignancies
| EpEX expression | EpICD expression | |||||||
|---|---|---|---|---|---|---|---|---|
| Tumour tissue | Number of tissue samples analysed (%) | |||||||
| Negative | Weak | Moderate | Strong | Negative | Weak | Moderate | Strong | |
| Pancreatic cancer (n=28) | 8 (28.6) | 10 (35.7) | 7 (25.0) | 3 (10.7) | 20 (71.4) | 6 (21.4) | 2 (7.1) | 0 (0) |
| Prostate cancer (n=45) | 3 (6.7) | 19 (42.2) | 16 (35.6) | 7 (15.5) | 17 (37.9) | 10 (22.2) | 11 (24.4) | 7 (15.5) |
| Oesophageal cancer (n=33) | 18 (54.5) | 8 (24.2) | 6 (18.2) | 1 (3.0) | 30 (90.9) | 2 (6.1) | 1 (3.0) | 0 (0) |
| Mesothelioma (n=17) | 13 (76.5) | 4 (23.5) | 0 (0) | 0 (0) | 17 (100) | 0 (0) | 0 (0) | 0 (0) |
| Gallbladder cancer (n=36) | 10 (27.8) | 14 (38.9) | 8 (22.2) | 4 (11.1) | 17 (47.2) | 12 (33.3) | 6 (16.7) | 1 (2.8) |
| Cervical cancer (n=13) | 8 (61.5) | 3 (23.1) | 2 (15.4) | 0 (0) | 10 (76.9) | 3 (23.1) | 0 (0) | 0 (0) |
| Laryngeal cancer (n=16) | 11 (68.8) | 3 (18.8) | 1 (6.2) | 1 (6.2) | 10 (62.6) | 4 (25.0) | 1 (6.2) | 1 (6.2) |
| Hepatocellular carcinoma (n=60) | 54 (90.0) | 5 (8.3) | 1 (1.7) | 0 (0) | 59 (98.3) | 1 (1.7) | 0 (0) | 0 (0) |
| Colon | ||||||||
| Low-grade dysplasia (n=38) | 1 (2.6) | 0 (0) | 0 (0) | 37 (97.4) | 0 (0) | 3 (7.9) | 14 (36.8) | 21 (55.3) |
| High-grade dysplasia (n=81) | 1 (1.2) | 0 (0) | 1 (1.2) | 79 (97.6) | 2 (2.4) | 12 (14.8) | 40 (49.4) | 27 (33.4) |
| Adenocarcinoma (n=33) | 0 (0) | 0 (0) | 3 (9.1) | 30 (90.9) | 1 (3.0) | 13 (39.4) | 10 (30.3) | 9 (27.3) |
| Stomach | ||||||||
| Intestinal cancer (n=36) | 1 (2.8) | 9 (25.0) | 6 (16.6) | 20 (55.6) | 20 (55.6) | 6 (16.6) | 3 (8.4) | 7 (19.4) |
| Diffuse cancer (n=12) | 3 (25.0) | 4 (33.3) | 3 (25.0) | 2 (16.7) | 10 (83.4) | 1 (8.3) | 1 (8.3) | 0 (0) |
| Mixed histology (n=3) | 0 (0) | 1 (33.3) | 1 (33.3) | 1 (33.4) | 3 (100) | 0 (0) | 0 (0) | 0 (0) |
| Small intestine cancer (n=14) | 0 (0) | 3 (21.4) | 4 (28.6) | 7 (50.0) | 6 (42.9) | 2 (14.3) | 1 (7.1) | 5 (35.7) |
| Endometrium | ||||||||
| Endometrioid cancer (n=38) | 6 (15.8) | 7 (18.4) | 8 (21.1) | 17 (44.7) | 29 (76.3) | 5 (13.2) | 4 (10.5) | 0 (0) |
| Serous cancer (n=25) | 1 (4.0) | 0 (0) | 11 (44.0) | 13 (52.0) | 10 (40.0) | 7 (28.0) | 5 (20.0) | 3 (12.0) |
| Urinary bladder cancer (n=48) | 25 (52.1) | 7 (14.6) | 10 (20.8) | 6 (12.5) | 42 (87.4) | 3 (6.3) | 0 (0) | 3 (6.3) |
| Pheochromocytoma (n=13) | 13 (100) | 0 (0) | 0 (0) | 0 (0) | 13 (100) | 0 (0) | 0 (0) | 0 (0) |
| Paraganglioma (n=5) | 5 (100) | 0 (0) | 0 (0) | 0 (0) | 5 (100) | 0 (0) | 0 (0) | 0 (0) |
| Lung | ||||||||
| Adenocarcinoma (n=50) | 1 (2.0) | 12 (24.0) | 24 (48.0) | 13 (26.0) | 17 (34.0) | 18 (36.0) | 13 (26.0) | 2 (4.0) |
| Bronchoalveolar carcinoma (n=6) | 1 (16.6) | 1 (16.7) | 1 (16.7) | 3 (50.0) | 3 (50.0) | 3 (50.0) | 0 (0) | 0 (0) |
| Squamous cell carcinoma (n=20) | 2 (10.0) | 4 (20.0) | 10 (50.0) | 4 (20.0) | 8 (40.0) | 9 (45.0) | 1 (5.0) | 2 (10.0) |
| Large cell carcinoma (n=11) | 3 (27.3) | 2 (18.2) | 5 (45.5) | 1 (9.0) | 4 (36.3) | 2 (18.2) | 5 (45.5) | 0 (0) |
| SCLC (n=33) | 9 (27.3) | 10 (30.3) | 10 (30.3) | 4 (12.1) | 13 (39.4) | 14 (42.4) | 4 (12.1) | 2 (6.1) |
| Ovarian | ||||||||
| Endometrioid cancer (n=27) | 0 (0) | 4 (14.8) | 13 (48.2) | 10 (37.0) | 3 (11.1) | 12 (44.5) | 10 (37.0) | 2 (7.4) |
| Serous cancer (n=27) | 0 (0) | 6 (22.2) | 13 (48.1) | 8 (29.7) | 8 (29.7) | 6 (22.2) | 9 (33.3) | 4 (14.8) |
| Mucinous cancer (n=11) | 1 (9.1) | 1 (9.1) | 5 (45.4) | 4 (36.4) | 2 (18.2) | 2 (18.2) | 5 (45.4) | 2 (18.2) |
| Breast | ||||||||
| Ductal cancer (n=19) | 8 (42.1) | 8 (42.1) | 2 (10.5) | 1 (5.3) | 9 (47.4) | 7 (36.8) | 3 (15.8) | 0 (0) |
| Mucinous cancer (n=7) | 0 (0) | 6 (85.7) | 1 (14.3) | 0 (0) | 2 (28.6) | 5 (71.4) | 0 (0) | 0 (0) |
| Tubular cancer (n=7) | 4 (57.1) | 3 (42.9) | 0 (0) | 0 (0) | 6 (85.7) | 1 (14.3) | 0 (0) | 0 (0) |
| Medullary cancer (n=38) | 7 (18.4) | 21 (55.3) | 7 (18.4) | 3 (7.9) | 18 (47.4) | 14 (36.8) | 4 (10.5) | 2 (5.3) |
| Lobular cancer (n=18) | 12 (66.7) | 5 (27.8) | 1 (5.5) | 0 (0) | 17 (94.5) | 1 (5.5) | 0 (0) | 0 (0) |
| Parotid gland | ||||||||
| Pleomorphic adenoma (n=31) | 17 (54.8) | 6 (19.4) | 8 (25.8) | 0 (0) | 27 (87.1) | 3 (9.7) | 1 (3.2) | 0 (0) |
| Adenolymphoma (n=21) | 4 (19.0) | 2 (9.5) | 12 (57.2) | 3 (14.3) | 14 (66.6) | 6 (28.6) | 1 (4.8) | 0 (0) |
| Cylindroma (n=27) | 16 (59.3) | 9 (33.3) | 2 (7.4) | 0 (0) | 21 (77.8) | 6 (22.2) | 0 (0) | 0 (0) |
| Well-differentiated NET (n=26) | 3 (11.5) | 6 (23.1) | 5 (19.2) | 12 (46.2) | 3 (11.5) | 7 (26.9) | 6 (23.1) | 10 (38.5) |
| Thymoma (n=34) | 25 (73.5) | 8 (23.6) | 1 (2.9) | 0 (0) | 34 (100) | 0 (0) | 0 (0) | 0 (0) |
Expression in lymphomas, soft tissue sarcomas and brain tumours are shown in the online supplementary table.
Table 2.
Expression of membranous full-length (EpCAMMF) EpCAM and the truncated variant (EpCAMMT) in EpEX-positive tumours
| Tumour entity | EpCAMMF | EpCAMMT | χ2 | ||
|---|---|---|---|---|---|
| n | % | n | % | ||
| Pancreatic cancer (n=20) | 13 | 65 | 7 | 35 | NA |
| Prostate cancer (n=42) | 41 | 97.6 | 1 | 2.4 | NA |
| Oesophageal cancer (n=15) | 10 | 66.7 | 5 | 33.3 | NA |
| Mesothelioma (n=4) | 4 | 100 | 0 | 0 | NA |
| Gallbladder cancer (n=26) | 22 | 84.6 | 4 | 15.4 | NA |
| Cervical cancer (n=5) | 4 | 80 | 1 | 20 | NA |
| Laryngeal cancer (n=5) | 5 | 100 | 0 | 0 | NA |
| Hepatocellular carcinoma (n=6) | 5 | 83.3 | 1 | 16.7 | NA |
| Colon | |||||
| Low-grade dysplasia (n=37) | 34 | 91.9 | 3 | 8.1 | 0.024 |
| High-grade dysplasia (n=80) | 66 | 82.5 | 14 | 17.5 | |
| Adenocarcinoma (n=33) | 22 | 66.7 | 11 | 33.3 | |
| Stomach | |||||
| Intestinal type cancer (n=35) | 19 | 54.3 | 16 | 45.7 | 0.180 |
| Diffuse cancer (n=9) | 6 | 66.7 | 3 | 33.3 | |
| Mixed histology (n=3) | 2 | 33.3 | 1 | 66.7 | |
| Small intestine cancer (n=14) | 9 | 64.3 | 5 | 35.7 | NA |
| Endometrium | |||||
| Endometrioid carcinoma (n=32) | 12 | 37.5 | 20 | 62.5 | 0.961 |
| Serous carcinoma (n=24) | 11 | 45.8 | 13 | 54.2 | |
| Urinary bladder cancer (n=23) | 11 | 47.8 | 12 | 52.2 | NA |
| Lung | |||||
| Adenocarcinoma (n=49) | 38 | 77.6 | 11 | 22.3 | 0.034 |
| Bronchoalveolar carcinoma (n=5) | 1 | 20 | 4 | 80 | |
| Squamous cell carcinoma (n=18) | 14 | 77.8 | 4 | 22.2 | |
| Large cell carcinoma (n=8) | 7 | 87.5 | 1 | 12.5 | |
| SCLC (n=24) | 20 | 83.3 | 4 | 16.7 | |
| Ovarian | |||||
| Endometrioid cancer (n=27) | 20 | 74.1 | 7 | 25.9 | 0.180 |
| Serous cancer (n=27) | 21 | 77.8 | 6 | 22.2 | |
| Mucinous cancer (n=10) | 10 | 100 | 0 | 0 | |
| Breast | |||||
| Ductal carcinoma (n=11) | 9 | 81.8 | 2 | 18.2 | 0.815 |
| Mucinous carcinoma (n=7) | 7 | 100 | 0 | 0 | |
| Tubular carcinoma (n=3) | 3 | 100 | 0 | 0 | |
| Medullary carcinoma (n=31) | 28 | 90.3 | 3 | 9.7 | |
| Lobular carcinoma (n=6) | 5 | 83.3 | 1 | 16.7 | |
| Parotid gland | |||||
| Pleomorphic adenoma (n=14) | 6 | 42.9 | 8 | 57.1 | 0.001 |
| Adenolymphoma (n=17) | 4 | 23.5 | 13 | 76.5 | |
| Cylindroma (n=11) | 10 | 90.9 | 1 | 9.1 | |
| Well-differentiated NET (n=23) | 19 | 82.6 | 4 | 17.4 | NA |
| Thymoma (n=9) | 8 | 88.9 | 1 | 10.1 | NA |
| Total (n=713) | 526 | 73.8 | 187 | 26.2 | |
NA, Not applicable.
Expression of EpCAMMF and EpCAMMT variants in human tissues
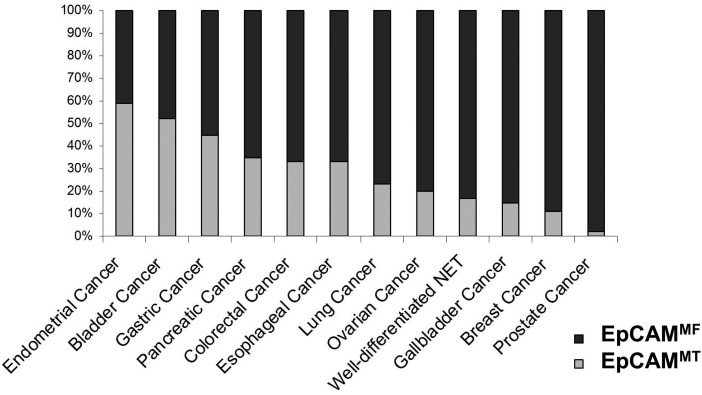
In total, 713 tumour samples were EpEX-positive and were further evaluated. Figure 3 shows gastric cancer samples expressing prevalently the EpCAMMF variant (figure 3A) as well as a tumour expressing the EpCAMMT variant (figure 3B), respectively. Altogether, the majority (73, 9%) of tissues analysed demonstrated a predominant expression of the EpCAMMF variant. However, the ratio between EpCAMMF and EpCAMMT changed significantly dependent on the tumour type. The prevalence of EpCAMMF and EpCAMMT expression in the most frequent EpEX-positive human malignancies is outlined in figure 4. We identified endometrial and bladder cancer as tumours with the highest proportion (>50%) of cases expressing the truncated EpCAM variant. Moreover, we observed tumour entities (ie, stomach, pancreatic, colorectal and oesophageal cancer) with an intermediate proportion (33%–50%) and tumours with a low proportion of expression of the EpCAMMT variant (<33%; ie, tumours of the lung, ovary, gallbladder, breast, prostate and well-differentiated neuroendocrine carcinomas), respectively. A detailed analysis of all tissue samples is shown in table 2. Taken together, these results support the view of a widespread loss of membranous EpICD expression in human epithelial carcinomas resulting in different degrees of the expression of the EpCAMMT and EpCAMMF variants. Of note, adenomas and adenolymphomas but not cylindromas of the parotid gland also showed a high prevalence of expression of the EpCAMMT variant (68%). Intriguingly, the expression of the EpCAMMT variant was observed in 3 (8.1%) of 37 samples of low-grade dysplasia (table 2). In contrast, EpCAMMT expression was observed in 14 (17.5%) out of 66 tissue samples of high-grade dysplasia and 11 (33.3%) out of 22 colon carcinomas (p=0.024). Finally, the EpCAMMT variants appear to be particularly frequent in bronchoalveolar carcinoma of the lung (80%, p=0.034).
Figure 3.
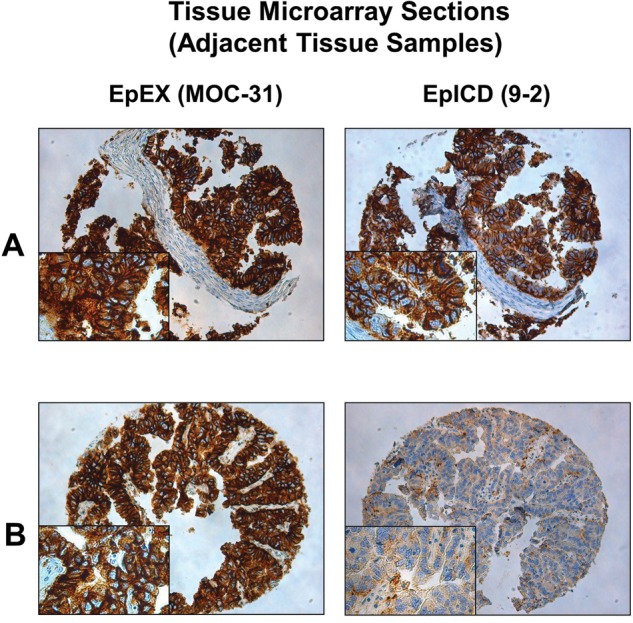
Staining of the EpEX antibody antibody MOC-31 and the EpICD antibody 9-2 using two tissue microarray (TMA) sections of the same series. (A) Two punches of the same gastric cancer sample with predominant EpCAMMF expression. (B) Two punches of the same gastric cancer sample with predominant EpCAMMT expression. Respective tissue samples (A and B) were situated in adjacent positions on the TMA.
Figure 4.
Expression of EpCAMMF and EpCAMMT in most frequent EpEX-positive human malignancies. Endometrial and bladder cancer demonstrated the highest proportion (>50%) of cases expressing the truncated EpCAM variant. Carcinomas of the stomach, pancreas, colon, rectum and oesophagus revealed an intermediate proportion (33%–50%) and tumours of the lung, ovary, gallbladder, breast, prostate and well-differentiated neuroendocrine carcinomas exhibited a low proportion (<33%) of expression of the EpCAMMT variant.
Discussion
In this study, we investigated for the first time the membranous expression pattern of two EpCAM variants in a large cohort including the major human malignancies by using two monoclonal antibodies targeting the ectodomain and endodomain of EpCAM. For the question addressed, we used the high-throughput technique of a multi-tissue array to minimise batch-to-batch variability and make the analyses most comparable. The application and validation of the microarray technology for biomarker assays compared with whole tissue sections have been sufficiently proven.16 Our data provide strong evidence that a different degree of expression of two membranous EpCAM variants occurs during carcinogenesis in the majority of epithelial cancers. Activation of EpCAM through cleavage of its intracellular domain EpICD seems to play an important role in the neoplastic transformation.
The regulated intramembrane proteolysis process occurs for an increasing number of membrane proteins and was first described for EpCAM by Maetzel et al3 who demonstrated that the cytoplasmic domain EpICD is cleaved by the two proteases, ADAM17 and y-secretase, producing a 5 kDa intracellular fragment with oncogenic potential. Since then, several studies reported significant alterations in the levels of the intracellular and extracellular domains of EpCAM as well as the accumulation of different EpCAM fragments within cellular compartments.17–19 Only recently, Schnell et al10 could demonstrate that EpCAM can be cleaved at multiple positions within its ectodomain resulting in various N-terminal proteolytic fragments. Importantly, in the present study the monoclonal antibody MOC-31 was used, which recognises the most N-terminal located epitope of the EpEX antigen thus detecting the full-length EpEX domain as described by Schnell et al. We postulate that combining MOC-31 with the EpICD antibody 9-2 detects the two most predominant variants of EpCAM in human cancer tissues.
A reciprocal loss of membrane EpEX but an increased nuclear and cytoplasmic accumulation of EpICD was first described in aggressive thyroid cancer depending on the histological subtype and differentiation grade of tumour cells.20 In papillary thyroid microcarcinoma, nuclear translocation of EpICD as well as loss of membranous EpEX was found to correlate with metastatic disease. In addition, the same authors could provide evidence that accumulation of nuclear and cytoplasmic EpICD is a frequent event in other epithelial cancers.9 Moreover, concurrent high EpICD and EpEX membrane expression was observed in some cancer cells suggesting the expression of the full-length protein (EpCAMMF) in a subset of tumours. Similar results have been reported by Lin et al18 who demonstrated an increased nuclear EpICD expression in colon cancer compared with normal tissue. Increased release of EpEX enhanced EpICD cleavage resulting in activation of reprogramming factors and epithelial-mesenchymal transition genes suggesting that EpICD participates in tumour initiation and progression. However, they could not detect a homogenous expression of soluble EpICD in cytoplasm or nucleus in any tumour cells assuming that cleavage of EpICD might be a dynamic process. We also could not detect a precise nuclear EpICD staining by using immunohistochemistry in the present study confirming the data by Schnell et al.10
The prognostic value of EpCAM is still a matter of debate. While we and others could demonstrate a poor clinical outcome in patients with tumours harbouring EpCAM overexpression,14 21–25 there is evidence for the same antigen to correlate with good prognosis in a variety of tumour entities.26 27 Our group could show that EpCAM can act both as tumour suppressor and tumour promotor28 29 in breast cancer cell lines and the dual role of EpCAM was confirmed by a recent study in breast cancer patients.30 Only recently, Eichelberg et al31 reported a positive EpCAM status to be significantly associated with a prolonged overall survival in renal cell carcinoma. Moreover, the prognostic value of cytoplasmic accumulation of EpCAM was demonstrated in patients with node positive breast cancer.17 However, none of the studies mentioned above considered the importance of incorporating different EpCAM variants within their assays. Notably, in pancreatic cancer we could show that loss of membranous EpICD expression predicts poor prognosis in patients with tumours harbouring the EpCAMMT variant12 suggesting a different biological function of these variants.
Most colorectal carcinomas undergo neoplastic progression through the adenoma–carcinoma sequence accompanied by an accumulation of successive genetic alterations.32 33 A focal loss of membranous EpEX expression was observed at the invasive margin in colorectal cancer, which was significantly associated with a higher extend of tumour budding and a more invasive phenotype.34 In our study, the majority of adenomas with low-grade dysplasia showed a predominant EpCAMMF expression, whereas a significant higher expression of the EpCAMMT variant was observed in high-grade dysplasia and invasive colon cancer specimens. Further studies on colorectal cancer samples are necessary to confirm these results. Moreover, subgroup analyses of different lung cancer types are necessary to clarify if differential expression of EpCAMMT variant in bronchoalveolar carcinoma is confirmed. Our preliminary results from EpCAM mRNA analyses in endometrial carcinoma rather suggest post-translational modification than EpCAM mutation in sporadic tumour development (data not shown). Additionally, the data from Lin et al18 emphasise the tumour-initiating abilities of EpCAM in colon cancer. We speculate that changes in expression of membranous EpCAM variants are associated with malignant transformation and contribute to the development in a subset of colon cancers. However, benign lesions of the parotid gland such as adenomas and adenolymphomas also showed a high prevalence of expression of the EpCAMMT variant. Future studies are necessary to clarify if EpCAM plays an important role in the development of these entities.
In conclusion, we show for the first time the differential expression of EpCAMMF and EpCAMMT variants in epithelial tumours. Additional studies investigating membranous expression of both EpEX and EpICD as well as the prognostic and predictive value of EpCAM variants might help to better elucidate the clinical relevance of EpCAM in carcinogenesis. Finally, a better understanding of the biological role of EpCAM will hopefully help to develop more effective anti-EpCAM strategies.
Take home messages.
At least two membranous variants of epithelial cell adhesion molecule (EpCAM) (EpCAMMT or EpCAMMF) are differentially expressed in human malignancies.
Malignancies with the highest degree of the truncated EpCAMMT variant are endometrial, urothelial, gastric, colorectal and pancreatic cancers.
Future studies evaluating the predictive and prognostic role of EpCAMMT and EpCAMMF variants with respect to treatment response to EpCAM-specific antibodies (catumaxomab, adecatumumab, etc) are necessary.
Supplementary Material
Footnotes
Contributors: DF, GG and GS planned the analysis and wrote the manuscript. GM, LT and AK performed immunohistochemical analysis and performed immunohistochemical reading. FL and AS developed and produced the 9-2 antibody clone.
Funding: This work was supported by the COMET Center ONCOTYROL and funded by the Federal Ministry for Transport Innovation and Technology (BMVIT) and the Federal Ministry of Economics and Labour/the Federal Ministry of Economy, Family and Youth (BMWA/BMWFJ), the Tiroler Zukunftsstiftung (TZS) and the State of Styria represented by the Styrian Business Promotion Agency (SFG) and supported by the Innsbruck Medical University, Fresenius Biotech GmbH and TILAK- Tiroler Landeskrankenanstalten GmbH (TILAK-Hospital Holding Company). This study was furthermore supported by the Austrian Cancer Society Tirol (grant to AS).
Competing interests: GS has served as consultant and advisory board member for Fresenius Biotech.
Patient consent: Obtained.
Ethics approval: Ethical Committee in Basel.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Balzar M, Winter MJ, de Boer CJ, et al. The biology of the 17-1A antigen (Ep-CAM). J Mol Med (Berl) 1999;77:699–712 [DOI] [PubMed] [Google Scholar]
- 2.Litvinov SV, Balzar M, Winter MJ, et al. Epithelial cell adhesion molecule (Ep-CAM) modulates cell-cell interactions mediated by classic cadherins. J Cell Biol 1997;139:1337–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maetzel D, Denzel S, Mack B, et al. Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell Biol 2009;11:162–71 [DOI] [PubMed] [Google Scholar]
- 4.Dontu G, Al-Hajj M, Abdallah WM, et al. Stem cells in normal breast development and breast cancer. Cell Prolif 2003;36:59–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Brien CA, Pollett A, Gallinger S, et al. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2007;445:106–10 [DOI] [PubMed] [Google Scholar]
- 6.Seimetz D, Lindhofer H, Bokemeyer C. Development and approval of the trifunctional antibody catumaxomab (anti-EpCAM x anti-CD3) as a targeted cancer immunotherapy. Cancer Treat Rev 2010;36:458–67 [DOI] [PubMed] [Google Scholar]
- 7.Niedzwiecki D, Bertagnolli MM, Warren RS, et al. Documenting the natural history of patients with resected stage II adenocarcinoma of the colon after random assignment to adjuvant treatment with edrecolomab or observation: results from CALGB 9581. J Clin Oncol 2011;29:3146–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpenter G, Red Brewer M. EpCAM: another surface-to-nucleus missile. Cancer Cell 2009;15:165–6 [DOI] [PubMed] [Google Scholar]
- 9.Ralhan R, He HC, So AK, et al. Nuclear and cytoplasmic accumulation of Ep-ICD is frequently detected in human epithelial cancers. PLoS One 2010;5:e14130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schnell U, Kuipers J, Giepmans BN. EpCAM proteolysis: new fragments with distinct functions? Biosci Rep 2013;33:e00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Went PT, Lugli A, Meier S, et al. Frequent EpCam protein expression in human carcinomas. Hum Pathol 2004;35:122–8 [DOI] [PubMed] [Google Scholar]
- 12.Fong D, Moser P, Kasal A, et al. Loss of membranous expression of the intracellular domain of EpCAM is a frequent event and predicts poor survival in patients with pancreatic cancer. Histolopathology 9 Oct 2013. Epub ahead of print. 10.1111/his.12307 [DOI] [PubMed] [Google Scholar]
- 13.Pai RK, West RB. MOC-31 exhibits superior reactivity compared with Ber-EP4 in invasive lobular and ductal carcinoma of the breast: a tissue microarray study. Appl Immunohistochem Mol Morphol 2009;17:202–6 [DOI] [PubMed] [Google Scholar]
- 14.Gastl G, Spizzo G, Obrist P, et al. Ep-CAM overexpression in breast cancer as a predictor of survival. Lancet 2000;356:1981–2 [DOI] [PubMed] [Google Scholar]
- 15.Spizzo G, Fong D, Wurm M, et al. EpCAM expression in primary tumour tissues and metastases: an immunohistochemical analysis. J Clin Pathol 2011;64:415–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang D, Salto-Tellez M, Putti TC, et al. Reliability of tissue microarrays in detecting protein expression and gene amplification in breast cancer. Mod Pathol 2003;16:79–84 [DOI] [PubMed] [Google Scholar]
- 17.Alberti S, Ambrogi F, Boracchi P, et al. Cytoplasmic Trop-1/Ep-CAM overexpression is associated with a favorable outcome in node-positive breast cancer. Jpn J Clin Oncol 2012;42:1128–37 [DOI] [PubMed] [Google Scholar]
- 18.Lin CW, Liao MY, Lin WW, et al. Epithelial cell adhesion molecule regulates tumor initiation and tumorigenesis via activating reprogramming factors and epithelial-mesenchymal transition gene expression in colon cancer. J Biol Chem 2012;287:39449–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunavisarut T, Kak I, Macmillan C, et al. Immunohistochemical analysis based Ep-ICD subcellular localization index (ESLI) is a novel marker for metastatic papillary thyroid microcarcinoma. BMC Cancer 2012;12:523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ralhan R, Cao J, Lim T, et al. EpCAM nuclear localization identifies aggressive thyroid cancer and is a marker for poor prognosis. BMC Cancer 2010;10:331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varga M, Obrist P, Schneeberger S, et al. Overexpression of epithelial cell adhesion molecule antigen in gallbladder carcinoma is an independent marker for poor survival. Clin Cancer Res 2004;10:3131–6 [DOI] [PubMed] [Google Scholar]
- 22.Spizzo G, Went P, Dirnhofer S, et al. Overexpression of epithelial cell adhesion molecule (Ep-CAM) is an independent prognostic marker for reduced survival of patients with epithelial ovarian cancer. Gynecol Oncol 2006;103:483–8 [DOI] [PubMed] [Google Scholar]
- 23.Benko G, Spajic B, Kruslin B, et al. Impact of the EpCAM expression on biochemical recurrence-free survival in clinically localized prostate cancer. Urol Oncol 2013;31:468–74 [DOI] [PubMed] [Google Scholar]
- 24.Stoecklein NH, Siegmund A, Scheunemann P, et al. Ep-CAM expression in squamous cell carcinoma of the esophagus: a potential therapeutic target and prognostic marker. BMC Cancer 2006;6:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fong D, Steurer M, Obrist P, et al. Ep-CAM expression in pancreatic and ampullary carcinomas: frequency and prognostic relevance. J Clin Pathol 2008;61:31–5 [DOI] [PubMed] [Google Scholar]
- 26.Seligson DB, Pantuck AJ, Liu X, et al. Epithelial cell adhesion molecule (KSA) expression: pathobiology and its role as an independent predictor of survival in renal cell carcinoma. Clin Cancer Res 2004;10:2659–69 [DOI] [PubMed] [Google Scholar]
- 27.Songun I, Litvinov SV, van de Velde CJ, et al. Loss of Ep-CAM (CO17–1A) expression predicts survival in patients with gastric cancer. Br J Cancer 2005;92:1767–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martowicz A, Spizzo G, Gastl G, et al. Phenotype-dependent effects of EpCAM expression on growth and invasion of human breast cancer cell lines. BMC Cancer 2012;12:501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gostner JM, Fong D, Wrulich OA, et al. Effects of EpCAM overexpression on human breast cancer cell lines. BMC Cancer 2011;11:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soysal SD, Muenst S, Barbie T, et al. EpCAM expression varies significantly and is differentially associated with prognosis in the luminal B HER2, basal-like, and HER2 intrinsic subtypes of breast cancer. Br J Cancer 2013;108:1480–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eichelberg C, Chun FK, Bedke J, et al. Epithelial cell adhesion molecule is an independent prognostic marker in clear cell renal carcinoma. Int J Cancer 2013;132:2948–55 [DOI] [PubMed] [Google Scholar]
- 32.Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. N Engl J Med 1988;319:525–32 [DOI] [PubMed] [Google Scholar]
- 33.Aytes A, Mollevi DG, Martinez-Iniesta M, et al. Stromal interaction molecule 2 (STIM2) is frequently overexpressed in colorectal tumors and confers a tumor cell growth suppressor phenotype. Mol Carcinog 2012;51:746–53 [DOI] [PubMed] [Google Scholar]
- 34.Gosens MJ, van Kempen LC, van de Velde CJ, et al. Loss of membranous Ep-CAM in budding colorectal carcinoma cells. Mod Pathol 2007;20:221–32 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.