Abstract
With genome sequence and composition highly analogous to human, rhesus macaque represents a unique reference for evolutionary studies of human biology. Here, we developed a comprehensive genomic framework of rhesus macaque, the RhesusBase2, for evolutionary interrogation of human genes and the associated regulations. A total of 1,667 next-generation sequencing (NGS) data sets were processed, integrated, and evaluated, generating 51.2 million new functional annotation records. With extensive NGS annotations, RhesusBase2 refined the fine-scale structures in 30% of the macaque Ensembl transcripts, reporting an accurate, up-to-date set of macaque gene models. On the basis of these annotations and accurate macaque gene models, we further developed an NGS-oriented Molecular Evolution Gateway to access and visualize macaque annotations in reference to human orthologous genes and associated regulations (www.rhesusbase.org/molEvo). We highlighted the application of this well-annotated genomic framework in generating hypothetical link of human-biased regulations to human-specific traits, by using mechanistic characterization of the DIEXF gene as an example that provides novel clues to the understanding of digestive system reduction in human evolution. On a global scale, we also identified a catalog of 9,295 human-biased regulatory events, which may represent novel elements that have a substantial impact on shaping human transcriptome and possibly underpin recent human phenotypic evolution. Taken together, we provide an NGS data-driven, information-rich framework that will broadly benefit genomics research in general and serves as an important resource for in-depth evolutionary studies of human biology.
Keywords: human evolution, rhesus macaque, human-specific trait, next-generation sequencing, human regulation, RhesusBase
Introduction
Rhesus macaque, with its genome sequence and composition highly analogous to human, is an emerging model organism that provides a unique perspective for evolutionary studies of human biology (Gibbs et al. 2007). Several recent studies have highlighted rhesus macaque as a unique model for defining and elucidating human-specific genes and functional networks (Knowles and McLysaght 2009; Toll-Riera et al. 2009; Li, Zhang, et al. 2010; Xie et al. 2012), and for further understanding, the evolutionary and functional relevance of human regulations individually and as a whole (Hudson and Snyder 2006; Wray 2007; Brawand et al. 2011; Barbosa-Morais et al. 2012; Shibata et al. 2012; Ramaswami et al. 2013). These lines of evidence lend support to the hypothesis that noncoding regulatory elements may contribute substantially to the phenotypic differences between humans and other primate species (Wray 2007). Despite these unique advantages, several unresolved issues have limited current use of the rhesus macaque model in evolutionary research—inadequate functional genomics annotations, error-prone gene models, and lack of a platform for visualizing and assessing high-throughput data generated by next-generation sequencing (NGS). A genomic context of rhesus macaque, equipped with comprehensive functional annotations, accurate macaque gene models, and NGS-oriented genomic framework for high-throughput data handling, would therefore provide a strong basis for the evolutionary studies of human biology.
To address these key questions, we previously reported the “RhesusBase,” the first knowledgebase for macaque functional genomics (Zhang et al. 2013). Through this database, we extensively refined macaque gene models and integrated macaque functional annotations at the genome-wide level (Zhang et al. 2013). Although RhesusBase has been successful in supporting research in this field, only limited NGS data sets were integrated into the database, and RNA-seq data from only one macaque animal were used for gene model revisions. Because of the rapid accumulation of high-throughput data derived from various NGS technologies, further incorporation of new data sets may significantly aid the refinement of rhesus macaque genome sequence and gene models and help compensate for individual variability in genomes and transcriptomes. In addition, the increasing attention to RhesusBase data brought to light the urgent need for a more user-friendly genomic framework that allows productive NGS data accession, visualization, and management.
Following the rapid pace of NGS technology development, here we report RhesusBase2 with the integration of 1,667 NGS data sets (>48.7 billion processed reads) and about 51.2 million new functional annotation records. With one order of magnitude increase in database size, we re-evaluated the macaque gene models in RhesusBase and released an accurate and up-to-date version. Particularly, we highlighted the unique evolutionary model of rhesus macaque for primate comparative analysis, by providing a user-friendly, NGS-oriented genomic framework to visualize and access macaque annotations for human orthologous genes and the associated regulations. In this macaque genomic framework, we identified a catalog of 9,295 human-biased regulatory events that have a substantial impact on shaping human transcriptome, supporting the hypothesis that novel regulatory elements may constitute an important part of human phenotypic evolution (King and Wilson 1975; Wray 2007; Carroll 2008; Shibata et al. 2012). Through establishing a unique and well-annotated genomic context of rhesus macaque, RhesusBase2 provides a comprehensive framework for in-depth evolutionary studies of human genes and the associated regulations.
Results
An NGS Data-Driven, Information-Rich Core Database for Rhesus Macaque
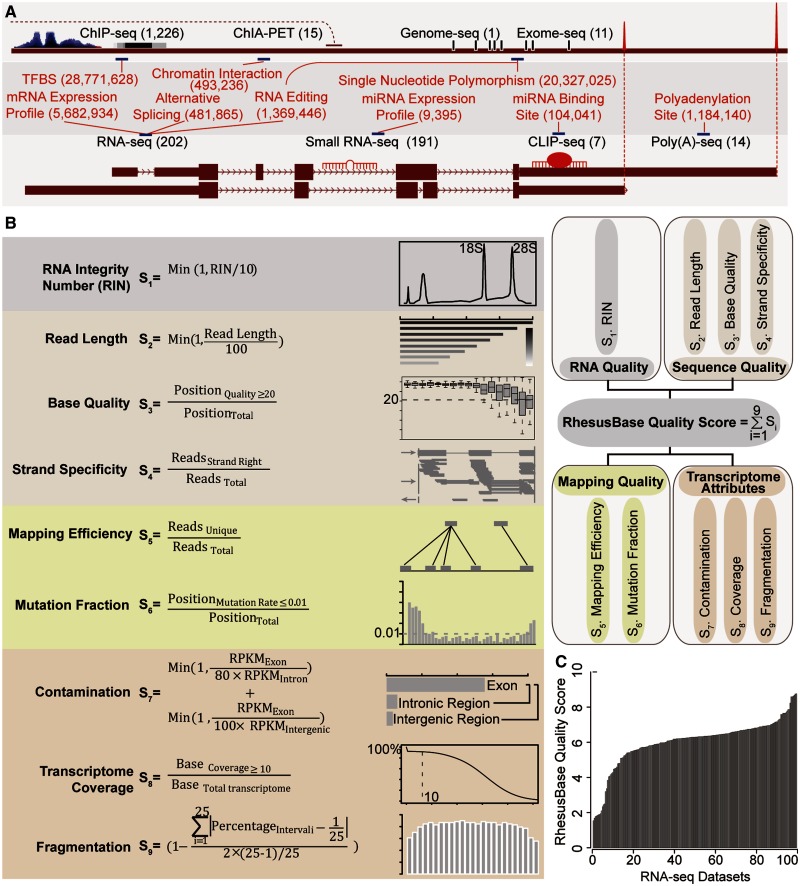
To expand the utility of rhesus macaque in primate comparative studies, we first set out to process and integrate 1,667 NGS data sets in human and rhesus macaque (fig. 1A, see Materials and Methods, supplementary table S1, Supplementary Material online). These data, aimed to provide in-depth physical and functional annotations of the rhesus macaque genome, represented studies that cover multiple gene regulatory mechanisms, such as genome or exome resequencing for population-wide DNA polymorphism, RNA-seq and poly(A)-seq for gene expression and structure, small RNA-seq for microRNA (miRNA) expression profile, as well as CLIP (cross-linking immunoprecipitation)-seq, ChIA-PET (chromatin interaction analysis paired-end tags), and ChIP (chromatin immunoprecipitation)-seq for gene expression regulation (table 1, supplementary table S1, Supplementary Material online). The sources and meta-data for all NGS data sets archived in the current version of RhesusBase are summarized in supplementary table S1, Supplementary Material online.
Fig. 1.
Integration, processing, and evaluation of NGS data sets. (A) Overview of the NGS data sets (shown in black) and corresponding annotations (red) processed and integrated into RhesusBase2. The numbers of NGS data sets and annotation entries are also shown. (B) The quality of RNA-seq data sets was assessed by standard evaluation steps, and a RhesusBase quality score was assigned to each data set according to multiple parameters as illustrated in the box. (C) The distribution of RhesusBase quality scores for RNA-seq data sets incorporated in RhesusBase2.
Table 1.
Statistics of NGS Data Processed in RhesusBase.
| Platforms | Data Sets |
Samples |
Total Reads (Million) |
|||
|---|---|---|---|---|---|---|
| v1 | v2 | v1 | v2 | v1 | v2 | |
| Genome-seq | 0 | 1 | 0 | Brain | 0 | 2,173 |
| Exome-seq | 0 | 11 | 0 | Blood | 0 | 1,054 |
| RNA-seq | 47 | 202 | 14 | 30a | 2,068 | 10,679 |
| Small RNA-seq | 0 | 191 | 0 | 66b | 0 | 1,970 |
| CLIP-seq | 7 | 7 | HEK293 | HEK293 | 34 | 34 |
| Poly(A)-seq | 0 | 14 | 0 | 6c | 0 | 206 |
| ChIP-seq | 0 | 1,226 | 0 | 103d | 0 | 28,660 |
| ChIA-PET | 0 | 15 | 0 | 5e | 0 | 3,885 |
| Sum | 54 | 1,667 | 15 | 182 | 2,102 | 48,661 |
Note.—Statistics of the NGS data sets archived in the previous (v1) and current version of RhesusBase (v2) is summarized.
aAdrenal, brain, breast, caudate nucleus, cerebellar cortex, cerebellum, colon, corneal endothelium, fat, frontal pole, heart, hippocampus, kidney, LCL, liver, lung, lymph node, muscle, neocortex, orbital cerebral cortex, ovary, prefrontal cortex, prostate, skinbone marrow, spleen, testis, thymus, thyroid, white blood cells, and mixtures of 16 tissues.
bABC158, ALL411, basal cells, Bjab103, blastocysts, BL115, BL134, BL510, brain, breast, cerebellum, CLLM633, CLLU626, columnar cells, EBV159, endometrium, epididymis, ESC, ES-RPE, cerebral cortex, fetal RPE, frontal cortex, GC40, GC136, GCB110, GCB385, H929, heart, HEK293, HeLa, HIV412, kidney, KMS12, L1236, L428, liver, lung, Ly3, MALT413, MCL112, MCL114, MM55, MM139, Mino122, Naïve-B-cell, ovary, parthenogenetic-activated blastocysts stem cell, principal and basal cells, peripheral blood mononuclear cells, PC44, peritubular, plasma, pigmented cluster, prostate, red blood cell, seminal vesicle, superior frontal gyrus, skeletal muscle, skin, splenic414, spermatozoa, seminal vesicle, testis, tonsil, U266, and uterus.
cBrain, ileum, kidney, liver, muscle, and testis.
dSee references Hudson and Snyder (2006), Euskirchen et al. (2007), Meyer et al. (2013) for details.
eK562, HCT-116, HeLa-S3, MCF-7, and NB4.
To provide quality assessment of these NGS data, each data set was processed and evaluated by following standardized computational pipelines and procedures, and subsequently assigned a RhesusBase quality score (ranging from 0 to 10) according to multiple criteria that consider overall workflow of data generation and processing (see Materials and Methods, supplementary table S1, Supplementary Material online). Taking RNA-seq data, for example, a data set with high RhesusBase quality score requires that 1) the RNA sample was prepared with optimal quality as indicated by the RNA Integrity Number; 2) sufficiently long reads were generated, with adequate base quality and strand specificity (for strand-specific RNA-seq); 3) there was a high rate for uniquely mapped reads and low mismatch rate across each base; 4) the RNA-seq assay was well performed with little contamination, as shown by enriched reads density at exonic regions, sufficient coverage of the whole transcriptome, and uniform read distribution across each transcript (fig. 1B, see Materials and Methods). Subscores of each evaluation were then summarized and normalized to generate the RhesusBase quality score for the NGS data set (fig. 1B and C, supplementary table S1, Supplementary Material online).
For other categories of NGS data sets integrated, the RhesusBase quality score was assigned to each data set according to a similar scoring system that considers attributes such as the sample quality, sequencing quality, mapping quality, and the degree to which the data set agrees with its known biological attributes (see Materials and Methods, supplementary fig. S1 and table S1, Supplementary Material online). Particularly, considering the differences of the sequencing platforms, experimental designs, and data qualities, we provided the percentile rank of the RhesusBase quality score among the same category of NGS data set in the same species, to allow RhesusBase users to set their own thresholds for NGS data quality control (ranging from 0 to 100, see Materials and Methods, supplementary table S1, Supplementary Material online). In general, NGS data sets with higher RhesusBase quality score and higher percentile rank are more reliable. RhesusBase thus offers a comprehensive quantitative evaluation system for multiple categories of NGS data sets (supplementary table S1, Supplementary Material online).
From these highly selected and processed NGS data sets, a total of 58,423,710 functional records were generated and incorporated in RhesusBase2, representing approximately one order of magnitude more NGS annotation entries compared with the previous version (fig. 1A, table 2). On the genome level, 14,028,737 macaque polymorphism sites (equivalent to 20,327,025 entries) were identified on the basis of public resources (Fang et al. 2011; Sayers et al. 2012), as well as in-house NGS data from macaque genome/exome resequencing (see Materials and Methods). On the transcriptome level, normalized mRNA expression profiles and alternative splicing events were compiled from RNA-seq data sets on a wide range of tissues and cell lines in rhesus macaque (see Materials and Methods). Combining information at these two levels, we also identified 14,118 potential RNA editing sites (equivalent to 1,369,446 entries) in rhesus macaque that introduce differences between RNA and its corresponding DNA sequence (see Materials and Methods). With respect to gene expression regulation, the expression profiles of more than 1,000 miRNAs were estimated based on 39 independent samples, and a list of 285,623 macaque polyadenylation (PA) sites were defined on the basis of the poly(A)-seq data to indicate the ends of macaque transcripts (see Materials and Methods). To broaden the application of RhesusBase, human annotations from 1,514 NGS data sets, including the recently released ENCODE data, were also archived in RhesusBase (table 1). On the basis of these human NGS data sets, normalized mRNA expression profiles in 105 independent samples, 28,771,628 transcription factors binding sites (TFBSs), 493,236 chromatin interactions, and 898,517 PA sites were identified, the expression profiles of miRNAs in 152 independent samples were estimated, and a total of 104,041 miRNA targets were determined (see Materials and Methods). These functional annotations in human were also mapped to the orthologous macaque regions to facilitate in-depth comparative transcriptome study of the primate species (table 2, see Materials and Methods).
Table 2.
Functional Annotations in RhesusBase.
| Entries |
|||
|---|---|---|---|
| Categories | v1a | v2 | |
| DNA | SNP | 5,682,738 | 20,327,025 |
| RNA | mRNA expression profile | 1,330,884 | 5,682,934 |
| PA site | 0 | 1,184,140 | |
| Alternative splicing | 0 | 481,865 | |
| RNA editing | 0 | 1,369,446 | |
| miRNA expression profile | 0 | 9,395 | |
| Regulation | TFBS | 174,805 | 28,771,628 |
| Chromatin interaction | 0 | 493,236 | |
| miRNA-binding site | 15,909 | 104,041 | |
| Sum | 7,204,336 | 58,423,710 | |
aStatistics of the functional annotations archived in the previous (v1) and current version of RhesusBase (v2) is summarized. From highly selected and processed NGS data sets, a total of 58,423,710 functional records were generated and incorporated in RhesusBase2 (v2), representing approximately one order of magnitude more NGS annotation entries compared with the previous version (v1).
Such a compendium of annotations provides comprehensive documentation of temporal and spatial regulations of the rhesus macaque genome. Particularly with respect to gene expression profile, one could easily retrieve the normalized expression profiles and splicing patterns of genes from hundreds of RNA-seq assays, together with the RhesusBase quality scores as additional quality control of particular NGS data sets. By circumventing the computational-intensive efforts of processing and evaluating raw data, RhesusBase2 provides a powerful platform for end users to fully take advantage of primate NGS data sets in extensive functional genomics studies.
Refinement of 30% of the Macaque Ensembl Transcripts (Release 68)
Considering that the majority of macaque genes annotated in Ensembl are inferred (by projection of human transcripts), for the previous version of RhesusBase, we addressed the issue of potentially incorrect macaque gene models through the use of ten RNA-seq data sets. This allowed us to revise the structure of 28% of the transcripts. As the newer RhesusBase2 comprised 97 macaque RNA-seq data sets, which correspond to 5.6 billion reads that cover 99% of the exons and 88% of the splice junctions annotated by Ensembl, we performed further evaluation and refinement of the macaque gene models.
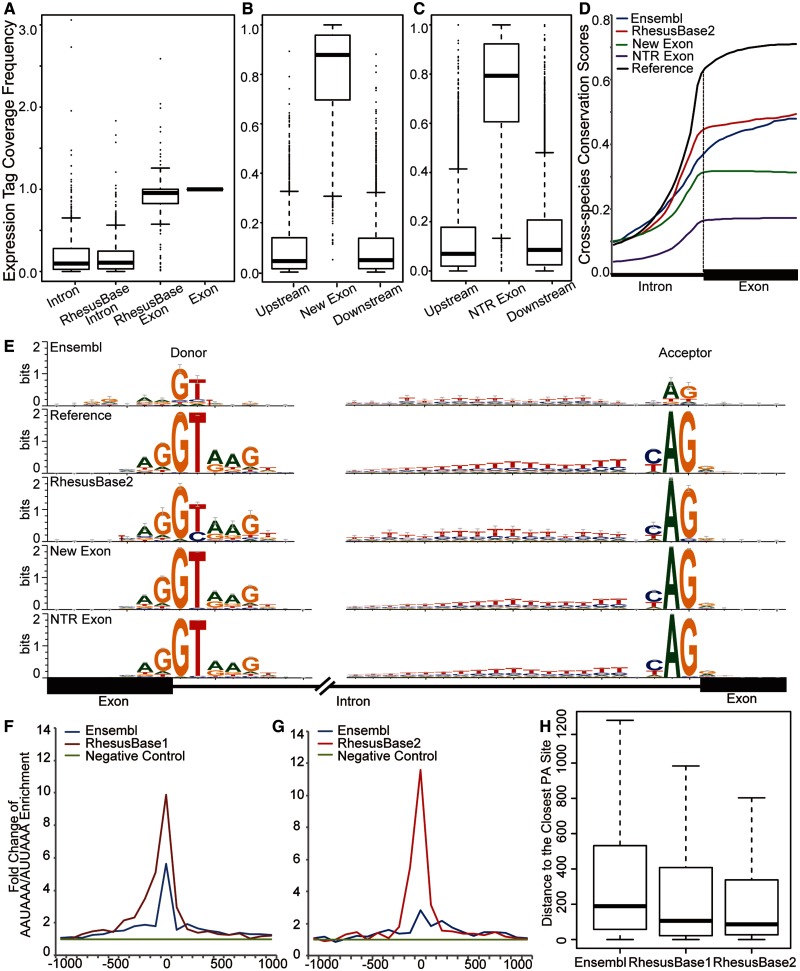
Interestingly, despite considerable expansion of the database, few additional mistakes were found at the annotated exon–intron boundaries (table 3, supplementary table S2, Supplementary Material online). Of the splice junctions previously revised by RhesusBase, 3,202 (or 79%) were supported by the new data. However, in some cases (520), the new data also supported the Ensembl gene models, which likely attributes previous variations to population differences in alternative splicing. Furthermore, with the additional NGS data sets, RhesusBase2 made further revisions of the Ensembl gene models on 909 junctions in 742 transcripts. We then evaluated these refined gene models in terms of the exon–intron distributions of the mRNA-seq expression tags, distributions of the cross-species conservation scores, and the sequence motif flanking the splice sites (see Materials and Methods) (Zhang et al. 2013). In a typical mRNA-seq assay, the distribution of expression tags should highly enrich in exonic compared with intronic regions. In addition, the cross-species conservation scores in exonic regions should be higher than those in intronic regions due to purifying selection (Wang et al. 2008). Comparing the revised gene models with the original Ensembl version, the expression tag coverage in exonic regions was markedly higher than that in intronic regions (fig. 2A–C, Mann–Whitney test, P value < 2.2e-16). The new gene models also exhibited a more predictable distribution of cross-species conservation scores between exons and introns (fig. 2D). In addition, more definite sequence motifs were detected flanking the revised splice junctions (fig. 2E) and further consistent with the motifs of known splice sites (fig. 2E, Reference). Taken together, these results indicate accurate refinements of the gene models and suggest that the extent of RNA-seq coverage provided by the current data assembly is sufficient for the accurate definition of most macaque splice junctions (table 3, supplementary table S2, Supplementary Material online).
Table 3.
Definite Refinement of the Macaque Transcripts by the RhesusBase (v2).
| Categories | Revision Eventsa |
Revised Transcripts |
||
|---|---|---|---|---|
| Confirmed | Novel | Confirmed | Novel | |
| Junctions | 3,202 | 909 | 2,374 | 742 |
| New exons | 2,203 | 5,053 | 1,441 | 2,904 |
| 5′-UTRs | 803 | 587 | 803 | 587 |
| 3′-UTRs | 2,781 | 3,619 | 2,781 | 3,619 |
| Sum | 19,157 | 12,201 | ||
aWith incorporation of new macaque NGS data sets, we performed further refinement of the macaque gene models and compared it with the revisions reported in previous version of RhesusBase. The number of previous gene model revisions confirmed by this study (confirmed) and new gene model revisions by this study (novel) is summarized.
Fig. 2.
Refinement and evaluation of macaque gene models. (A) Accurate refinement of the splice junctions in macaque genes is illustrated by the exon–intron distribution patterns of the RNA-seq expression tag coverage. Exon: exonic regions defined by both gene models; intron: intronic regions defined by both gene models; RhesusBase exon: previously intronic regions now defined by revised gene models as exonic regions; RhesusBase intron: previously exonic regions now defined by revised gene models as intronic regions. (B and C) Normalized RNA-seq expression tag coverage in exonic regions, upstream and downstream intronic regions, for previously missed exons (B) or transcripts (C). (D) Intron–exon distributions of cross-species conservation score. Reference: splice junction supported by both gene models; Ensembl: splice junction defined by Ensembl; RhesusBase2: refined splice junction in this study; new exon: new exons not annotated by Ensembl; NTR exon: exons in NTRs identified in this study. (E) Sequence motifs flanking the splice junctions calculated on the basis of previous gene models (Ensembl), revised gene models (RhesusBase), or the splice junctions for new exons (new exon) and NTRs (NTR exon). Reference: distribution calculated using splice junctions supported by both gene models. (F and G) Enrichments of AAUAAA/AUUAAA hexamers near the end of the 3′-UTRs were calculated based on gene models of Ensembl (release 68) (Ensembl), the previous version of RhesusBase (RhesusBase1), the current version of RhesusBase (RhesusBase2), and 5′-UTR sequences as the negative control (negative control). (H) The distributions of the distance between the PA sites estimated from poly(A)-seq data and the 3′-end of the transcripts, as defined by Ensembl (release 68) (Ensembl), the previous version of RhesusBase (RhesusBase1), and the current version of RhesusBase (RhesusBase2).
Furthermore, with clustered and junction-supporting RNA-seq reads, 5,053 additional new exons were identified in 2,904 transcripts (table 3, supplementary table S2, Supplementary Material online). We also identified 28,223 novel transcriptional regions (NTRs) located in the intergenic regions defined by Ensembl (release 68, see Materials and Methods). A full list of the NTRs could be downloaded in the batch mode from the website of RhesusBase (http://www.rhesusbase.org/download/download.jsp, last accessed March 8, 2014). Attributes such as RNA-seq expression tag coverage (fig. 2B and C), cross-species conservation (fig. 2D), and sequence motifs near the splice junctions (fig. 2E) all corroborated that these are bona fide exons and NTRs. Compared with the previous version of RhesusBase, 4.9-fold more exons and 3.5-fold more NTRs were annotated by the RNA-seq data incorporated in RhesusBase2, which is in line with the notion that deeper sequencing raises sensitivity in identifying novel transcripts or isoforms with low abundance.
Finally, as the ends of transcripts are typically underrepresented in RNA-seq assays due to mRNA degradation and NGS-associated technical issues (Wang et al. 2009; Miura et al. 2013), it is possible that the coverage of RNA-seq would have significant impact on resolving 5′- or 3′-UTRs of transcripts. Indeed, 4,206 UTRs were further revised by RhesusBase2 (table 3, supplementary table S2, Supplementary Material online). These modified gene models were substantiated by the enriched AAUAAA/AUUAAA hexamers of the poly(A) signal sequences near the end of the revised 3′-UTRs (fig. 2G), as well as the smaller distance between the 3′-end of the transcript models and the putative PA sites as estimated from the poly(A)-seq data (fig. 2H, Mann–Whitney test, P value < 2.2e-16 for RhesusBase2 vs. Ensembl, see Materials and Methods). Of note, through the distribution of PA sites defined on the basis of the poly(A)-seq data, we found that 2,174 previously extended 3′-UTRs by the previous version of RhesusBase actually represented alternative variants of the genes annotated by Ensembl; this may be the reason for the unexpected peak of AAUAAA/AUUAAA signal sequences nearby the “incorrect 3′-terminal defined by Ensembl,” as denoted by the previous version of RhesusBase (fig. 2F).
Overall, the fine-scale structures in 30% of the macaque Ensembl transcripts (release 68) were refined in RhesusBase2 (table 3), representing an accurate, up-to-date, and near-complete set of macaque gene model annotations.
Evolutionary Interrogation of Human Genes and Regulations in NGS-Oriented Genomic Framework of Rhesus Macaque
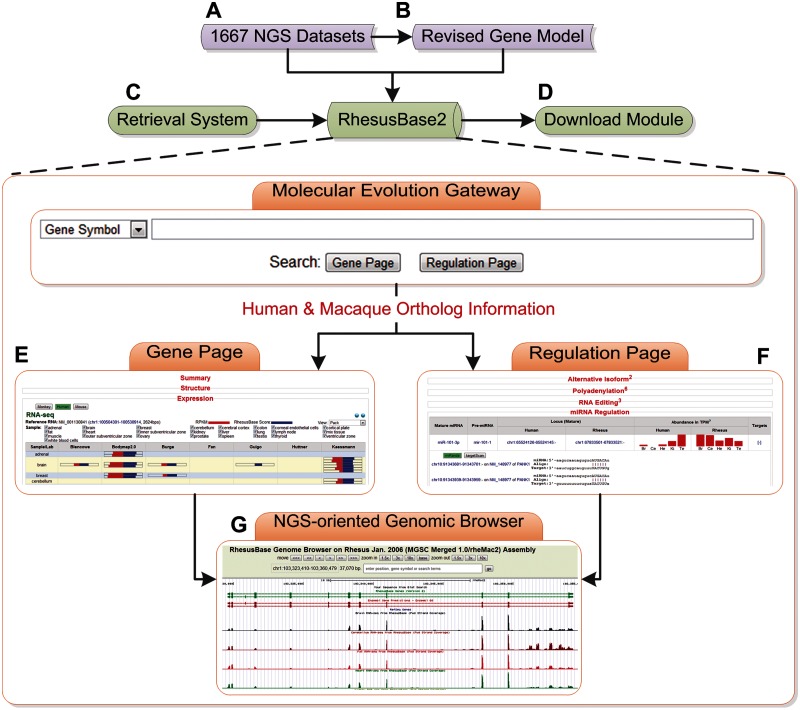
On the basis of comprehensive macaque annotations and accurate gene models, we further developed RhesusBase Molecular Evolution Gateway with multiple NGS-oriented genomic interfaces to enhance data visualization and analysis and to facilitate comparative studies in a user-friendly manner (www.rhesusbase.org/molEvo, last accessed March 8, 2014, fig. 3). To this end, for each human gene and its associated regulations, we designed a gene page (fig. 3E) and a regulation page (fig. 3F) to visualize the corresponding annotations of its macaque ortholog. Briefly, detailed functional annotations in different categories are available at the gene page assigned to the orthologous gene in rhesus macaque, such as gene information, expression profile, regulation, variation and repeats, phenotypes and diseases, function, and drug development (fig. 3E). RhesusBase2 also provides detailed illustrations of macaque annotations for each human regulation on the orthologous human gene, such as alternative splicing, alternative cleavage, and PA, RNA editing and miRNA regulation (fig. 3F). Position-related annotations in these gene-centric interfaces were further hyperlinked to a newly implemented, position-centric genome browser to facilitate efficient visualization of more than eight functional categories associated with the genomic regions of this gene, such as the revised gene models, expressed sequence tags and RNA-seq, splice junctions, RNA-editing sites, transcription regulations, cross-species conservation scores, and variation and repeats (fig. 3G). Specifically, we highlighted the improved macaque gene models on these interfaces to facilitate examination of these revisions in a user-friendly manner. Briefly, on the Gene Page, we included the sequences of the transcripts annotated by the Ensembl gene models, as well as the new RhesusBase2 gene models, with revised sequences highlighted in red. We also included a functional track on the RhesusBase Genome Browser to visualize these changes. A full list of the revised gene models could also be downloaded in the batch mode from the website of RhesusBase (http://www.rhesusbase.org/download/download.jsp, last accessed March 8, 2014). These efforts provide a well-annotated genomic context for comparative studies of genes and regulations in primate species, which may serve as an important resource for in-depth evolutionary studies of human genes and the associated transcriptional regulation (Wray et al. 2003).
Fig. 3.
NGS-oriented genomic interfaces of macaque genomics. (A–G) Overview of database management system and interactive user interfaces in RhesusBase2. RhesusBase2 core database was developed on the basis of 1,667 NGS data sets (A) and accurate macaque gene models (B). Equipped with keywords, location, and sequence-based information retrieval systems (C), as well as BioMart and batch-download interfaces (D), RhesusBase2 allows productive data accession and management. In particular, for each human gene and its associated regulations, one gene page (E) and one regulation page (F) were designed to visualize the corresponding annotations of its macaque ortholog in a user-friendly manner. All position-related annotations were hyperlinked to a newly implemented, position-centric genome browser for efficient visualization of position-based annotations in human and rhesus macaque (G).
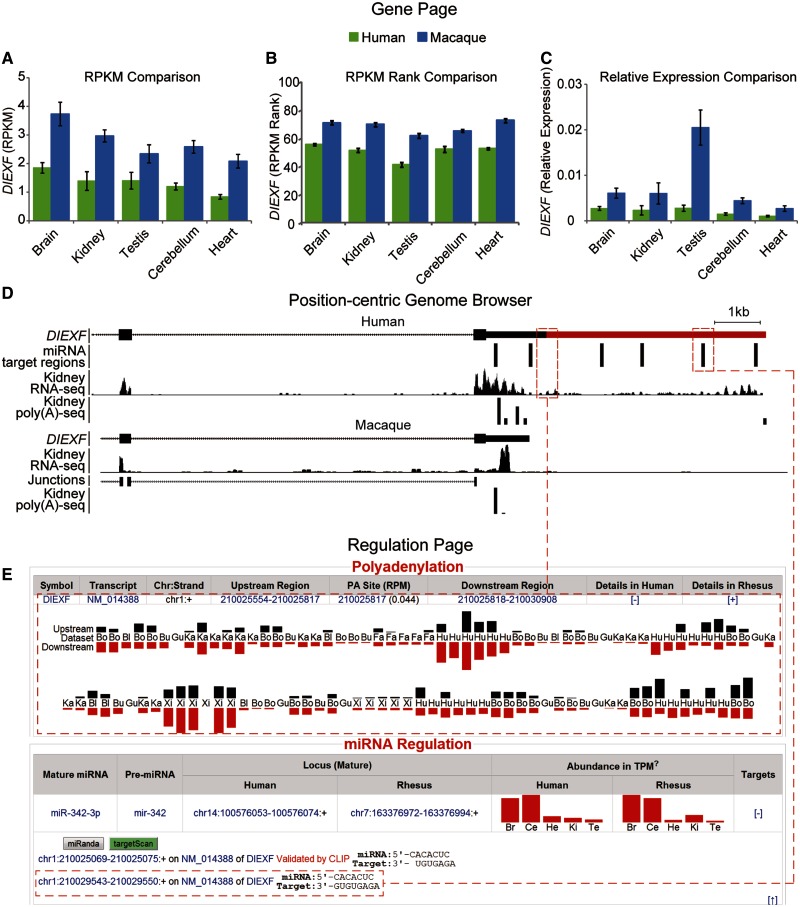
One example of using RhesusBase Molecular Evolution Gateway in generation of hypothetical link of human-biased regulations to human-specific traits is shown in figure 4. DIEXF (digestive organ expansion factor homolog) reportedly regulates the p53 pathway to control the expansion growth of digestive organs (Chen et al. 2005), whereas the increased metabolic requirements of human brain were considered to be balanced by the reduction of digestive system in human evolution (Aiello and Wheeler 1995; Babbitt et al. 2011). It is thus interesting to investigate whether this gene is differentially expressed between human and rhesus macaque and whether some human-biased regulations might contribute to the changes. From the RhesusBase Gene Page assigned to this gene (www.rhesusbase.org/genePage.jsp?id=712825, last accessed March 8, 2014), generally lower mRNA expressions were detected in human tissues than those in rhesus macaque, as indicated by RPKM (Reads Per Kilobase of exon model per Million mapped reads) scores estimated by mRNA-seq across paired tissues in human and rhesus macaque (two-way analysis of variance, P value = 1.53e-9, fig. 4A, see Materials and Methods). Given that the RPKM scores of human genes are largely comparable with their macaque orthologs in the same tissue (Pearson correlation coefficients ranging from 0.72 to 0.84, supplementary fig. S2, Supplementary Material online), such a difference likely did not arise from false positives due to platform difference and technological noise. Besides direct RPKM comparisons, the percentile rank of the expression level of this gene in each RNA-seq assay, also shown on the Gene Page, further confirmed the differential expression of DIEXF between human and rhesus macaque (two-way analysis of variance, P value < 2e-16, fig. 4B, see Materials and Methods). Specifically, from the Gene Page, we also provided the RPKM score of extensively used housekeeping gene for individual RNA-seq data set for an alternative normalization approach of calculating the relative expression levels, as in the real-time PCR assays (see Materials and Methods). In this case, cross-species comparisons using relative expression levels normalized to GAPDH also revealed differential expression of DIEXF between human and rhesus macaque (two-way analysis of variance, P value = 2.37e-06, fig. 4C).
Fig. 4.
Application of RhesusBase in evolutionary and mechanistic characterization of one human candidate gene DIEXF. (A–C) mRNA expression profiles for DIEXF in human and its macaque ortholog are available on the Gene Page (binned by tissue types), indicating higher expression in rhesus macaque across multiple tissues. Gene expression levels across the five tissues in human and rhesus macaque are shown in RPKM values (A), the percentile rank of the expression levels in the associated NGS assay (B), and the relative expression levels normalized to GAPDH as the internal control (C). Data are shown in mean ± SEM (standard error of the mean). Additional survey of the DIEXF genomic regions on the Position-centric Genome Browser (D) and the Regulation Page (E) revealed a human-specific extension of 3′-UTR region (highlighted by red bar in D), as indicated by the RhesusBase2-archived poly(A)-seq and RNA-seq annotations in human and rhesus macaque (D). This human-specific 3′-UTR region was further predicted to harbor multiple miRNA target sites (E).
Additional survey of the DIEXF genomic regions in the position-centric RhesusBase Genome Browser revealed a human-specific extension of the 3′-UTR length, illustrated by the RhesusBase2-archived poly(A)-seq and RNA-seq annotations in human and rhesus macaque (fig. 4D). Interestingly, from the regulation page for DIEXF (www.rhesusbase.org/molEvo/hrRegu.jsp?type=symbol&value=DIEXF, last accessed March 8, 2014), we further found that this human-specific 3′-UTR region harbors multiple predicted miRNA target sites (fig. 4E). Collectively, RhesusBase2 interfaces facilitate comprehensive assessment of interlinked gene regulations across primate species and, in the case of DIEXF, provide additional clues on its down-regulation in human and its possible evolutionary implications in digestive system reduction in human as proposed by the “expensive tissue hypothesis” (Aiello and Wheeler 1995; Babbitt et al. 2011).
Human-Biased Regulations Shape the Unique Transcriptome in Human
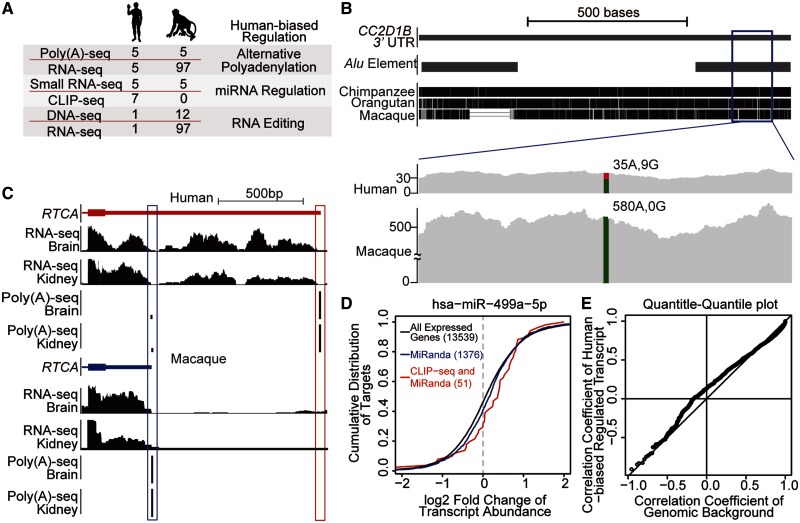
On a global scale, as proof-of-concept demonstration of RhesusBase2 in evolutionary interrogation of human biology, we identified a catalog of 9,295 human-biased regulatory events using RhesusBase2-archived macaque annotations as a reference (fig. 5A, table 4). First, by combining RNA-seq data with the corresponding genome resequencing data, we identified 1,069 human-specific RNA-editing sites (see Materials and Methods, fig. 5B). Second, through integrating RNA-seq and poly(A)-seq data for characterizing transcript ends, we identified 55 human-specific alternative cleavage and PA events that create longer transcripts (see Materials and Methods, fig. 5C). Third, small RNA-seq and CLIP-seq together pinpointed 8,171 human-biased miRNA regulatory events (see Materials and Methods, fig. 5D). In all these cases (supplementary table S3, Supplementary Material online), the human-biased regulatory events were defined with statistical confidence, given that the average genome and transcriptome coverage in rhesus macaque was usually deeper than that in the human (fig. 5A).
Fig. 5.
Human-biased regulatory events contribute substantially to the transcriptomic difference between human and macaque. (A) Identification of human-biased regulation using RhesusBase2-archived NGS data sets. The numbers of NGS data sets used to identify each type of human-biased regulation are shown. (B) A human-specific A-to-G RNA editing site located in an Alu element located in 3′-UTR of CC2D1B (chr1: 52,589,179 in NCBI36). Extent of sequencing coverage flanking the editing site is shown for both human and rhesus macaque data. At the focal editing site, the A allele is highlighted in green and the G allele in red. (C) Compared with the transcript structure of macaque RTCA gene (in blue) and the corresponding poly(A)-seq signals (highlighted in the blue box), the transcript structure for human ortholog is shown with human-specific 3′-terminal (in red), indicated by poly(A)-seq signals (highlighted in the red box) and RNA-seq read densities in human and rhesus macaque data. (D) The graph shows a cumulative distribution of the log 2 fold changes of mRNA abundance between human and rhesus macaque for different sets of mRNAs: targets of the hsa-miR-499a-5p identified by MiRanda prediction (blue line) or by both CLIP-seq and MiRanda prediction (red line), and genomic background with all expressed genes (black line). Displacement of the curve to the right reveals decreased mRNA abundance in human, which is indicative of human-biased mRNA repression in the presence of the human-biased miRNA. (E) Quantile–quantile plot shows the distribution of the correlation coefficients of tissue expression profiles, for genes targeted by human-biased regulatory events and the genomic background.
Table 4.
A Catalog of 9,295 Human-Biased Regulatory Events.
| Categoriesa | Events | Transcripts | Genes |
|---|---|---|---|
| Alternative PA | 55 | 44 | 44 |
| RNA editing | 1,069 | 687 | 330 |
| miRNA regulation | 8,171 | 3,436 | 2,531 |
| Sum | 9,295 | 4,167 | 2,905 |
aA catalog of 9,295 human-biased regulatory events was identified using RhesusBase2-archived macaque annotations as a reference. The number of genes and transcripts with human-biased regulatory events is summarized.
Although most protein-coding genes show highly conserved tissue expression profiles across primate species, as illustrated by high correlation coefficients (Barbosa-Morais et al. 2012), genes targeted by these human-biased regulatory events exhibited significantly lower correlation in tissue-specific expression between human and macaque, when compared with the genomic background (fig. 5E). These more recently evolved regulatory events thus may have a substantial contribution in shaping the human transcriptome, providing additional evidence that noncoding regulatory elements may contribute substantially to the phenotypic differences between humans and other primate species (Wray 2007).
Discussion
Functional Features of RhesusBase2 in Annotating Macaque Genome and Gene Models
Despite the unique advantages of rhesus macaque in evolutionary studies of human genes and regulations, inadequate functional genomics annotations and error-prone gene models limited current use of this model. Prior to RhesusBase, Ensembl, National Center for Biotechnology Information (NCBI), and the University of California–Santa Cruz (UCSC) Genome Browser were the three main resources for macaque annotations, the majority of which were putatively predicted due to the limited functional genomic data for rhesus macaque. With the development of the first rhesus macaque database, and now the significantly expanded second version, RhesusBase2 represents the most comprehensive resource to date for the macaque genomic annotations (fig. 1, table 2). Particularly with respect to gene expression profiles, RhesusBase2 extensively compiled high-throughput transcriptome sequencing data from 97 macaque RNA-seq experiments covering 18 tissues in 38 individuals, as well as 105 human RNA-seq experiments covering 25 tissues (fig. 1, table 2). Functional genomics information such as microarray data from BioGPS (Wu et al. 2009) and in situ hybridization data from Allen Brain Atlas (Jones et al. 2009) were also merged and integrated, which are largely missing in other online resources. RhesusBase2 thus broadens the scope of understanding of the macaque genome structure and functions.
In contrast to the Ensembl gene models, which mainly rely on ab initio or comparative genomics-guided predictions, RhesusBase, and now the significantly expanded second version, applied in-depth experimental data (e.g., RNA-seq) for significant refinement of rhesus macaque gene models (fig. 2, table 3). Consequently, we found that about 30% of the macaque transcripts were previously misannotated by Ensembl (release 68). Although it remains a formal possibility that yet more NGS data are still needed for complete delineation of transcriptome components, RhesusBase2 represents an accurate, up-to-date, and near-complete set of macaque gene model annotations (fig. 2, table 3).
A Comparative Genomic Framework for Evolutionary Interrogation of Human Genes and Regulations
Although public databases such as Ensembl, NCBI, and UCSC Genome Browser have established web servers to access and analyze macaque data, they are not tailored specifically for any particular species, especially in terms of NGS data processing and visualization for cross-species comparative studies. On the basis of comprehensive macaque annotations and accurate gene models archived in RhesusBase2, we developed an NGS-oriented genomic framework to facilitate the evolutionary studies of human genes and regulations in a comparative evolutionary context (fig. 3). Through newly implemented gene pages, regulation pages, and a position-centric genome browser, we provide detailed genomic annotations for each human gene and its macaque ortholog in a user-friendly manner (fig. 3). As a proof-of-concept application of this comparative framework, we present novel clues to the understanding of digestive system reduction in human evolution through mechanistic characterization of the DIEXF gene (fig. 4). On a global scale, we also identified a catalog of 9,295 human-biased regulatory events that have a substantial impact on shaping human transcriptome, possibly representing novel regulatory elements underpinning recent human phenotypic evolution (fig. 5). Overall, the framework we developed provides a well-annotated genomic context for comparative studies of genes and regulations in primates, which may serve as an important resource for in-depth evolutionary studies of human genes and the associated transcriptional regulation.
When comparing RhesusBase2 with the previous version, we found that inclusion of more NGS data provided further depth for the precise and comprehensive definition of macaque transcripts and regulations, especially for those with low or context-dependent expression. Therefore, as more deep sequencing data are available, genome structure and its functional attributes will become progressively better defined. RhesusBase is built with the power and flexibility for efficient data incorporation, processing, and retrieval. It is thus designed to best meet the rapid pace of NGS technology development, data collection, and presentation. As a primate center built according to international AAALAC standards (Association for Assessment and Accreditation of Laboratory Animal Care International), we will continue to maintain and support RhesusBase through generating and integrating NGS data, providing an up-to-date, user-friendly, “one-stop” resource for the community.
Materials and Methods
Integration and Computational Processing of NGS Data
In-house functional genomics data on rhesus macaque, as well as public data either retrieved through the PubMed keywords query “(high-throughput sequencing) AND (rhesus macaque)” or directly downloaded from public databases, such as GEO or SRA, were processed and integrated (supplementary table S1, Supplementary Material online). A total of 1,514 human NGS data sets, including the recently released ENCODE data, were also incorporated to facilitate in-depth comparative analyses in primate species (table 1, supplementary table S1, Supplementary Material online).
NGS data sets were then processed according to standardized pipelines as reported previously (Hafner et al. 2010; Shen et al. 2011; Derti et al. 2012; Feng et al. 2012; Li et al. 2012; Zhang et al. 2013; Chen et al. 2014): 1) For macaque genome resequencing and exome-sequencing data sets, the raw sequencing reads were aligned to the rhesus macaque genome (rheMac2) with BWA (v0.5.9-r16) (Li and Durbin 2009), and only uniquely mapped reads were retained. 2) RNA-seq reads were mapped to the genomes of rhesus macaque (rheMac2) or human (NCBI36/hg18) by TopHat (v1.2.0) (Trapnell et al. 2009), with ambiguously mapped reads discarded. 3) For small RNA-seq data sets in rhesus macaque and human, clean reads with the adaptor trimmed by cutadapt (v1.0) were mapped to the corresponding orthologous precursor miRNA (miRBase Release 18) with Bowtie (v0.12.8) (Langmead et al. 2009). Only sequences perfectly matching the reference, with a length of more than 15 nucleotides, were retained to estimate the expression levels of miRNAs. 4) Reads generated by ChIP-seq assays were mapped to human genome (GRCh37/hg19) with Bowtie (v0.12.5) (Langmead et al. 2009) or BWA (Li and Durbin 2009). ChIP-seq peaks were then called using MACS (v1.3.7) (Zhang et al. 2008) following previous published pipelines (Feng et al. 2012). 5) Chromatin interaction data identified by ChIA-PET (Li, Fullwood, et al. 2010) were directly downloaded from UCSC Genome Browser (Meyer et al. 2013). 6) Seven AGO PAR-CLIP sequencing data sets of human HEK293 cell line were downloaded from GEO database and subjected to PAR-CLIP sequencing analysis protocol to identify AGO crosslink-centered regions (Hafner et al. 2010). Briefly, after adapter removal, short (<20 nt) or repetitive reads were discarded and the remaining reads were mapped to human genome (NCBI36/hg18) by GMAP (Wu and Watanabe 2005). Only uniquely mapped reads with less than two mismatch were used to generate PAR-CLIP clusters, and the specific T to C mutations within these clusters were used as indicators for reliable protein-binding sites (Hafner et al. 2010). 7) Poly(A)-seq data in human and rhesus macaque were downloaded and aligned to the reference genomes following the original report (Derti et al. 2012). After a filtering protocol to remove false positives due to internal priming events, the 3′-ends of uniquely mapped reads (PA tags) located within 30 bp on the same strand were clustered. A PA site was then defined by its location at the peak of the PA-tag cluster (Derti et al. 2012).
RhesusBase Quality Score: Comprehensive Evaluation on NGS Data Sets
To facilitate NGS data assessment and comparison, FastQC (v0.10.0), Samtools (v0.1.16) (Li et al. 2009), Bamtools (v0.1.0.2) (Barnett et al. 2011), and Bedtools (v2.16.2) (Quinlan and Hall 2010) were used to process the reads mapping results, and a series of Perl (v5.12.4), Python (v2.7.3), and R (v2.15.3) scripts were implemented to evaluate the quality of the NGS data and the processing procedures. Subsequently, a RhesusBase quality score (ranging from 0 to 10) was assigned to each NGS data set according to multiple criteria that consider overall workflow of data generation associated with the original study and the downstream computational processing (fig. 1B, supplementary fig. S1, Supplementary Material online).
For the 202 mRNA-seq data integrated in RhesusBase (supplementary table S1, Supplementary Material online), nine parameters were considered in the evaluation and scoring of NGS data sets (fig. 1B), including 1) normalized RNA integrity number as an indicator for the quality of RNA samples; 2) normalized lengths of RNA-seq reads; 3) normalized percentage of high-quality positions (the 25th percentile Phred quality score ≥ 20) across all reads; 4) the quality of strand specificity, estimated on the basis of the percentage of reads with correct strand assignment. For RNA-seq data sets with nonstrand-specific design, this subscore was set to zero; 5) the quality of reads mapping efficiency, estimated on the basis of the percentage of uniquely mapped reads to the reference genome; 6) the mutation rates across the reads, estimated by the percentage of sites with low mutation rate (≤0.01) across the reads; 7) degrees of potential contamination. To ensure the RNA-seq reads were generated from mature mRNAs, the RPKM of exonic, intronic, and intergenic regions were calculated and compared, and the ratios of RPKMexonic/RPKMintronic and RPKMexonic/RPKMintergenic were normalized and summarized to estimate the degrees of contamination for individual RNA-seq study; 8) the coverage of the transcriptome, estimated by the percentage of sites with high reads coverage (≥ 10) across the whole transcriptome; and 9) the quality of fragmentation as indicated by the distribution of reads across the transcript. Each transcript was equally divided into 25 intervals. For an ideal RNA-seq performed with perfect RNA fragmentation, the fraction of RNA-seq reads mapped to each interval is expected to 0.04. When combining all transcripts across the transcriptome, the absolute deviations from 0.04 for each interval were summed and normalized to indicate the quality of fragmentation for one RNA-seq study (Xie et al. 2012).
For the 191 small RNA-seq data sets (supplementary table S1, Supplementary Material online), RhesusBase quality score was assigned to each NGS data set considering the normalized RNA integrity number, the percentage of high-quality positions (the 25th percentile Phred quality score ≥ 20) across all reads, the percentage of clean reads after adapter removal, the percentage of mappable reads to premiRNAs, and the coverage of the known miRNA transcriptome (Shen et al. 2011) (supplementary fig. S1, Supplementary Material online). We also introduced a similar scoring system to evaluate seven CLIP-seq data sets (supplementary table S1 and fig. S1, Supplementary Material online), except that the percentage of mappable reads to mature miRNAs and mRNA regions, instead of premiRNAs, was considered (Hafner et al. 2010).
For genome/exome resequencing data sets, RhesusBase quality scores was assigned to each NGS data set by summarizing and normalizing five parameters: the lengths of the reads, the percentage of high-quality sites (Phred quality score ≥ 20) across all bases of reads, the quality of reads mapping efficiency, the mutation rates across the reads, and the coverage of the whole genome or exome (Chen et al. 2014) (supplementary fig. S1, Supplementary Material online). For ChIA-PET data sets, RhesusBase quality score was assigned to each NGS data set according to the percentage of high-quality positions (the 25th percentile Phred quality score ≥ 20) across all reads and the quality of reads mapping efficiency (supplementary fig. S1, Supplementary Material online).
For the 1,226 ChIP-seq data sets (supplementary table S1, Supplementary Material online), multiple parameters were considered to evaluate the quality of the original NGS study and the downstream computational processing, including 1) nonredundant fraction to indicate the percentage of nonredundant reads in one ChIP-seq study; 2) PCR bottlenecking coefficient1 to indicate the complexity of the ChIP-seq library, defined by the ratio of the number of genomic locations where exactly one read maps uniquely to the number of distinct genomic locations to which at least one read maps uniquely; 3) normalized strand coefficient and relative strand coefficient as measurements for assessing signal-to-noise ratio (Landt et al. 2012); and 4) self-consistent peaks and replicate-consistent peaks to estimate the consistency of replicates (Landt et al. 2012). For poly(A)-seq data sets, five parameters were used to evaluate the quality of NGS data: the percentage of high-quality positions (the 25th percentile Phred quality score ≥ 20) across all reads, the quality of reads mapping efficiency, the percentage of poly(A)-seq peaks with peak width ≤ 30, the percentage of reads within narrow peaks (peak width ≤ 30) among all uniquely mapped reads, and the percentage of reads within known 3′-UTR among all uniquely mapped reads (Derti et al. 2012) (supplementary fig. S1, Supplementary Material online).
For each category of NGS data, subscores of each evaluation were summarized and normalized to generate the RhesusBase quality score for the NGS data set (ranging from 0 to 10, fig. 1B, supplementary fig. S1 and table S1, Supplementary Material online). We also provided the percentile rank of the RhesusBase quality score among the same category of NGS data sets in the same species, defined as the percentage of scores in the same category that are equal or lower, to allow RhesusBase users to set their own thresholds for NGS data quality control (ranging from 0 to 100, supplementary table S1, Supplementary Material online). Generally, NGS data sets with higher RhesusBase quality score or higher percentile rank are more reliable. For advanced RhesusBase users, each subscore is also informative in meta-analyses; for example, a RNA-seq data set with low quality of strand specificity should be used with caution when analyzing overlapping genes (such as cis-natural antisense transcript). One case study for the interpretation of RhesusBase quality score is available online on the website of RhesusBase (http://www.rhesusbase.org/help/FAQ/SQ1.jsp, last accessed March 8, 2014). The subscores of RhesusBase quality score for each NGS data set are also available on RhesusBase website (http://www.rhesusbase.org/RhesusBaseScore.jsp, last accessed March 8, 2014).
Generation and Incorporation of Functional Annotations from NGS Data
On the genome level, macaque polymorphism sites were identified on the basis of the NGS data from macaque whole-genome or whole-exome resequencing (supplementary table S1, Supplementary Material online, table 2). After basic computational processing of these NGS data as described in the previous section, variant calling was performed with Samtools (v0.1.16) pipelines using parameters as previously described (Li et al. 2009). Single-nucleotide variations were further subject to a stringent filtering protocol and only sites with 1) one mismatch type only, 2) ≥5 supported reads, of which ≥3 reads with high PHRED quality (≥25), 3) no significant strand-biased distribution of the detected mutations (Fisher’s exact test, P value < 0.05), and 4) ≥2 supporting reads located on either strand were retained.
On the transcriptome level, normalized mRNA expression profiles and alternative splicing events were identified from RNA-seq data sets integrated in RhesusBase2 (supplementary table S1, Supplementary Material online). Briefly, after reads mapping and quality controls, mRNA expression levels were calculated in terms of RPKMs for different tissues, from which the expression level of each gene was estimated by normalizing the read counts to the length of the transcript, as well as the total uniquely mapped reads generated by this given NGS study (Mortazavi et al. 2008). On the basis of these RNA-seq data sets, alternative splicing events were also identified following the protocols of JuncBase (v0.4) with default parameters (Brooks et al. 2011). The small RNA-seq data sets of rhesus macaque and human were also processed (supplementary table S1, Supplementary Material online), with the expression levels of miRNAs estimated in terms of tags per million (TPM).
Combining genome and transcriptome sequencing, we also identified 14,118 potential RNA editing sites in rhesus macaque that introduce differences between RNA and its corresponding DNA sequence, using a similar approach as we demonstrated in another study (Chen et al. 2014). Briefly, stringent criteria were used to control for false positives, considering the poor genome assembly (Zhang et al. 2012) and error-prone gene structures in rhesus macaque (Zhang et al. 2013). All NGS assays were performed on macaque tissues derived from the same animal to exclude individual differences in the genome and transcriptome. In addition, more stringent definition of “uniquely mapped reads” was used, in which one cDNA read was considered to be uniquely mapped only if it had no second-best hit or the second-best hit included at least three additional sequence alignment mismatches, when considering both the genome and the transcriptome mapping models. Only RNA site with a homozygous genotype were included in an initial list of RNA-editing sites, which were subject to additional filtering protocol to remove candidate sites with 1) low read coverage (<5), 2) poor base-calling quality, 3) multiple types of variation, 4) strand-biased cDNA read distributions (Fisher’s exact test, P value < 0.05), and 5) location in repeat genomic regions or misannotated gene structures (Chen et al. 2014). Human RNA editing sites identified previously were also integrated for subsequent comparative editome analyses (Peng et al. 2012; Ramaswami et al. 2012). For the human set, sites in the initial list were subject to further computational pipelines to exclude sites corresponding to genomic polymorphisms deposited in dbSNP (Peng et al. 2012; Ramaswami et al. 2012).
With respect to gene expression regulations, ChIP-seq data sets (supplementary table S1, Supplementary Material online) were processed for identifying the TFBSs using MACS with default parameters (v1.3.7) (Zhang et al. 2008). A list of miRNA targets were identified by MiRanda and TargetScan prediction (Lewis et al. 2003; John et al. 2004) or by a combination of AGO PAR-CLIP sequencing data (supplementary table S1, Supplementary Material online) and MiRanda and TargetScan prediction (Lewis et al. 2003; John et al. 2004; Hafner et al. 2010). According to the poly(A)-seq data, a list of PA sites were also defined in human and rhesus macaque following the original report (Derti et al. 2012).
Genome-Wide Refinement and Assessment of Macaque Gene Models
A series of Perl (v5.12.4) scripts were implemented to refine the fine-scale transcript structures in rhesus macaque, based on the macaque mRNA-seq and poly(A)-seq data. Exon–intron boundary was revised according to computational pipelines previously established by this laboratory (Zhang et al. 2013). To control for false positives in splice junction definition, only splice junctions supported by at least 30% of the total reads covering this region were included in the revisions. Using mRNA reads collectively derived from 37 strand-specific RNA-seq data sets (Merkin et al. 2012; Zhang et al. 2013), we also extended the 3′-UTRs of the earlier gene models to new terminal sites. As the read coverage of genes is dependent on the gene expression levels and the total reads generated by the given NGS assay, it might be inadequate to use a fixed reads coverage cutoff to perform the transcript extension. We thus introduced a two-step 3′-UTR extension approach similar to that reported previously (Miura et al. 2013). First, when investigating the read coverage of the extended regions in sliding windows, if the density of the expression tags in one 260-bp sliding window dropped by at least 2-fold compared with the upstream region of previous terminal site or dropped by at least 1.2-fold compared with the previous sliding window (windows are separated by 36 bp), the position of the end of this sliding window was marked as the pooled terminal site (only extensions with length of more than 100 bp were included). Second, a pooled terminal site was further adjusted by separately examining the 37 strand-specific RNA-seq data sets, through which the 3′-UTRs could be further extended according to a cutoff of reads coverage estimated by the RNA-seq data (at least 80 positions in 100 bp window with coverage corresponding to ≥ 1 RPKM), following a previously published pipeline (Miura et al. 2013). On the one hand, the pooled terminal site was extended to the most downstream site when at least five data sets supported the extension (only extensions with ≥ additional 250 bp were included); on the other hand, if the pooled terminal site in itself was not supported by at least five independent data sets, the original terminal site defined by Ensembl (release 68) would instead be used. Refinement of 5′-UTRs was done with a similar approach, except for the definition of pooled terminal sites. We also identified potential new exons and NTRs missed in previous annotations, by performing Cufflink (v2.0.2) pipelines (Trapnell et al. 2012) with a similar approach as our previous report (Zhang et al. 2013). A full list of the revised gene models and NTRs could be downloaded in the batch mode from the website of RhesusBase (http://www.rhesusbase.org/download/download.jsp, last accessed March 8, 2014).
Gene model revisions were then evaluated on the basis of three parameters: the distribution of the RNA-seq expression tags, cross-species conservation scores, and the sequence motif nearby the splice junctions or the transcript termini, as described in detail in previous study (Zhang et al. 2013). The distribution of the RNA-seq expression tags was generated using in-house Perl (v5.12.4) and R scripts. Cross-species conservation scores were calculated according to the previous guideline (Meyer et al. 2013). The sequence motifs flanking the donor/acceptor splice sites were deduced by WebLogo (v3.2), and the distribution of AAUAAA/AUUAAA hexamers of the poly(A) signal sequences near the ends of the transcripts was determined and shown (fig. 2F and 2G, supplementary fig. S3, Supplementary Material online). For well-annotated gene models, such as Ensembl gene models in human, the majority of poly(A) sites identified by poly(A)-seq agree with known 3′-end of the transcript models to single-base precision (Derti et al. 2012). We thus further evaluated the 3′-UTR extensions by calculating the distance between the 3′-end of the transcript models and the poly(A) sites estimated from poly(A)-seq data deposited in RhesusBase (supplementary fig. S1, Supplementary Material online). Because of the limited coverage of the poly(A)-seq data on the macaque transcriptome, the poly(A)-seq tags were used only in evaluating the completeness of the transcripts in this study, rather than defining the 3′-end of the transcripts.
Development of RhesusBase2 Interactive User Interfaces
We developed RhesusBase2 interfaces using Apache-based web development technologies to manage the deposited meta-data with a MySQL relational schema. Aside from the existing interfaces of the previous version, the updated RhesusBase2 interface was equipped with functional components of user-friendly information retrieval and download systems, the Molecular Evolution Gateway with gene and regulation pages to visualize functional annotations in human and rhesus macaque, and a new RhesusBase Genome Browser, with which the users can easily access the sequences and functional annotations of selected macaque genes in the UCSC Genome Browser format. A demonstration of the use of the RhesusBase Genome Browser in candidate gene study is available online at the website of RhesusBase (http://www.rhesusbase.org/help/FAQ/SQ2.jsp, last accessed March 8, 2014).
Currently, the Gene Page is built on the basis of the NCBI Entrez Gene system, and genes without corresponding Entrez Gene annotation are therefore not archived. In such instances, we highly recommend the users to view the annotations (such as the revised gene models) in the RhesusBase Genome Browser, which was designed for intuitively displaying position-based annotations.
Several different normalization approaches were introduced in the comparison of gene expression between human and rhesus macaque, including direct RPKM comparisons (fig. 4A), comparisons of the percentile rank of the RPKM scores in different NGS studies (fig. 4B), and comparisons of relative expression levels using housekeeping gene GAPDH as the internal control (fig. 4C). All annotations and database schema in RhesusBase are freely accessible at www.rhesusbase.org (last accessed March 8, 2014).
Identification of Human-Biased Regulatory Events Using RhesusBase Annotations
A list of human RNA-editing sites from a previous report (Peng et al. 2012) was integrated and mapped to macaque syntenic regions using LiftOver with default parameters to identify the orthologous loci in rhesus macaque. The 97 RNA-seq data sets for rhesus macaque were then processed to examine the incidence of RNA-editing in rhesus macaque. A human-specific RNA editing site was defined when the macaque orthologous site satisfies either of two criteria: 1) for a focal site of RNA editing in human, the DNA sequence in rhesus macaque is divergent from the human reference or edited nucleotide; 2) the DNA sequence in rhesus macaque is consistent with that in human and the orthologous site was covered by at least 149 RNA-seq reads, but without any editing signal. In this regard, the minimal RNA-seq read coverage of the orthologous site was set to be 149 to efficiently distinguish true absence of editing regulation in rhesus macaque from the failure of detection, as under the assumption of binomial distribution, the detection power of editing sites with an editing frequency of as low as 2% is still more than 95%.
Genomic loci, sequence information, and structure annotations of human miRNAs were downloaded from miRBase (v19). Macaque orthologous miRNAs were identified when no more than two mismatches were found in mature sequences between human and rhesus macaque, based on pairwise whole-genome alignment between the two species (Meyer et al. 2013). RhesusBase2-archived small RNA-seq data for five tissues (brain, cerebellum, heart, kidney, and testis) from both human and rhesus macaque were then processed to identify miRNAs with human-biased expression. Briefly, clean reads with the adaptor trimmed were mapped to the corresponding orthologous precursor miRNA. Only sequences perfectly matching the reference, with a length of more than 15 nucleotides, were retained. The miRNA expression level was then estimated in terms of TPM. For miRNAs with five-tissue TPM sums of more than 250 in both human and rhesus macaque, those exhibiting significantly differential expression between the two species (fold change > 10, Fisher’s exact test P value < 0.01) were defined as human-biased miRNAs. Finally, 8,171 human-biased miRNA regulatory events were identified, with miRNA targets identified by MiRanda prediction (John et al. 2004) or by a combination of CLIP-seq and MiRanda prediction (John et al. 2004; Hafner et al. 2010).
Poly(A)-seq data from multiple tissues of human and rhesus macaque (brain, liver, kidney, testis, muscle, and ileum) were cross-compared to identify human-specific PA (Derti et al. 2012). Briefly, PA tags within 30 bp were clustered, and a PA site was defined by its location at the peak of the PA-tag cluster. Human PA sites were mapped to macaque syntenic regions using LiftOver with default parameters, and a human-specific PA site was defined when macaque PA signals were undetectable across the 100-bp window flanking the focal syntenic site. A total of 3,262 human RefSeq transcripts were then identified as candidate transcripts targeted by human-specific PA regulatory events, with at least one human-specific PA site downstream of the last human-macaque shared PA site. To further control for false positives, mRNA-seq data across six tissues (brain, cerebellum, heart, kidney, liver, and testis) in human and rhesus macaque (Brawand et al. 2011) were introduced to verify these candidates. This was done by comparing the RNA-seq read densities in common 3′-UTR regions shared by human and rhesus macaque (from stop codon to the last shared PA site), as well as in human-specific 3′-UTR regions (from the shared PA site to the human-specific PA site). The macaque genomic regions syntenic to human regions were defined using LiftOver (-minMatch 0.8). Transcribed regions with evident expression (RPKM > 0.5) were subjected to Fisher’s exact test with P value of 0.05 used to indicate a significant difference in read distributions between human and rhesus macaque. Finally, 44 human transcripts were identified as targets of human-specific PA regulations—these showed significantly different human-macaque distributions of reads flanking the last shared PA site in all tests (at least two independent tests were needed), as well as low expression signals (RPKM < 1.0) in macaque genomic regions that are syntenic to putative human-specific extension regions, as indicated by 97 RhesusBase2-archived mRNA-seq data sets.
For genes targeted by human-biased regulations, the tissue expression profiles in human and rhesus macaque were determined as described previously (Xie et al. 2012). The Pearson correlation coefficients between the RPKM scores in the corresponding tissues were calculated, and the distribution of correlation coefficients was visualized using R (v2.15.3) scripts.
Supplementary Material
Supplementary tables S1–S3 and figures S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
C.-Y.L. conceived the idea and designed the study. S.-J.Z., C.-J.L., and P.Y. performed most of the experiments. X.Z., J.-Y.C., X.Y., J.P., S.Y., C.W., J.X., Y.E.Z., and X.Z. performed part of the experiments. S.-J.Z., C.-J.L., P.Y., and X.Z. analyzed the data and performed the statistical analyses. C.-Y.L. and B.C.-M.T. wrote the paper. All authors read and approved the final manuscript. The authors thank Dr Heping Cheng at Peking University and Dr Qing-Rong Liu at the National Institutes of Health (USA) for insightful suggestions. This work was supported by grants from the National Key Basic Research Program of China (2013CB531200, 2012CB518004), Beijing Joint Research Program of Scientific Research and Graduate Student Training, the National Natural Science Foundation of China (31171269, 31221002) , the National Science Council (NSC102-2321-B-182-007), and the National Health Research Institute (NHRI-EX103-10321SI).
References
- Aiello LC, Wheeler P. The expensive-tissue hypothesis—the brain and the digestive system in human and primate evolution. Curr Anthropol. 1995;36:199–221. [Google Scholar]
- Babbitt CC, Warner LR, Fedrigo O, Wall CE, Wray GA. Genomic signatures of diet-related shifts during human origins. Proc Biol Sci. 2011;278:961–969. doi: 10.1098/rspb.2010.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- Barnett DW, Garrison EK, Quinlan AR, Stromberg MP, Marth GT. BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics. 2011;27:1691–1692. doi: 10.1093/bioinformatics/btr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D, Soumillon M, Necsulea A, Julien P, Csardi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- Brooks AN, Yang L, Duff MO, Hansen KD, Park JW, Dudoit S, Brenner SE, Graveley BR. Conservation of an RNA regulatory map between Drosophila and mammals. Genome Res. 2011;21:193–202. doi: 10.1101/gr.108662.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Chen J, Ruan H, Ng SM, Gao C, Soo HM, Wu W, Zhang Z, Wen Z, Lane DP, Peng J. Loss of function of def selectively up-regulates Delta113p53 expression to arrest expansion growth of digestive organs in zebrafish. Genes Dev. 2005;19:2900–2911. doi: 10.1101/gad.1366405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JY, Peng Z, Zhang R, Yang XZ, Bertrand CT, Fang H, Liu CJ, Shi M, Ye ZQ, Zhang YE, et al. RNA editome in rhesus macaque shaped by purifying selection. PLoS Genet. 2014;10(4):e1004274. doi: 10.1371/journal.pgen.1004274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, Babak T. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22:1173–1183. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen GM, Rozowsky JS, Wei CL, Lee WH, Zhang ZD, Hartman S, Emanuelsson O, Stolc V, Weissman S, Gerstein MB, et al. Mapping of transcription factor binding regions in mammalian cells by ChIP: comparison of array- and sequencing-based technologies. Genome Res. 2007;17:898–909. doi: 10.1101/gr.5583007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Zhang Y, Zhang R, Yang L, Li M, Ye K, Guo X, Wang J, Su B. Genome sequence and global sequence variation map with 5.5 million SNPs in Chinese rhesus macaque. Genome Biol. 2011;12:R63. doi: 10.1186/gb-2011-12-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, Remington KA, Strausberg RL, Venter JC, Wilson RK, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson ME, Snyder M. High-throughput methods of regulatory element discovery. Biotechniques. 2006;41:673, 675, 677. doi: 10.2144/000112322. passim. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AR, Overly CC, Sunkin SM. The Allen Brain Atlas: 5 years and beyond. Nat Rev Neurosci. 2009;10:821–828. doi: 10.1038/nrn2722. [DOI] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Knowles DG, McLysaght A. Recent de novo origin of human protein-coding genes. Genome Res. 2009;19:1752–1759. doi: 10.1101/gr.095026.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Li CY, Zhang Y, Wang Z, Cao C, Zhang PW, Lu SJ, Li XM, Yu Q, Zheng X, Du Q, et al. A human-specific de novo protein-coding gene associated with human brain functions. PLoS Comput Biol. 2010;6:e1000734. doi: 10.1371/journal.pcbi.1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Fullwood MJ, Xu H, Mulawadi FH, Velkov S, Vega V, Ariyaratne PN, Mohamed YB, Ooi HS, Tennakoon C, et al. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol. 2010;11:R22. doi: 10.1186/gb-2010-11-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012;338:1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41:D64–D69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura P, Shenker S, Andreu-Agullo C, Westholm JO, Lai EC. Widespread and extensive lengthening of 3' UTRs in the mammalian brain. Genome Res. 2013;23:812–825. doi: 10.1101/gr.146886.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Peng Z, Cheng Y, Tan BC, Kang L, Tian Z, Zhu Y, Zhang W, Liang Y, Hu X, Tan X, et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat Biotechnol. 2012;30:253–260. doi: 10.1038/nbt.2122. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G, Lin W, Piskol R, Tan MH, Davis C, Li JB. Accurate identification of human Alu and non-Alu RNA editing sites. Nat Methods. 2012;9:579–581. doi: 10.1038/nmeth.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G, Zhang R, Piskol R, Keegan LP, Deng P, O'Connell MA, Li JB. Identifying RNA editing sites using RNA sequencing data alone. Nat Methods. 2013;10:128–132. doi: 10.1038/nmeth.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2012;40:D13–D25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Lv Y, Huang L, Liu W, Wen M, Tang T, Zhang R, Hungate E, Shi S, Wu CI. Testing hypotheses on the rate of molecular evolution in relation to gene expression using microRNAs. Proc Natl Acad Sci U S A. 2011;108:15942–15947. doi: 10.1073/pnas.1110098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Sheffield NC, Fedrigo O, Babbitt CC, Wortham M, Tewari AK, London D, Song L, Lee BK, Iyer VR, et al. Extensive evolutionary changes in regulatory element activity during human origins are associated with altered gene expression and positive selection. PLoS Genet. 2012;8:e1002789. doi: 10.1371/journal.pgen.1002789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toll-Riera M, Bosch N, Bellora N, Castelo R, Armengol L, Estivill X, Alba MM. Origin of primate orphan genes: a comparative genomics approach. Mol Biol Evol. 2009;26:603–612. doi: 10.1093/molbev/msn281. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, 3rd, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- Xie C, Zhang YE, Chen JY, Liu CJ, Zhou WZ, Li Y, Zhang M, Zhang R, Wei L, Li CY. Hominoid-specific de novo protein-coding genes originating from long non-coding RNAs. PLoS Genet. 2012;8:e1002942. doi: 10.1371/journal.pgen.1002942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SJ, Liu CJ, Shi M, Kong L, Chen JY, Zhou WZ, Zhu X, Yu P, Wang J, Yang X, et al. RhesusBase: a knowledgebase for the monkey research community. Nucleic Acids Res. 2013;41:D892–D905. doi: 10.1093/nar/gks835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Goodsell J, Norgren RB., Jr Limitations of the rhesus macaque draft genome assembly and annotation. BMC Genomics. 2012;13:206. doi: 10.1186/1471-2164-13-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.