Abstract
Studies of microbial evolutionary dynamics are being transformed by the availability of affordable high-throughput sequencing technologies, which allow whole-genome sequencing of hundreds of related taxa in a single study. Reconstructing a phylogenetic tree of these taxa is generally a crucial step in any evolutionary analysis. Instead of constructing genome assemblies for all taxa, annotating these assemblies, and aligning orthologous genes, many recent studies 1) directly map raw sequencing reads to a single reference sequence, 2) extract single nucleotide polymorphisms (SNPs), and 3) infer the phylogenetic tree using maximum likelihood methods from the aligned SNP positions. However, here we show that, when using such methods to reconstruct phylogenies from sets of simulated sequences, both the exclusion of nonpolymorphic positions and the alignment to a single reference genome, introduce systematic biases and errors in phylogeny reconstruction. To address these problems, we developed a new method that combines alignments from mappings to multiple reference sequences and show that this successfully removes biases from the reconstructed phylogenies. We implemented this method as a web server named REALPHY (Reference sequence Alignment-based Phylogeny builder), which fully automates phylogenetic reconstruction from raw sequencing reads.
Keywords: next-generation sequencing, Escherichia coli, Pseudomonas syringae
Introduction
One of the unifying goals across fields as diverse as evolutionary biology, epidemiology, and ecology is to understand the evolutionary relationships between different taxa (Preston et al. 1998; Gill et al. 2005; Ishii et al. 2006; Chun et al. 2009; Ogura et al. 2009; Harris et al. 2010), which are typically quantified by constructing phylogenetic trees (Nei and Kumar 2000). Recently, our ability to resolve such trees has greatly improved due to the rate at which sequence data can be generated via high-throughput sequencing methods. However, using high-throughput sequencing data to precisely determine phylogenetic relationships between taxa is not trivial.
Traditionally, phylogenies are reconstructed from whole-genome sequence data by 1) assembling sequence reads into contigs; 2) annotating open reading frames; 3) identifying orthologous open reading frames across all genomes; 4) aligning orthologous coding regions; and 5) reconstructing a phylogenetic tree from these multiple alignments (Touchon et al. 2009; Luo et al. 2011; Rodriguez-R et al. 2012). Subsequently, a phylogenetic tree is then reconstructed from the alignments, typically by applying maximum likelihood methods such as RAxML (Stamatakis 2006) or PhyML (Guindon et al. 2010), or Bayesian methods such as PhyloBayes (Lartillot et al. 2009) or MrBayes (Huelsenbeck and Ronquist 2001).
Although it is generally accepted that this method allows accurate reconstruction of phylogenetic trees (Rosenberg and Kumar 2003), the series of steps involved is not only time consuming but requires a sophisticated combination of bioinformatic methods.
Recently, an alternative method that is simpler and less time consuming has been applied in several large-scale microbial studies (Harris et al. 2010; Epstein et al. 2012; Harris et al. 2012; McCann et al. 2013; Cui et al. 2013). In this method, raw short-sequence reads from each taxon are directly mapped to the genome sequence of a single reference. Homologous sites from all taxa (and in some studies only those sites containing single nucleotide polymorphisms [SNPs]) are then concatenated into a multiple sequence alignment from which the phylogenetic tree is reconstructed.
There are reasons to suspect that such reference-mapping-based phylogeny reconstruction methods might introduce systematic errors. First, multiple alignments are traditionally constructed progressively, that is, starting by aligning the most closely related pairs and iteratively aligning these subalignments (Notredame et al. 2000; Chenna et al. 2003). Aligning all sequences instead to a single reference is likely to introduce biases. For example, reads with more SNPs are less likely to successfully and unambiguously align to the reference sequence, as is common in alignments of more distantly related taxa. This mapping asymmetry between strains that are closely and distantly related to the reference sequence may affect the inferred phylogeny, and this has indeed been observed (Spencer et al. 2007). Second, as maximum likelihood methods explicitly estimate branch lengths, including only alignment columns that contain SNPs and excluding (typically many) columns that are nonpolymorphic, may also affect the topology of the inferred phylogeny. This effect has been described before for morphological traits (Lewis 2001) and is one reason long-branch attraction can be alleviated with maximum likelihood methods when nonpolymorphic sites are included in the alignment (Felsenstein 1981). Furthermore, the more general issue of selectively leaving out data from multiple sequence alignments has been studied recently and found to affect tree topology (Shavit Grievink et al. 2013).
By simulating sequence evolution across small phylogenies of known topology, we identify parameter regimes where the combination of single-taxon reference mapping and SNP extraction generally leads to severe errors in phylogeny reconstruction. These simulations also show that even when including nonpolymorphic sites in an alignment, the effect of mapping to a single reference can lead to systematic errors. In particular, we find that when some taxa are diverged by more than 5–10% from the reference, the distance to the reference is systematically underestimated. This can generate incorrect tree topologies, especially when other branches in the tree are short. Moreover, using data from a set of 21 Escherichia coli genomes, a set of 19 Pseudomonas syringae genomes, and a set of 32 Sinorhizobium meliloti genomes, we show that biases due to mapping to a single reference and exclusion of nonpolymorphic sites significantly affect the inferred phylogenetic trees for realistic data sets.
To alleviate these problems, we present a method that combines alignments obtained by mapping reads to not one but to multiple reference sequences. Applying this method to both the simulated and real data sets suggests that, by combining sequence mappings to multiple references, mapping biases can be avoided and accurate phylogenies can be reconstructed when each taxon is close (i.e., <5% divergence) to at least one of the reference sequences. To make this phylogeny reconstruction procedure available to researchers, including experimental biologists without specific expertise in bioinformatics, we have implemented this method as a web server called Reference sequence Alignment-based Phylogeny (REALPHY) builder (available at http://realphy.unibas.ch, last accessed March 13, 2014). REALPHY takes as input raw short-sequence read data sets and reconstructs phylogenies by aligning the reads to one or more reference sequences.
Results and Discussion
The inference of phylogenetic trees from collections of polymorphic sites identified by mapping short-sequence reads from multiple genomes to a single reference genome is an increasingly common practice (Harris et al. 2010; Epstein et al. 2012; Holt et al. 2012; McAdam et al. 2012; Okoro et al. 2012; Cui et al. 2013). However, as indicated in the Introduction, there are several reasons to suspect that this method may introduce systematic errors.
To test in what situations this method may result in incorrect tree reconstruction, we simulated sequence evolution along known phylogenies, systematically varying both topology (i.e., the placement of the reference genome) and branch lengths. For each data set, we then compared the true tree topology with the tree topologies inferred from 1) the correct and complete alignment of the evolved sequences; 2) the alignment obtained after mapping short reads and retaining only SNP positions; and 3) the alignment after mapping short reads and retaining both SNPs and nonpolymorphic sites.
Sequence Simulation
Tree Shapes and Branch Lengths
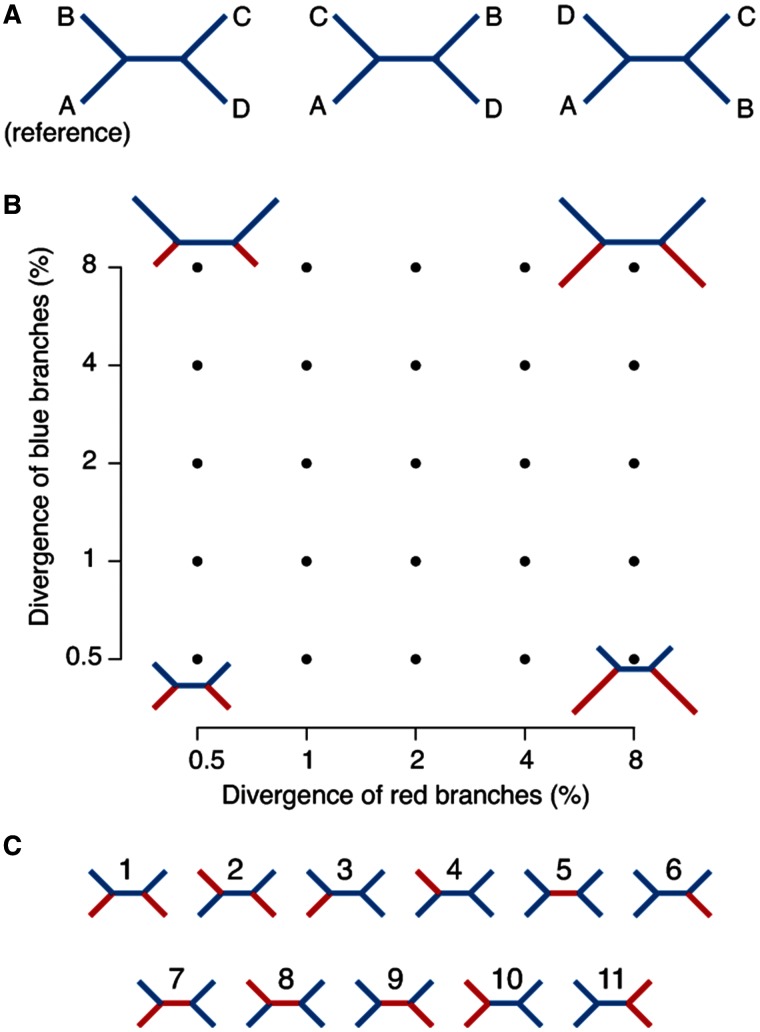
To allow a systematic exploration of parameter space, we restricted our analysis to unrooted four-taxon trees, which have only five branches and only three possible topologies: (A,B),(C,D); (A,C),(B,D); and (A,D),(B,C) (fig. 1A). Throughout, we use taxon A (fig. 1A) as the reference sequence to which short-sequence reads from all other taxa are mapped. To understand the effects of differences in branch length on tree reconstruction, we considered all ways of partitioning the five branches into two subsets (the red and blue branches in fig. 1C). Because our four-taxon tree is asymmetric (the sequence from one taxon is designated as the reference sequence), there are 11 possible groupings of the branches, which we call tree shapes. For each tree shape, we varied the branch lengths in these two groups over a range of values (0.5%, 1%, 2%, 4%, and 8% divergence). This gave rise to 25 possible branch length combinations, shown as grid points in figure 1B. Varying tree shape and branch lengths in this manner gave rise to a total of 275 (25 × 11) different trees.
Fig. 1.
Tree shapes and branch lengths used to simulate sequence evolution. (A) The three possible topologies in a four-taxon tree. (B) The sample space of tree topologies. Each axis indicates the divergence along one set of branches: the divergence of the red branches is indicated along the x-axis and the divergence of the blue branches is indicated along the y-axis. We sampled at five points along each axis, that is, at 0.5, 1, 2, 4, and 8% divergence, for a total of 25 different combinations of branch lengths. (C) All possible tree shapes are considered in the analyses. There are 11 total tree shapes in a four-taxon tree that divide the branches into two types (shown here as red and blue). In all our analyses, the reference node is the lower left node of the tree.
To refer to any of these trees individually, we specify each of the parameters varied above: first, the tree shape (1–11), followed by the divergence level of the majority set of branches (i.e., the blue branches in fig. 1C), and finally the divergence level of the minority set of branches (i.e., the red branches in fig. 1C). We represent the results for different branch length combinations in a matrix, for example, trees with a divergence of 0.5% over blue branches and 8% over red branches correspond to the bottom-right corner of figure 1B.
Recombination
Recombination (gene conversion) occurs frequently in bacterial species (Didelot and Maiden 2010). Thus, in addition to varying tree shape and branch lengths, we investigated the effect of short-read mapping on phylogeny reconstruction in the presence of recombination. To simulate this process, 10% of the nucleotides in the reference sequence were replaced with the orthologous nucleotides from the sequence of its cousin taxon (taxon D in fig. 1; using taxon C would yield identical results). Thus, with the inclusion of recombination, we simulated sequence evolution over a total of 550 trees (2 × 275).
The Impact of SNP Extraction and Read Mapping Bias on Tree Topology
Accurate Phylogenetic Reconstruction When Using the True Alignment
We first tested whether the correct tree topology could be reliably recovered from the true alignment (the evolution of 100,000 nucleotides simulated along a four-taxon tree). We found that when there was no recombination, all tree topologies were reconstructed correctly by PhyML (Guindon et al. 2010), a maximum likelihood tree inference program. Not surprisingly, when a sufficient amount of recombination was incorporated, phylogeny reconstruction was no longer error-free (supplementary fig. S1, Supplementary Material online).
Phylogenies Reconstructed Using Only SNP Positions Are Unreliable
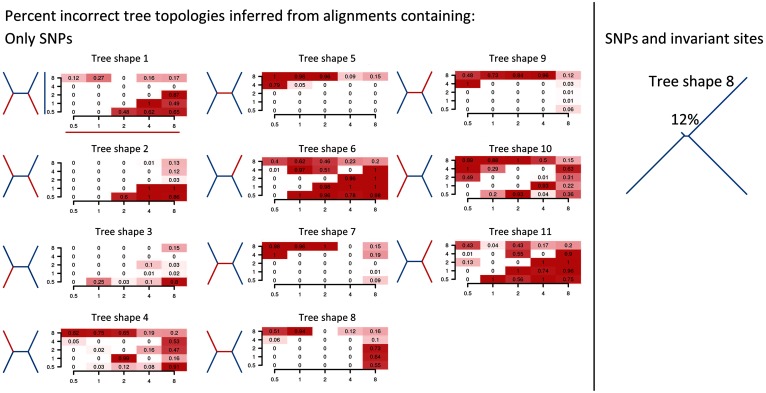
We then tested for parameter regimes that led to incorrect phylogenies when mapping to a single reference, extracting SNP positions only, and reconstructing a phylogeny using maximum likelihood. We identified 131 different parameter settings for which incorrect topologies were inferred for a fraction of data sets, even in the absence of recombination (fig. 2). Up to 100% of all inferred tree topologies were incorrect for some parameter sets (e.g., for tree shape 1 at 1% and 4% divergence; fig. 2). This contrasted strongly with the results using the true alignment, for which no incorrect topologies were inferred for any of the parameter settings.
Fig. 2.
Parameter combinations for which incorrect topologies were inferred from mapped alignments excluding (left) and including (right) nonpolymorphic sites (without recombination). Left: Fraction of incorrectly reconstructed trees from a total of 100 replicates for all parameter combinations, inferred only on extracted SNPs. Each panel shows data for a different tree shape. Tree shapes are indicated on the left of each panel. Each heatmap shows the divergence (in percent) of red branches across the x-axis and divergences across blue branches on the y-axis. Right: Tree shape 8 with a divergence of 0.5% along the short branches and 8% along the long branches is shown. The percentage above the tree indicates the proportion of trees (out of 100) for which the incorrect topology was inferred.
When recombination was included, the reliability of phylogenetic reconstruction using only SNP positions decreased further. There were 140 parameter sets for which incorrect topologies were inferred, and the number of incorrectly inferred trees increased from 6,641 to 8,871 out of a total of 27,500 data sets.
Importantly, we also found that the choice of the reference taxon affected error rates (supplementary fig. S1, Supplementary Material online). For example, although tree shapes three and four have identical branch lengths and differ only in the position of the reference sequence, the accuracy of tree reconstruction differed considerably. When the reference taxon was on a short branch (0.5% divergence) and all other branches were long (8% divergence), no errors were made in inferring the topology. In contrast, when the sister taxon of the reference was on a short branch and all other branches were long, errors were made in 82% of all cases.
Including Nonpolymorphic Sites Improves Reliability
The above analyses were performed on alignments containing only SNP positions. When nonpolymorphic positions were included in the alignments (i.e., all nonpolymorphic positions that were successfully mapped to the reference genome), the accuracy of phylogenetic inference improved. Erroneous topologies were reconstructed for only a single parameter set, in tree shape eight: when the branch of the reference’s sister taxon and the internal branch were short (0.5%) and all other branches were long (8%), the incorrect topology was inferred in 12% of all simulations (fig. 2). When recombination was included, the accuracy again decreased strongly for five parameter combinations compared with the true alignments (supplementary fig. S1, Supplementary Material online).
Thus, when aligning short reads to a single reference genome, there were still some parameter sets for which trees could not be reliably reconstructed. However, for the same parameter sets, no inaccuracies arose when trees were inferred without reference mapping (i.e., using the correct and complete alignment). This demonstrates that the inaccuracy in phylogenetic reconstruction was due to biases that arose in mapping short reads to a single reference sequence.
It is likely that the inaccurately inferred tree topologies are caused primarily by a combination of two factors: 1) Short-read aligners such as Bowtie2 can only map sequences closely related to the reference, such that sequences with too many mismatches are discarded and 2) the relative distance to the reference is important, as regions that are on average more closely related to the reference are less likely to be discarded than regions that are more distant to the reference. Figure 3 qualitatively illustrates how biases are introduced. Assuming that the alignment algorithm only allows a single mismatch between the query and reference sequence within a short region, only a single mutation in the branch leading to the reference would be allowed, and any additional mutations in the region would cause the alignments to be discarded (fig. 3A). In contrast, three separate mutations would be allowed in the terminal branches that lead to the other leaves in the tree (fig. 3C). As a consequence, the fraction of columns having identical nucleotides in all taxa would be inflated, whereas the fraction of columns in which all nucleotides would be equal except for the nucleotide in the reference is underestimated (supplementary fig. S2, Supplementary Material online). As shown in supplementary figure S3, Supplementary Material online, such biases decrease the extent to which the likelihood function supports the correct phylogeny over incorrect alternative topologies, and this is most dramatic for the problematic tree shape 8 (supplementary fig. S3B, Supplementary Material online).
Fig. 3.
Mapping to a single reference introduces alignment biases. Assuming, for illustrative purposes, that the alignment algorithm allows only one mismatch between query and reference within a 21-bp region, each panel shows the maximal number of mutations allowed in order for successful mapping of all orthologous fragments to occur, as a function of the positions in the tree where mutations occur. (A) If a single mutation occurs on the reference branch, then the distance from the reference to all other sequences reaches one immediately, and no further mutations are allowed. (B) One mutation on the internal branch as well as one mutation on the sister branch are allowed before all three query sequences reach a distance of one to the reference. (C) Three independent mutations on each of the external branches are allowed before all query sequences reach a distance of one to the reference.
Branch Lengths Are Highly Inaccurate When Using SNP Positions Only
To analyze how these biases affect branch lengths, we quantified branch lengths from all phylogenetic trees that were correctly reconstructed: 1) from the true alignments; 2) from alignments obtained after short-read mapping and SNP extraction (without nonpolymorphic sites); and 3) from the full alignments obtained after short-read mapping (including all nonpolymorphic sites). Because we obviously expected to infer longer overall branch lengths when including SNP positions alone, we assessed the accuracy of the inferred relative branch lengths instead of total branch lengths. We defined the relative length of a branch as its length divided by the sum of all branch lengths within the tree. To quantify the effects of reference mapping and SNP extraction on tree reconstruction, we determined, for each branch in the tree, the ratio of its relative length after mapping and SNP extraction, to the relative length of the branch inferred from the true and complete alignment.
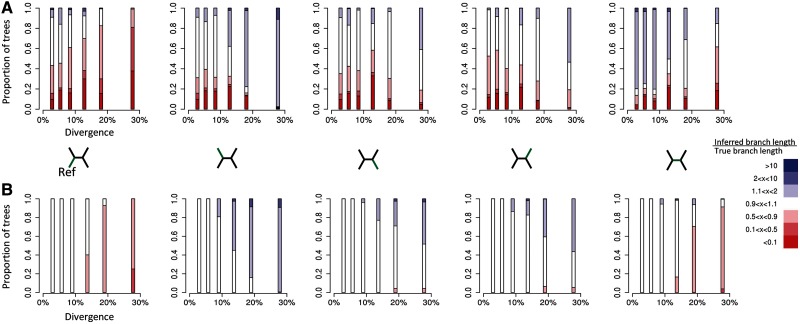
We found again that accuracy was low when using single reference-mapped alignments containing SNP positions alone. The inferred branch lengths in these phylogenies differed considerably from the true branch lengths (fig. 4A), and we found that even at relatively low levels of divergence (5.9%), in at least 13% of all reconstructions, each of the five branches was estimated to be less than one-tenth of their true relative length.
Fig. 4.
Deviation of relative branch lengths, as inferred from mapped sequence alignments, from the true relative branch lengths for (A) phylogenies inferred using SNP positions only and (B) phylogenies inferred using all positions. For each branch in our simulated four-taxon trees, the figure shows the proportion of trees in which the estimated relative branch length deviated from the true relative branch length to a certain degree (color).The trees were subdivided into six equally sized bins based on the overall divergence level (proportion of columns within the original multiple sequence alignment that contain SNPs) and the branch length ratios were calculated for each divergence class (position on the x-axis). The proportion of trees inferred from mapped sequence alignments that contain relative branch lengths that are more than ten times greater than those from the true tree are shown in dark blue. Relative branch lengths that are more than ten times shorter are shown in dark red. Relative branch lengths that are within 10% of the true branch length are shown in white (see legend). The figure shows one plot for each of the five branches within the tree (this branch is indicated in green in the four-taxon trees between A and B). The reference sequence is always the taxon on the bottom left of the tree. Trees were only included in the statistics if the mapped tree topology matched the true (known) tree topology.
By including both nonpolymorphic and SNP positions, the tree reconstruction accuracy increased considerably (fig. 4B). When total divergence across the true tree was less than 10%, branch length estimation was generally accurate, and branch lengths were only rarely under- or overestimated by more than 10%. However, at higher divergence levels, accuracy decreased rapidly. This decrease in accuracy also exposed a consistent bias, in which the lengths of the interior branch and the branch leading to the reference were consistently underestimated, while the lengths of the other branches in the tree were consistently overestimated. This confirmed our qualitative considerations above regarding the systematic biases that mapping to a single reference introduces.
Combining Alignments from Mappings to Multiple Reference Taxa Allows for Accurate and Unbiased Phylogeny Reconstruction
Although the inclusion of nonpolymorphic sites in the alignment considerably improved the accuracy of tree reconstruction, there were parameter regimes where topologies could still not be reconstructed correctly. Furthermore, relative branch lengths were inferred to be up to two times longer/shorter than they ought to be (fig. 4B). Earlier, we have argued that this bias is caused by the relative position of the reference to the other sequences in the phylogeny. This suggests that this bias may be overcome by using multiple references that are more evenly distributed across the tree. However, as detailed in Materials and Methods, care has to be taken that no other systematic biases are introduced when combining alignments from mappings to multiple references. We therefore developed an iterative procedure for merging alignment columns from mappings to different references into a final nonredundant alignment, ensuring that each genomic position from each reference occurs in at most one column of the final alignment and that conflicts between the mappings using different references are resolved.
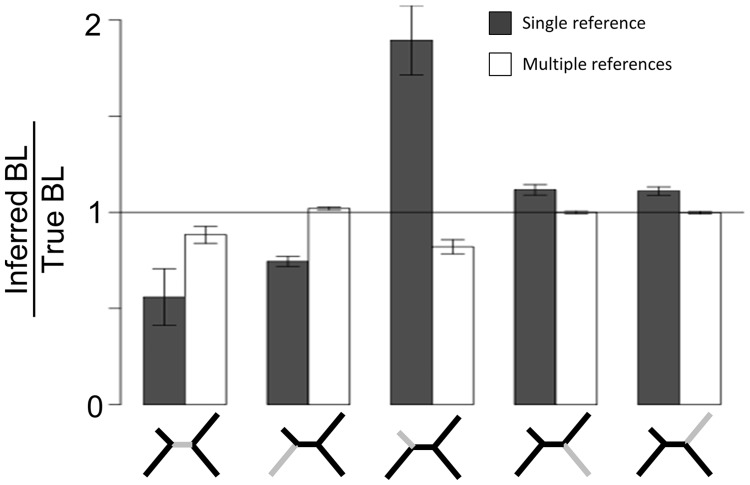
To test whether this strategy allowed us to create more accurate phylogenies, we focused on the parameter setting that caused incorrect topologies even when nonpolymorphic sites are included (tree shape 8 with 0.5% and 8% divergence) and built four separate alignments by using each of the taxa as a reference. After merging the alignments, the correct phylogeny was reconstructed in 100% of the cases. Furthermore, whereas relative branch lengths differed by up to 2-fold from the true alignment when using a single reference and when reconstructing the phylogeny from the merged alignment, relative branch lengths differed by at most 18% (fig. 5). This demonstrated that mapping sequences to multiple reference taxa allows for much more accurate tree reconstruction, even for substantially divergent sequences.
Fig. 5.
Accuracy of estimated relative branch lengths when inferring a phylogeny from a single reference alignment (gray bars) and from a merged alignment of all four references (white bars). The relative branch length (BL) of a particular branch is defined as the length of the branch divided by the sum of all BLs in the tree. The BL ratio is the ratio of the estimated BL and the BL of the true tree. The bars show the BL ratios for each of the five branches (indicated at the bottom) of the trees inferred in 88 independent trials (all correctly reconstructed topologies) of alignments from tree shape eight with divergences of 0.5% and 8%. Note that the closer the bars are to one, the more similar the estimated tree is to the true tree.
We have now implemented this new method as a web server, REALPHY, to make this resource widely available.
Application to Bacterial Genome Sequences
The simulations presented above show under what conditions mapping to a single reference and inferring phylogenetic trees from SNP positions can lead to errors even for simple four-taxon trees. Here we show that these errors do typically occur in realistic data sets and that by merging alignments from multiple references, REALPHY avoids such errors. We analyzed three published data sets with sequences from E. coli (Touchon et al. 2009), P. syringae (Baltrus et al. 2011), and S. meliloti (Epstein et al. 2012). The first two data sets demonstrate how biases from mapping to a single reference can affect the inferred phylogeny. In addition, they allow us to compare phylogenies constructed by REALPHY with those using classical alignment methods (Touchon et al. 2009; Baltrus et al. 2011) and demonstrate that, as a consequence of the larger number of sites that are included in REALPHY analyses, we obtain more accurate phylogenies. The S. meliloti data set illustrates how the use of only SNP positions can lead to errors in the reconstructed tree.
Data from E. coli
Touchon et al. (2009) determined the phylogeny of 20 fully sequenced E. coli and Shigella strains as well as one E. fergusonii strain using classical methods. They used whole-genome data and reciprocal best BlastP to identify 1,878 genes that were present in all genomes. These orthologs were aligned (covering ∼40% of the shortest genome’s length), and a distance matrix based on this alignment was calculated and used to build a neighbor-joining tree.
We applied REALPHY with default parameters to the same data set, performing 21 separate runs using each of the 21 taxa as a single reference sequence. The topologies of the inferred trees were almost identical. We identified only one branch point at which not all 21 phylogenies agreed. This branch point concerned the subclade containing E. coli APEC O1, UTI89 and S88 (fig. 6). Here, in 9 out of the 21 cases, instead of APEC O1 clustering with S88, APEC O1 clustered with UTI89.
Fig. 6.
Comparison of REALPHY phylogenies to phylogenies inferred in previous publications. Both REALPHY trees (green) were built using PhyML, with the general time-reversible (GTR) model of nucleotide evolution and gamma distributed rate variation. The annotation on the branch points in black denotes the bootstrap support for the branch points from a total of 100 bootstrap experiments (only shown if <100) for REALPHY trees, Bayesian probabilities for the Baltrus tree (shown if <0.95) and bootstrap values out of 1,000 for the Touchon tree (shown if <1,000). Annotations in gray show the number of REALPHY single-reference trees that support the particular branch points (only shown if <21 for E. coli and <3 for P. syringae). Boxed parts of the trees contain differences to the previously published corresponding tree. (A) E. coli phylogeny reconstructed by Touchon et al. (2009) (left) compared with a phylogeny reconstructed from all 21 merged reference alignments produced by REALPHY. The differences between the two trees are the placements of E. coli 536 and S88. (B) P. syringae phylogeny reconstructed by Baltrus et al. (2011) (left) compared with a phylogeny based on mappings to the three fully sequenced P. syringae strains: P. syringae B728a, P. syringae pv. phaseolicola 1448a and P. syringae pv. tomato DC3000. Right: The root of the tree was arbitrarily selected to facilitate comparison between the two topologies. When inferring trees from single reference genome alignments, two branch points are not supported by all three trees (annotated on the corresponding branches). These branch points concern the placement of Cit7 (P. syringae B728a as reference) and Pae (P. syringae pv. phaseolicola 1448a as reference).
We also merged all reference alignments using our merging procedure (1,896,194 bp total length; 170,886 SNP positions, covering 43% of the shortest genome's length) and inferred a tree for the combined alignment. The tree inferred for this merged alignment was identical to the consensus tree obtained with 12 of the 21 different references.
We found that REALPHY's tree differed at two branch points (both in clade B2) from the tree calculated by Touchon et al. (2009) (fig. 6A). The first branch point concerned the aforementioned UTI89, APEC O1, and S88 clade. The fact that this branch point was only supported by 12 of the 21 reference alignments suggests the data is indeed not completely unambiguous, and this is substantiated by a bootstrap experiment with 100 repeats showing a support of 88% for this branch, whereas all other branches have 100% support. The second branch point that differed between ours and the Touchon tree concerned the placement of E. coli 536, which was placed at the root of the B2 clade in our reconstruction but clustered with CFT073 and ED1a in the tree reconstructed by Touchon et al. (2009). Notably, Touchon et al. also presented a second tree based on a MAUVE alignment. In this case, 536 was placed as outgroup to the B2 subclade, as our consensus and merged-data tree did. Furthermore, Touchon et al. showed that the support for this branch is only about 92% compared with 100% for the rest of the tree.
The facts that REALPHY uses a larger number of sites (43% of the smallest E. coli genome compared with 40% for the Touchon et al. [2009] data) and that REALPHY’s tree matches the consensus tree of all reference alignments suggest that REALPHY's tree may be more accurate. To investigate this further, we selected only the seven strains from the B2 clade and reran REALPHY on this data set. Because of the much higher similarity of this subset of sequences, the reference alignments included a much larger number of sites (covering about 76% of the shortest B2 genome). We found that the tree inferred by REALPHY for the merged alignment was identical to the trees inferred for all seven reference alignments (supplementary fig. S4, Supplementary Material online). Moreover, this tree supported all REALPHY’s branches from the tree of all 21 taxa, strongly supporting that REALPHY’s tree was more accurate than the tree constructed by Touchon et al. Interestingly, the tree built from the B2 clade differs from both REALPHY’s and the Touchon et al. tree in the placement of the CFT073 strain, demonstrating that phylogenetic trees can often be further refined by analyzing sequences from subclades separately.
In summary, the analysis of the E. coli data showed that the resulting tree can be biased by the reference strain and that usage of merged alignments from multiple references avoids this bias in this case. It also indicated that REALPHY performs at least as well as classical methods that are more complex and time consuming and can even outperform these methods when it is using a larger number of sites.
Data from P. syringae
In our second analysis, we studied a published P. syringae data set (Baltrus et al. 2011), consisting of three fully sequenced genomes and 16 draft genomes in FASTA format. This sequence set was considerably more divergent than the above E. coli data set (∼9% compared with ∼14%, respectively). As discussed earlier, we expect the effects of reference mapping bias to increase as sequence divergence increases. Indeed this bias becomes apparent when comparing the reference alignment lengths from P. syringae to those of E. coli. Although for the E. coli data approximately 43% of the genomes was covered by the REALPHY alignments, for the P. syringae genomes, this coverage ranges from 17.6% to 18.9%. As may be expected, this alignment bias is significant enough to affect the inferred P. syringae topology. When we used P. syringae B728a as the reference sequence, it was placed as most basal taxon in the group II clade instead of Cit7; and when we used P. phaseolicola 1448a as reference, P. phaseolicola 1448a was placed as the most basal taxon in group III instead of Pae (fig. 6B). As both differences concern the clade in which the reference strain is present, it is probable that these differences are the result of a mapping bias to the reference sequence. This bias was removed when we constructed a merged alignment obtained from all three reference genomes. This alignment contained a total of 1,403,205 bp (236,228 SNPs, covering 23% of the smallest reference) and the inferred tree agreed completely with the consensus tree of the three individual reference phylogenies.
Notably, there were some disagreements between the topology of our tree and the multilocus sequence typing (MLST) tree inferred by Baltrus et al. (2011). As the MLST tree is inferred from only a small number of sites, Baltrus et al. inferred another tree based on a concatenated alignment of 324 proteins (corresponding to roughly 6% of the shortest P. syringae genome’s length), and this phylogeny is more similar to the one inferred by REALPHY. In this case, Pma and Por_1_6 clustered together, as well as Pto_DC3000 and Pla106, agreeing with our inferred topology. As our phylogeny is based on an alignment that contains far more sites than both the protein alignment and the MLST alignment, this suggests that our phylogeny is likely to model the evolutionary relationships between P. syringae strains more accurately than the phylogenies presented by Baltrus et al.
This example further confirms that usage of a single reference can significantly bias the resulting topology and that REALPHY's inferred phylogenies are often more accurate than phylogenies constructed from a smaller number of selected sites.
Data from S. meliloti
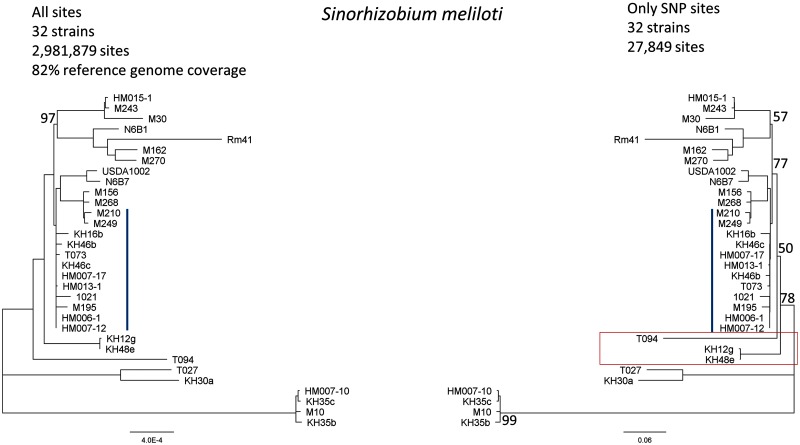
In the previous examples, the phylogenies were constructed from all alignment sites, that is, both SNP sites and nonpolymorphic sites. To illustrate reconstruction errors that result from using only SNP sites, we applied REALPHY to a set of S. meliloti strains (Epstein et al. 2012). Because this data set consists of very closely related strains that differ only by a maximum of ∼1%, we do not expect to observe significant reference alignment bias (trees inferred from the two references Rm41 and 1021 were identical). However, the usage of SNP sites only may affect the inferred phylogeny. To test this, we inferred a phylogeny using PhyML from a complete alignment and an alignment containing only SNP sites (fig. 7). We found that there is one significant difference between the resulting tree topologies, affecting the placement of T094. In addition, the relative branch lengths of the tree inferred from SNP sites only changed significantly (supplementary fig. S5, Supplementary Material online). Interestingly, for the S. meliloti data set, relative branch lengths changed more severely than for the E. coli and P. syringae data sets, despite the fact that both the E. coli and the P. syringae data set are more diverged than the S. meliloti data set. These results further highlight the importance of including nonpolymorphic sites in alignments from which phylogenies are inferred using maximum likelihood methods.
Fig. 7.
Comparison between two phylogenies inferred from a REALPHY alignment of Sinorhizobium meliloti strains (Epstein et al. 2012) including (left) and excluding (right) nonpolymorphic alignment sites. The alignments were created by merging the reference alignments from S. meliloti Rm41 and 1021. The red box highlights differing branch points. Bootstrap support is indicated if below 100%, except for the blue clade where the support is low.
Conclusion
In recent years, numerous studies (e.g., Harris et al. 2010; Croucher et al. 2011; Mutreja et al. 2011; Holt et al. 2012; McAdam et al. 2012) have reconstructed phylogenetic trees for large numbers of closely related bacterial strains by mapping short-sequence reads to a reference genome sequence. Here, we have analyzed the performance of such methods on simulated and real sequence data and have shown that there are two primary pitfalls to this approach. The most readily apparent is that when SNP alignments are used to construct trees with maximum likelihood methods, it can lead to incorrect tree topologies and inaccuracy in the inferred branch lengths. Furthermore, when query sequences are sufficiently divergent from the reference sequence, the most divergent regions of the genome may fail to map, and this mapping bias may lead to incorrect branch lengths and topologies. Notably, the simulations that we have presented here did not include any variation in mutation rates across the genome; biases in transitions or transversions; or clustering of mutations due to selection; each of these could serve to exacerbate the problem of biased sequence mapping.
To address these pitfalls, we have presented a new method, REALPHY, which can successfully avoid biases from mapping to a single reference by implementing a procedure for merging alignments obtained by mapping to multiple reference genomes into a single nonredundant alignment.
REALPHY was mainly designed to reconstruct phylogenies for microbial genomes, that is, bacterial genomes and single-celled eukaryotes such as fungi, but it can in principle be equally applied to data from higher eukaryotic organisms. However, such applications have not been tested yet and, as described in Materials and Methods, the computational resources that are required increase with the size of the input genomic data and may become prohibitive for large eukaryotic genomes that contain many repetitive sequences.
To make this method available to a large community of researchers, including pure biologists without bioinformatics expertise, we provide REALPHY through a web server, allowing the fast and automated generation of multiple sequence alignments from a variety of genome sequence data formats (e.g., Illumina sequence reads, contigs, draft genomes, fully sequenced genomes), and the automatic reconstruction of phylogenies from these alignments.
Materials and Methods
REALPHY Implementation
A flowchart of the REALPHY implementation is presented in figure 8.
Fig. 8.
Illustration of the individual steps in the REALPHY pipeline (running from top to bottom). All fully sequenced or assembled genomes (FASTA and GenBank files) are divided into all overlapping 50-bp subsequences. Short sequences are aligned to individual reference sequences with Bowtie2. Alignment columns are created from all pairwise mappings to the references. Individual reference alignments are merged into a single multiple sequence alignment. A phylogeny is reconstructed from merged and individual reference alignments via PhyML.
Pipeline Requirements
The REALPHY pipeline requires the user to provide a set of DNA sequences for each taxon to be included in the phylogenetic tree. This set will typically consist of short-sequence reads but may also include larger sequences, such as fully or partially assembled genomes. In addition, REALPHY requires one or more reference sequence sets to which all sequences will be aligned. Each reference sequence set should consist of a whole-genome sequence, a set of chromosome sequences, or a set of contigs.
Alignment
Sequence reads from each query genome provided as FASTQ formatted files are directly mapped to each of the reference sequences using Bowtie2. Assembled genomes provided in FASTA or GenBank format are divided into all possible subsequences of 50 bp (default) to be able to efficiently map these sequences to a reference genome with Bowtie2. REALPHY calls Bowtie2 with the default k-mer length of 22, allowing one mismatch within the k-mers to maximize sensitivity. For each short sequence, only the best mappings are retained, that is, when there are n >1 “best” mappings; each of the mappings is assigned a weight 1/n. For each reference sequence, the short-read mappings for all query genomes are combined into a multiple alignment containing all orthologous positions that can be reliably identified across the reference and query genomes.
It is possible that paralogous fragments from a query genome may map to the same position as an ortholog in a reference sequence. If these paralogous fragments have diverged, reads from the same query genome may report differing nucleotides aligned to the same position of the reference genome. To avoid such inconsistent mappings, only unambiguous positions are included in the final alignment. Unambiguous position assignment results if the weighted sum of mappings from the query genome is ≥10, and ≥95% of the mappings show the same nucleotide. This percentage was chosen to make it unlikely that paralogous mappings would pass the cutoff but would reduce false negatives due to sequencing errors, which are relatively common in high-throughput sequencing data (Nakamura et al. 2011). By default, only those alignment columns are retained in which a nucleotide from each of the taxa is present.
In some cases, a small number of genomes may be highly diverged from all reference sequences in some genomic regions, resulting in no successful alignments. In other cases, some genomic regions may be missing entirely. This may be due to their absence in the sequencing data set due to uneven sequence coverage or due to gene deletions. Even if only a few strains are affected by these problems, these regions will be missing from the final alignment, as by default, REALPHY only includes regions of the genome for which all strains are present in the alignment. Although such situations are by definition problematic and may lead to inaccurate phylogenies, the user can choose to override the default parameters and include columns in the alignment in which either all or a specified proportion of genomes can have ambiguous or missing nucleotides. These missing nucleotides will be represented by gaps. Importantly, it has been shown that under certain circumstances, phylogenetic trees reconstructed from such alignments can be more reliable than trees reconstructed from alignments in which gapped positions are omitted (Shavit Grievink et al. 2013).
Combining Alignments from Mapping to Different Reference Sequences
The results of the short-read mappings consist of a collection of alignment columns where mappings for all taxa exist. The easiest procedure for combining alignment columns that result from mapping to different references would be to collect the union of all alignment columns and apply a phylogenetic reconstruction method to this data set. However, such a data set would be highly redundant, with a given position from a given reference occurring multiple times, that is, once for each reference to which it was mapped. More importantly, certain positions may be represented more frequently than others in a full collection of alignment columns, which is likely to introduce biases in phylogeny reconstruction. For example, it is well known that substitution rates vary over several orders of magnitude for different genes within a genome (as reviewed in Rocha 2006). As a consequence, positions from slowly evolving genes may be reliably mapped to distal reference genomes, whereas positions from fast evolving genes can only be mapped to the closest reference genomes. Consequently, positions from slowly evolving genes are likely to be overrepresented in the full collection of alignment columns.
To avoid such biases, REALPHY combines alignment columns from different references into a final set of alignment columns using the following procedure (supplementary fig. S6, Supplementary Material online). Alignment columns from all alignments are pooled and then iteratively processed as follows: 1) Randomly select an alignment (column C) from the pool. This column will contain both nucleotides for aligned nonreference genomes (e.g., short-sequence read data) as well as nucleotides derived from positions xr in each of the other reference genomes r. 2) For each of these positions xr occurring in column C, we also select the alignment column Cr of nucleotides mapped to position xr in the reference r (if this column Cr is present in the pool). 3) All selected columns, that is, C and the Cr for all other references, are then removed from the pool, and a consensus column is calculated by applying a simple majority rule. 4) This consensus column is then added to the collection of final alignment columns. We continue to select random columns from the pool until there are no columns left. This ensures that each reference genome position occurs in only one of the final alignment columns and that possible disagreements about which nucleotide from a given taxon should be aligned to a given reference position are resolved through a simple majority rule.
Tree Building
Based on the final set of DNA sequence alignment columns, the pipeline determines a phylogenetic tree by applying PhyML (Guindon et al. 2010, default parameters) or Dnapars (a maximum parsimony method; Felsenstein 2005). We chose PhyML as it is optimized for speed in terms of handling large numbers of taxa as well as long sequence alignments. The maximum likelihood method PhyML is run with the general time-reversible (GTR) model of nucleotide evolution and gamma distributed rate variation by default. Dnapars from the Phylip program suite is run with its default settings.
Output
For each reference genome the output consists of a FASTA and a PHYLIP formatted file that contain an aligned set of orthologous sites (SNPs as well as nonpolymorphic sites), a tree file in Newick format, and multiple tab-delimited files (one for each query genome) containing the positions on the reference genome to which the identified SNPs were aligned.
Computational Resources
The resources REALPHY requires depend mainly on the genome length, the number of genomes, and the number of references. The disk space required (∼60 MB per Mbase × number of genomes × number of references) and the computing time (∼2 min per Mbase × number of genomes × number of references) are linearly dependent on these three factors. Furthermore, the amount of RAM required depends primarily on the sequence length and the number of genomes (∼250 MB per Mbase × number of genomes). The computing time required for mapping (which is performed by Bowtie2) will be affected by the repetitiveness of the genomes. As we have not yet tested REALPHY on data from large eukaryotic genomes with many repetitive regions, we currently cannot meaningfully estimate how computational times will scale for such large genomes.
Implementation
The pipeline has been fully automated and is provided as a web server at http://realphy.unibas.ch (last accessed March 18, 2014). In addition, a stand-alone implementation in Java can be downloaded from the same website.
Sequence Simulation
We simulated sequence evolution in a custom-made Java program along four-taxon trees in which branch lengths were systematically varied between 0.5% and 8% divergence (fig. 1). These sequences were 100,000-bp long, with a GC content of 50%. Evolution occurred with identical transition and transversion rates, that is, using the elementary Jukes–Cantor model (Jukes and Cantor 1969). For each parameter combination (i.e., the combination of branch lengths in the tree), we repeated the simulation 100 times.
Phylogenetic Analyses
Multiple sequence alignments were built as described for the REALPHY algorithm. From these alignments as well as the true alignments, phylogenies were reconstructed using the maximum likelihood method PhyML with a GTR substitution matrix and a gamma-distributed rate heterogeneity model (Guindon et al. 2010).
Supplementary Material
Acknowledgment
The authors thank Chris Field for helpful discussions. This work was supported by the Royal Society of New Zealand with a James Cook Research Fellowship to P.B.R.; and the Swiss National Science Foundation with an Ambizione Fellowship (grant number PZ00P3-144605) to O.K.S., and a Swiss National Science Foundation Grant (grant number 31003A_135397) to E.v.N. Funding for open access charge: Swiss National Science Foundation Grant (grant number 31003A_135397).
Supplementary Material
Supplementary figures S1–S6 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
References
- Baltrus DA, Nishimura MT, Romanchuk A, Chang JH, Mukhtar MS, Cherkis K, Roach J, Grant SR, Jones CD, Dangl JL. Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog. 2011;7(7):e1002132. doi: 10.1371/journal.ppat.1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31(13):3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun J, Grim CJ, Hasan NA, Lee JH, Choi SY, Haley BJ, Taviani E, Jeon YS, Kim DW, Lee JH, et al. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci U S A. 2009;106(36):15442–15447. doi: 10.1073/pnas.0907787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331(6016):430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Yu C, Yan Y, Li D, Li Y, Jombart T, Weinert LA, Wang Z, Guo Z, Xu L, et al. Historical variations in mutation rate in an epidemic pathogen. Yersinia pestis. Proc Natl Acad Sci U S A. 2013;110(2):577–582. doi: 10.1073/pnas.1205750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Maiden MCJ. Impact of recombination on bacterial evolution. Trends Microbiol. 2010;18(7):315–322. doi: 10.1016/j.tim.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein B, Branca A, Mudge J, Bharti AK, Briskine R, Farmer AD, Sugawara M, Young ND, Sadowsky MJ, Tiffin P, et al. Population genomics of the facultatively mutualistic bacteria Sinorhizobium meliloti and S. medicae. PLoS Genet. 2012;8(8):e1002868. doi: 10.1371/journal.pgen.1002868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17(6):368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 2005 PHYLIP (phylogeny inference package). Version 3.69 [cited 2014 Mar 18]. Available from: http://evolution.genetics.washington.edu/phylip.html. [Google Scholar]
- Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, et al. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol. 2005;187(7):2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Harris SR, Clarke IN, Seth-Smith HMB, Solomon AW, Cutcliffe LT, Marsh P, Skilton RJ, Holland MJ, Mabey D, Peeling RW, et al. Whole-genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nat Genet. 2012;44(4):413–419, S1. doi: 10.1038/ng.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SR, Feil EJ, Holden MTG, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327(5964):469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt KE, Baker S, Weill FX, Holmes EC, Kitchen A, Yu J, Sangal V, Brown DJ, Coia JE, Kim DW, et al. Shigella sonnei genome sequencing and phylogenetic analysis indicate recent global dissemination from Europe. Nat Genet. 2012;44(9):1056–1059. doi: 10.1038/ng.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Ishii S, Ksoll WB, Hicks RE, Sadowsky MJ. Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Appl Environ Microbiol. 2006;72(1):612–621. doi: 10.1128/AEM.72.1.612-621.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro MN, editor. Mammalian protein metabolism. Vol. 3. New York: Academic Press; 1969. pp. 21–132. [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25(17):2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- Lewis PO. A likelihood approach to estimating phylogeny from discrete morphological character data. Syst Biol. 2001;50(6):913–925. doi: 10.1080/106351501753462876. [DOI] [PubMed] [Google Scholar]
- Luo C, Walk ST, Gordon DM, Feldgarden M, Tiedje JM, Konstantinidis KT. Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proc Natl Acad Sci U S A. 2011;108(17):7200–7205. doi: 10.1073/pnas.1015622108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdam PR, Templeton KE, Edwards GF, Holden MTG, Feil EJ, Aanensen DM, Bargawi HJA, Spratt BG, Bentley SD, Parkhill J, et al. Molecular tracing of the emergence, adaptation, and transmission of hospital-associated methicillin-resistant. Staphylococcus aureus. Proc Natl Acad Sci U S A. 2012;109(23):9107–9112. doi: 10.1073/pnas.1202869109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann HC, Rikkerink EHA, Bertels F, Fiers M, Lu A, Rees-George J, Andersen MT, Gleave AP, Haubold B, Wohlers MW, et al. Genomic analysis of the kiwifruit pathogen Pseudomonas syringae pv. actinidiae provides insight into the origins of an emergent plant disease. PLoS Pathog. 2013;9(7):e1003503. doi: 10.1371/journal.ppat.1003503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutreja A, Kim DW, Thomson NR, Connor TR, Lee JH, Kariuki S, Croucher NJ, Choi SY, Harris SR, Lebens M, et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature. 2011;477(7365):462–465. doi: 10.1038/nature10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Oshima T, Morimoto T, Ikeda S, Yoshikawa H, Shiwa Y, Ishikawa S, Linak MC, Hirai A, Takahashi H, et al. Sequence-specific error profile of Illumina sequencers. Nucleic Acids Res. 2011;39(13):e90. doi: 10.1093/nar/gkr344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S. Oxford: Oxford University Press; 2000. Molecular evolution and phylogenetics. [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302(1):205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Ogura Y, Ooka T, Iguchi A, Toh H, Asadulghani M, Oshima K, Kodama T, Abe H, Nakayama K, Kurokawa K, et al. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc Natl Acad Sci U S A. 2009;106(42):17939–17944. doi: 10.1073/pnas.0903585106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoro CK, Kingsley RA, Connor TR, Harris SR, Parry CM, Al-Mashhadani MN, Kariuki S, Msefula CL, Gordon MA, de Pinna E, et al. Intracontinental spread of human invasive Salmonella Typhimurium pathovariants in sub-Saharan Africa. Nat Genet. 2012;44(11):1215–1221. doi: 10.1038/ng.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston GM, Haubold B, Rainey PB. Bacterial genomics and adaptation to life on plants: implications for the evolution of pathogenicity and symbiosis. Curr Opin Microbiol. 1998;1(5):589–597. doi: 10.1016/s1369-5274(98)80094-5. [DOI] [PubMed] [Google Scholar]
- Rocha EPC. The quest for the universals of protein evolution. Trends Genet. 2006;22(8):412–416. doi: 10.1016/j.tig.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Rodriguez-R LM, Grajales A, Arrieta-Ortiz M, Salazar C, Restrepo S, Bernal A. Genomes-based phylogeny of the genus Xanthomonas. BMC Microbiol. 2012;12(1):43. doi: 10.1186/1471-2180-12-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg MS, Kumar S. Taxon sampling, bioinformatics, and phylogenomics. Syst Biol. 2003;52(1):119–124. doi: 10.1080/10635150390132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shavit Grievink L, Penny D, Holland BR. Missing data and influential sites: choice of sites for phylogenetic analysis can be as important as taxon sampling and model choice. Genome Biol Evol. 2013;5(4):681–687. doi: 10.1093/gbe/evt032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer M, Bryant D, Susko E. Conditioned genome reconstruction: how to avoid choosing the conditioning genome. Syst Biol. 2007;56(1):25–43. doi: 10.1080/10635150601156313. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, et al. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 2009;5(1):e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.