The INO80 and SWR1 complexes belong to the INO80 subfamily of ATP-dependent chromatin remodeling complexes. These complexes interact with nucleosomes and remodel chromatin by either sliding nucleosomes along the DNA or exchanging histones within nucleosomes. In doing so, the INO80 and SWR1 complexes promote recruitment of regulatory factors to DNA.
Distinct from other subfamilies of chromatin remodeling complexes, INO80 and SWR1 contain core ATPases with a unique split ATPase domain, as well as Rvb proteins, which share limited homology to bacterial RuvB, the Holliday Junction DNA Helicase. They bind to histone H2A variants (H2AX, H2A.Z) and have been implicated in the alteration of chromatin structure during transcription, replication, and DNA repair. This SnapShot depicts these two remodeling complexes, their mechanism of action and their biological functions.
Composition of the INO80 and SWR1 Complexes
The budding yeast INO80 complex is composed of the core ATPase, Ino80 (INOsitol requiring), Rvb1 (RuVB-like), Rvb2, Act1 (ACTin), Arp4 (Actin-Related Protein), Arp5, Arp8, Nhp10 (Non-Histone Protein), Taf14 (TATA binding protein-Associated Factor), Ies1 (Ino Eighty Subunit), Ies2, Ies3, Ies4, Ies5 and Ies6.
The budding yeast SWR1 complex contains fourteen polypeptides: the core ATPase, Swr1 (Sick With Rat8 ts), Rvb1, Rvb2, Act1, Arp4, Arp6, Swc2 (SWr Complex), Swc3, Swc4, Swc5, Swc6, Swc7, Yaf9 (Yeast homolog of the human leukemogenic protein AF9), and Bdf1 (BromoDomain Factor).
Remodeling Mechanisms
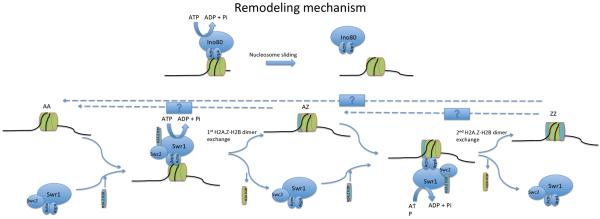
Nucleosome is the basic unit of chromatin, and is composed of 147 bp of DNA wrapped in 1.65 turns of left-handed superhelix around an octamer containing two copies each of the four core histone proteins (H2A, H2B, H3 and H4). The interactions between the remodeling complexes (INO80 and SWR1) and nucleosomes likely involve actin and the Arp subunits, which associate with the conserved HSA (helicase-SANT-associated) domain of the core ATPase in each complex. The chromatin substrates of the INO80 subfamily include histone H2A variants (H2AX and H2A.Z). Specifically, the interaction between INO80 complex and phosphorylated H2AX (γ-H2AX, in yeast H2A is also H2AX) within the nucleosome is mediated by Nhp10 and Arp4 (SWR1 may also associate with γ-H2AX via Arp4); while the interaction between SWR1 complex and the free H2A.Z-H2B dimer is mediated by the Swc2 subunit and the N-terminus of the Swr1 subunit. The INO80 complex has ATP-dependent DNA helicase activity and preferentially binds to four-way junction or three-way junction DNA structures in vitro, consistent with its functions in homologous recombination and DNA replication. The INO80 complex remodels chromatin by mobilizing (sliding) nucleosomes on the chromatin template in an ATP-dependent fashion as demonstrated by in vitro studies. The detailed steps of INO80 chromatin remodeling and its connection with INO80 helicase activity remain unknown. INO80 is capable of moving a mononucleosome from an end position to a more central position along a piece of DNA and may serve as a nucleosome spacing factor. Furthermore, INO80 has also been implicated in nucleosome eviction in vivo.
The SWR1 complex uses the energy of ATP hydrolysis to replace canonical H2A in nucleosomes with the H2A.Z variant in a stepwise and unidirectional fashion, as demonstrated by in vitro studies. SWR1 first binds to one side of AA nucleosomes (which contain two H2A-H2B dimers) and a free H2A.Z-H2B dimer, leading to hyperstimulation of the SWR1 ATPase activity, eviction of nucleosomal H2A-H2B, and deposition of H2A.Z-H2B. The AZ nucleosome (containing one H2A-H2B dimer and one H2A.Z-H2B dimer) subsequently dissociates from SWR1 to form an intermediate product. SWR1 can specifically bind to the A side of the AZ nucleosome, leading to a second histone replacement reaction that forms the ZZ nucleosome (containing 2 H2A.Z-H2B dimers). The mechanisms for the reverse reactions (ZZ to AZ, AZ to AA, and/or ZZ to AA) remain unclear. In budding yeast, the SWR1 complex shares 4 subunits (Act1, Arp4, Yaf9, and Swc4) with the NuA4 complex (a histone acetyltransferase complex). These two complexes function together to regulate the deposition of H2A.Z into nucleosomes. Current models suggest that SWR1 recruitment to chromatin depends on those shared subunits between the SWR1 and NuA4 complexes. Recruitment also depends on Bdf1, which recognizes acetylated histone H4, most likely modified by the NuA4 complex. Once recruited, SWR1 replaces conventional histone H2A with the H2A.Z variant, which may then undergo further acetylation by NuA4.
Biological Functions of the INO80 Complex
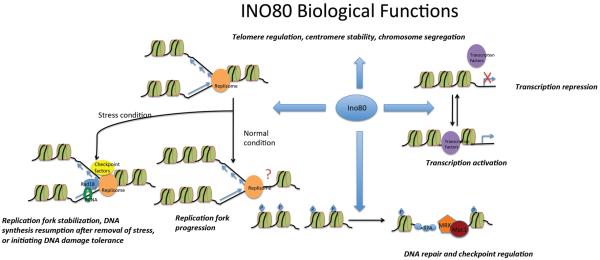
The transcription of roughly 20% of yeast genes is regulated, either positively or negatively, by the INO80 complex. For example, binding of Pho4 (a transcription factor) to the PHO5 promoter initiates recruitment of INO80, SWI/SNF (SWItch/Sucrose NonFermentable, a nucleosome remodeling complex), and SAGA (Spt/Ada/Gcn5/acetyltransferase, a histone acetyltransferase complex). These complexes work together to remodel nucleosomes on the promoter in order to increase DNA accessibility for the transcription machinery.
Yeast INO80 binds to sites of DNA double strand breaks via an interaction with γ-H2AX. It promotes the eviction of nucleosomes near double strand breaks, leading to efficient recruitment of the DNA repair machinery, which may include complexes such as MRX (Mre11-Rad50-Xrs2) and the checkpoint kinase Mec1. INO80 is also implicated in UV (Ultraviolet) damage repair.
Yeast INO80 contributes to the efficient progression of the replication fork under normal conditions, perhaps by sliding or eviction nucleosomes in the path of replication fork. Under conditions of replication stress, the INO80 complex promotes stabilization of the replication fork, resumption of DNA synthesis and tolerance to DNA damage possibly through remodeling of nucleosomes at stressed replication fork. Phosphorylation of the Ies4 subunit of INO80 regulates the replication checkpoint. The INO80 complex is also involved in telomere regulation, centromere stability, and chromosome segregation.
Biological Functions of the SWR1 Complex
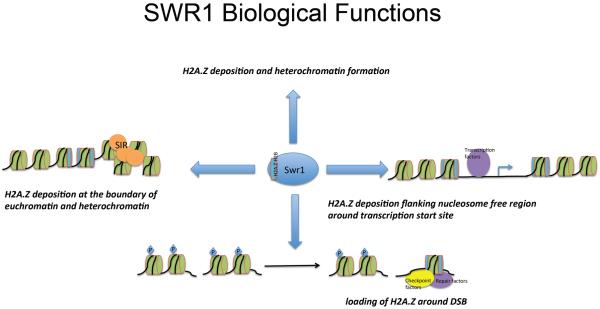
SWR1 deposits H2A.Z into nucleosomes flanking the nucleosome-free region around promoters. The incorporation of H2A.Z into the nucleosome may change nucleosome stability and dynamics, and also facilitate or inhibit recruitment of other factors. Thus, The incorporation of H2A.Z may regulate transcription both positively and negatively.
Emerging data suggest that the SWR1 complex functions in concert with the NuA4 complex at DSB lesions to exchange γ-H2AX with H2A.Z, which may facilitate recruitment of repair factors and checkpoint factors.
SWR1 exchanges H2A.Z into genes near subtelomeric regions, antagonizes the spreading of heterochromatin into euchromatic regions. Thus, H2A.Z deposition may protect genes from SIR (Silent Information Regulator) dependent telomeric silencing. In higher eukaryotic cells, H2A.Z is also implicated in gene silencing and heterochromatin formation.
Figure 1–4.
ACKNOWLEDGMENTS
Work in the authors' laboratory is supported by grants from NIGMS to X. S. Y. B. is supported by the Epigenetic Scholar Program at MD Anderson Cancer Center.
REFERENCES
- Bao Y, Shen X. INO80 subfamily of chromatin remodeling complexes. Mutat Res. 2007;618:18–29. doi: 10.1016/j.mrfmmm.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway RC, Conaway JW. The INO80 chromatin remodeling complex in transcription, replication and repair. Trends Biochem Sci. 2009;34:71–77. doi: 10.1016/j.tibs.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Lu PY, Levesque N, Kobor MS. NuA4 and SWR1-C: two chromatin-modifying complexes with overlapping functions and components. Biochem Cell Biol. 2009;87:799–815. doi: 10.1139/O09-062. [DOI] [PubMed] [Google Scholar]
- Luk E, Ranjan A, Fitzgerald PC, Mizuguchi G, Huang Y, Wei D, Wu C. Stepwise Histone Replacement by SWR1 Requires Dual Activation with Histone H2A.Z and Canonical Nucleosome. Cell. 2010;143:725–736. doi: 10.1016/j.cell.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison AJ, Shen X. Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat Rev Mol Cell Biol. 2009;10:373–384. doi: 10.1038/nrm2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altaf M, Auger A, Covic M, Cote J. Biochem Cell Biol. 2009;87(1):35–50. doi: 10.1139/O08-140. [DOI] [PubMed] [Google Scholar]
- Bao Y, Shen X. Cell. 2007 May 4;129(3):632. doi: 10.1016/j.cell.2007.04.018. 2007. [DOI] [PubMed] [Google Scholar]
- van Attikum H, Gasser SM. Trends Cell Biol. 2009;19(5):207–17. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Osley MA, Shen X. Altering nucleosomes during DNA double-strand break repair in yeast. Trends Genet. 2006;22:671–677. doi: 10.1016/j.tig.2006.09.007. [DOI] [PubMed] [Google Scholar]