Abstract
cAMP responsive element-binding protein H (CREBH) is an endoplasmic reticulum (ER) anchored transcription factor that is highly expressed in the liver and small intestine and implicated in nutrient metabolism and proinflammatory response. ApoA-IV is a glycoprotein secreted primarily by the intestine and to a lesser degree by the liver. ApoA-IV expression is suppressed in CREBH-deficient mice and strongly induced by enforced expression of the constitutively active form of CREBH, indicating that CREBH is the major transcription factor regulating Apoa4 gene expression. Here, we show that CREBH directly controls Apoa4 expression through two tandem CREBH binding sites (5′-CCACGTTG-3′) located on the promoter, which are conserved between human and mouse. Chromatin immunoprecipitation and electrophoretic mobility-shift assays demonstrated specific association of CREBH with the CREBH binding sites. We also demonstrated that a substantial amount of CREBH protein was basally processed to the active nuclear form in normal mouse liver, which was further increased in steatosis induced by high-fat diet or fasting, increasing apoA-IV expression. However, we failed to find significant activation of CREBH in response to ER stress, arguing against the critical role of CREBH in ER stress response.
Keywords: cAMP responsive element-binding protein H, ER stress, hepatic steatosis
cAMP responsive element-binding protein H (CREBH) is an endoplasmic reticulum (ER)-bound bZiP transcription factor that is highly and specifically expressed in the liver and small intestine (1, 2). It is activated by proteolytic cleavage in the transmembrane domain by site 1 (S1P) and site 2 (S2P) proteases, which release the N-terminal portion of the protein that enters the nucleus to act as a transcription factor (3, 4). CREBH was initially identified as a liver-enriched transcription factor that could bind to cAMP response element (CRE) and activate the transcription driven by CRE-containing promoters, such as rat phosphoenolpyruvate carboxykinase (PEPCK) promoter (1, 2).
Recent studies employing genetic ablation of CREBH or in vivo delivery of sequence-specific shRNA revealed that CREBH is involved in a variety of physiological functions of the liver. It has been shown that CREBH is proteolytically activated by ER stress (5), induced acute phase response genes (5), and the iron metabolism regulator, hepcidin (6). It has also been demonstrated that CREBH is induced in the liver of fasted mice (7), and promotes the transcription of genes involved in gluconeogenesis (8) and plasma TG clearance (9).
While CREBH controls the expression of a variety of target genes, the upstream signal that activates CREBH and the promoter element mediating the CREBH function remain poorly understood. Despite the postulated roles for CREBH in ER stress-mediated inflammatory response and hepcidin expression, it remains controversial whether CREBH is indeed activated by ER stress. Zhang et al. (5) first reported that CREBH was activated by ER stress, in a manner similar to ATF6α. However, subsequent studies by several other groups failed to detect proteolytic activation of CREBH by ER stress inducers in stable cell lines expressing exogenous CREBH (10, 11). On the other hand, hepatic CREBH mRNA is induced by fasting and suppressed by refeeding, which appears to be mediated by glucocorticoid receptor and PPARα that bind to peroxisome proliferator responsive element (PPRE) and glucocorticoid transcriptional response element on the CREBH promoter (7, 8, 12).
Comparison of gene expression profiles of WT and CREBH-deficient mice identified Apoa4 as one of the genes that are tightly controlled by CREBH, both in the liver and small intestine (9). ApoA-IV is produced and secreted by enterocytes in the small intestine and at a lesser level by the liver (13, 14), and is mainly associated with HDL particles in normal human serum (15). ApoA-IV mRNA is induced in the liver of fasted mice in a CREBH-dependent manner (9, 16). Transcription factor hepatic nuclear factor 4α (HNF-4α) was also shown to play a role in apoA-IV mRNA expression in mouse liver (16).
In the current study, we investigated the mechanism by which CREBH activates the Apoa4 gene promoter, and evaluated the effect of ER stress on CREBH activation in mouse liver. We demonstrate that CREBH directly activates Apoa4 gene expression in mice and humans through tandem CREBH binding sites on its promoter. Liver steatosis induced hepatic apoA-IV expression, which was dependent on CREBH. We also found that a significant amount of CREBH protein was spontaneously processed in normal mouse liver to the active nuclear form, and tunicamycin, a commonly used chemical ER stress inducer, suppressed CREBH processing.
MATERIALS AND METHODS
Animals
Creb3l3−/− mice have been described (9, 17). Creb3l3−/− mice were backcrossed more than 10 generations to C57BL/6 mice (Taconic Farms). Mice were fed either a standard chow diet containing 13.2% fat, 24.6% protein, and 62.1% carbohydrate (kcal/100 kcal) (#5053, LabDiet), a high-fat and high-cholesterol Western diet containing 42.0% fat, 15.2% protein, 42.7% carbohydrate, and 0.2% cholesterol (TD 88137, Harlan Teklad), or a ketogenic diet consisting of 93.4% fat, 4.7% protein, and 1.8% carbohydrate (F-3666, Bio-Serv). Mice were housed in a 12 h light/dark cycle with ad libitum access to food and water, unless indicated otherwise, and euthanized by CO2 between 9:00 AM and 11:00 AM. For some experiments, mice were fasted overnight for 16 h with free access to water. Male mice around 2 months old with body weights of 24 ± 1 g were used exclusively. Plasma was collected from EDTA-treated whole blood by centrifuging at 3,000 rpm for 15 min. Tissues were resected and snap-frozen on dry ice and stored at −80°C for further analysis. All procedures involving animals were in accordance with the Weill Cornell Medical College Institutional Animal Care and Use Committee.
Plasmid constructs
The N-terminal fragment of CREBH [CREBH(N)] (amino acids 1-332) was amplified from a human cDNA clone (Open BioSystems, BC101504) and inserted into pcDNA3.1 vector using the EcoRI and XbaI sites. Fragments of mouse Apoa4 promoter were isolated by PCR using RP24-302M3 BAC plasmid (BACPAC Resources Center, CHORI) containing the Apoa4 gene as a template, and cloned into pGL3-basic vector (Promega) using MluI and XhoI restriction enzymes. The primers used were: 5′-ggacgcGTAGCTAGCTGCTTCTAGGGAT-3′ for 5.9 kb; 5′-ggacgcgtCACTCTGCATGGCTCTTGCATATG-3′ for 0.9 kb; 5′-ggacgcGTTCTCTCAGACTGGCACAG-3′ for 0.33 kb; 5′-ggacgcGTTTCTGGCTATCCTTCCCA-3′ for 0.23 kb; 5′-ggacgcGTCAGCTTCCACGTTGTCTT-3′ for 0.15 kb; 5′-ggacgcGTCACACTGGGGAGGAGG-3′ for 0.07 kb proximal promoter fragments; and 5′-ctcGAGTCAGAAGAAACGGTGTACC-3′ for common reverse primer (lowercase letters represent additional nucleotides for cloning). Site-directed mutagenesis was performed using the QuikChange II XL site-directed mutagenesis kit (Stratagene) with primers 5′-TTACGCGTCAGCTTCCtttcTGTCTTAGGGCC-3′ for site A, and 5′-GTGTGTCACCTTCCAtttcGGAGTCACACTG-3′ for site B (mutated nucleotides are shown in lowercase letters). UPRE-luciferase reporter was described previously (18).
Cell culture and transfection
HEK-293T, Hepa1-6, HepG2, and Huh7 cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. Cells were transfected with 25 ng luciferase reporter, 25 ng of effector, and 5 ng of pRL/CMV Renilla plasmids using Lipofectamine 2000 (Invitrogen). Cells were harvested 24 h later for luciferase assays using a Dual-Luciferase reporter assay kit (Promega). In some experiments, cells were treated with tunicamycin (1 μg/ml) for 16 h before harvest. Transfection efficiency was normalized to the Renilla activity.
Adenoviral transduction of cells
Recombinant adenovirus expressing mouse CREBH(N) was generated as described elsewhere (9). For adenovirus infection experiments, primary mouse hepatocytes and HepG2 cells were plated on 60 mm dishes at 1 × 106 cells/dish and infected with either recombinant adenovirus Ad-GFP or Ad-CREBH(N). Cells were harvested 24 h after infection for RNA isolation and real-time PCR analysis.
RNA isolation and real-time PCR
Total RNAs were isolated using TRIzol reagent (Invitrogen) and used for the synthesis of cDNA using a high capacity cDNA reverse transcription kit (Applied Biosystems). The abundance of each cDNA was quantified by SYBR real-time PCR using the Mx3005P™ system (Stratagene). The following primers were used: mouse Apoa4, Actb (9); human APOA4, 5′-CCAAGATCGACCAGAACGTGG-3′ and 5′-GTCCTGAGCATAGGGAGCCA-3′; human ACTB, 5′-GCGAGAAGATGACCCAGATC-3′ and 5′-CCAGTGGTACGGCCAGAGG-3′.
Western blot analysis
Liver nuclear extracts and microsomal fractions were prepared and subjected to Western blotting analysis, as described previously (9). The following primary antibodies were used in this study: mouse monoclonal antibodies against apoB (19), lamin B1 (Santa Cruz, sc-56145), rat monoclonal antibody against hemagglutinin (HA) tag (Roche, 3F10), goat polyclonal antibody against apoA-IV (Santa Cruz, sc-19036), monoclonal antibody for human apoA-IV (Cell Signaling, #5700), rabbit polyclonal antibodies against apoE (Meridian Life Sciences, K23100R), ATF6α (20), calnexin (Enzo, ADI-SPA-865), and CREBH (9).
Electrophoretic mobility shift assay
Electrophoretic mobility shift assay (EMSA) was carried out using 32P-labeled oligonucleotides and CREBH(N) proteins prepared using in vitro TNT® quick coupled transcription/translation system (Promega). Double stranded probes containing the CREBH binding site from the mouse Apoa4 promoter were synthesized based on the following sequences: WT, 5′-TTACGCGTCAGCTTCCACGTTGTCTTAGGGCC-3′; Mut, 5′-TTACGCGTCAGCTTCCtttcTGTCTTAGGGCC-3′ (mutated nucleotides are in lowercase). Probes were end-labeled with [γ-32]ATP using the T4 polynucleotide kinase, annealed, and purified through a Sephadex G25 spun column. A 50 fmol annealed probe was incubated with 1.5 μl of the in vitro translation reaction mixture for CREBH(N) for 20 min in EMSA buffer [10 mM Tris (pH 7.6), 50 mM NaCl, 1 mM EDTA, 5% glycerol, 50 μg/ml poly(deoxyinosinic-deoxycytidylic) acid]. For competition assays, 5-, 25-, or 125-fold excess unlabeled probes or 0.3 μg antibodies were added to the reaction mixture. Samples were electrophoresed on a 6% polyacrylamide gel in 0.5× TBE buffer [40 mM Tris, 45 mM boric acid, 1 mM EDTA (pH 8.0)]. Gels were dried and analyzed using a Personal Molecular Imager FX™ system (Bio-Rad).
Chromatin immunoprecipitation assay
Hepa1-6 cells were transfected with HA-CREBH(N) or control plasmids. Forty-eight hours later, cells were used for chromatin immunoprecipitation (ChIP) experiments as described elsewhere (21). Immunoprecipitation was performed using anti-HA (Roche, 3F10) or anti-Flag antibody (Biolegend, L5). Eluted DNA was subjected to real-time PCR analysis.
Quantification of hepatic TG
Total lipids were extracted from the liver according to the method of Bligh and Dyer (22). Briefly, lipids were extracted from homogenized tissues with chloroform-methanol (2:1, v/v). After centrifugation, the organic phase was collected, dried under nitrogen, and then dissolved in 60% butanol, 40% of a 2:1 mixture of Triton-X114, and methanol. Aliquots were assayed for TG levels using a colorimetric TG assay kit (Sigma).
Statistical analysis
All data are expressed as mean ± SEM for animal experiments. Two-tailed Student's t-tests were used to evaluate statistical differences between groups.
RESULTS
CREBH is sufficient to induce apoA-IV mRNA in hepatocytes
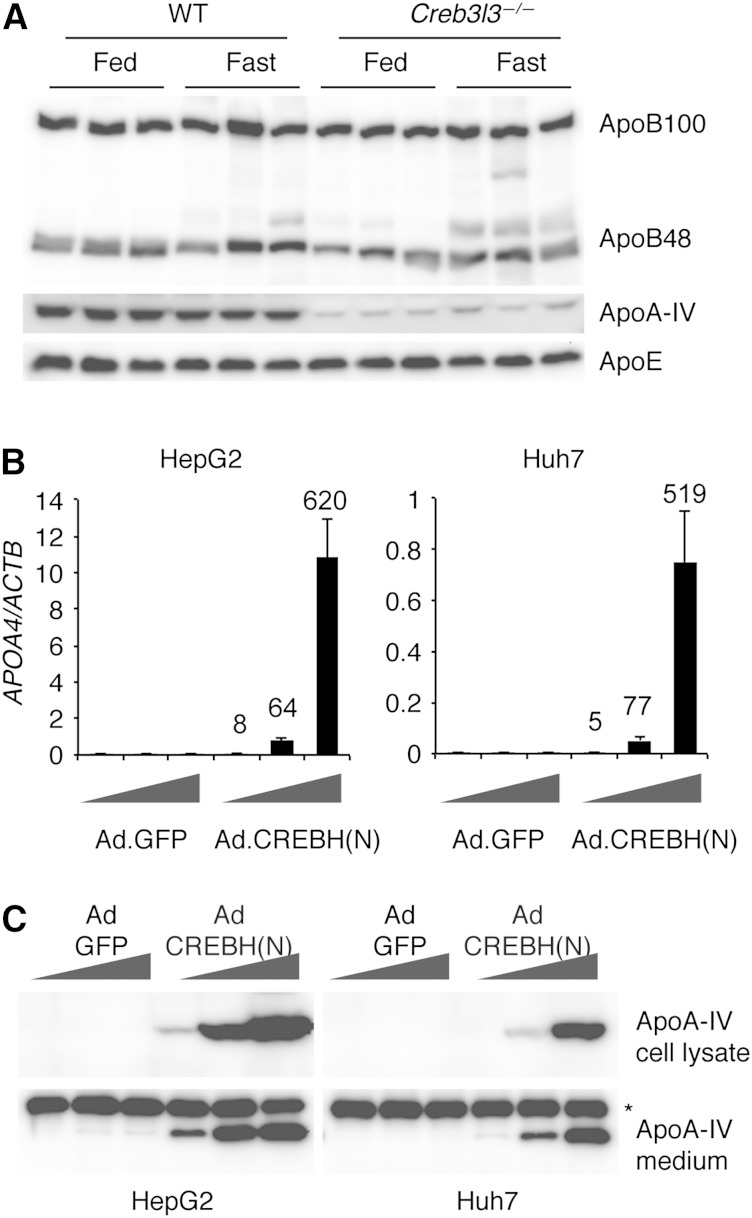
In both humans and rodents, many apolipoprotein genes are present as gene clusters on the chromosomes. APOE/C1/C4/C2 and APOA1/C3/A4/A5 gene clusters are located on chromosomes 19 and 11 in humans, and chromosomes 7 and 9 in mice, respectively. To determine the role of CREBH in the transcription of genes within these clusters, we tested the effects of loss of CREBH or overexpression of a constitutively active CREBH(N) on the mRNA levels of these genes. Consistent with our previous report (9), apoA-IV and apoC-II mRNAs were markedly decreased both in the liver and small intestine of CREBH-deficient mice (Table 1). Conversely, overexpression of CREBH(N) using a recombinant adenoviral vector strongly induced apoA-IV and apoC-II mRNAs in primary hepatocytes and mouse liver (Table 1), suggesting that CREBH directly activates the transcription of Apoa4 and Apoc2 genes. ApoB and several other apolipoprotein mRNAs were also modestly decreased in CREBH knockout mice, but they were not induced by CREBH(N) overexpression, suggesting that they might be indirectly regulated by CREBH. Western blot analysis of plasma samples revealed that the plasma apoA-IV level was markedly decreased in CREBH knockout mice, while apoB and apoE were preserved (Fig. 1A).
TABLE 1.
Expression profile of apolipoprotein genes
| Liver |
Jejunum |
Liver |
Primary Hepatocytes |
|||||
| WT | Creb3l3−/− | WT | Creb3l3−/− | Ad.GFP | Ad.CREBH(N) | Ad.GFP | Ad.CREBH(N) | |
| Apoe/c1/c4/c2 cluster | ||||||||
| Apoe | 1.00 ± 0.09 | 0.55 ± 0.04c | ND | ND | 1.00 ± 0.01 | 0.86 ± 0.12 | 1.00 ± 0.20 | 1.61 ± 0.10 |
| Apoc1 | 1.00 ± 0.05 | 0.78 ± 0.07 | ND | ND | 1.00 ± 0.03 | 0.51 ± 0.23 | 1.00 ± 0.19 | 3.14 ± 0.79 |
| Aopoc4 | 1.00 ± 0.02 | 0.67 ± 0.06c | ND | ND | 1.00 ± 0.03 | 1.59 ± 0.35 | 1.00 ± 0.16 | 2.95 ± 0.37a |
| Apoc2 | 1.00 ± 0.07 | 0.24 ± 0.02c | 1.00 ± 0.04 | 0.29 ± 0.04c | 1.00 ± 0.18 | 19.23 ± 1.44b | 1.00 ± 0.11 | 11.58 ± 1.14a |
| Apoa1/c3/a4/a5 cluster | ||||||||
| Apoa1 | 1.00 ± 0.10 | 0.33 ± 0.04c | ND | ND | 1.00 ± 0.06 | 1.48 ± 0.13 | 1.00 ± 0.42 | 16.16 ± 8.66 |
| Apoc3 | 1.00 ± 0.05 | 0.99 ± 0.07 | 1.00 ± 0.08 | 1.26 ± 0.18 | 1.00 ± 0.02 | 1.47 ± 0.29 | 1.00 ± 0.32 | 5.76 ± 1.78 |
| Apoa4 | 1.00 ± 0.30 | 0.02 ± 0.00b | 1.00 ± 0.14 | 0.09 ± 0.04c | 1.00 ± 0.16 | 298.6 ± 32.9b | 1.00 ± 0.12 | 40.33 ± 8.40a |
| Apoa5 | 1.00 ± 0.09 | 0.47 ± 0.06c | ND | ND | 1.00 ± 0.02 | 1.53 ± 0.33 | 1.00 ± 0.47 | 16.03 ± 2.21a |
| Apob | 1.00 ± 0.03 | 0.67 ± 0.04c | 1.00 ± 0.10 | 0.38 ± 0.08b | 1.00 ± 0.07 | 1.28 ± 0.16 | 1.00 ± 0.05 | 1.91 ± 0.22 |
Values represent fold changes of mRNA levels relative to those of WT mice or Ad.GFP infected samples. ND, not determined.
P < 0.05.
P < 0.01.
P < 0.001.
Fig. 1.
CREBH regulates apoA-IV expression. A: Plasma samples were collected from WT and Creb3l3−/− mice in the fed state or after a 16 h fast, and subjected to Western blotting. B: HepG2 and Huh7 cells were infected with increasing doses of CREBH(N) or the control GFP adenoviruses. Forty-eight hours later, cells were harvested to measure apoA-IV mRNA levels by real-time PCR. Values were normalized to β-actin. Data are given as means ± SD. Numbers on the bars represent fold changes in mRNA levels relative to Ad.GFP controls. C: Western blot analysis of intracellular and secreted apoA-IV. ApoA-IV in the conditioned medium was enriched by immunoprecipitation before Western blotting. *Nonspecific band.
To determine whether the human APOA4 gene was also regulated by CREBH, we infected HepG2 and Huh7 cells with recombinant adenovirus expressing CREBH(N). ApoA-IV mRNA was strongly induced by CREBH(N) overexpression in both human liver cell lines in a dose-dependent manner. Western blotting analysis of cell lysates and conditioned media confirmed the induction of apoA-IV protein by CREBH(N) overexpression (Fig. 1B, C), indicating that CREBH regulates apoA-IV transcription in human hepatocytes as well.
CREBH is the major transcription factor activating Apoa4 transcription
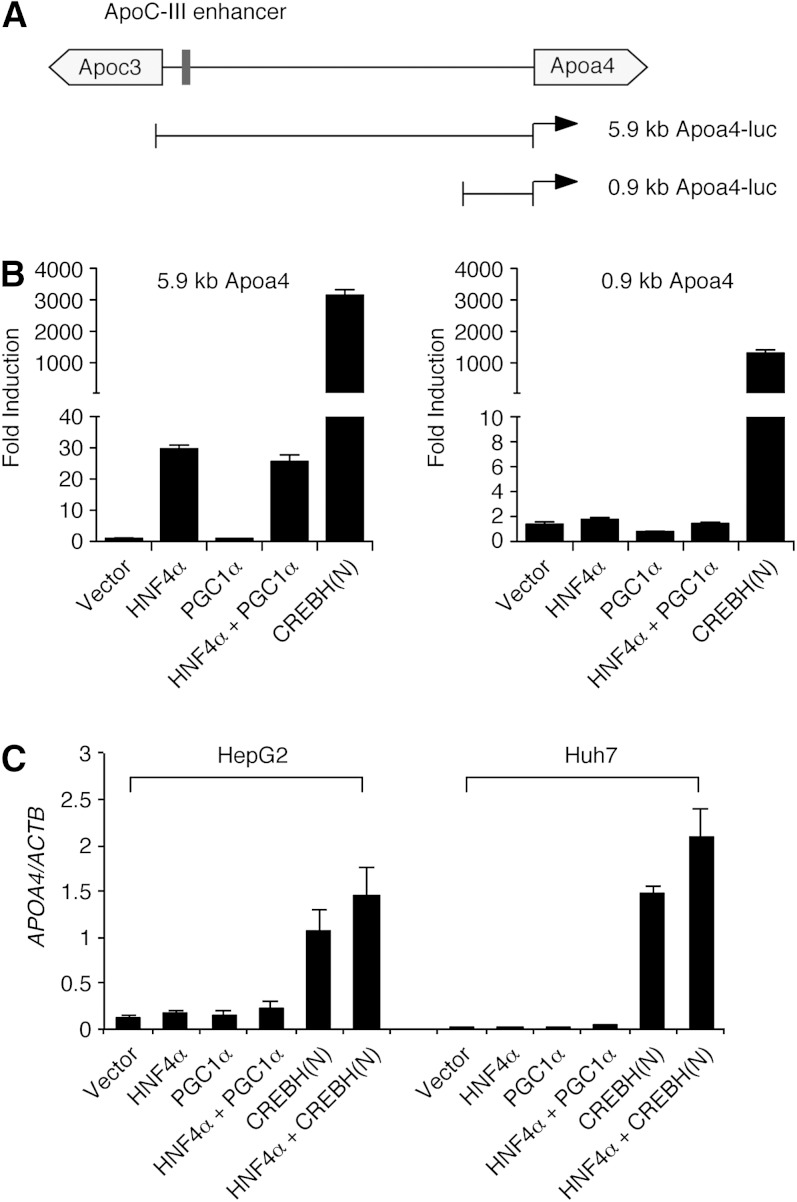
Apoa4 and Apoc3 genes are separated by a 5.8 kb intergenic region and transcribed in the opposite direction (Fig. 2A). Earlier studies demonstrated that nuclear hormone receptors such as HNF-4α and estrogen receptor α (ERRα) activate apoA-IV transcription through the apoC-III enhancer, which is located near the Apoc3 gene (16, 23–26). To investigate the roles of different transcription factors in Apoa4 transcription, luciferase reporter constructs containing 5.9 kb or 0.9 kb fragments of Apoa4 promoter were generated. Cotransfection of the HNF-4α construct with the 5.9 kb Apoa4-luciferase reporter increased the luciferase activity by ∼30-fold in 293T cells (Fig. 2B). PPARγ coactivator-1α (PGC1α) did not further activate the 5.9 kb Apoa4-luciferase reporter, despite it being known as a coactivator of HNF-4α (16, 25). HNF-4α failed to activate the 0.9 kb Apoa4-luciferase reporter, consistent with the requirement for apoCIII enhancer in HNF-4α-mediated apoA-IV activation (25). CREBH(N) cotransfection dramatically increased the luciferase activity of both 5.9 kb Apoa4-luciferase and 0.9 kb Apoa4-luciferase reporters by more than 1,000-fold (Fig. 2B). Together with the marked reduction of apoA-IV mRNA in CREBH knockout mice, these data suggest that CREBH is the major transcription factor that activates Apoa4 gene expression through the proximal promoter region. Consistent with the luciferase assay data, CREBH(N) transfection strongly induced the endogenous apoA-IV mRNA in HepG2 and Huh7 cells, while HNF-4α and PGC1α showed only weak effects (Fig. 2C).
Fig. 2.
CREBH activates the Apoa4 promoter. A: Schematic diagram of the apoC-III/A-IV intergenic region and Apoa4-luciferase reporter constructs. B: Apoa4-luciferase reporter constructs containing 5.9 kb or 0.9 kb Apoa4 promoter fragments were cotransfected into 293T cells with indicated plasmids. Twenty-four hours later, luciferase activities in cell lysates were measured and normalized to the Renilla activity. Values represent fold changes compared with luciferase reporter only samples. C: HepG2 and Huh7 cells were transfected with indicated constructs. Forty-eight hours later, cells were harvested for real-time PCR analysis. ApoA-IV mRNA levels were normalized to β-actin. Data are given as means ± SD.
Identification of CREBH recognition sites on the Apoa4 promoter
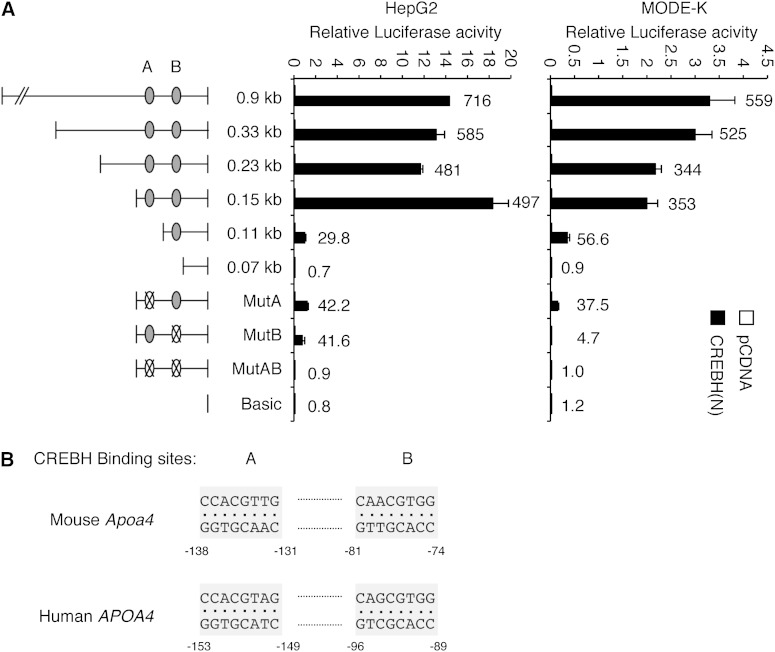
In order to identify the cis-acting elements required for the activation of the Apoa4 promoter by CREBH, we generated a series of reporter plasmids containing various lengths of mouse Apoa4 promoter, and performed luciferase assays following cotransfection with CREBH(N) plasmid using HepG2 and a mouse intestinal epithelial cell line, MODE-K (Fig. 3A). Serial deletion of the Apoa4 promoter from the 5′ end revealed that the 0.15 kb proximal fragment is crucial for the activation of the Apoa4 promoter by CREBH in both HepG2 and MODE-K cells. Further deletion to the 0.07 kb upstream of the transcription start site completely abolished the transactivation of the reporter by the cotransfected CREBH(N). We found two potential CREBH binding sites, site A (5′-CCACGTTG-3′) and site B (5′-CAACGTGG-3′), between nucleotides −138 and −74 of the mouse Apoa4 promoter, which have identical sequences in opposite orientations. Notably, the potential CREBH binding sites are well-conserved in human and mouse APOA4 promoters (Fig. 3B). Disruption of either site A or B by deletion (0.11 kb) or point mutations (MutA and MutB) partially reduced the fold induction of the luciferase activity by CREBH(N) (Fig. 3A). Interestingly, disruption of site B (MutB) had a more severe impact on CREBH-induced reporter activity in MODE-K than in HepG2 cells. Further studies will determine whether CREBH regulates Apoa4 gene differently in the intestine and liver. Disruption of both sites completely abolished the transactivation of the Apoa4 promoter by CREBH, indicating that these sequences are required for the activation of Apoa4 promoter by CREBH (Fig. 3A).
Fig. 3.
Identification of CREBH binding sites on the Apoa4 gene promoter. A: Luciferase reporter constructs containing various fragments of the Apoa4 gene promoter with site-specific mutations were generated and tested for CREBH transactivation in HepG2 and MODE-K cells. Dual-luciferase assays were performed 24 h after transfection. Values represent means ± SD. Numbers on the bars represent fold induction of the luciferase activity by CREBH(N) cotransfection. B: Alignment of the putative CREBH binding sites on human and mouse Apoa4 gene promoter.
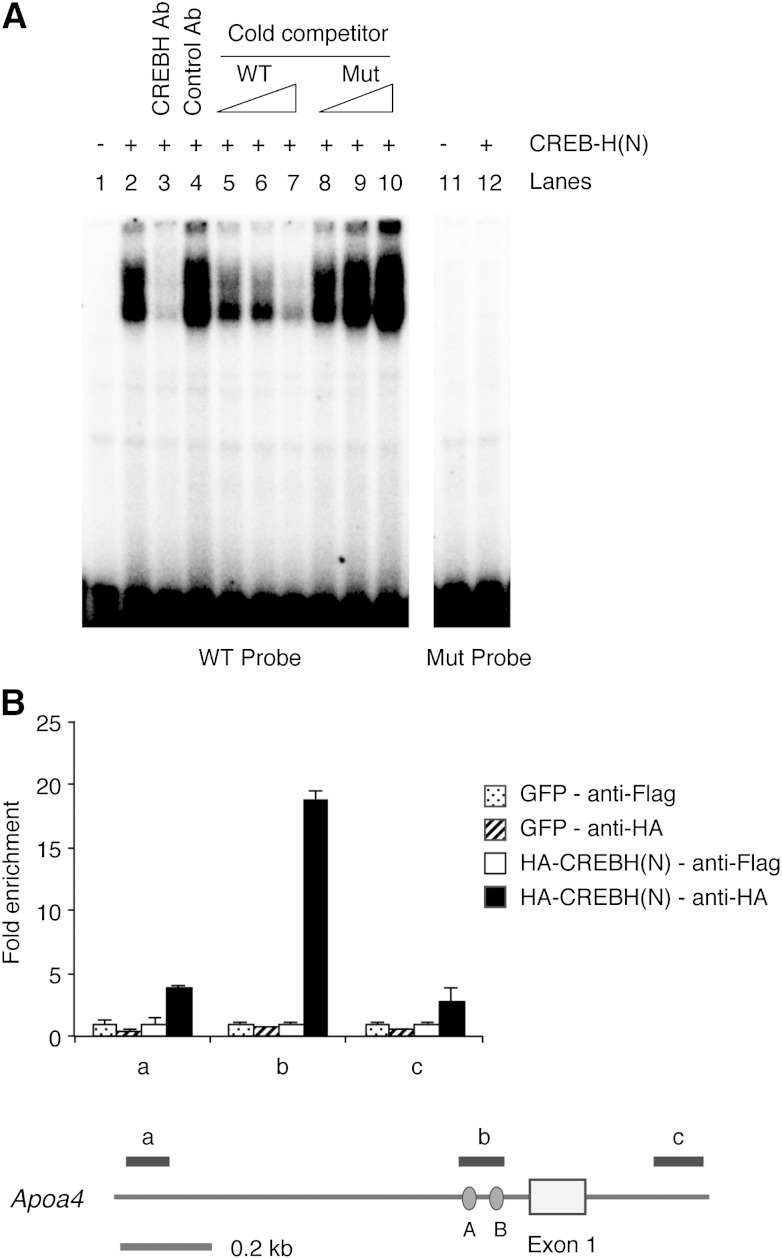
To test if CREBH can directly bind to these cis-acting elements, EMSA was performed with in vitro translated CREBH(N) proteins (Fig. 4A). The CREBH(N) protein formed a complex with a radiolabeled oligonucleotide probe containing site A sequences (5′-ttacgcgtcagcttCCACGTTGTcttagggcc-3′, uppercase letters represent the CREBH binding site) (lane 2). CREBH(N)-probe complex formation was inhibited by either a specific antibody (lane 3) or the excess unlabeled WT probe (lanes 5–7). A mutant probe with the disrupted ACGT core (5′-ttacgcgtcagcttCCtttcTGTcttagggcc-3′) did not bind to CREBH(N) (lane 12) and failed to compete for CREBH(N) binding with the WT probe (lanes 8–10). These data indicate that CREBH directly binds to the Apoa4 gene promoter and activates its transcription.
Fig. 4.
Direct binding of CREBH to the Apoa4 gene promoter. A: Double strand oligonucleotides containing the putative CREBH binding site were used for EMSA with in vitro translated CREBH(N) protein. Control or CREBH-specific antibodies were added in reactions for lanes 3 and 4. Increasing amounts (5×, 25×, 125×) of WT or Mut probe with disrupted ACGT core were used as cold competitors in reactions for lanes 5–10. Mut, mutant. B: Hepa1-6 cells infected with HA-CREBH(N) or GFP control adenoviruses were subjected to ChIP analysis. The recovered chromatin samples were analyzed by quantitative real-time PCR with primer pairs amplifying the Apoa4 promoter or the intronic region as shown in the schematic diagram. Values represent fold enrichment relative to anti-Flag immunoprecipitation samples.
To confirm the direct binding of CREBH to the Apoa4 promoter within the cell, we performed ChIP assays. Our CREBH antibody was not suitable for ChIP because the major epitope is in the bZiP domain involved in dimerization and DNA binding. Therefore, we overexpressed HA-tagged CREBH(N) in Hepa1-6 cells by adenoviral infection. Immunoprecipitation of chromatin isolated from HA-CREBH(N) expressing cells with HA antibody markedly enriched the Apoa4 promoter region containing CREBH recognition sites but not neighboring regions, indicating the sequence-specific binding of CREBH to the Apoa4 promoter (Fig. 4B).
CREBH is essential for the increased expression of apoA-IV in steatosis
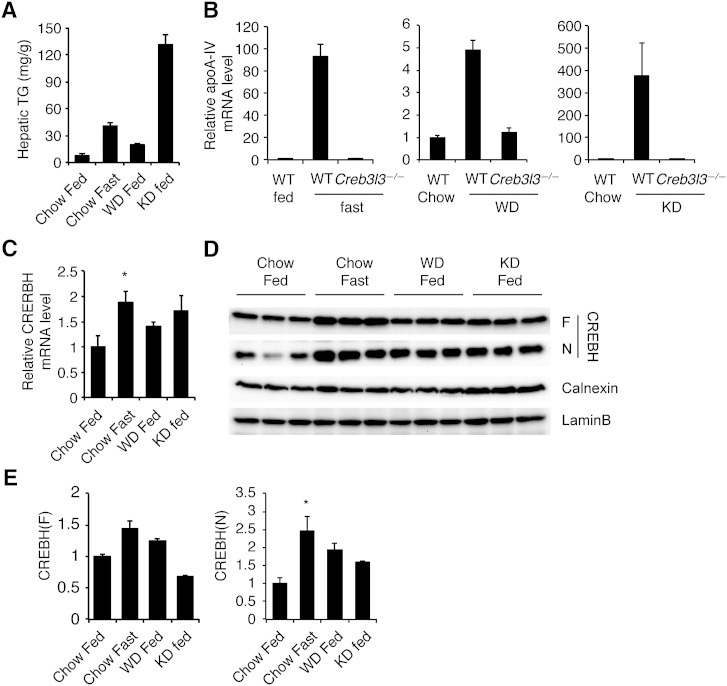
It has been shown that hepatic apoA-IV expression is increased in various animal models of liver steatosis (16, 27, 28). Fasting also induces hepatic steatosis in mice, which likely leads to the induction of apoA-IV (16, 27). To determine whether CREBH plays a role in apoA-IV expression in a steatotic liver, we fed WT and CREBH-deficient mice a Western diet containing high fat and high cholesterol or a ketogenic diet containing high fat and very low carbohydrate. These diets, along with fasting, caused hepatic steatosis to varying degrees of severity and increased the abundance of hepatic apoA-IV mRNA (Fig. 5A, B), as previously reported (27, 28). Interestingly, the abundance of apoA-IV mRNA correlated well with the hepatic TG level, which is consistent with a previous report (27). Loss of CREBH completely abolished the induction of hepatic apoA-IV mRNA in these models of hepatic steatosis (Fig. 5B), indicating the critical role of CREBH in hepatic apoA-IV expression. Notably, loss of CREBH increased hepatic TG content (data not shown), excluding the possibility that reduced TG content accounted for diminished apoA-IV mRNA in CREBH-deficient liver.
Fig. 5.
CREBH is essential for apoA-IV expression in steatosis. A: Hepatic TG content of mice fasted (n = 5), or fed normal chow (n = 3), Western diet (WD) for 10 weeks (n = 5), or ketogenic diet (KD) for 4 days (n = 5). ApoA-IV (B) and CREBH (C) mRNA level compared with the chow-fed group (n = 3–5 mice/group). D: ER and nuclear fractions were subjected to Western blotting. F, full length CREBH precursor; N, CREBH(N). Calnexin and lamin B levels were measured in ER and nuclear fractions, respectively. E: Quantification of Western blots of CREBH(F) to calnexin, and CREBH(N) to lamin B. *P < 0.05.
We next asked whether CREBH transcription or processing is activated by Western or ketogenic diets. As we demonstrated previously (9), both CREBH mRNA and processed CREBH(N) protein were increased in fasted mice (Fig. 5C–E). Similarly, Western or ketogenic diet-fed mice exhibited increased CREBH(N) protein levels, although the differences did not reach statistical significance (Fig. 5D, E). The abundance of CREBH mRNA and the precursor form of the protein were not increased by Western or ketogenic diets (Fig. 5C, E). Curiously, modest induction of CREBH mRNA and protein by fasting or ketogenic diet administration was underwhelming compared with the robust apoA-IV induction. Furthermore, the apoA-IV mRNA level was only modestly increased by Western diet compared with fasting or ketogenic diet, despite comparable CREBH(N) levels between these groups. Collectively, these data suggest that hepatic apoA-IV mRNA expression is determined not only by CREBH(N) abundance, but also by other mechanisms involving CREBH.
CREBH processing is not increased by ER stress
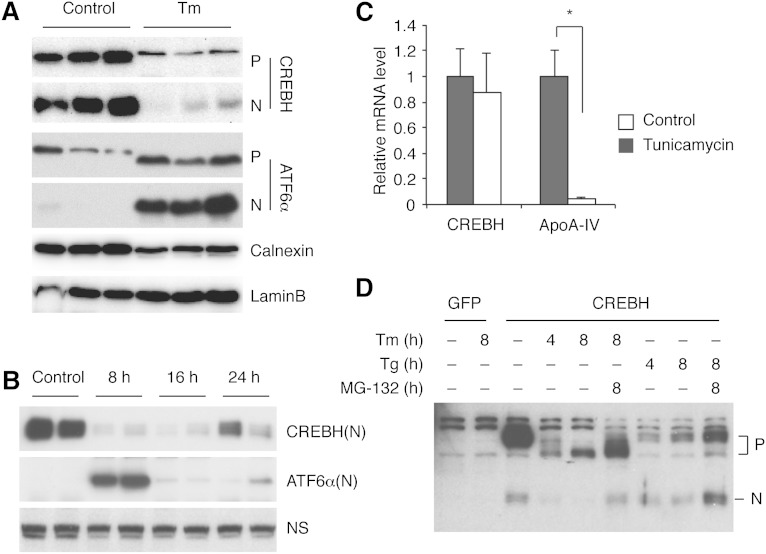
CREBH was initially suggested to be activated by ER stress in a manner analogous to ATF6α activation (5). However, other independent studies failed to demonstrate the proteolytic activation of CREBH by ER stress, raising the concern of whether or not CREBH is involved in ER stress response (10, 11). Because the CREBH protein is not produced in most hepatoma cell lines at a measurable level, these studies examined the effect of ER stress-inducing agents on overexpressed CREBH. To evaluate the effect of ER stress on the endogenous CREBH in vivo, we injected mice with tunicamycin and examined the abundance of CREBH protein in the ER and nucleus by Western blotting analysis. ATF6α was used as a control, which was minimally processed in untreated liver, but strongly activated by tunicamycin treatment (Fig. 6A). CREBH(N) was readily detected in nuclear extracts from untreated mouse liver, indicating that a substantial amount of CREBH protein is basally processed in normal mouse liver (Fig. 6A). Tunicamycin treatment dramatically reduced both membrane bound precursor CREBH(F) and the nuclear CREBH(N). CREBH(N) levels remained low during the prolonged treatment with tunicamycin for up to 24 h (Fig. 6B). Consistently, the hepatic apoA-IV mRNA level was decreased by tunicamycin injection (Fig. 6C). Tunicamycin had no effect on CREBH mRNA levels (Fig. 6C). These results are in agreement with a recent report that CREBH protein is N-glycosylated, which is required for optimal proteolytic activation of CREBH (10).
Fig. 6.
CREBH processing is not increased by ER stress in mouse liver. A: Mice were injected with tunicamycin (1.5 mg/kg) or 150 mM dextrose as control intraperitoneally, and euthanized 6 h later. Liver microsomal and nuclear fractions were subjected to Western blotting. Calnexin and lamin B were used as loading controls for ER and nuclear proteins, respectively. P, full-length CREBH precursor; N, CREBH(N). B: Mice were injected with tunicamycin and euthanized at indicated time points. CREBH(N) proteins in the liver were revealed by Western blot. ATF6α (N) levels were also measured as a control. NS, nonspecific band. C: Mice were injected with tunicamycin (1.5 mg/kg) intraperitoneally, deprived of food, and euthanized 16 h later. Hepatic CREBH and apoA-IV mRNA levels were measured by quantitative RT-PCR. D: Hepa 1-6 cells stably expressing GFP or full-length CREBH were treated with tunicamycin (1 μg/ml), thapsigargin (1 μM), and/or 10 μM MG-132. CREBH protein level was measured by Western blot.
To further investigate the effects of ER stress on CREBH activation, we stably overexpressed the full-length CREBH in Hepa1-6 cells. CREBH was basally processed to CREBH(N), which was suppressed by tunicamycin treatment (Fig. 6D). Importantly, neither tunicamycin nor thapsigargin (an ER Ca2+-ATPase inhibitor) increased CREBH processing, arguing against the notion that CREBH is activated by ER stress. MG-132 substantially increased both the CREBH precursor and the nuclear form, suggesting that they are degraded by the proteasome.
DISCUSSION
CREBH is expressed in the liver and small intestine and regulates a group of genes that are involved in TG and lipoprotein metabolism (9). Apoa4 is one of the genes that is downregulated in CREBH-deficient mice both in the liver and small intestine (9). Hepatic apoA-IV expression was highly increased in steatosis caused by high-fat diets or fasting, which was abrogated by the loss of CREBH. We found that apoA-IV mRNA was dramatically induced by the overexpression of constitutively active CREBH(N) in mouse liver and cultured hepatocytes, identifying CREBH as the major transcription factor regulating Apoa4 gene expression. We also identified two cis-acting elements on the Apoa4 gene promoter that are required for activation by CREBH. The CREBH binding sites (5′-CCACGTTG-3′) were distinct from CRE (5′-TGACGTCA-3′) or UPRE sequences (TGACGTGG-3′). It would be of interest to test whether other CREBH target genes also possess CREBH binding sites similar to those present on the Apoa4 promoter. Genome-wide ChIP followed by high-throughput sequencing would be useful for identifying the common CREBH binding site on its target genes.
Hepatic apoA-IV expression is strongly induced in mouse models of steatosis, implicating a potential role of apoA-IV in TG homeostasis (16, 27, 28). Although apoA-IV is mainly expressed in the small intestine of humans (29, 30), recent genome-wide expression profiling studies revealed that hepatic apoA-IV expression is increased in steatosis, as well as in alcoholic and nonalcoholic steatohepatitis (supplementary Fig. I, GEO database, accession numbers GSE33814, GSE48452, GSE37031, and GSE28619) (31–34). ApoA-IV is a 46 kDa glycoprotein (43 kDa for mouse) that has been implicated in multiple metabolic functions (14, 35). It can be transferred from chylomicrons and VLDLs to HDLs in exchange for apoCs and thereby facilitates lipolysis of TG-rich lipoproteins by LPL (36, 37). ApoA-IV also appears to play a role in reverse cholesterol transport and protection from atherosclerosis (38). ApoA-IV has been implicated in fat absorption in the small intestine (39–41), the central regulation of food intake (42), and the regulation of insulin secretion from β-cells (43). A recent study demonstrated that TG accumulation induces apoA-IV expression in mouse liver, which in turn promotes TG secretion to reduce hepatic TG burden (27). Given the critical role of CREBH in Apoa4 gene expression, it will be interesting to investigate the role of CREBH in these metabolic functions ascribed to apoA-IV.
Fasting increases CREBH mRNA as well as nuclear CREBH(N) protein in the liver (7). Glucocorticoid receptor and PPARα have been implicated in the transcriptional activation of CREBH in response to fasting (7, 8). In contrast, little is known about the regulation of proteolytic activation of CREBH. ER transmembrane transcription factors including CREBH have to be transported to the Golgi apparatus for proteolytic activation (3). Thus, Golgi transport is a critical regulatory step in the activation of ER transmembrane transcription factors. For example, ATF6α and the SREBP-SREBP cleavage-activating protein (SCAP) complex are retained in the ER by the association with BiP and Insig, respectively (44, 45). ER stress and sterol depletion dissociate ATF6α and the SREBP-SCAP complex from the ER resident proteins, triggering the Golgi transport and the subsequent proteolytic activation. CREBH was relatively highly processed in normal mouse liver, which contained a substantial amount of the nuclear CREBH(N). The abundance of CREBH(N) was further increased in hepatic steatosis induced by fasting or various diets. The molecular mechanism for the Golgi transport of CREBH remains to be determined.
Although CREBH was essential for hepatic apoA-IV expression in various mouse models of steatosis, the abundance of CREBH(N) did not precisely correlate with apoA-IV levels. For example, the CREBH(N) level was increased by no more than 2-fold in ketogenic diet-fed mice compared with the chow diet group, contrasting to >300-fold induction of apoA-IV mRNA. Western diet increased apoA-IV mRNA only modestly (∼5-fold), despite the CREBH(N) level being comparable between the Western and ketogenic diet groups. We speculated that CREBH activity might also be regulated by TG accumulation. It is possible that CREBH undergoes posttranslational modifications or interacts with transcriptional coactivators in response to TG accumulation, increasing the transcriptional activity of CREBH. In this regard, a recent study demonstrated that CREB3, another transmembrane bZIP transcription factor closely related to CREBH, activates apoA-IV transcription upon overexpression (46). It would be interesting to investigate whether CREB3 also plays a role in apoA-IV expression in the liver and if so, whether CREB3 and CREBH physically and functionally interact with each other to cooperatively regulate Apoa4 gene expression.
CREBH is structurally similar to ATF6α that is activated by ER stress and can activate the UPRE-luciferase reporter, suggesting that CREBH might be involved in UPR gene induction (3, 5). UPRE was originally described as a consensus DNA binding sequence for ATF6α, which was selected from random oligonucleotide pools by affinity maturation using recombinant ATF6α protein (47). UPRE is identical to the XBP1 consensus sequence that was also identified by in vitro affinity maturation (48). Accordingly, the UPRE reporter is strongly activated under ER stress conditions by XBP1 and ATF6α (18, 49) and therefore has been used as an indicator of UPR activation. However, despite the strong induction of UPRE reporter activity by CREBH(N) cotranscfection, CREBH(N) overexpression had little effect on the expression of canonical UPR genes such as Hspa5 (BiP), Ddit3 (CHOP), and Dnajb9 (ERdj4), suggesting that the function of CREBH is distinct from ER stress response (data not shown).
It has been shown that CREBH is activated by ER stress and regulates the expression of hepcidin and proinflammatory and acute phase response genes, linking ER stress to inflammation and iron metabolism (6, 17, 50). However, subsequent studies by other investigators failed to demonstrate the activation of CREBH processing by ER stress (10, 11). One caveat of these studies is that CREBH processing was examined after overexpression in cultured cells. Here, we confirmed that CREBH processing is not increased by chemical ER stress-inducers both in vitro and in vivo. Tunicamycin rather decreased both precursor and mature forms of CREBH, likely by inducing the degradation of unglycosylated CREBH protein. Given that the role of CREBH in the inflammatory and the acute phase responses has been explored in the setting of ER stress, our findings warrant the reevaluation of the mechanisms by which CREBH controls these host responses.
Supplementary Material
Acknowledgments
The authors thank Michael Park for critical reading of the manuscript, B. Spiegelman for Flag-PGC1α, and S. Duncan for Flag-HNF4α plasmids.
Footnotes
Abbreviations:
- ChIP
- chromatin immunoprecipitation
- CRE
- cAMP response element
- CREBH
- cAMP responsive element-binding protein H
- CREBH(N)
- N-terminal fragment of cAMP responsive element-binding protein H
- EMSA
- electrophoretic mobility shift assay
- ER
- endoplasmic reticulum
- HA
- hemagglutinin
- HNF-4α
- hepatic nuclear factor 4α PGC1α, PPARγ coactivator-1α
This study was supported by National Institutes of Health Grant R01DK089211.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one figure.
REFERENCES
- 1.Chin K. T., Zhou H. J., Wong C. M., Lee J. M., Chan C. P., Qiang B. Q., Yuan J. G., Ng I. O., Jin D. Y. 2005. The liver-enriched transcription factor CREB-H is a growth suppressor protein underexpressed in hepatocellular carcinoma. Nucleic Acids Res. 33: 1859–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Omori Y., Imai J., Watanabe M., Komatsu T., Suzuki Y., Kataoka K., Watanabe S., Tanigami A., Sugano S. 2001. CREB-H: a novel mammalian transcription factor belonging to the CREB/ATF family and functioning via the box-B element with a liver-specific expression. Nucleic Acids Res. 29: 2154–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asada R., Kanemoto S., Kondo S., Saito A., Imaizumi K. 2011. The signalling from endoplasmic reticulum-resident bZIP transcription factors involved in diverse cellular physiology. J. Biochem. 149: 507–518. [DOI] [PubMed] [Google Scholar]
- 4.Lee A. H. 2012. The role of CREB-H transcription factor in triglyceride metabolism. Curr. Opin. Lipidol. 23: 141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang K., Shen X., Wu J., Sakaki K., Saunders T., Rutkowski D. T., Back S. H., Kaufman R. J. 2006. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 124: 587–599. [DOI] [PubMed] [Google Scholar]
- 6.Vecchi C., Montosi G., Zhang K., Lamberti I., Duncan S. A., Kaufman R. J., Pietrangelo A. 2009. ER stress controls iron metabolism through induction of hepcidin. Science. 325: 877–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danno H., Ishii K. A., Nakagawa Y., Mikami M., Yamamoto T., Yabe S., Furusawa M., Kumadaki S., Watanabe K., Shimizu H., et al. 2010. The liver-enriched transcription factor CREBH is nutritionally regulated and activated by fatty acids and PPARalpha. Biochem. Biophys. Res. Commun. 391: 1222–1227. [DOI] [PubMed] [Google Scholar]
- 8.Lee M. W., Chanda D., Yang J., Oh H., Kim S. S., Yoon Y. S., Hong S., Park K. G., Lee I. K., Choi C. S., et al. 2010. Regulation of hepatic gluconeogenesis by an ER-bound transcription factor, CREBH. Cell Metab. 11: 331–339. [DOI] [PubMed] [Google Scholar]
- 9.Lee J. H., Giannikopoulos P., Duncan S. A., Wang J., Johansen C. T., Brown J. D., Plutzky J., Hegele R. A., Glimcher L. H., Lee A. H. 2011. The transcription factor cyclic AMP-responsive element-binding protein H regulates triglyceride metabolism. Nat. Med. 17: 812–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan C. P., Mak T. Y., Chin K. T., Ng I. O., Jin D. Y. 2010. N-linked glycosylation is required for optimal proteolytic activation of membrane-bound transcription factor CREB-H. J. Cell Sci. 123: 1438–1448. [DOI] [PubMed] [Google Scholar]
- 11.Bailey D., Barreca C., O'Hare P. 2007. Trafficking of the bZIP transmembrane transcription factor CREB-H into alternate pathways of ERAD and stress-regulated intramembrane proteolysis. Traffic. 8: 1796–1814. [DOI] [PubMed] [Google Scholar]
- 12.Gentile C. L., Wang D., Pfaffenbach K. T., Cox R., Wei Y., Pagliassotti M. J. 2010. Fatty acids regulate CREBh via transcriptional mechanisms that are dependent on proteasome activity and insulin. Mol. Cell. Biochem. 344: 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stan S., Delvin E., Lambert M., Seidman E., Levy E. 2003. Apo A-IV: an update on regulation and physiologic functions. Biochim. Biophys. Acta. 1631: 177–187. [DOI] [PubMed] [Google Scholar]
- 14.Tso P., Liu M. 2004. Apolipoprotein A-IV, food intake, and obesity. Physiol. Behav. 83: 631–643. [DOI] [PubMed] [Google Scholar]
- 15.Lagrost L., Gambert P., Boquillon M., Lallemant C. 1989. Evidence for high density lipoproteins as the major apolipoprotein A-IV-containing fraction in normal human serum. J. Lipid Res. 30: 1525–1534. [PubMed] [Google Scholar]
- 16.Hanniman E. A., Lambert G., Inoue Y., Gonzalez F. J., Sinal C. J. 2006. Apolipoprotein A-IV is regulated by nutritional and metabolic stress: involvement of glucocorticoids, HNF-4 alpha, and PGC-1 alpha. J. Lipid Res. 47: 2503–2514. [DOI] [PubMed] [Google Scholar]
- 17.Luebke-Wheeler J., Zhang K., Battle M., Si-Tayeb K., Garrison W., Chhinder S., Li J., Kaufman R. J., Duncan S. A. 2008. Hepatocyte nuclear factor 4alpha is implicated in endoplasmic reticulum stress-induced acute phase response by regulating expression of cyclic adenosine monophosphate responsive element binding protein H. Hepatology. 48: 1242–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee A. H., Iwakoshi N. N., Glimcher L. H. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell. Biol. 23: 7448–7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen A. T., Braschi S., Geoffrion M., Fong L. G., Crooke R. M., Graham M. J., Young S. G., Milne R. 2006. A mouse monoclonal antibody specific for mouse apoB48 and apoB100 produced by immunizing “apoB39-only” mice with mouse apoB48. Biochim. Biophys. Acta. 1761: 182–185. [DOI] [PubMed] [Google Scholar]
- 20.Hur K. Y., So J. S., Ruda V., Frank-Kamenetsky M., Fitzgerald K., Koteliansky V., Iwawaki T., Glimcher L. H., Lee A. H. 2012. IRE1alpha activation protects mice against acetaminophen-induced hepatotoxicity. J. Exp. Med. 209: 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson J. D., Denisenko O., Bomsztyk K. 2006. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 1: 179–185. [DOI] [PubMed] [Google Scholar]
- 22.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 23.Carrier J. C., Deblois G., Champigny C., Levy E., Giguere V. 2004. Estrogen-related receptor alpha (ERRalpha) is a transcriptional regulator of apolipoprotein A-IV and controls lipid handling in the intestine. J. Biol. Chem. 279: 52052–52058. [DOI] [PubMed] [Google Scholar]
- 24.Carrière V., Vidal R., Lazou K., Lacasa M., Delers F., Ribeiro A., Rousset M., Chambaz J., Lacorte J. M. 2005. HNF-4-dependent induction of apolipoprotein A-IV gene transcription by an apical supply of lipid micelles in intestinal cells. J. Biol. Chem. 280: 5406–5413. [DOI] [PubMed] [Google Scholar]
- 25.Rhee J., Ge H., Yang W., Fan M., Handschin C., Cooper M., Lin J., Li C., Spiegelman B. M. 2006. Partnership of PGC-1alpha and HNF4alpha in the regulation of lipoprotein metabolism. J. Biol. Chem. 281: 14683–14690. [DOI] [PubMed] [Google Scholar]
- 26.Sauvaget D., Chauffeton V., Citadelle D., Chatelet F. P., Cywiner-Golenzer C., Chambaz J., Pincon-Raymond M., Cardot P., Le Beyec J., Ribeiro A. 2002. Restriction of apolipoprotein A-IV gene expression to the intestine villus depends on a hormone-responsive element and parallels differential expression of the hepatic nuclear factor 4alpha and gamma isoforms. J. Biol. Chem. 277: 34540–34548. [DOI] [PubMed] [Google Scholar]
- 27.VerHague M. A., Cheng D., Weinberg R. B., Shelness G. S. 2013. Apolipoprotein A-IV expression in mouse liver enhances triglyceride secretion and reduces hepatic lipid content by promoting very low density lipoprotein particle expansion. Arterioscler. Thromb. Vasc. Biol. 33: 2501–2508. [DOI] [PubMed] [Google Scholar]
- 28.Sanderson L. M., Boekschoten M. V., Desvergne B., Muller M., Kersten S. 2010. Transcriptional profiling reveals divergent roles of PPARalpha and PPARbeta/delta in regulation of gene expression in mouse liver. Physiol. Genomics. 41: 42–52. [DOI] [PubMed] [Google Scholar]
- 29.Elshourbagy N. A., Walker D. W., Boguski M. S., Gordon J. I., Taylor J. M. 1986. The nucleotide and derived amino acid sequence of human apolipoprotein A-IV mRNA and the close linkage of its gene to the genes of apolipoproteins A-I and C–III. J. Biol. Chem. 261: 1998–2002. [PubMed] [Google Scholar]
- 30.Karathanasis S. K., Yunis I., Zannis V. I. 1986. Structure, evolution, and tissue-specific synthesis of human apolipoprotein AIV. Biochemistry. 25: 3962–3970. [DOI] [PubMed] [Google Scholar]
- 31.López-Vicario C., González-Périz A., Rius B., Morán-Salvador E., Garcia-Alonso V., Lozano J. J., Bataller R., Cofán M., Kang J. X., Arroyo V., et al. 2014. Molecular interplay between Δ5/Δ6 desaturases and long-chain fatty acids in the pathogenesis of non-alcoholic steatohepatitis. Gut. 63: 344–355. [DOI] [PubMed] [Google Scholar]
- 32.Ahrens M., Ammerpohl O., von Schonfels W., Kolarova J., Bens S., Itzel T., Teufel A., Herrmann A., Brosch M., Hinrichsen H., et al. 2013. DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 18: 296–302. [DOI] [PubMed] [Google Scholar]
- 33.Starmann J., Falth M., Spindelbock W., Lanz K. L., Lackner C., Zatloukal K., Trauner M., Sultmann H. 2012. Gene expression profiling unravels cancer-related hepatic molecular signatures in steatohepatitis but not in steatosis. PLoS ONE. 7: e46584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Affò S., Dominguez M., Lozano J. J., Sancho-Bru P., Rodrigo-Torres D., Morales-Ibanez O., Moreno M., Millán C., Loaeza-del-Castillo A., Altamirano J., et al. 2013. Transcriptome analysis identifies TNF superfamily receptors as potential therapeutic targets in alcoholic hepatitis. Gut. 62: 452–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sato W., Horie Y., Kataoka E., Ohshima S., Dohmen T., Iizuka M., Sasaki J., Sasaki T., Hamada K., Kishimoto H., et al. 2006. Hepatic gene expression in hepatocyte-specific Pten deficient mice showing steatohepatitis without ethanol challenge. Hepatol. Res. 34: 256–265. [DOI] [PubMed] [Google Scholar]
- 36.Weinberg R. B., Spector M. S. 1985. Human apolipoprotein A-IV: displacement from the surface of triglyceride-rich particles by HDL2-associated C-apoproteins. J. Lipid Res. 26: 26–37. [PubMed] [Google Scholar]
- 37.Goldberg I. J., Scheraldi C. A., Yacoub L. K., Saxena U., Bisgaier C. L. 1990. Lipoprotein ApoC-II activation of lipoprotein lipase. Modulation by apolipoprotein A-IV. J. Biol. Chem. 265: 4266–4272. [PubMed] [Google Scholar]
- 38.Duverger N., Tremp G., Caillaud J. M., Emmanuel F., Castro G., Fruchart J. C., Steinmetz A., Denefle P. 1996. Protection against atherogenesis in mice mediated by human apolipoprotein A-IV. Science. 273: 966–968. [DOI] [PubMed] [Google Scholar]
- 39.Lu S., Yao Y., Meng S., Cheng X., Black D. D. 2002. Overexpression of apolipoprotein A-IV enhances lipid transport in newborn swine intestinal epithelial cells. J. Biol. Chem. 277: 31929–31937. [DOI] [PubMed] [Google Scholar]
- 40.Yao Y., Lu S., Huang Y., Beeman-Black C. C., Lu R., Pan X., Hussain M. M., Black D. D. 2011. Regulation of microsomal triglyceride transfer protein by apolipoprotein A-IV in newborn swine intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 300: G357–G363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan X., Munshi M. K., Iqbal J., Queiroz J., Sirwi A. A., Shah S., Younus A., Hussain M. M. 2013. Circadian regulation of intestinal lipid absorption by apolipoprotein AIV involves forkhead transcription factors A2 and O1 and microsomal triglyceride transfer protein. J. Biol. Chem. 288: 20464–20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujimoto K., Fukagawa K., Sakata T., Tso P. 1993. Suppression of food intake by apolipoprotein A-IV is mediated through the central nervous system in rats. J. Clin. Invest. 91: 1830–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang F., Kohan A. B., Kindel T. L., Corbin K. L., Nunemaker C. S., Obici S., Woods S. C., Davidson W. S., Tso P. 2012. Apolipoprotein A-IV improves glucose homeostasis by enhancing insulin secretion. Proc. Natl. Acad. Sci. USA. 109: 9641–9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen J., Chen X., Hendershot L., Prywes R. 2002. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell. 3: 99–111. [DOI] [PubMed] [Google Scholar]
- 45.Ye J., DeBose-Boyd R. A. 2011. Regulation of cholesterol and fatty acid synthesis. Cold Spring Harb. Perspect. Biol. 3: a004754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanecka A., Ansems M., van Hout-Kuijer M. A., Looman M. W., Prosser A. C., Welten S., Gilissen C., Sama I. E., Huynen M. A., Veltman J. A., et al. 2012. Analysis of genes regulated by the transcription factor LUMAN identifies ApoA4 as a target gene in dendritic cells. Mol. Immunol. 50: 66–73. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y., Shen J., Arenzana N., Tirasophon W., Kaufman R. J., Prywes R. 2000. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J. Biol. Chem. 275: 27013–27020. [DOI] [PubMed] [Google Scholar]
- 48.Clauss I. M., Chu M., Zhao J. L., Glimcher L. H. 1996. The basic domain/leucine zipper protein hXBP-1 preferentially binds to and transactivates CRE-like sequences containing an ACGT core. Nucleic Acids Res. 24: 1855–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamamoto K., Sato T., Matsui T., Sato M., Okada T., Yoshida H., Harada A., Mori K. 2007. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev. Cell. 13: 365–376. [DOI] [PubMed] [Google Scholar]
- 50.Shin D. Y., Chung J., Joe Y., Pae H. O., Chang K. C., Cho G. J., Ryter S. W., Chung H. T. 2012. Pretreatment with CO-releasing molecules suppresses hepcidin expression during inflammation and endoplasmic reticulum stress through inhibition of the STAT3 and CREBH pathways. Blood. 119: 2523–2532. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.