Abstract
PPARs regulate the expression of genes for energy metabolism in a ligand-dependent manner. PPARs can influence fatty acid oxidation, the level of circulating triglycerides, glucose uptake and insulin sensitivity. Here, we demonstrate that 5-hydroxyeicosapentaenoic acid (HEPE), 8-HEPE, 9-HEPE, 12-HEPE and 18-HEPE (hydroxylation products of EPA) obtained from methanol extracts of Pacific krill (Euphausia pacifica) can act as PPAR ligands. Two of these products, 8-HEPE and 9-HEPE, enhanced the transcription levels of GAL4-PPARs to a significantly greater extent than 5-HEPE, 12-HEPE, 18-HEPE, EPA, and EPA ethyl-ester. 8-HEPE also activated significantly higher transcription of GAL4-PPARα, GAL4-PPARγ, and GAL4-PPARδ than EPA at concentrations greater than 4, 64, and 64 μM, respectively. We also demonstrated that 8-HEPE increased the expression levels of genes regulated by PPARs in FaO, 3T3-F442A, and C2C12 cells. Furthermore, 8-HEPE enhanced adipogenesis and glucose uptake. By contrast, at the same concentrations, EPA showed weak or little effect, indicating that 8-HEPE was the more potent inducer of physiological effects.
Keywords: peroxisome proliferator-acitivated receptor, fatty acid oxidation, adipogenesis, glucose uptake
PPARs are members of a nuclear receptor superfamily that play critical roles in the regulation of storage and catabolism of lipids (1). They contribute to these regulation processes by activating gene expression in a ligand-dependent manner that involves recognition of and binding to peroxisome proliferator response elements composed of TGACCT-related direct repeats separated by one nucleotide (2, 3). PPARs form heterodimers with peroxisome proliferator response elements via the retinoid-X receptor, which is a receptor for 9-cis-retinoic acid (4, 5). Three types of PPAR have been identified, namely α, γ, and δ. PPARα is expressed at high levels in the liver where it promotes fatty acid oxidation, ketogenesis, lipid transport, and gluconeogenesis (6, 7). PPARα responds to the concentration of fatty acids in the liver and enhances fatty acid breakdown by upregulating genes encoding β-oxidation enzymes (8–10). PPARγ is highly expressed in adipose tissue, where it serves as an essential regulator for adipocyte differentiation. PPARγ promotes lipid storage in mature adipocytes by increasing the expression of several key genes (11, 12). PPARδ (also known as PPARβ) is widely expressed, but with relatively higher levels in the brain, colon, and skin (13–15); PPARδ is the predominant PPAR isoform in skeletal muscle. Studies using transgenic mice showed that targeted expression of activated PPARδ increases the numbers of oxidative type 1 muscle fibers, which enhance whole-body insulin sensitivity and exercise endurance capacity (16).
The identification of unsaturated fatty acids as PPAR ligands provided firm evidence that the direct interaction of nuclear receptors with these fatty acids is required for some PPAR-dependent transcription activity (4, 8, 17–19). Unsaturated fatty acids can bind to all three types of PPAR, with PPARα exhibiting the highest affinity for concentrations equivalent to circulating blood levels (8, 20). In contrast, the long-chain fatty acid, erucic acid (C22:1), is a weak ligand that appears to have more affinity for PPARδ (21). Overall, saturated fatty acids are poor PPAR ligands compared with unsaturated fatty acids (4, 8, 19). Eicosanoids, a class of fatty acids, are mainly derived from arachidonic acid (AA) either via the lipoxygenase pathway, leading to the formation of leukotrienes and hydroxyeicosatetraenoic acids (HETEs), or via the cyclooxygenase pathway that produces prostaglandins and thromboxanes. They can act as activators for different PPARs; for example, the prostaglandin D2 derivative, 15-deoxy-Δ12,14-PGJ2, is a ligand for PPARγ (17, 22); 8-HETE, a compound associated with phorbol ester-induced inflammation, is a ligand for PPARα (8, 19); and leukotriene B4, a chemotactic inflammation mediator, binds to PPARα (19, 23).
Increasing levels of triglycerides are associated with obesity and are indicators of progressive development of insulin resistance, hypertension, and hyperlipidemia (24, 25). Thus, normalization of triglyceride levels has been investigated as a possible means of prevention for these conditions. It is well-known that dietary PUFAs can ameliorate some of the deleterious effects of these disorders (26, 27). This property may reflect both activation of PPAR-dependent β- and ω-oxidation pathways (28), as well as PUFA-dependent suppression of lipogenic and glycolytic enzymes (29). Krill are a source of PUFAs, as their oil is rich in EPA and docosahexaenoic acid. In a similar manner to fish oil, krill oil has been shown to have anti-inflammatory effects and to lower the levels of triglycerides and cholesterol in the plasma (30, 31). Although lipids are major bioactive compounds in krill oil, we found in a previous study that a water-soluble extract from Pacific krill could suppress weight gain in mice caused by a high fat diet. The levels of triglycerides in the livers of mice given a water-soluble extract from Pacific krill decreased (32).
Here, we investigated krill extracts and found that hydroxyeicosapentaenoic acids (HEPEs) (5-, 8-, 9-, 12-, and 18-HEPE) displayed PPAR ligand activities. Interestingly, the PPAR ligand activities of 8-HEPE and 9-HEPE were significantly greater than EPA. These results indicate that hydroxylation of EPA at the C-8 or C-9 position increased ligand activity for PPARs.
MATERIAL AND METHODS
Reagents
HEPEs (purity >98%) and EPA (purity >98%) were purchased from Cayman Chemical (Ann Arbor, MI). EPA ethyl-ester (EPA-Et) (purity >99%), the PPAR agonists GW7647 and GW1929, and the PPARα antagonist MK-886 were purchased from Wako Pure Chemical Industries (Osaka, Japan). 2-Deoxy-d-glucose, the PPARγ antagonist GSK0660, and the PPARδ antagonist T0070907 were purchased from Sigma-Aldrich (St. Louis, MO). The PPAR agonist GW501516 was purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Extraction and isolation from Pacific krill
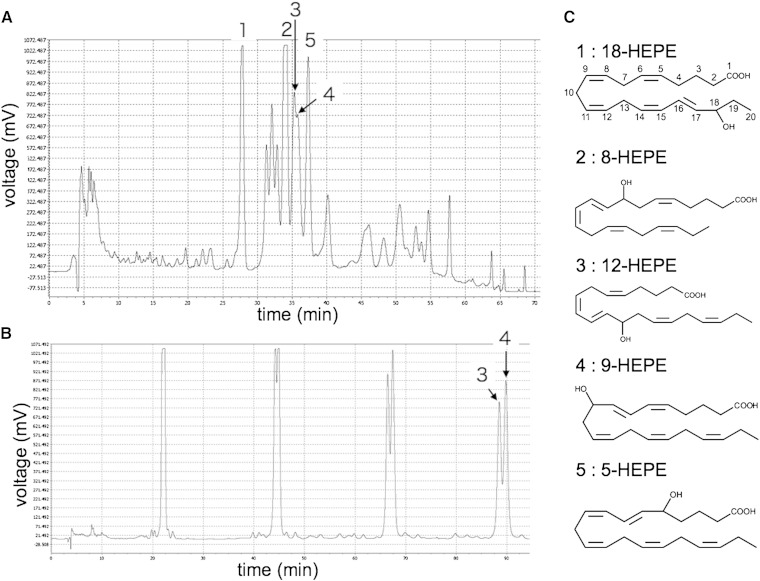
We used dried Pacific krill purchased from Kawashu (Iwate, Japan). The krill were powdered and then extracted with methanol under reflux for 4 h. The methanol extract was diluted with distilled water (1:4 by volume) and subjected to column chromatography using Diaion HP-20 (Mitsubishi Chemical, Tokyo, Japan). A fraction was eluted with 80% methanol from the HP-20. This fraction was diluted with distilled water (1:3 by volume) and applied to an InertSep C18 (GL Science Inc., Tokyo, Japan). The fraction was eluted with 100% methanol. HEPEs were separated on an InertSustain ODS-3 column (20.0 mm diameter × 250 mm; GL Science Inc.) with isocratic elution of acetonitrile/water/formic acid (55/45/0.1) at a flow rate of 15 ml/min and detection at 235 nm (Fig. 2A). The fraction that contained compounds 3 and 4 was subjected to recycle-HPLC, and peaks 3 and 4 were separated on an InertSustain ODS-3 column (20.0 mm diameter × 250 mm; GL Science Inc.) with isocratic elution of acetonitrile/water/formic acid (60/40/0.1) at a flow rate of 15 ml/min and detection at 235 nm (Fig. 2B).
Fig. 2.
HPLC chromatograms. A: Extract purified using HP-20 and InertSep C18 (20 mg). Five compounds that activate GAL4-PPARα transcription are present. B: Chromatogram from the recycle-HPLC separation of compounds 3 and 4. LC-TOFMS identified the following compounds: 1, 18-HEPE; 2, 8-HEPE; 3, 12-HEPE; 4, 9-HEPE; and 5, 5-HEPE. C: The structures of the different HEPEs. The numbers on 18-HEPE indicate carbon positions.
Identification of HEPEs
The compounds purified from Pacific krill were identified as 5-, 8-, 9-, 12-, and 18-HEPE by LC-TOFMS (Agilent Technologies, Palo Alto, CA) analysis. The HEPEs were separated on an InertSustain ODS-3 column (2.0 mm diameter × 250 mm; GL Science Inc.) with isocratic elution of acetonitrile/water/formic acid (40/60/0.5) at a flow rate of 0.3 ml/min. Physicochemical properties, such as monoisotopic mass, molecular formula, UV spectrum, and retention time in the HPLC, were identical with reference standard compounds (supplementary Tables II, III; supplementary Fig. III).
Cell culture
3T3-F442A (DS Pharma Biomedical Co., Ltd., Osaka, Japan), NIH-3T3, and C2C12 cells (RIKEN BioResource Center, Ibaraki, Japan) were cultured in DMEM containing 10% FBS (Invitrogen, Carlsbad, CA) and antibiotic antimycotic solution (Sigma-Aldrich). FaO cells (DS Pharma Biomedical Co.) were cultured in Ham's F12 containing 10% FBS and antibiotic antimycotic solution. Myogenesis was induced in C2C12 cells using differentiation medium (DMEM with 2% horse serum).
Luciferase reporter assay
The ligand-dependent transcriptional activities of PPARs were assayed using luciferase reporter systems. The ligand binding domains of mouse PPARs (PPARα, amino acids 201–468; PPARγ, amino acids 238–506; PPARδ, amino acids 171–441) were cloned into the pFN26A (BIND) hRluc-neo Flexi Vector (Promega, Tokyo, Japan). NIH-3T3 cells were seeded in 24-well culture plates (1 × 105 cells/well) and cultured overnight. pFN26A vector carrying an inserted PPAR ligand binding domain and the pGL4.35 vector (Promega) were cotransfected using Lipofectamine 2000 (Invitrogen) into NIH-3T3 cells; the transfected cells were cultured for 36 h. The cells were cultured with HEPE, EPA, EPA-Et, GW7647, GW1929, or GW501516 for 24 h and then harvested using passive lysis buffer (Promega). Renilla and firefly luciferase activities were determined using a dual luciferase assay system (Promega).
Quantitative real-time PCR
FaO, C2C12, and 3T3-F442A cells were seeded in 24-well culture plates (1 × 105 cells/well) and cultured overnight. 8-HEPE, EPA, GW7647, GW1929, or GW501516 was then added to the culture for 6 (FaO and C2C12) or 24 h (3T3-F442A). Total RNAs were extracted with an RNeasy kit (QIAGEN, Tokyo, Japan) and used to synthesize cDNAs using a PrimeScript RT reagent kit (Takara, Shiga, Japan); all kits were used according to the manufacturers’ recommendations. Quantitative real-time PCR was performed with the gene-specific primers listed in supplementary Table I and Fast SYBR Green master mix (Applied Biosystems, Foster City, CA).
Triglyceride staining and quantification
3T3-F442A cells were seeded in 24-well culture plates (1 × 105 cells/well) and cultured overnight. 8-HEPE or EPA was then added to the culture for 7 days at which time the cells were stained with Oil Red O. Stained cells were examined using a Nikon microscope (×400). An adipogenesis assay kit (Bio Vision, Mountain View, CA) was used to measure triglyceride content.
2-Deoxyglucose uptake measurement
C2C12 cells were seeded in 24-well culture plates (2 × 104 cells/well) and cultured overnight. The medium was then replaced with DMEM containing 2% horse serum and antibiotic antimycotic solution and the cells were cultured for a further 5 days. Following this treatment to induce myogenesis, they were cultured in DMEM containing 8-HEPE, EPA, or GW501516 for 6 h. A 2-deoxyglucose uptake measurement kit (Cosmo Bio, Tokyo, Japan) was used to quantify glucose uptake into the C2C12 cells.
Statistical analysis
Statistically significant differences between the experimental groups were identified using one-way ANOVA and Tukey's post hoc tests. Data are shown as means ± SD.
RESULTS
Activation of transcription of GAL4-PPARs by Pacific krill extracts
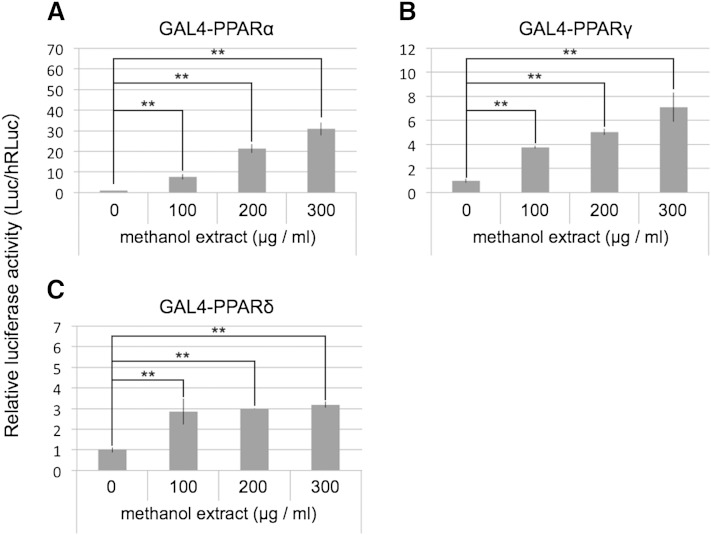
To examine methanol extracts from Pacific krill for possible PPAR ligand activity, we created fusion proteins in which the yeast GAL4 DNA-binding domain was linked to the ligand-binding domain of PPARα, PPARγ, or PPARδ (GAL4-PPARα, GAL4-PPARγ, or GAL4-PPARδ). When a reporter containing a GAL4 upstream activating sequence is utilized, these chimeric receptors allow PPAR activity to be assayed independently of endogenous receptors. We confirmed the specificity of the GAL4-PPARα, GAL4-PPARγ, and GAL4-PPARδ reporter systems using PPAR agonists (supplementary Fig. I). Using this reporter approach, we found that Pacific krill extracts activated transcription of GAL4-PPARα, GAL4-PPARγ, and GAL4-PPARδ in a dose-dependent manner (Fig. 1).
Fig. 1.
GAL4-PPAR assays of a methanol extract from Pacific krill. NIH-3T3 cells were transfected with reporter plasmids and cultured with the extract for 24 h. Then, firefly luciferase (Luc) activities were measured, and the values were normalized against Renilla luciferase (hRLuc) activity. Plotted values represent the mean ± SD from four independent cultures. **P < 0.01.
HEPEs activated transcription of GAL4-PPARs
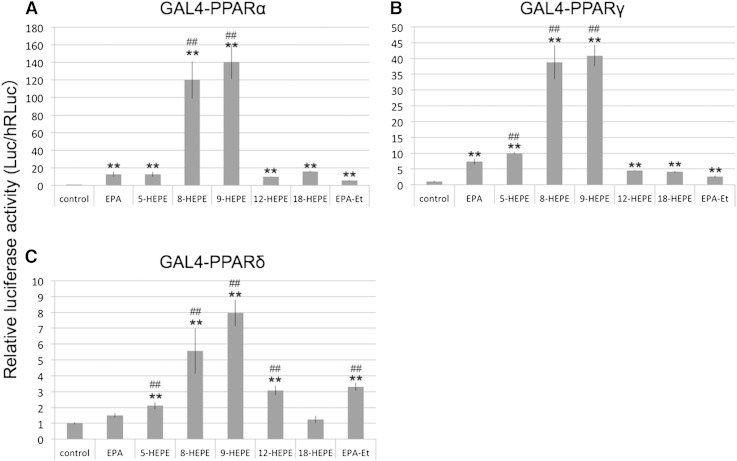
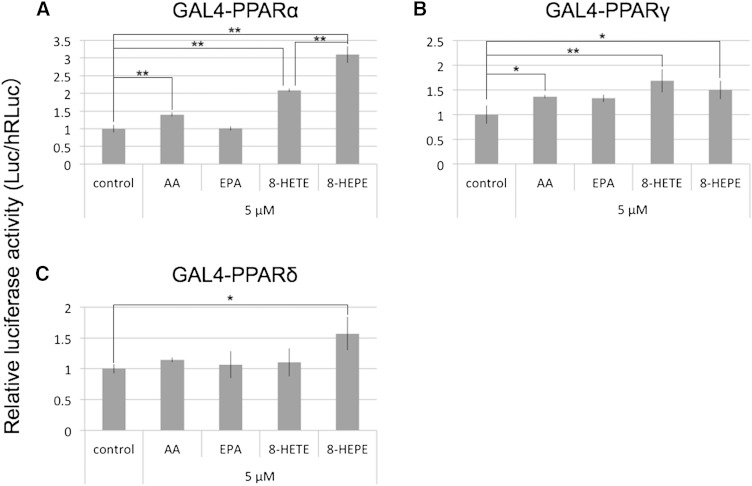
We purified the compounds in the Pacific krill extracts that activated transcription of GAL4-PPARα using an activity-guided fractionation method (Fig. 2; supplementary Fig. II). LC-TOFMS identified the compounds in the active fractions as HEPEs (supplementary Tables II, III; supplementary Fig. III). Transcription of GAL4-PPARα and GAL4-PPARγ was activated by 5-, 8-, 9-, 12-, and 18-HEPE (Fig. 3A, B), while transcription of GAL4-PPARδ was activated by 5-, 8-, 9-, and 12-HEPE (Fig. 3C). Surprisingly, 8- and 9-HEPE showed significantly greater transcriptional activation of GAL4-PPARs than 5-, 12-, 18-HEPE, EPA, and EPA-Et. 8-HEPE is known to be able to activate PPARα (8); however, no reports have previously been made on whether it could activate PPARγ or PPARδ, nor has its activity for PPARs been compared with EPA. We therefore compared the relative PPAR ligand activities of 8-HEPE and EPA at a range of concentrations (4, 16, 64, and 128 μM; Table 1). With regard to GAL4-PPARα, 8-HEPE had a significantly greater effect on transcription activation at all concentrations tested (Table 1). For GAL4-PPARγ and GAL4-PPARδ, 8-HEPE showed a significantly greater effect at the highest tested concentrations (64 and 128 μM; Table 1). Forman, Chen, and Evans (8) showed that 8-HETE also activates PPARα. Here, we compared the relative activities of 8-HEPE and 8-HETE as ligands for PPARs. We found that AA, 8-HETE, and 8-HEPE activated transcription of GAL4-PPARα and GAL4-PPARγ at a concentration of 5 μM (Fig. 4A, B), while 8-HEPE activated transcription of GAL4-PPARδ (Fig. 4C). We also found that 8-HEPE showed a greater effect than 8-HETE on the transcription levels of GAL4-PPARα and GAL4-PPARδ.
Fig. 3.
GAL4-PPAR assays of HEPEs. NIH-3T3 cells were transfected with reporter plasmids and cultured with 128 μM EPA, EPA-Et, or HEPEs for 24 h. Then firefly luciferase (Luc) activities were measured, and the values were normalized against Renilla luciferase (hRLuc) activity. Plotted values represent the mean ± SD from four independent cultures. Significant differences from the control are indicated by **P < 0.01. Significant differences from EPA are indicated by ##P < 0.01.
TABLE 1.
Activities of EPA and 8-HEPE in the induction of GAL4-PPARs transcription
| Compound | GAL4-PPARα | GAL4-PPARγ | GAL4-PPARδ |
| EPA | |||
| 4 μM | 1.20 ± 0.18 | 1.58 ± 0.23 | 1.39 ± 0.39 |
| 16 μM | 2.80 ± 0.92a | 2.23 ± 0.32a | 1.42 ± 0.08a |
| 64 μM | 7.10 ± 1.32a | 4.37 ± 0.47a | 1.43 ± 0.23 |
| 128 μM | 12.65 ± 3.20a | 7.35 ± 0.88a | 1.50 ± 0.15a |
| 8-HEPE | |||
| 4 μM | 7.28 ± 0.87ab | 1.49 ± 0.28 | 0.90 ± 0.09 |
| 16 μM | 31.69 ± 3.50ab | 2.53 ± 0.35a | 1.48 ± 0.11a |
| 64 μM | 95.60 ± 10.13ab | 6.02 ± 0.42ab | 1.90 ± 0.08ab |
| 128 μM | 119.90 ± 21.13ab | 38.78 ± 5.40ab | 5.55 ± 1.43ab |
Values represent the mean ± SD from four independent cultures.
Significant differences from the control are indicated by P < 0.01.
Significant differences from EPA are indicated by P < 0.01.
Fig. 4.
GAL4-PPAR assays of AA, EPA, 8-HETE, and 8-HEPE. NIH-3T3 cells were transfected with reporter plasmids and cultured with 5 μM AA, EPA, 8-HETE, or 8-HEPE for 24 h. Then firefly luciferase (Luc) activities were measured, and the values were normalized against Renilla luciferase (hRLuc) activity. Plotted values represent the mean ± SD from four independent cultures. 8-HETE and 8-HEPE standards were used. *P < 0.05, **P < 0.01.
Here, we demonstrate that HEPEs from Pacific krill can act as PPARα activators (Fig. 2; supplementary Figs. II, III). Moreover, through use of Diaion HP-20 column chromatography to increase concentrations, we found that their activities varied in a concentration-dependent manner (supplementary Fig. IIB). Measurements of HEPEs and EPA indicated that HP-20 chromatography was sufficient to concentrate HEPEs (supplementary Table IV) and that InertSep C18 chromatography removed EPA. Methanol extract of Pacific krill contained 0.72 μg/mg 8-HEPE, 0.24 μg/mg 9-HEPE, and 7.1 μg/mg EPA (supplementary Table IV). A cocktail that contained the same relative amounts of 8-HEPE and 9-HEPE, as in the methanol extract, showed about 60% of the activity of the methanol extract. However, addition of EPA to the cocktail did not increase activity (supplementary Fig. IV). These results indicate that 8-HEPE and 9-HEPE are the major PPARα activators in Pacific krill extracts.
8-HEPE increases expression of genes related to fatty acid oxidation in mitochondrial and peroxisomal pathways
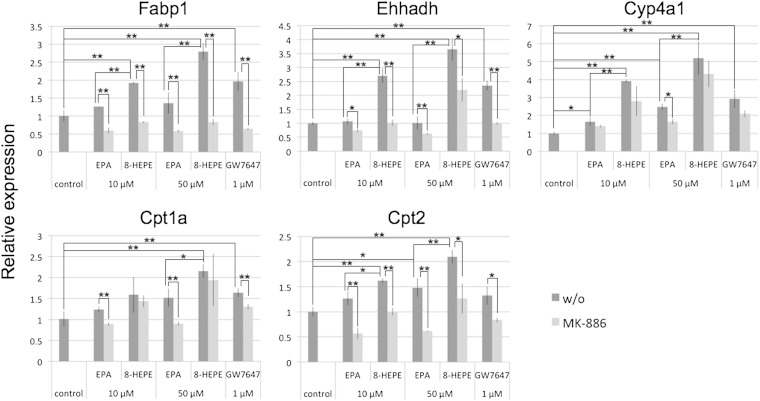
PPARα regulates the expression of genes encoding enzymes and proteins responsible for fatty acid oxidation. To examine the activity of 8-HEPE as a PPARα ligand, we investigated whether 8-HEPE increased the expression of genes regulated by PPARα. Previous studies showed that Wy14,643, an activator of PPARα, upregulated expression of liver fatty acid-binding protein (Fabp1), enoyl-CoA hydratase/3-hydroxyacyl CoA dehydrogenase (Ehhadh), cytochrome P450 CYP4A enzyme (Cyp4a1) and carnitine palmitoyltransferase (Cpt1a, Cpt2) in mouse liver cells (10) and in the rat hepatoma cell line, FaO (33). We cultured FaO cells with EPA, 8-HEPE, or GW7647, a PPARα-specific agonist, for 6 h. Expression of Fabp1, Ehhadh, and Cpt1a was increased by 8-HEPE, but not by EPA; expression of Cyp4a1 and Cpt2 was increased by EPA and 8-HEPE. Overall, 8-HEPE showed a greater effect on gene expression levels than EPA (Fig. 5). MK-886, a PPARα-specific antagonist, inhibited 8-HEPE-induced expression of Fabp1, Ehhadh, and Cpt2 (Fig. 5).
Fig. 5.
Gene expression after 8-HEPE activation of PPARα in FaO cells. The cells were cultured with EPA, 8-HEPE, or GW7647 for 6 h. The PPARα antagonist MK-886 (50 μM) was added to the media at the same time as EPA, 8-HEPE, or GW7647. Gene expression levels were measured by real-time PCR and normalized against expression of Actb. Plotted values represent the mean ± SD from three independent cultures. 8-HEPE purified from Pacific krill was used. *P < 0.05, **P < 0.01. w/o, without.
8-HEPE induces adipogenesis in mouse preadipocyte cells
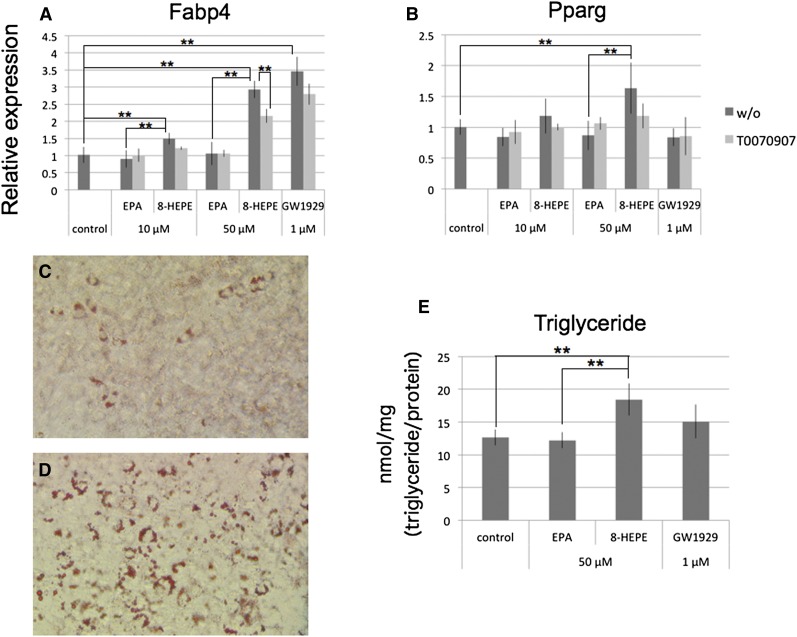
PPARγ plays a central role in regulation of gene expression and differentiation in adipocytes (11). We therefore examined whether 8-HEPE could act as a PPARγ ligand in these cells. We cultured 3T3-F442A cells, a mouse preadipocyte cell line, with EPA, 8-HEPE, or GW1929, a PPARγ-specific agonist, for 24 h. We found that expression of Fabp4 and Pparg was increased by 8-HEPE, but not by EPA (Fig. 6A, B). Both 8-HEPE and GW1929 increased the level of Fabp4 expression (Fig. 6A), but only 8-HEPE had an effect on Pparg expression (Fig. 6B). T0070907, a PPARγ-specific antagonist, inhibited 8-HEPE-induced expression of Fabp4 (Fig. 6A).
Fig. 6.
The effect of 8-HEPE treatment on adipogenesis in 3T3-F442A cells. A, B: 3T3-F442A cells were cultured with EPA, 8-HEPE, or GW1929 for 24 h. The PPARγ antagonist T0070907 (5 μM) was added to the media at the same time as EPA, 8-HEPE, or GW1929. Expression of Fabp4 and Pparg was measured by real-time PCR and normalized against expression of Rplp0. C–E: 3T3-F442A cells were cultured with 50 μM of EPA or 8-HEPE, or 1 μM GW1929 for 7 days. Cells cultured with EPA (C) or 8-HEPE (D) were stained by Oil Red O. E: Triglyceride contents were measured and normalized against protein concentration. Plotted values represent the mean ± SD from four independent cultures. 8-HEPE purified from Pacific krill was used. **P < 0.01. w/o, without.
Next, we cultured 3T3-F442A cells with 50 μM EPA or 8-HEPE, or 1 μM GW1929 for 7 days, and measured triglyceride accumulation to evaluate adipocyte differentiation. Compared with the control or EPA-treated cells, a greater level of triglyceride accumulation was observed in 8-HEPE-treated cells (Fig. 6C–E).
8-HEPE increases Angptl4 expression and enhances glucose uptake in mouse myoblast cells
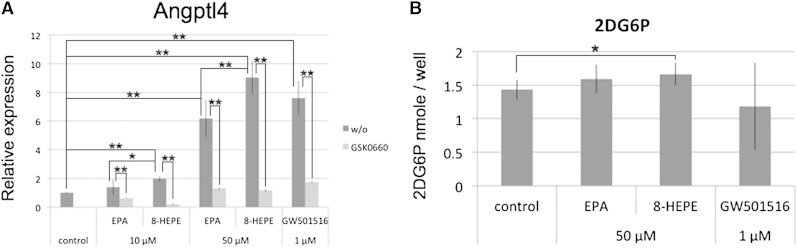
PPARδ is the predominant isoform found in skeletal muscle; the agonist GW501516, which is PPARδ-specific, increases the expression of angiopoietin-like protein 4 (Angptl4) (34, 35) and enhances glucose uptake (36) in C2C12 cells, a mouse myoblast cell line. Angptl4 is one of the major targets of PPARδ, and Angptl4 is the gene that is most highly induced by long-chain fatty acids in human myotubes (34). Here, we examined the relative level of Angptl4 expression by real-time PCR. We cultured C2C12 cells with EPA 8-HEPE or GW501516 for 6 h and found that expression of Angptl4 was increased to a greater extent by 8-HEPE than EPA (Fig. 7A). GSK0660, a PPARδ-specific antagonist, inhibited 8-HEPE-induced expression of Angptl4 (Fig. 7A).
Fig. 7.
The effect of 8-HEPE treatment on Angptl4 expression and glucose uptake in C2C12 cells. A: C2C12 cells were cultured with EPA, 8-HEPE, or GW501516 for 6 h. The PPARδ antagonist GSK0660 (25 μM) was added to the media at the same time as EPA, 8-HEPE, or GW501516. Expression of Angptl4 was measured by real-time PCR and normalized against expression of Actb. B: C2C12 cells, in which myogenesis had been induced, were cultured with 50 μM of EPA or 8-HEPE, or 1 μM GW501516 for 6 h. Glucose uptake was measured in the cells using 2-deoxyglucose. 8-HEPE purified from Pacific krill was used. *P < 0.05, **P < 0.01. 2DG6P, 2-deoxyglucose-6-phosphate. w/o, without.
We then measured glucose uptake in C2C12 cells that had been induced to undergo myotube differentiation for 5 days. 8-HEPE enhanced the uptake of glucose in these cells (Fig. 6B); the level of 8-HEPE-enhanced glucose uptake was higher, albeit nonsignificantly, than that induced by EPA (Fig. 6B). These results indicate that 8-HEPE acts as a PPARδ ligand. Overall, our analyses show that although 8-HEPE has a greater activity than EPA in upregulating Angptl4 expression, its effect on glucose uptake is comparable to EPA.
DISCUSSION
Our analyses here demonstrate that 8-HEPE and 9-HEPE are PPAR ligands and that their activities are greater than those of 5-HEPE, 12-HEPE, 18-HEPE, EPA, and EPA-Et (Fig. 3). These results indicate that the position of hydroxylation on EPA is correlated with the strength of ligand activity for PPARs and that hydroxylation at the C-8 or C-9 positions produces more effective ligands. A similar phenomenon has been found in HETEs in which the position of hydroxylation influences ligand activity for PPARα (20). It was also reported that 8-HETE has a significantly greater ability to activate PPARα compared with 5-HETE, 11-HETE, 12-HETE, and 15-HETE (20). This report and the present results show that hydroxylation at the C-8 position is important for activation of PPARα in both HETEs and HEPEs. However, 8-HETE and 8-HEPE activities with regard to PPARγ and PPARδ have not previously been identified (8, 20). The likely reason is that the concentrations of 8-HETE and 8-HEPE tested in the experiments were too low to activate PPARγ or PPARδ. For example, earlier studies used 3 μM 8-HETE and 1 μM 8-HEPE (8, 20), whereas, in the present study, activation of PPARγ and PPARδ was observed at concentrations greater than 5 μM 8-HEPE (Fig. 4). For PPARα and PPARδ, 8-HEPE showed a greater effect than 8-HETE, while both 8-HETE and 8-HEPE were similarly active with regard to for PPARγ (Fig. 4). Previous studies did not compare the relative ligand activity of 8-HEPE and EPA for PPARs. As shown here, 8-HEPE had a greater ability than EPA to activate PPARα, PPARγ, and PPARδ at concentrations of more than 4 μM, 64 μM, and 64 μM, respectively (Table 1). The concentration of total fatty acids in serum is about 700 μM (37), while serum concentrations of specific fatty acids that are abundant in the diet, such as linoleic acid and AA, generally fall into the range 25–30 μM (8). Physiologically, most free fatty acids in serum are bound and buffered with albumin. The concentration of unbound free fatty acids is in the nanomolar range (38).
PPARs regulate the expression of numerous genes related to energy metabolism, and the activation of PPARs can have beneficial effects in obesity and diabetes (1, 10, 39–41). PPARα activators regulate obesity by increasing hepatic fatty acid oxidation and decreasing the levels of the circulating triglycerides responsible for adipose cell hypertrophy and hyperplasia (42–44); PPARγ activators induce adipocyte gene expression and improve insulin sensitivity (11, 45); and, PPARδ activators induce fatty acid oxidation in skeletal muscle (46) and activate glucose transport in myotubes (36). Our analyses of the effects of 8-HEPE in FaO, 3T3-F442A, and C2C12 cells showed that it induced expression of genes known to be regulated by PPARα (Fig. 5), PPARγ (Fig. 6A), and PPARδ (Fig. 7A); this 8-HEPE-induced upregulation of gene expression was suppressed by PPAR antagonists. 8-HEPE also induced adipogenesis (Fig. 6B–E) and enhanced glucose uptake (Fig. 7B). These results indicate that 8-HEPE acts as a ligand for intrinsic PPARα, PPARγ, and PPARδ.
Eicosanoids are a family of biologically active oxygenated derivatives of C20 PUFAs. Two pathways are principally involved in the production of eicosanoids. The lipoxygenase pathway transforms PUFAs into lipoxins, leukotrienes, and monohydroxy fatty acids, whereas the cyclooxygenase pathway produces prostaglandins and thromboxanes. Production of HEPEs has been observed in platelets (47), macrophages (48, 49), and lung cells (50); and Tomio et al. (51) showed that 12-HEPE and 15-HEPE are decreased in 12/15-lipoxygenase knockout mice. In general, the physiological effects of HEPEs are unclear; however, it has been suggested that 12-HEPE and 15-HEPE have a suppressive effect on the development of endometriotic lesions (51). There have been comparatively few investigations of the biochemical or physiological properties of HEPEs, including 8-HEPE. It was shown that Escherichia coli hemolysin induced production of 8-HEPE in rabbit macrophages (48). However, no enzyme with the capability of transforming EPA into 8-HEPE has been identified to date. In 8-HETE, 8(S)-lipoxygenase can transform AA into 8(S)-HETE (52), a compound that can stereoselectively activate PPARα (8, 20). Future studies will be need to address the questions of 8-HEPE stereoselectivity and the identity of the enzyme that can transform EPA into 8-HEPE. Our study indicates that 8-HEPE and 9-HEPE induce fatty acid oxidation, adipogenesis, and glucose uptake via activation of PPARs in vivo. These metabolic responses are very interesting areas of research for the regulation of obesity and diabetes. The content of 8-HEPE in krill is more than 10 times higher than in the Japanese pilchard (Sardinops melanostictus) or the Pacific saury (Cololabis saira), species that are generally regarded as possessing high EPA contents (supplementary Table V). Our study shows that Pacific krill will be a valuable source of HEPEs for addressing the physiological functions of HEPEs.
Supplementary Material
Footnotes
Abbreviations:
- AA
- arachidonic acid
- Angptl4
- angiopoietin-like protein 4
- Cpt
- carnitine palmitoyltransferase
- CYP4A
- cytochrome P450 CYP4A enzyme
- EPA-Et
- EPA ethyl-ester
- Ehhadh
- enoyl-CoA hydratase/3-hydroxyacyl CoA
- Fabp
- fatty acid-binding protein
- HEPE
- hydroxyeicosapentaenoic acid
This work was supported by funds from the Basic Biotechnology Project of Iwate Prefecture, Japan, Grants-in-Aid from the Sanriku Foundation and Center for Revitalization Promotion, Japan Science and Technology Agency.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of four figures and five tables.
REFERENCES
- 1.Schoonjans K., Staels B., Auwerx J. 1996. The peroxisome proliferator activated receptors (PPARS) and their effects on lipid metabolism and adipocyte differentiation. Biochim. Biophys. Acta. 1302: 93–109. [DOI] [PubMed] [Google Scholar]
- 2.Tugwood J. D., Issemann I., Anderson R. G., Bundell K. R., McPheat W. L., Green S. 1992. The mouse peroxisome proliferator activated receptor recognizes a response element in the 5′ flanking sequence of the rat acyl CoA oxidase gene. EMBO J. 11: 433–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang B., Marcus S. L., Sajjadi F. G., Alvares K., Reddy J. K., Subramani S., Rachubinski R. A., Capone J. P. 1992. Identification of a peroxisome proliferator-responsive element upstream of the gene encoding rat peroxisomal enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase. Proc. Natl. Acad. Sci. USA. 89: 7541–7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kliewer S. A., Sundseth S. S., Jones S. A., Brown P. J., Wisely G. B., Koble C. S., Devchand P., Wahli W., Willson T. M., Lenhard J. M., et al. 1997. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors alpha and gamma. Proc. Natl. Acad. Sci. USA. 94: 4318–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marcus S. L., Miyata K. S., Zhang B., Subramani S., Rachubinski R. A., Capone J. P. 1993. Diverse peroxisome proliferator-activated receptors bind to the peroxisome proliferator-responsive elements of the rat hydratase/dehydrogenase and fatty acyl-CoA oxidase genes but differentially induce expression. Proc. Natl. Acad. Sci. USA. 90: 5723–5727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernal-Mizrachi C., Weng S., Feng C., Finck B. N., Knutsen R. H., Leone T. C., Coleman T., Mecham R. P., Kelly D. P., Semenkovich C. F. 2003. Dexamethasone induction of hypertension and diabetes is PPAR-alpha dependent in LDL receptor-null mice. Nat. Med. 9: 1069–1075. [DOI] [PubMed] [Google Scholar]
- 7.Reddy J. K., Hashimoto T. 2001. Peroxisomal beta-oxidation and peroxisome proliferator-activated receptor alpha: an adaptive metabolic system. Annu. Rev. Nutr. 21: 193–230. [DOI] [PubMed] [Google Scholar]
- 8.Forman B. M., Chen J., Evans R. M. 1997. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc. Natl. Acad. Sci. USA. 94: 4312–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desvergne B., Wahli W. 1999. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr. Rev. 20: 649–688. [DOI] [PubMed] [Google Scholar]
- 10.Lee S. S., Pineau T., Drago J., Lee E. J., Owens J. W., Kroetz D. L., Fernandez-Salguero P. M., Westphal H., Gonzalez F. J. 1995. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol. Cell. Biol. 15: 3012–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tontonoz P., Hu E., Graves R. A., Budavari A. I., Spiegelman B. M. 1994. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 8: 1224–1234. [DOI] [PubMed] [Google Scholar]
- 12.Rosen E. D., Spiegelman B. M. 2001. PPARgamma: a nuclear regulator of metabolism, differentiation, and cell growth. J. Biol. Chem. 276: 37731–37734. [DOI] [PubMed] [Google Scholar]
- 13.Kliewer S. A., Forman B. M., Blumberg B., Ong E. S., Borgmeyer U., Mangelsdorf D. J., Umesono K., Evans R. M. 1994. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proc. Natl. Acad. Sci. USA. 91: 7355–7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amri E. Z., Bonino F., Ailhaud G., Abumrad N. A., Grimaldi P. A. 1995. Cloning of a protein that mediates transcriptional effects of fatty acids in preadipocytes. Homology to peroxisome proliferator-activated receptors. J. Biol. Chem. 270: 2367–2371. [DOI] [PubMed] [Google Scholar]
- 15.Braissant O., Foufelle F., Scotto C., Dauça M., Wahli W. 1996. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinology. 137: 354–366. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y-X., Zhang C-L., Yu R. T., Cho H. K., Nelson M. C., Bayuga-Ocampo C. R., Ham J., Kang H., Evans R. M. 2004. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol. 2: e294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forman B. M., Tontonoz P., Chen J., Brun R. P., Spiegelman B. M., Evans R. M. 1995. 15-Deoxy-delta 12, 14-prostaglandin J2 is a ligand for the adipocyte determination factor PPAR gamma. Cell. 83: 803–812. [DOI] [PubMed] [Google Scholar]
- 18.Dowell P., Peterson V. J., Zabriskie T. M., Leid M. 1997. Ligand-induced peroxisome proliferator-activated receptor alpha conformational change. J. Biol. Chem. 272: 2013–2020. [DOI] [PubMed] [Google Scholar]
- 19.Krey G., Braissant O., L'Horset F., Kalkhoven E., Perroud M., Parker M. G., Wahli W. 1997. Fatty acids, eicosanoids, and hypolipidemic agents identified as ligands of peroxisome proliferator-activated receptors by coactivator-dependent receptor ligand assay. Mol. Endocrinol. 11: 779–791. [DOI] [PubMed] [Google Scholar]
- 20.Yu K., Bayona W., Kallen C. B., Harding H. P., Ravera C. P., McMahon G., Brown M., Lazar M. A. 1995. Differential activation of peroxisome proliferator-activated receptors by eicosanoids. J. Biol. Chem. 270: 23975–23983. [DOI] [PubMed] [Google Scholar]
- 21.Johnson T. E., Holloway M. K., Vogel R., Rutledge S. J., Perkins J. J., Rodan G. A., Schmidt A. 1997. Structural requirements and cell-type specificity for ligand activation of peroxisome proliferator-activated receptors. J. Steroid Biochem. Mol. Biol. 63: 1–8. [DOI] [PubMed] [Google Scholar]
- 22.Kliewer S. A., Lenhard J. M., Willson T. M., Patel I., Morris D. C., Lehmann J. M. 1995. A prostaglandin J2 metabolite binds peroxisome proliferator-activated receptor gamma and promotes adipocyte differentiation. Cell. 83: 813–819. [DOI] [PubMed] [Google Scholar]
- 23.Devchand P. R., Keller H., Peters J. M., Vazquez M., Gonzalez F. J., Wahli W. 1996. The PPARalpha-leukotriene B4 pathway to inflammation control. Nature. 384: 39–43. [DOI] [PubMed] [Google Scholar]
- 24.Durrington P. N. 1993. Diabetes, hypertension and hyperlipidaemia. Postgrad. Med. J. 69(Suppl 1): S18–S25; discussion S25–S29. [PubMed] [Google Scholar]
- 25.Reaven G. M. 1994. Syndrome X: 6 years later. J. Intern. Med. Suppl. 736: 13–22. [PubMed] [Google Scholar]
- 26.Willumsen N., Skorve J., Hexeberg S., Rustan A. C., Berge R. K. 1993. The hypotriglyceridemic effect of eicosapentaenoic acid in rats is reflected in increased mitochondrial fatty acid oxidation followed by diminished lipogenesis. Lipids. 28: 683–690. [DOI] [PubMed] [Google Scholar]
- 27.Spady D. K., Woollett L. A., Dietschy J. M. 1993. Regulation of plasma LDL-cholesterol levels by dietary cholesterol and fatty acids. Annu. Rev. Nutr. 13: 355–381. [DOI] [PubMed] [Google Scholar]
- 28.Green S. 1995. PPAR: a mediator of peroxisome proliferator action. Mutat. Res. 333: 101–109. [DOI] [PubMed] [Google Scholar]
- 29.Jump D. B., Clarke S. D., Thelen A., Liimatta M. 1994. Coordinate regulation of glycolytic and lipogenic gene expression by polyunsaturated fatty acids. J. Lipid Res. 35: 1076–1084. [PubMed] [Google Scholar]
- 30.Vigerust N. F., Bjørndal B., Bohov P., Brattelid T., Svardal A., Berge R. K. 2013. Krill oil versus fish oil in modulation of inflammation and lipid metabolism in mice transgenic for TNF-α. Eur. J. Nutr. 52: 1315–1325. [DOI] [PubMed] [Google Scholar]
- 31.Ierna M., Kerr A., Scales H., Berge K., Griinari M. 2010. Supplementation of diet with krill oil protects against experimental rheumatoid arthritis. BMC Musculoskelet. Disord. 11: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sadzuka Y., Sugiyama I., Miyashita M., Ueda T., Kikuchi S., Oshiro E., Yano A., Yamada H. 2012. Beneficial effects by intake of Euphausiacea pacifica on high-fat diet-induced obesity. Biol. Pharm. Bull. 35: 568–572. [DOI] [PubMed] [Google Scholar]
- 33.Vanden Heuvel J. P., Kreder D., Belda B., Hannon D. B., Nugent C. A., Burns K. A., Taylor M. J. 2003. Comprehensive analysis of gene expression in rat and human hepatoma cells exposed to the peroxisome proliferator WY14,643. Toxicol. Appl. Pharmacol. 188: 185–198. [DOI] [PubMed] [Google Scholar]
- 34.Staiger H., Haas C., Machann J., Werner R., Weisser M., Schick F., Machicao F., Stefan N., Fritsche A., Häring H-U. 2009. Muscle-derived angiopoietin-like protein 4 is induced by fatty acids via peroxisome proliferator-activated receptor (PPAR)-delta and is of metabolic relevance in humans. Diabetes. 58: 579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robciuc M. R., Skrobuk P., Anisimov A., Olkkonen V. M., Alitalo K., Eckel R. H., Koistinen H. A., Jauhiainen M., Ehnholm C. 2012. Angiopoietin-like 4 mediates PPAR delta effect on lipoprotein lipase-dependent fatty acid uptake but not on beta-oxidation in myotubes. PLoS ONE. 7: e46212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krämer D. K., Al-Khalili L., Perrini S., Skogsberg J., Wretenberg P., Kannisto K., Wallberg-Henriksson H., Ehrenborg E., Zierath J. R., Krook A. 2005. Direct activation of glucose transport in primary human myotubes after activation of peroxisome proliferator-activated receptor delta. Diabetes. 54: 1157–1163. [DOI] [PubMed] [Google Scholar]
- 37.Groop L. C., Bonadonna R. C., Simonson D. C., Petrides A. S., Shank M., DeFronzo R. A. 1992. Effect of insulin on oxidative and nonoxidative pathways of free fatty acid metabolism in human obesity. Am. J. Physiol. 263: E79–E84. [DOI] [PubMed] [Google Scholar]
- 38.Richieri G. V, Kleinfeld A. M. 1995. Unbound free fatty acid levels in human serum. J. Lipid Res. 36: 229–240. [PubMed] [Google Scholar]
- 39.Kubota N., Terauchi Y., Miki H., Tamemoto H., Yamauchi T., Komeda K., Satoh S., Nakano R., Ishii C., Sugiyama T., et al. 1999. PPAR gamma mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance. Mol. Cell. 4: 597–609. [DOI] [PubMed] [Google Scholar]
- 40.Tsuchida A., Yamauchi T., Takekawa S., Hada Y., Ito Y., Maki T., Kadowaki T. 2005. Peroxisome proliferator-activated receptor (PPAR)alpha activation increases adiponectin receptors and reduces obesity-related inflammation in adipose tissue: comparison of activation of PPARalpha, PPARgamma, and their combination. Diabetes. 54: 3358–3370. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y-X., Lee C-H., Tiep S., Yu R. T., Ham J., Kang H., Evans R. M. 2003. Peroxisome-proliferator-activated receptor delta activates fat metabolism to prevent obesity. Cell. 113: 159–170. [DOI] [PubMed] [Google Scholar]
- 42.Rosenson R. S. 2008. Fenofibrate: treatment of hyperlipidemia and beyond. Expert Rev. Cardiovasc. Ther. 6: 1319–1330. [DOI] [PubMed] [Google Scholar]
- 43.Jeong S., Yoon M. 2009. Fenofibrate inhibits adipocyte hypertrophy and insulin resistance by activating adipose PPARalpha in high fat diet-induced obese mice. Exp. Mol. Med. 41: 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guerre-Millo M., Gervois P., Raspé E., Madsen L., Poulain P., Derudas B., Herbert J. M., Winegar D. A., Willson T. M., Fruchart J. C., et al. 2000. Peroxisome proliferator-activated receptor alpha activators improve insulin sensitivity and reduce adiposity. J. Biol. Chem. 275: 16638–16642. [DOI] [PubMed] [Google Scholar]
- 45.Brown K. K., Henke B. R., Blanchard S. G., Cobb J. E., Mook R., Kaldor I., Kliewer S. A., Lehmann J. M., Lenhard J. M., Harrington W. W., et al. 1999. A novel N-aryl tyrosine activator of peroxisome proliferator-activated receptor-gamma reverses the diabetic phenotype of the Zucker diabetic fatty rat. Diabetes. 48: 1415–1424. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka T., Yamamoto J., Iwasaki S., Asaba H., Hamura H., Ikeda Y., Watanabe M., Magoori K., Ioka R. X., Tachibana K., et al. 2003. Activation of peroxisome proliferator-activated receptor delta induces fatty acid beta-oxidation in skeletal muscle and attenuates metabolic syndrome. Proc. Natl. Acad. Sci. USA. 100: 15924–15929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morita I., Takahashi R., Saito Y., Murota S. 1983. Stimulation of eicosapentaenoic acid metabolism in washed human platelets by 12-hydroperoxyeicosatetraenoic acid. J. Biol. Chem. 258: 10197–10199. [PubMed] [Google Scholar]
- 48.Rose F., Kiss L., Grimminger F., Mayer K., Grandel U., Seeger W., Bieniek E., Sibelius U. 2000. E. coli hemolysin-induced lipid mediator metabolism in alveolar macrophages: impact of eicosapentaenoic acid. Am. J. Physiol. Lung Cell. Mol. Physiol. 279: L100–L109. [DOI] [PubMed] [Google Scholar]
- 49.Dioszeghy V., Rosas M., Maskrey B. H., Colmont C., Topley N., Chaitidis P., Kühn H., Jones S. A., Taylor P. R., O'Donnell V. B. 2008. 12/15-Lipoxygenase regulates the inflammatory response to bacterial products in vivo. J. Immunol. 181: 6514–6524. [DOI] [PubMed] [Google Scholar]
- 50.Hwang D. H., Boudreau M., Chanmugam P. 1988. Dietary linolenic acid and longer-chain n-3 fatty acids: comparison of effects on arachidonic acid metabolism in rats. J. Nutr. 118: 427–437. [DOI] [PubMed] [Google Scholar]
- 51.Tomio K., Kawana K., Taguchi A., Isobe Y., Iwamoto R., Yamashita A., Kojima S., Mori M., Nagamatsu T., Arimoto T., et al. 2013. Omega-3 polyunsaturated fatty acids suppress the cystic lesion formation of peritoneal endometriosis in transgenic mouse models. PLoS ONE. 8: e73085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fürstenberger G., Marks F., Krieg P. 2002. Arachidonate 8(S)-lipoxygenase. Prostaglandins Other Lipid Mediat. 68–69: 235–243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.