Abstract
Sterols are an essential class of lipids in eukaryotes, where they serve as structural components of membranes and play important roles as signaling molecules. Sterols are also of high pharmacological significance: cholesterol-lowering drugs are blockbusters in human health, and inhibitors of ergosterol biosynthesis are widely used as antifungals. Inhibitors of ergosterol synthesis are also being developed for Chagas’s disease, caused by Trypanosoma cruzi. Here we develop an in silico pipeline to globally evaluate sterol metabolism and perform comparative genomics. We generate a library of hidden Markov model-based profiles for 42 sterol biosynthetic enzymes, which allows expressing the genomic makeup of a given species as a numerical vector. Hierarchical clustering of these vectors functionally groups eukaryote proteomes and reveals convergent evolution, in particular metabolic reduction in obligate endoparasites. We experimentally explore sterol metabolism by testing a set of sterol biosynthesis inhibitors against trypanosomatids, Plasmodium falciparum, Giardia, and mammalian cells, and by quantifying the expression levels of sterol biosynthetic genes during the different life stages of T. cruzi and Trypanosoma brucei. The phenotypic data correlate with genomic makeup for simvastatin, which showed activity against trypanosomatids. Other findings, such as the activity of terbinafine against Giardia, are not in agreement with the genotypic profile.
Keywords: sterol metabolism, sterol biosynthesis inhibitor, comparative genomics, hidden Markov model, hierarchical clustering, Trypanosoma, Giardia
Sterols are an intriguing class of metabolites, ubiquitous in eukaryotes but largely absent from archaea and bacteria. Sterols in eukaryotes serve multiple physiological functions associated with two main purposes: as structural components of membranes (bulk function) and as precursors to signaling molecules that regulate growth and development (sparking function). The sterol biosynthetic pathways encompass phylum-specific branches that yield distinctive end products. Cholesterol is the most abundant sterol in vertebrates, where it is an essential component of cell membranes and acts as a precursor to vitamin D and steroid hormones. Invertebrates such as Caenorhabditis elegans or Drosophila melanogaster are sterol auxotrophs that rely on uptake of exogenous sterols (1–3). Fungi synthesize ergosterol, which has a greater degree of unsaturation than cholesterol and an additional methyl group at C-24. Plants make a large variety of phytosterols, such as the 24-alkylated campesterol, sitosterol, or stigmasterol. Sterol diversity may be equally high in the protozoa, involving cholesterol and stigmasterol in Paramecium (4), ergosterol and stigmasterols in Acanthamoeba (5), cycloartenol and cyclolaudenol in Dicytostelium (6), and ergosterol in trypanosomatids (7, 8).
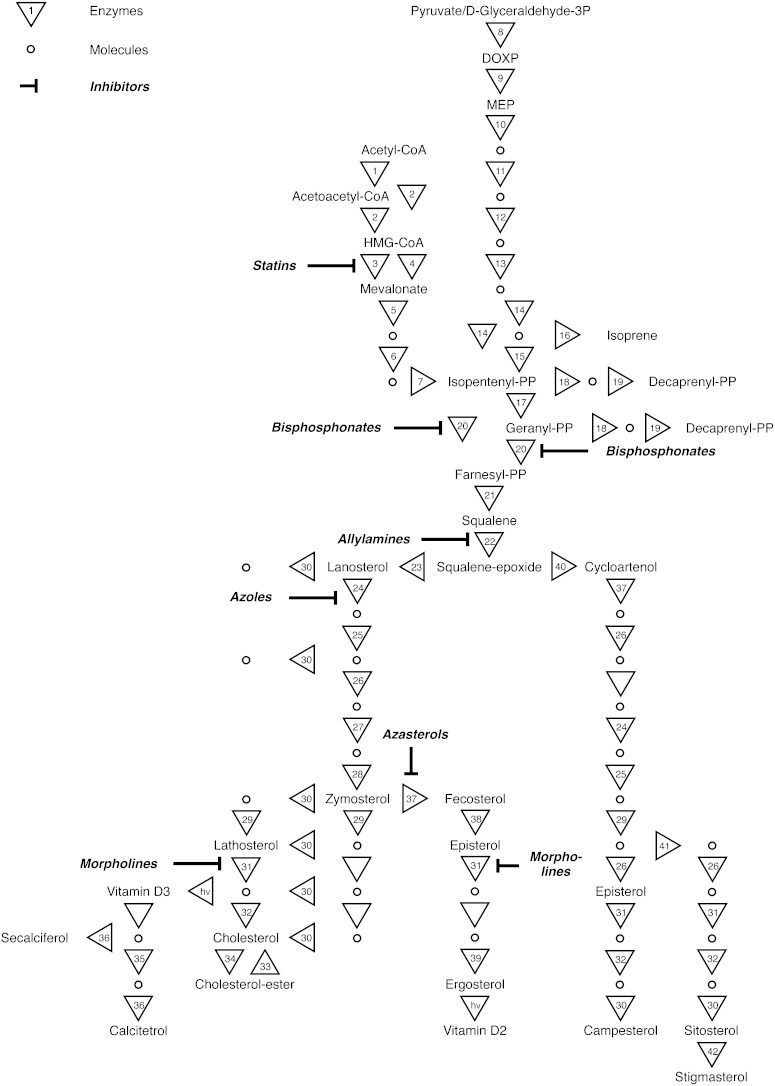
The sterol biosynthetic pathways involve more than 40 enzymes (Table 1). Isopentenyl diphosphate, the building block of isoprenoids, is made either from acetyl-CoA via the MEV pathway or from pyruvate plus glyceraldehyde 3-phosphate via the deoxyxylulose 5-phosphate (DOXP) pathway (Fig. 1). The condensation of two molecules of farnesyl diphosphate to produce squalene is the committing step of sterol synthesis. From there, the synthesis proceeds via lanosterol and zymosterol to cholesterol and ergosterol derivatives, or via cycloartenol to the various kinds of phytosterols (Fig. 1). The enzymes that mediate sterol biosynthesis are of dual interest: First, there is a pharmacological interest because sterol biosynthesis inhibitors (SBIs) are widely deployed as chemotherapeutics. As indicated in Fig. 1, human HMG-CoA reductase serves as the target of cholesterol-lowering statins such as simvastatin (9). Human farnesyl diphosphate synthase is the target of bisphosphonates (e.g., tiludronate), used against osteoporosis (10). Squalene epoxidase and sterol 24-methyltransferase are antifungal targets, inhibited by allylamines (e.g., terbinafine) and azasterols, respectively (11–14). A particularly promising target is sterol 14-demethylase (CYP51). Azole inhibitors of CYP51 (e.g., ketoconazole) are widely used for fungal infections and, because trypanosomatid parasites also make ergosterol (15–17), lend themselves for a piggyback approach toward the urgently required new drugs for Chagas's disease. The latest antifungal approved by the U.S. Food and Drug Administration, posaconazole, was shown to be highly active against Trypanosoma cruzi in culture and in vivo (18–20), and it is currently undergoing phase 2 clinical trials for the treatment of Chagas's disease. Second, sterol biosynthetic enzymes are of phylogenetic importance. Sterols were proposed to hold a key to understanding eukaryote phylogeny because the evolution of eukaryotes is thought to be interlinked with that of the sterols (21).
TABLE 1.
Sterol biosynthetic enzymes
| # | EC | KEGG | Enzyme Name |
| 1 | 2.3.1.9 | K00626 | Acetyl-CoA C-acetyltransferase |
| 2 | 2.3.3.10 | K01641 | Hydroxymethylglutaryl-CoA synthase |
| 3 | 1.1.1.34 | K00021 | Hydroxymethylglutaryl-CoA reductase (NADPH) |
| 4 | 1.1.1.88 | K00054 | Hydroxymethylglutaryl-CoA reductase |
| 5 | 2.7.1.36 | K00869 | Mevalonate (MEV) kinase |
| 6 | 2.7.4.2 | K00938 | Phospho-MEV kinase |
| 7 | 4.1.1.33 | K01597 | Diphospho-MEV decarboxylase |
| 8 | 2.2.1.7 | K01662 | 1-Deoxy-d-xylulose-5-phosphate synthase |
| 9 | 1.1.1.267 | K00099 | 1-Deoxy-d-xylulose-5-phosphate reductoisomerase |
| 10 | 2.7.7.60 | K00991 | 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase |
| 11 | 2.7.1.148 | K00919 | 4-Diphosphocytidyl-2-C-methyl-d-erythritol kinase |
| 12 | 4.6.1.12 | K01770 | 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase |
| 13 | 1.17.7.1 | K03526 | (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase |
| 14 | 1.17.1.2 | K03527 | 4-Hydroxy-3-methylbut-2-enyl diphosphate reductase |
| 15 | 5.3.3.2 | K01823 | Isopentenyl-diphosphate delta-isomerase |
| 16 | 4.2.3.27 | K12742 | Isoprene synthase |
| 17 | 2.5.1.1 | K00787 | Geranyl diphosphate synthase |
| 18 | 2.5.1.68 | K12503 | Short-chain Z-isoprenyl diphosphate synthase |
| 19 | 2.5.1.86 | K14215 | Trans,polycis-decaprenyl diphosphate synthase |
| 20 | 2.5.1.10 | K00787 | Farnesyl diphosphate synthase |
| 21 | 2.5.1.21 | K00801 | Farnesyl-diphosphate farnesyltransferase |
| 22 | 1.14.13.132 | K00511 | Squalene monooxygenase |
| 23 | 5.4.99.7 | K01852 | Lanosterol synthase |
| 24 | 1.14.13.70 | K05917 | Cytochrome P450, family 51 (CYP51; sterol 14-demethylase) |
| 25 | 1.3.1.70 | K00222 | Delta14-sterol reductase |
| 26 | 1.14.13.72 | K07750 | Methylsterol monooxygenase |
| 27 | 1.1.1.170 | K07748 | Sterol-4α-carboxylate 3-dehydrogenase (decarboxylating) |
| 28 | 1.1.1.270 | K09827 | 3-Keto steroid reductase |
| 29 | 5.3.3.5 | K01824 | Cholestenol delta-isomerase |
| 30 | 1.3.1.72 | K09828 | Delta24-sterol reductase |
| 31 | 1.14.21.6 | K00227 | Lathosterol oxidase |
| 32 | 1.3.1.21 | K00213 | 7-Dehydrocholesterol reductase |
| 33 | 3.1.1.13 | K01052 | Lysosomal acid lipase/cholesteryl ester hydrolase |
| 34 | 2.3.1.26 | K00637 | Sterol O-acyltransferase |
| 35 | 1.14.13.13 | K07438 | Calcidiol 1-monooxygenase |
| 36 | 1.14.13.126 | K07436 | Vitamin D3 24-hydroxylase |
| 37 | 2.1.1.41 | K00559 | Sterol 24-C-methyltransferase |
| 38 | 5.-.-.- | K09829 | C-8 sterol isomerase |
| 39 | 1.3.1.71 | K00223 | Delta24[24 (1)]-sterol reductase |
| 40 | 5.4.99.8 | K01853 | Cycloartenol synthase |
| 41 | 2.1.1.143 | K08242 | 24-Methylenesterol C-methyltransferase |
| 42 | n.a. | K09832 | Cytochrome P450, family 710, subfamily A |
EC, Enzyme Commission number; KEGG, Kyoto Encyclopedia of Genes and Genomes identifier.
Fig. 1.
Overview on sterol biosynthesis. Enzymes are represented by triangles, metabolites by circles. Inhibitors are indicated in bold. Key metabolites are spelled out. See Table 1 for enzyme name EC classifiers.
Here we present an in silico pipeline for comparative genomics of eukaryotes based on prediction of sterol biosynthetic enzymes. Our aim is to shed light on the evolutionary relationships of these enzymes and to identify new antiparasitic drug target candidates. Focusing on trypanosomatids, we further compare the stage-specificity of expression of the sterol biosynthetic enzymes, and we probe the potential of selected SBIs against a panel of parasites.
MATERIALS AND METHODS
Sequences
Proteome files were downloaded from Uniprot (22), EuPathDB (23), and Integr8 (24). The predicted proteomes were tested for completeness against the 100 most conserved proteins of the Core Eukaryotic Genes Mapping Approach database (25), which we had determined based on HMMer 3.0 profile (26, 27) searches of eukaryote reference proteomes (C. elegans, Chlamydomonas reinhardtii, D. discoideum, D. melanogaster, D. rerio, E. cuniculi, Entamoeba histolytica, G. lamblia, Homo sapiens, K. lactis, L. major, M. musculus, Plasmodium falciparum, S. cerevisiae, S. pombe, Trypanosoma brucei, T. cruzi, T. parva, and T. vaginalis). Additional proteomes were included only if hmmscan of HMMer 3.0 returned a hit for at least 99 profiles with an expectancy (E) value ≤10−30. Redundancy-reduced sets of reference sequences for the enzymes of interest (Table 1) were downloaded from UniRef90 (28).
Chemicals
Simvastatin (S6196-25MG), tiludronate disodium salt hydrate (T4580-10MG), terbinafine hydrochloride (T8826-100MG), and ketoconazole (K1003-100MG) were purchased from Sigma-Aldrich. Fenpropimorph was kindly offered by M. Witschel from BASF. The test compounds were dissolved in dimethyl sulfoxide at 10 mg/ml and stored at −20°C. Resazurin sodium salt (Alamar Blue) was purchased from Sigma-Aldrich (R7017-1G), and Chlorophenol red galactopyranoside (CPRG) from Roche (10884308001).
Cell lines
The cell lines used for in vitro drug sensitivity determination were rat L6 myoblasts, T. brucei rhodesiense STIB 900, T. cruzi Tulahuen C2C4 (expressing Escherichia coli β-galactosidase), Leishmania donovani MHOM-ET-67/L82 axenic amastigotes, Giardia intestinalis strain G1, and P. falciparum strain K1.
Bioinformatics
Multiple alignments were performed with ClustalW 2.0.10 (29). All profile constructions and searches were carried out with HMMer 3.0 (26). The heat map was produced with the R library gplots (30). Hierarchical clustering was performed with the R library Pvclust, using Canberra distance and the McQuitty algorithm (31). Pvclust assesses uncertainty in hierarchical cluster analysis by implementing multiscale bootstrap resampling (n = 10,000) to estimate “approximately unbiased” (au) errors, where P = (100 – au)/100. For principal component (PC) analysis we used basic R functions and ggplot2. The analyses were automated with Unix shell scripts and with Perl scripts. The phylogenetic trees were constructed from amino acid sequences with Mega 5, using the neighbor-joining algorithm and Jones-Taylor-Thornton substitution model. The number of bootstrap replications was 1,000.
Gene expression
Tag counts of the sterol biosynthesis enzymes were extracted from three published T. brucei short-read libraries (long slender bloodstream forms, short stumpy bloodstream forms, and procyclic tsetse fly midgut forms) and from four T. cruzi short-read libraries (intracellular amastigotes, trypomastigotes, epimastigotes, and metacyclics). The libraries had been produced by Illumina sequencing using the spliced-leader trapping (SLT) protocol (32). Numbers of reads were normalized by using the DESeq (33) bioconductor package. The gene expression data were modeled with prcomp as a numerical matrix of M genes times N libraries. Eigenvalues and orthogonal eigenvectors were computed based on the square-symmetrical correlation matrix.
In vitro drug sensitivity
In vitro drug sensitivity assays were performed as described (34–37). The tests were done over 72 h of incubation, except for the T. cruzi assay, which lasted 96 h. For L6 cells, L. donovani, T. brucei, and G. intestinalis, the redox-sensitive dye resazurin (Alamar Blue) served as an indicator of cell viability. For P. falciparum, incorporation of 3H-hypoxanthine was used. For T. cruzi, β-galactosidase activity was quantified with the substrate CPRG/Nonidet. IC50 values were estimated by linear interpolation based on the semilogarithmic dose-response curves.
RESULTS AND DISCUSSION
Sterol metabolic profiling of eukaryote genomes
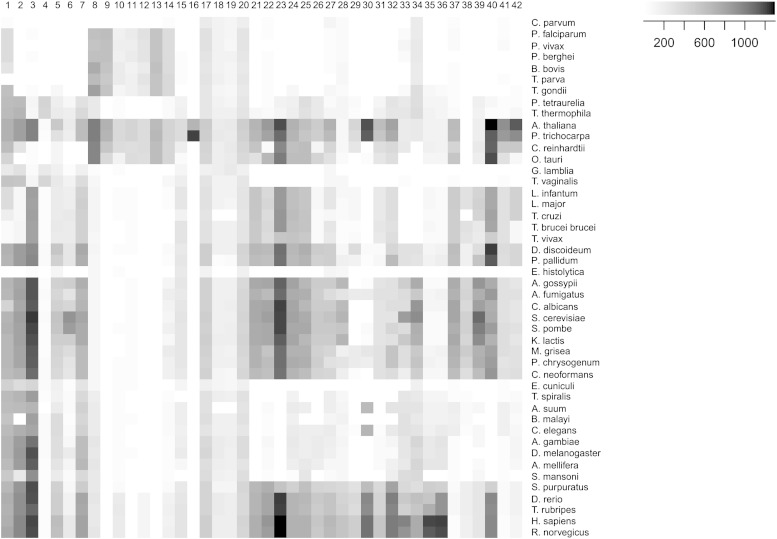
Aiming for a broad overview on sterol metabolism, we assembled a list of 42 relevant enzymes ranging from terpenoid backbone synthesis over squalene synthase to the formation of the different sterols and vitamin D derivatives as outlined in Fig. 1. For each enzyme, all the amino acid sequences that had been annotated in the manually curated section of UniProt with the corresponding EC number were retrieved (Table 1). Each of these sequence sets was redundancy reduced to 90%. Then a ClustalW multiple alignment was performed and converted to a position-dependent scoring matrix with hmmbuild of HMMer 3.0. The resultant 42 profiles were concatenated to a hidden Markov model (HMM) library for terpenoid backbone and sterol biosynthetic enzymes. This library was used for an in silico screen of eukaryote proteomes. For each proteome and each enzyme, we retrieved the profile-alignment score of the protein that had returned the lowest expectancy (E) value. This approach allowed organizing the data in an unbiased, quantitative way, by plotting the obtained high scores as a two-dimensional heat map where the horizontal cross-sections represent the “sterol biosynthetic profile” of a given organism (Fig. 2).
Fig. 2.
Sterol biosynthetic profiles. The heat map denotes the best scores achieved by each proteome (rows, n = 46) against each profile for a sterol biosynthetic enzyme (columns, n = 42). See Table 1 for enzyme name and Fig. 1 for its position in the pathway.
The known differences among the analyzed eukaryotes were obvious. The absence of the MEV pathway (enzyme nos. 2 to 7 of Table 1) and the presence of the MEP/DOXP pathway (enzyme nos. 8 to 14) were apparent in most apicomplexan species, with the exception of Cryptosporidium parvum, which lacked either pathway (Fig. 2, top). This is in agreement with C. parvum having lost the plastid genome in the course of evolution (38). It was interesting to note that E. histolytica was also deficient in either pathway, a finding that, to our knowledge, had not been reported before (39). Entamoeba was reported to be capable of limited cholesterol synthesis (40). Our data, however, support earlier reports suggesting an absolute requirement for cholesterol by Entamoeba (41). The vascular plants possessed both pathways, but the green algae C. reinhardtii and Ostreococcus tauri only had the MEP/DOXP arm, a fact that had been elegantly demonstrated for C. reinhardtii and other green algae by feeding experiments with radiolabeled glucose (42).
As expected, the sterol 24-C-methyltransferases SMT1 and SMT2 (enzyme nos. 37 and 41), characteristic of ergosterol synthesis, were absent in all vertebrates. The genome-wide screen also confirmed the presence of a set of sterol synthetic enzymes, including 24-C-methyltransferases, in trypanosomatids. Surprisingly, though, the trypanosomatids lacked sterol O-acyltransferase as well as cholesteryl ester hydrolase (enzyme nos. 34 and 33, respectively), and yet, T. brucei had been shown to be capable of sterol esterification (43), suggesting that trypanosomatids might use unusual enzymes for ester synthesis and hydrolysis.
Only 2 of the 42 sterol metabolic enzymes were present in all the analyzed proteomes: geranyl/farnesyl diphosphate synthase (enzyme nos. 17 and 20, respectively). Thus, our proteome-wide profiling approach indicates that the synthesis of farnesylpyrophosphate is essential to all eukaryotes and that farnesylpyrophosphate is a metabolite of central importance to organisms, whether they use the MEV or the MEP/DOXP pathway. A possible explanation is that farnesylation and geranylation of proteins is essential to eukaryotes. This hypothesis is in agreement with the fact that farnesyltransferases (EC 2.5.1.29, 2.5.1.58, and 2.5.1.60) occur in all the analyzed eukaryotes as determined with a profile search analogous to those for sterol biosynthetic enzymes (data not shown).
Hierarchical clustering reveals convergent evolution
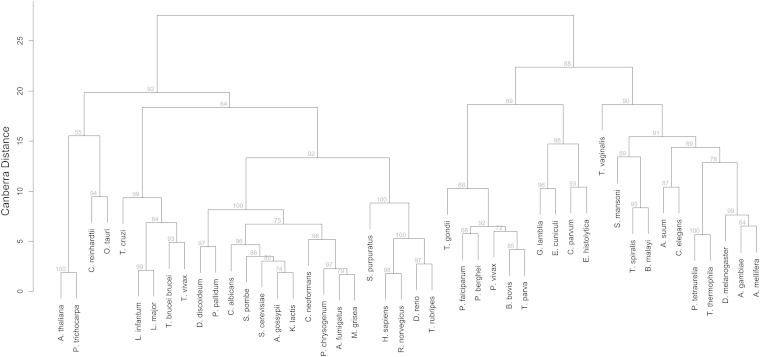
To detect less obvious and possibly new relationships regarding sterol metabolism of eukaryotes, we subjected the sterol metabolic profiles (Fig. 2) to hierarchical clustering. Every analyzed species was represented by a 42-tuple vector consisting of the best scores of the respective proteome to each profile. Hierarchical clustering of these vectors produced the “sterol biosynthesis tree” shown in Fig. 3, which basically subdivided the eukaryotes into species that make their own sterols (sterol prototrophs, left side) and species that do not (sterol auxotrophs, right side). The tree locally mirrored the phylogeny of the analyzed eukaryotes. The green plants cosegregated, as did the trypanosomatids, the protostomes, and the deuterostomes where the sea urchin Strongylocentrotus purpuratus clearly clustered with the chordates. The fungi formed a cluster except for E. cuniculi, and the apicomplexans formed a cluster except for C. parvum. These two outliers segregated with G. lamblia and E. histolytica in a phylogenetically diverse branch of amitochondriates (Fig. 3). The analyzed ciliates clustered with the insects; however, this association was not significant, and clearly, a larger number of ciliate genomes would be desirable to better resolve their position in the sterol biosynthesis landscape.
Fig. 3.
Sterol biosynthetic tree of eukaryotes. The numerical vectors (rows of Fig. 2) were clustered based on Canberra distance and the McQuitty algorithm. Gray numbers are “approximately unbiased” errors (au), where P = (100 − au)/100.
We interpret cases where clustering based on sterol biosynthesis enzymes does not coincide with eukaryote phylogeny as indicative of convergent evolution. Thus, the amoebazoon Entamoeba and the fungus Encephalitozoon, both obligate endoparasites, clustered together on the sterol-auxotrophic branch of the tree, whereas the free-living amoebazoa D. discoideum and P. palladium segregate on the sterol-prototrophic branch together with the free-living, or facultative parasitic, fungi (Fig. 3). A likely explanation of this clustering is that the eukaryote progenitor synthesized sterols de novo, whereas the obligate endoparasites independently lost the corresponding genes in adaptation to a parasitic lifestyle. The trypanosomatids are a notable exception: of all the included obligate endoparasites, they were the only species located on the sterol prototrophic arm of the tree (Fig. 3).
What is the origin of trypanosomatid ergosterol synthesis?
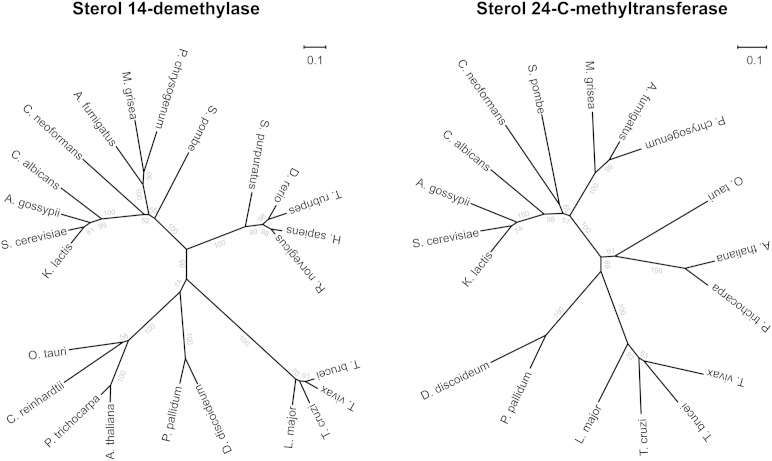
The sterol biosynthesis tree (Fig. 3) is based on functional predictions and cannot serve for phylogenetic models. To investigate the evolutionary origin of the sterol biosynthetic genes in trypanosomatids, we constructed phylogenetic trees for two key enzymes of ergosterol synthesis: lanosterol 14α-demethylase (enzyme no. 24 in Table 1), the target of azoles, and sterol 24-C-methyltransferase (enzyme no. 37), the dedicative enzyme for ergosterol synthesis. Both trees had a similar topology (except that sterol 24-C-methyltransferase does not occur in animals) with distinct branches for the major groups of eukaryotes and a highly significant separate branch for the included trypanosomatid sequences (Fig. 4). It was impossible to root the trees because there are no suitable outgroups for the two enzymes such as orthologs from bacteria. Acquisition of foreign genes by trypanosomes from plants has been suggested for glycosomal enzymes of T. brucei (44, 45). The phylogenetic trees of lanosterol 14α-demethylase and sterol 24-C-methyltransferase clearly do not support such a scenario for the sterol biosynthetic enzymes. Based on this bioinformatic analysis, one would exclude horizontal transfer as the evolutionary origin of trypanosomatid ergosterol synthesis.
Fig. 4.
Phylogenetic trees of key enzymes. Neighbor-joining trees of sterol 14-demethylase (enzyme no. 24), the target of azoles (A), and sterol 24-C-methyltransferase (enzyme no. 37), the dedicative enzyme of ergosterol synthesis (B). The scale bars indicate number of substitutions per site.
Susceptibility of parasites to SBIs
Given the antichagasic potential of azoles, we tested a panel of further known inhibitors of sterol biosynthesis, as indicated in Fig. 1, against parasites (P. falciparum, L. donovani, T. cruzi, T. brucei, and G. lamblia) and mammalian cells (rat L6 myoblasts). IC50 values were determined in vitro (Table 2). As expected, the mammalian cells were rather tolerant to most of the tested drugs, except for simvastatin, which had an IC50 of 0.70 µM against the L6 cells. This result was surprising because simvastatin is widely used as a cholesterol-lowering agent. We also tested mouse embryonic fibroblast and also against lovastatin, with the same result: IC50 values clearly below 1 µM. Apparently, cholesterol synthesis was essential under our test conditions. Tiludronate was inactive against all the tested cells. The most potent compound against T. cruzi was ketoconazole, followed by simvastatin, which also showed a moderate activity against T. brucei (IC50 = 4.6 µM) and L. donovani (IC50 = 4.7 µM). The activity against T. cruzi was not conclusive due to the toxicity of simvastatin to L6 host cells (hence the parentheses in Table 2). Terbinafine, ketoconazole, and fenpropimorph were moderately active against P. falciparum with IC50 below 10 µM. Terbinafine was the only tested compound that showed a considerable effect on Giardia (IC50 = 0.62 µM). However, terbinafine inhibits squalene monooxygenase (enzyme no. 22), an enzyme that is absent in Giardia (Fig. 2). Comparing the in silico data of Fig. 2 with the in vitro data of Table 2, we would expect a negative correlation between the score of the profile search for a given enzyme and species and the IC50 of a known inhibitor of the same enzyme. As shown in Fig. 5, this was the case with simvastatin (Spearman rank order correlation coefficient rS = −0.89, P < 0.05). For other compound-target pairs, there was either no correlation (ketoconazole and sterol 14-demethylase, rS = 0.06) or a positive, albeit nonsignificant, correlation (tiludronate and farnesyl diphosphate synthase, rS = 0.31; fenpropimorph and lathosterol oxidase, rS = 0.64). For terbinafine and its presumed target squalene epoxidase (enzyme no. 22), there was even a significant positive correlation between profile score and IC50 (rS = 0.89, P < 0.05; Fig. 5).
TABLE 2.
| L6 | Pfa | Ldo | Tcr | Tbr | Gla | |
| Simvastatin | 0.70 | 28.43 | 4.66 | (1.78) | 4.58 | 45.20 |
| Tiludronate | 204.00 | 21.04 | 119.07 | 169.82 | 120.66 | 64.22 |
| Terbinafine | 40.59 | 6.07 | 15.95 | 21.32 | 88.45 | 0.62 |
| Ketoconazole | 22.25 | 4.01 | 28.93 | 0.01 | 26.28 | 10.67 |
| Fenpropimorph | 33.59 | 3.22 | 35.60 | 8.95 | 25.18 | 4.88 |
IC50 in µM.
L6 rat skeletal muscle cells.
Gla, G. lamblia, Ldo, L. donovani; Pfa, P. falciparum; Tbr, T. brucei; Tcr, T. cruzi.
Fig. 5.
Correlation of genotype and phenotype. For each species, the in vitro IC50 (in µM) of the drugs simvastatin (A) and terbinafine (B) is plotted versus the highest score attained of the species’ proteome against the HMM profile of the presumed target of that drug. Gla, G. lamblia; L6, rat L6 myoblasts; Ldo, L. donovani; Pfa, P. falciparum; Tbr, T. brucei; Tcr, T. cruzi.
Stage-specific regulation of sterol biosynthetic enzymes in trypanosomes
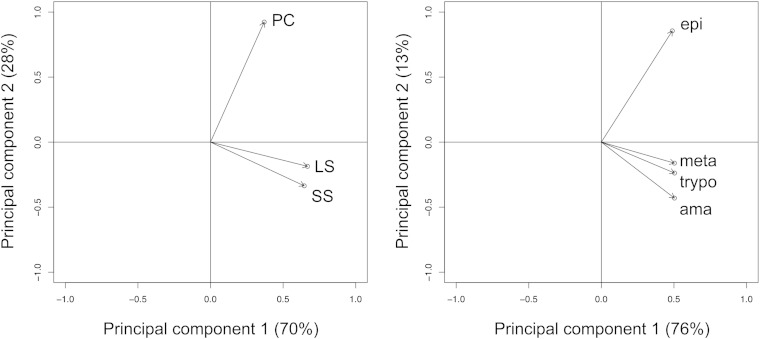
A possible reason for the lack of correlation between genomic makeup and drug susceptibility is that a particular target enzyme may be differently expressed across the life-cycle stages of a parasite. We investigated the expression of sterol biosynthetic enzymes at the mRNA level in the trypanosomatids T. brucei and T. cruzi using previously published SLT data. SLT takes advantage of the conserved miniexon that is spliced in trans to all trypanosomal mRNA (46). We analyzed data from different stages of T. brucei (slender bloodstream form, stumpy bloodstream form, and procyclic tsetse-midgut form) and T. cruzi (intracellular amastigote form, trypomastigote form, and epimastigote triatomine-midgut form) (32, 47). For both species, we found marked differences between the life-cycle stages regarding the steady-state expression levels of sterol biosynthetic genes. Generally, we detected higher mRNA levels of genes involved in sterol biosynthesis in the insect stages than in the mammalian stages of the organisms. This is in good agreement with the availability of sterols for the parasite in a mammalian host. PC analysis was performed for genes with orthologs in both species (n = 31). Plotting the first two PCs revealed the proliferating insect stages of both T. brucei and T. cruzi to more closely align with PC-2 than with PC-1, and to positively correlate with either PC (Fig. 6). In contrast, the proliferating mammalian stages, as well as all the nonproliferating stages, more closely aligned with PC-1 and oppositely correlated with PC-2 (Fig. 6). Taken together, PC analysis of the steady-state expression levels of sterol biosynthetic genes singled out the insect stages of T. brucei as well as T. cruzi (Fig. 6), demonstrating parallel metabolic adaptations in African and South American trypanosomes.
Fig. 6.
Expression profiles in Trypanosoma spp. Loading plots based on PC-1 and PC-2 of the steady-state mRNA levels of sterol biosynthetic genes expressed in count per million. A: T. brucei procyclic forms (PC) and long slender (LS) and short stumpy (SS) bloodstream forms. B: T. cruzi epimastigotes (epi), metacyclics (meta), trypomastigotes (trypo), and amastigotes (ama).
CONCLUSION
Starting from the assumption that sterol synthesis is intrinsically linked to the evolution of eukaryotes, we performed comparative genomics based on the profiling of sterol biosynthetic enzymes (Fig. 1). The aim was to investigate convergent evolution in eukaryotes and possibly link this to chemotherapeutic strategies against parasites. We refrained from functionally annotating the analyzed proteomes because our in silico pipeline did not furnish proof of function. It provided quantitative scores for all enzymes and proteomes (Fig. 2), which lent itself for clustering. The resulting tree's principal subdivision was between the sterol-prototrophs and the sterol-auxotrophs (Fig. 3). The latter included all of the analyzed protostomes, the former the deuterostomes (i.e., the vertebrates plus the sea urchin S. purpuratus).
The interesting branches of the sterol metabolic tree were those where the grouping deviated from evolutionary descent. This was observed for unicellular obligate endoparasites, which appeared to have independently lost genes for sterol biosynthetic enzymes, presumably in adaptation to a parasitic lifestyle. Thus, the microsporidian Encephalitozoon did not group with the free-living and facultative parasitic fungi but with parasitic protozoa such as Giardia. The same was observed for Entamoeba, which did not group with the free-living amoebozoa but with the apicomplexan Cryptosporidium. In fact, both Entamoeba and Cryptosporidium exhibited extreme cases of metabolic reduction, lacking the sterol biosynthetic enzymes as well as either pathway, MEV or non-MEV, for isoprenoid synthesis. The only enzymes present in all the analyzed proteomes were the farnesyl/geranyl diphosphate synthases and the protein farnesyl transferase complex, indicating that protein prenylation is indispensable to all eukaryotes.
The only obligate endoparasites that possessed sterol biosynthetic genes were the trypanosomatids, Trypanosoma spp. and Leishmania spp. (Fig. 2). Ergosterol has long been known to occur in trypanosomatids (48), and all the analyzed species scored positive for the key enzyme in ergosterol synthesis, sterol 24-C-methyltransferase (enzyme no. 37 in Figs. 1 and 2 and Table 1). In the sterol metabolic tree of Fig. 3, all the trypanosomatids grouped together, sister to the slime molds Dictyostelium and Polysphondylium, which belong to the amoebozoa but have been proposed to have algal ancestry based on their capability to synthesize cycloartenol (6). An unrooted phylogenetic tree of sterol 24-C-methyltransferase (Fig. 4) did not shed light on the ancestry of the enzyme (it was not possible to root the tree due to the lack of an ortholog from prokaryotes to use as an outgroup). The main conclusion from the phylogenetic analysis is that there is no evidence for horizontal transfer as the origin of sterol biosynthetic genes in trypanosomatids.
Apart from their enigmatic history, a major question brought forward by the sterol biosynthetic enzymes of trypanosomatids is to what extent they are exploitable for chemotherapy. Azoles are in development as antichagasic agents because they exhibit selective activity against T. cruzi. Their target is sterol 14-demethylase (enzyme no. 24), and in a phylogenetic tree of the enzyme, the trypanosomatid orthologs form a clearly distinct branch (Fig. 4). We tested other known inhibitors of sterol biosynthesis against trypanosomatids and other parasites (Table 2). In general, the in vitro activity of the inhibitors did not correlate with the presence of their presumed target enzyme (Fig. 5). Although the activity of ketoconazole against T. cruzi was unmatched, simvastatin also showed activity against trypanosomatids. However, the results were not conclusive because, surprisingly, simvastatin was toxic to mammalian cell lines even though it is widely used as a cholesterol-lowering drug. Furthermore, simvastatin was successfully used for the treatment of mouse (49) and dog (50) models of Chagas's disease.
Comparing the stage-specificity of expression of the sterol biosynthetic enzymes in T. brucei and T. cruzi revealed interesting parallels. The sterol metabolic enzymes were differentially regulated across the different life stages, and the expression patterns were similar for both species. The bloodstream forms correlated positively with each other and negatively with the insect forms (Fig. 6). The expression levels of the sterol biosynthetic genes were generally higher in the insect stages than in the mammalian stages of both T. cruzi and T. brucei. Sterol 24-C-methyltransferase (enzyme no. 37) might be a good drug target because it is highly expressed and thus probably essential in all the life-cycle stages. Finally, the trypanosomatids lacked the bona fide genes for sterol esterification (Fig. 2, enzyme nos. 33 and 34), and yet they had been shown to build sterol esters (43). Thus, we hypothesize that trypanosomatids possess atypical sterol ester synthase and esterase, which represent another possible point for chemotherapeutic intervention. In summary, we conclude that sterol metabolism offers further potential drug targets for selective inhibition of trypanosomes.
Acknowledgments
The author thanks Monica Cal, Sibylle Sax, and Christina Kunz-Renggli for technical assistance and drug sensitivity testing; Matthias Witschel for fenpropimorph; and Marcel Tanner for initiating and continuously supporting this line of research.
Footnotes
Abbreviations:
- DOXP
- deoxyxylulose 5-phosphate
- MEP
- methylerythritol 4-phosphate
- MEV
- mevalonate
- PC
- principal component
- SBI
- sterol biosynthesis inhibitor
- SLT
- spliced-leader trapping
This work was supported by the Swiss TPH.
REFERENCES
- 1.Hieb W. F., Rothstein M. 1968. Sterol requirement for reproduction of a free-living nematode. Science. 160: 778–780. [DOI] [PubMed] [Google Scholar]
- 2.Carvalho M., Schwudke D., Sampaio J. L., Palm W., Riezman I., Dey G., Gupta G. D., Mayor S., Riezman H., Shevchenko A., et al. 2010. Survival strategies of a sterol auxotroph. Development. 137: 3675–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurzchalia T. V., Ward S. 2003. Why do worms need cholesterol? Nat. Cell Biol. 5: 684–688. [DOI] [PubMed] [Google Scholar]
- 4.Kaneshiro E. S. 1987. Lipids of Paramecium. J. Lipid Res. 28: 1241–1258. [PubMed] [Google Scholar]
- 5.Smith F. R., Korn E. D. 1968. 7-Dehydrostigmasterol and ergosterol: the major sterols of an amoeba. J. Lipid Res. 9: 405–408. [PubMed] [Google Scholar]
- 6.Nes W. D., Norton R. A., Crumley F. G., Madigan S. J., Katz E. R. 1990. Sterol phylogenesis and algal evolution. Proc. Natl. Acad. Sci. USA. 87: 7565–7569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunt R. C., Ellar D. J. 1974. Isolation of the plasma membrane of a trypanosomatid flagellate: general characterisation and lipid composition. Biochim. Biophys. Acta. 339: 173–189. [DOI] [PubMed] [Google Scholar]
- 8.Urbina J. A., Vivas J., Ramos H., Larralde G., Aguilar Z., Avilan L. 1988. Alteration of lipid order profile and permeability of plasma membranes from Trypanosoma cruzi epimastigotes grown in the presence of ketoconazole. Mol. Biochem. Parasitol. 30: 185–195. [DOI] [PubMed] [Google Scholar]
- 9.Goldstein J. L., Brown M. S. 1990. Regulation of the mevalonate pathway. Nature. 343: 425–430. [DOI] [PubMed] [Google Scholar]
- 10.Kavanagh K. L., Guo K., Dunford J. E., Wu X., Knapp S., Ebetino F. H., Rogers M. J., Russell R. G., Oppermann U. 2006. The molecular mechanism of nitrogen-containing bisphosphonates as antiosteoporosis drugs. Proc. Natl. Acad. Sci. USA. 103: 7829–7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gigante F., Kaiser M., Brun R., Gilbert I. H. 2010. Design and preparation of sterol mimetics as potential antiparasitics. Bioorg. Med. Chem. 18: 7291–7301. [DOI] [PubMed] [Google Scholar]
- 12.Gros L., Castillo-Acosta V. M., Jimenez Jimenez C., Sealey-Cardona M., Vargas S., Manuel Estevez A., Yardley V., Rattray L., Croft S. L., Ruiz-Perez L. M., et al. 2006. New azasterols against Trypanosoma brucei: role of 24-sterol methyltransferase in inhibitor action. Antimicrob. Agents Chemother. 50: 2595–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorente S. O., Rodrigues J. C., Jimenez Jimenez C., Joyce-Menekse M., Rodrigues C., Croft S. L., Yardley V., de Luca-Fradley K., Ruiz-Perez L. M., Urbina J., et al. 2004. Novel azasterols as potential agents for treatment of leishmaniasis and trypanosomiasis. Antimicrob. Agents Chemother. 48: 2937–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magaraci F., Jimenez C. J., Rodrigues C., Rodrigues J. C., Braga M. V., Yardley V., de Luca-Fradley K., Croft S. L., de Souza W., Ruiz-Perez L. M., et al. 2003. Azasterols as inhibitors of sterol 24-methyltransferase in Leishmania species and Trypanosoma cruzi. J. Med. Chem. 46: 4714–4727. [DOI] [PubMed] [Google Scholar]
- 15.Nes C. R., Singha U. K., Liu J., Ganapathy K., Villalta F., Waterman M. R., Lepesheva G. I., Chaudhuri M., Nes W. D. 2012. Novel sterol metabolic network of Trypanosoma brucei procyclic and bloodstream forms. Biochem. J. 443: 267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urbina J. A. 2009. Ergosterol biosynthesis and drug development for Chagas disease. Mem. Inst. Oswaldo Cruz. 104 (Suppl. 1): 311–318. [DOI] [PubMed] [Google Scholar]
- 17.Singh N., Kumar M., Singh R. K. 2012. Leishmaniasis: current status of available drugs and new potential drug targets. Asian Pac. J. Trop. Med. 5: 485–497. [DOI] [PubMed] [Google Scholar]
- 18.Urbina J. A., Payares G., Contreras L. M., Liendo A., Sanoja C., Molina J., Piras M., Piras R., Perez N., Wincker P., et al. 1998. Antiproliferative effects and mechanism of action of SCH 56592 against Trypanosoma (Schizotrypanum) cruzi: in vitro and in vivo studies. Antimicrob. Agents Chemother. 42: 1771–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferraz M. L., Gazzinelli R. T., Alves R. O., Urbina J. A., Romanha A. J. 2007. The anti-Trypanosoma cruzi activity of posaconazole in a murine model of acute Chagas’ disease is less dependent on gamma interferon than that of benznidazole. Antimicrob. Agents Chemother. 51: 1359–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pinazo M. J., Espinosa G., Gallego M., Lopez-Chejade P. L., Urbina J. A., Gascon J. 2010. Successful treatment with posaconazole of a patient with chronic Chagas disease and systemic lupus erythematosus. Am. J. Trop. Med. Hyg. 82: 583–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L. L., Wang G. Z., Zhang H. Y. 2007. Sterol biosynthesis and prokaryotes-to-eukaryotes evolution. Biochem. Biophys. Res. Commun. 363: 885–888. [DOI] [PubMed] [Google Scholar]
- 22.Magrane M.; UniProt Consortium. 2011. UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford). 2011: bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aurrecoechea C., Barreto A., Brestelli J., Brunk B. P., Cade S., Doherty R., Fischer S., Gajria B., Gao X., Gingle A., et al. 2013. EuPathDB: the eukaryotic pathogen database. Nucleic Acids Res. 41: D684–D691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kersey P., Bower L., Morris L., Horne A., Petryszak R., Kanz C., Kanapin A., Das U., Michoud K., Phan I., et al. 2005. Integr8 and Genome Reviews: integrated views of complete genomes and proteomes. Nucleic Acids Res. 33: D297–D302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parra G., Bradnam K., Korf I. 2007. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 23: 1061–1067. [DOI] [PubMed] [Google Scholar]
- 26.Eddy S. R. 1998. Profile hidden Markov models. Bioinformatics. 14: 755–763. [DOI] [PubMed] [Google Scholar]
- 27.Eddy S. R. 2009. A new generation of homology search tools based on probabilistic inference. Genome Inform. 23: 205–211. [PubMed] [Google Scholar]
- 28.Suzek B. E., Huang H., McGarvey P., Mazumder R., Wu C. H. 2007. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 23: 1282–1288. [DOI] [PubMed] [Google Scholar]
- 29.Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.R-Core-Team. 2013. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 31.Suzuki R., Shimodaira H. 2006. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 22: 1540–1542. [DOI] [PubMed] [Google Scholar]
- 32.Nilsson D., Gunasekera K., Mani J., Osteras M., Farinelli L., Baerlocher L., Roditi I., Ochsenreiter T. 2010. Spliced leader trapping reveals widespread alternative splicing patterns in the highly dynamic transcriptome of Trypanosoma brucei. PLoS Pathog. 6: e1001037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anders S., Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Räz B., Iten M., Grether-Bühler Y., Kaminsky R., Brun R. 1997. The Alamar Blue assay to determine drug sensitivity of African trypanosomes in vitro. Acta Trop. 68: 139–147. [DOI] [PubMed] [Google Scholar]
- 35.Desjardins R. E., Canfield C. J., Haynes J. D., Chulay J. D. 1979. Quantitative assessment of antimalarial activity in vitro by a semiautomated microdilution technique. Antimicrob. Agents Chemother. 16: 710–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snyder C., Chollet J., Santo-Tomas J., Scheurer C., Wittlin S. 2007. In vitro and in vivo interaction of synthetic peroxide RBx11160 (OZ277) with piperaquine in Plasmodium models. Exp. Parasitol. 115: 296–300. [DOI] [PubMed] [Google Scholar]
- 37.Buckner F. S., Verlinde C. L., La Flamme A. C., Van Voorhis W. C. 1996. Efficient technique for screening drugs for activity against Trypanosoma cruzi using parasites expressing beta-galactosidase. Antimicrob. Agents Chemother. 40: 2592–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abrahamsen M. S., Templeton T. J., Enomoto S., Abrahante J. E., Zhu G., Lancto C. A., Deng M., Liu C., Widmer G., Tzipori S., et al. 2004. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 304: 441–445. [DOI] [PubMed] [Google Scholar]
- 39.Lorenzi H. A., Puiu D., Miller J. R., Brinkac L. M., Amedeo P., Hall N., Caler E. V. 2010. New assembly, reannotation and analysis of the Entamoeba histolytica genome reveal new genomic features and protein content information. PLoS Negl. Trop. Dis. 4: e716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lujan H. D., Diamond L. S. 1997. Cholesterol requirement and metabolism in Entamoeba histolytica. Arch. Med. Res. 28 (Spec. no.): 96–97. [PubMed] [Google Scholar]
- 41.Diamond L. S., Cunnick C. C. 1991. A serum-free, partly defined medium, PDM-805, for axenic cultivation of Entamoeba histolytica Schaudinn, 1903 and other Entamoeba. J. Protozool. 38: 211–216. [DOI] [PubMed] [Google Scholar]
- 42.Disch A., Schwender J., Muller C., Lichtenthaler H. K., Rohmer M. 1998. Distribution of the mevalonate and glyceraldehyde phosphate/pyruvate pathways for isoprenoid biosynthesis in unicellular algae and the cyanobacterium Synechocystis PCC 6714. Biochem. J. 333: 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coppens I., Levade T., Courtoy P. J. 1995. Host plasma low density lipoprotein particles as an essential source of lipids for the bloodstream forms of Trypanosoma brucei. J. Biol. Chem. 270: 5736–5741. [DOI] [PubMed] [Google Scholar]
- 44.Michels P. A., Marchand M., Kohl L., Allert S., Wierenga R. K., Opperdoes F. R. 1991. The cytosolic and glycosomal isoenzymes of glyceraldehyde-3-phosphate dehydrogenase in Trypanosoma brucei have a distant evolutionary relationship. Eur. J. Biochem. 198: 421–428. [DOI] [PubMed] [Google Scholar]
- 45.Hannaert V., Saavedra E., Duffieux F., Szikora J. P., Rigden D. J., Michels P. A., Opperdoes F. R. 2003. Plant-like traits associated with metabolism of Trypanosoma parasites. Proc. Natl. Acad. Sci. USA. 100: 1067–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cornelissen A. W. C. A., Verspieren M. P., Toulme J., Swinkels B. W., Borst P. 1986. The common 5′ terminal sequence on trypanosome mRNAs: a target for anti-messenger oligodeoxynucleotides. Nucleic Acids Res. 14: 5605–5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ekanayake D. K., Minning T., Weatherly B., Gunasekera K., Nilsson D., Tarleton R., Ochsenreiter T., Sabatini R. 2011. Epigenetic regulation of transcription and virulence in Trypanosoma cruzi by O-linked thymine glucosylation of DNA. Mol. Cell. Biol. 31: 1690–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Halevy S., Sarel S. 1965. Isolation of ergosterol from the trypanosomatid Leptomonas culicidarum. J. Protozool. 12: 293–296. [DOI] [PubMed] [Google Scholar]
- 49.Silva R. R., Shrestha-Bajracharya D., Almeida-Leite C. M., Leite R., Bahia M. T., Talvani A. 2012. Short-term therapy with simvastatin reduces inflammatory mediators and heart inflammation during the acute phase of experimental Chagas disease. Mem. Inst. Oswaldo Cruz. 107: 513–521. [DOI] [PubMed] [Google Scholar]
- 50.Melo L., Caldas I. S., Azevedo M. A., Goncalves K. R., da Silva do Nascimento A. F., Figueiredo V. P., de Figueiredo Diniz L., de Lima W. G., Torres R. M., Bahia M. T., et al. 2011. Low doses of simvastatin therapy ameliorate cardiac inflammatory remodeling in Trypanosoma cruzi-infected dogs. Am. J. Trop. Med. Hyg. 84: 325–331. [DOI] [PMC free article] [PubMed] [Google Scholar]