Abstract
Plasma lipidome is now increasingly recognized as a potentially important marker of chronic diseases, but the exact extent of its contribution to the interindividual phenotypic variability in family studies is unknown. Here, we used the rich data from the ongoing San Antonio Family Heart Study (SAFHS) and developed a novel statistical approach to quantify the independent and additive value of the plasma lipidome in explaining metabolic syndrome (MS) variability in Mexican American families recruited in the SAFHS. Our analytical approach included two preprocessing steps: principal components analysis of the high-resolution plasma lipidomics data and construction of a subject-subject lipidomic similarity matrix. We then used the Sequential Oligogenic Linkage Analysis Routines software to model the complex family relationships, lipidomic similarities, and other important covariates in a variance components framework. Our results suggested that even after accounting for the shared genetic influences, indicators of lipemic status (total serum cholesterol, TGs, and HDL cholesterol), and obesity, the plasma lipidome independently explained 22% of variability in the homeostatic model of assessment-insulin resistance trait and 16% to 22% variability in glucose, insulin, and waist circumference. Our results demonstrate that plasma lipidomic studies can additively contribute to an understanding of the interindividual variability in MS.
Keywords: lipidomics, obesity, lipids, genetics, insulin resistance, plasma lipidomics, variance components
The full spectrum of lipid molecules in human plasma and their potential role in human health and disease are areas of intense interest (1–9). This interest owes substantially to the sophisticated technologies that make it possible to accurately capture and quantify the human lipidome (8, 10). Associative evidence gleaned from plasma lipidomic studies promises vital contributions to biomarker research—an important mainstay of the continued efforts for chronic disease prevention. The plasma lipidomic profile of humans exhibits a wide range of diversity (11, 12) and is associated with several conditions including obesity (13, 14), hypertension (14–16), disorders of glucose metabolism (13, 17), metabolic syndrome (MS) (10, 18), cardiovascular diseases (19), cystic fibrosis (20), nicotine consumption (21), and response to antilipid treatment (22).
Lipidomic association studies conducted in the context of families have an important consideration that the concentrations of the plasma lipid species might be phenotypically as well as genetically correlated with each other (23) especially in related individuals. In that case, it is important to dissect out the genetic basis of the phenotypic traits and the potential contribution of the lipidome over and beyond the genetic basis. In this regard, although the diversity of the human lipidome is established (11, 12), the extent to which the lipidome can independently explain the interindividual variability in the phenotypic expression of disease states is currently unknown. In family studies, the contribution of polygenes to a trait is usually quantified as heritability (24–26) and provides a reasonable first-pass estimate of the likelihood of finding genetic variants that may contribute to the trait in question. Similarly, identification and quantification of a lipidomic variance component (VC) can provide a clue to the likelihood of unveiling potential lipidomic biomarkers without being confounded by the genetic basis of the trait.
In this study, our aim was to determine the proportion of variability of MS traits that is explained by the plasma lipidome independently of the known confounders. For these analyses, we used data from the ongoing San Antonio Family Heart Study (SAFHS) in Mexican Americans (27). These data and samples provide an appropriate opportunity for the aforementioned investigation for the following reasons: i) MS is very common in Mexican Americans (28); ii) the SAFHS recruited 42 large and extended pedigrees that help delineate the potential contribution of genetics to MS (29, 30); iii) high-resolution plasma lipidomic studies have been conducted on a large number of SAFHS participants (15); and iv) using these data, we have previously demonstrated that specific lipid species are associated with hypertension (15), central obesity (31), and type 2 diabetes (32).
Here, we used novel VC methods to measure the variability in phenotypic traits related to MS that is explained by the plasma lipidome. Using these methods, we addressed the following three research questions: First, what is the degree of variability in MS-related traits that can be ascribed to the plasma lipidome in Mexican American families? Second, is this association independent of and additive to the known association of MS with clinically used measures of lipemic status like total serum cholesterol, serum TGs, and serum HDL chlolesterol (HDL-C)? Third, is the association of plasma lipidome with MS independent of obesity?
METHODS
Study subjects
Data for this study came from Mexican American families recruited in the ongoing SAFHS. The recruitment and ascertainment procedures used in the SAFHS have been described in details elsewhere (27, 29, 30). Briefly, the study has now recruited more than 1,600 subjects from 42 large and extended families with a majority of these subjects having completed up to three additional follow-up visits spaced ∼5 years apart. For this study, we used the data and samples collected during the first visit only. This study therefore only uses cross-sectional data from the SAFHS cohort. Complete lipidomic and other phenotypic data were available for 1,206 subjects (from 42 families). Informed consent was obtained from all participants before collection of samples. The Institutional Review Board of the University of Texas Health Sciences Center at San Antonio approved the study.
Lipidomic studies
We analyzed the plasma samples at the Metabolomics Laboratory, Baker IDI Heart and Diabetes Institute, Melbourne, Australia. The lipid extraction and LC/MS methods used in these analyses have recently been described in detail (23). We included a total of 319 lipid species representing 23 lipid classes.
Phenotypic traits
We used data on a total of 14 phenotypic traits related to various components of MS. These included fasting and postglucose load plasma levels of glucose and insulin (assessed through 2 h oral glucose tolerance test); two homeostasis model of assessment (HOMA) measures (i.e., HOMA-IR, representing insulin resistance, and HOMA-β, representing β-cell function); three measures of obesity [i.e., BMI, waist circumference, and waist-hip ratio (WHR)]; systolic blood pressure (SBP) and diastolic blood pressure (DBP); and three serum lipid measures (i.e., total cholesterol, TGs, and HDL-C). The methods of assessment for these traits in the SAFHS participants have been extensively described previously (27, 30).
VC approach
In the VC approach, the total phenotypic variance (Ω) of a trait is analytically decomposed into components that reflect different characteristics (33). The three VC models most relevant to the analyses of the lipidomics data are shown in Table 1. The models are the polygenic (P), lipidomic (L), and polygenic-lipidomic (PL) models. In these models, Ω is represented as a sum of components, each of which is a product of a similarity matrix and its corresponding VC. In a polygenic model, the VCs are σ2G and σ2E, which represent the genetic and environmental components, respectively. The corresponding similarity matrices (dimension n × n, where n represents the sample size) for these two components are ϕ and I (i.e., the kinship and identity matrix, respectively). The elements of a kinship matrix (ϕ) indicate the genetic similarity (kinship coefficient) denoted by relationships between each pair of the study subjects. For example, the routinely used kinship coefficients for different relationships are as follows: identical twins, 1; parent-offspring or sibling, 0.5; and grandparent-grandchild, avuncular, half-siblings, or double first cousins, 0.25. Further, the kinship coefficients for third-, fourth-, fifth-, and sixth-degree relatives are 0.0078, 0.0020, 0.0005, and 0.0001, respectively. The elements of the identity matrix (I) are 1 for diagonals and 0 for off-diagonals.
TABLE 1.
Models used for estimating polygenic and lipidomic VCs
| No. | Model | Specification | Parameters Estimated |
| 1 | Polygenic (P) | Ω = 2 ϕ σ2G + I σ2R | h2r = σ2G/Ω, e2 = σ2R/Ω |
| 2 | Lipidomic (L) | Ω = λ σ2L + I σ2R | L2 = σ2L/Ω, r2 = σ2R/Ω |
| 3 | Polygenic-lipidomic (PL) | Ω = 2 ϕ σ2G + λ σ2L + I σ2R | h2r = σ2G/Ω, L2 = σ2L/Ω, e2 = σ2R/Ω |
Ω, total phenotypic variance; ϕ, kinship coefficient matrix; λ, lipidomic similarity matrix; σ2G, polygenic VC; σ2L, lipidomic VC; σ2R, residual VC; e2, residual environmental contribution; h2r, narrow-sense heritability; I, identity matrix; L2, lipidomic contribution.
The flexibility of the VC approach to analyses of complex pedigrees permits additional and independent VCs by designing and defining similarity matrices based on various other measures of interest. We exploited this feature of VCs by including a term based on the plasma lipidomic similarity between pairs of individuals and the corresponding lipidomic VC. The inclusion of this term alone or in addition to the polygenic component described previously was referred to as the L or the PL model (Table 1). Detailed subsequently are the methods used to generate the lipidomic similarity matrix essential for these analyses.
Lipidomic similarity matrix (λ)
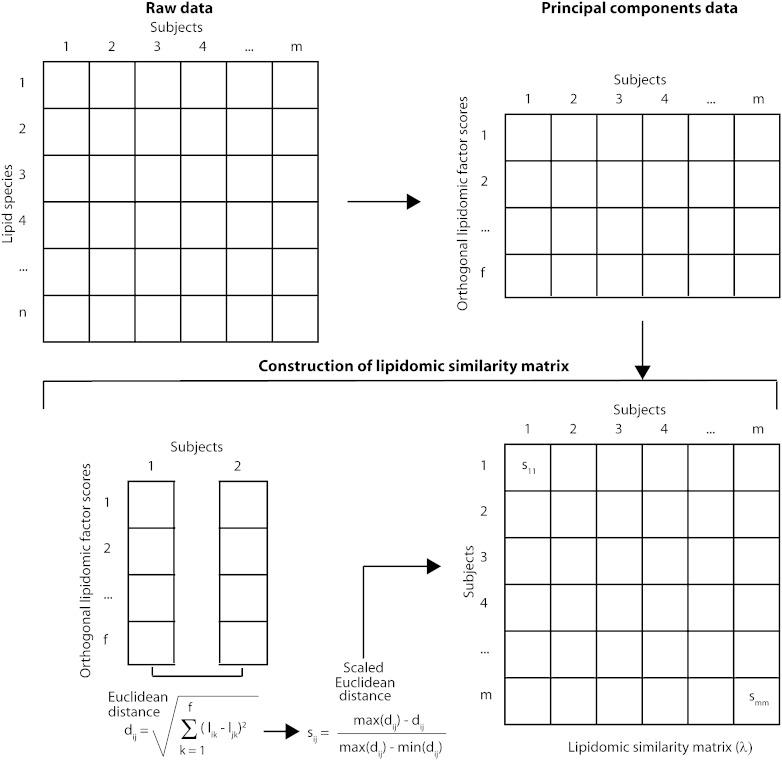
Our goal was to express the similarity between a pair of individuals based on the concentrations of 319 plasma lipid species (Fig. 1). To reduce the dimensionality of the plasma lipidomic data and to ensure that the reduced dimensions are orthogonal to each other, we conducted principal components analyses and extracted all the factors with an eigenvalue exceeding unity (described hereafter as significant factors). This cutoff was chosen because, in the context of principal components analyses, eigenvalues below 1 tend to indicate variables that are noncontributory to the variance of the principal components [also known as Kaiser's criterion (34)]. We then rotated this factor solution using a varimax rotation and obtained factor scores for all the significant factors for each individual. Next, we estimated the Euclidean distance between a pair of individuals i and j as follows:
where l indicates the score for the kth of the f significant factors. We then scaled this Euclidean distance, as shown in Fig. 1, for two reasons: first, this distance conceptually reflects the dissimilarity between two individuals, whereas the elements of λ need to quantify the similarity; and second, the elements of the λ matrix are expected to lie in the range (0, 1).
Fig. 1.
Analytical approach. For details, see text.
Statistical analyses
Principal components analyses were conducted using Stata 12.0 (Stata Corp., College Station, TX) software package. Contribution of the factors to the explanation of the between-subject variability was assessed by ANOVA. All regression models additionally included age, age2, sex, age × sex interaction, age2 × sex interaction, and use of antidiabetic, antihypertensive, or antilipid drugs as additional covariates for adjustment. For running the polygenic, lipidomic, and PL models, we used the Sequential Oligogenic Linkage Analysis Routines software package (35). In these models, the phenotypic traits were first inverse-normalized before subjecting them to analyses. Statistical significance of the estimated parameters (shown in Table 1) was determined by constraining the parameter of interest to 0 and then estimating Chi-square (1 degree of freedom) as −2(LLunconstrained model – LLconstrained model), where LL represents the log-likelihood. Correction for multiple tests was done using Bonferroni's method.
RESULTS
Study participants
The mean age of the study participants was ∼40 years, and the study sample was 60% female. The clinical characteristics of the study subjects are detailed in Table 2. Our study subjects had a high prevalence of type 2 diabetes (∼15%), central obesity (∼48%), and hypertriglyceridemia (∼41%). The prevalence of hypertension (SBP > 140 mm Hg or DBP > 90 mm Hg or history of antihypertensive treatment) was only 13.44%. More than 40% of the study participants had MS, indicating that the families of Mexican Americans included in this study represented a high-risk population for MS in the United States.
TABLE 2.
Clinical characteristics of study participants
| Characteristic | Value |
| Age [mean (SD)], years | 39.55 (16.93) |
| Females [n (%)] | 729 (60.45) |
| Fasting glucose [mean (SD)], mM | 5.58 (2.48) |
| Two-hour postglucose load glucose [mean (SD)], mM | 7.31 (5.03) |
| Fasting insulin [mean (SD)], μU/ml | 16.21 (20.22) |
| Two-hour postglucose load insulin [mean (SD)], μU/ml | 76.37 (73.35) |
| HOMA-IR [mean (SD)] | 4.44 (7.49) |
| HOMA-β [mean (SD)] | 17.19 (56.67) |
| Waist circumference [mean (SD)], cm | 95.08 (17.31) |
| BMI [mean (SD)], kg/m2 | 29.27 (6.63) |
| WHR [mean (SD)] | 0.90 (0.10) |
| SBP [mean (SD)], mm Hg | 120.57 (18.79) |
| DBP [mean (SD)], mm Hg | 70.67 (10.21) |
| Total serum cholesterol [mean (SD)], mg/dl | 189.63 (38.93) |
| Serum TGs [mean (SD)], mg/dl | 145.62 (103.09) |
| HDL-C [mean (SD)], mg/dl | 50.08 (12.89) |
| Subjects with type 2 diabetes [n (%)] | 180 (14.92) |
| Subjects with central obesity [n (%)] | 577 (47.84) |
| Subjects with raised serum TGs [n (%)] | 495 (41.04) |
| Subjects with low serum HDL-C [n (%)] | 260 (21.56) |
| Subjects with high blood pressure [n (%)] | 988 (81.92) |
| Subjects with MS [n (%)] | 490 (40.63) |
Principal components analysis of the plasma lipidome
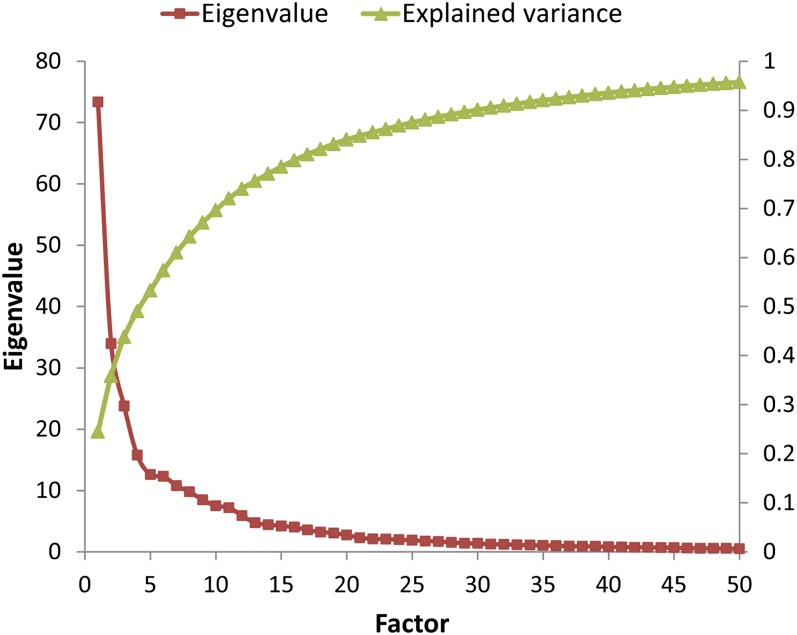
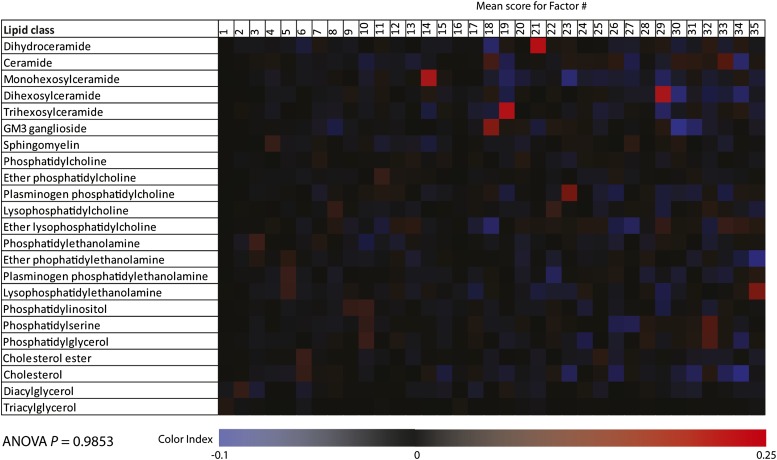
The results of principal components analysis of the 319 lipid species are shown in Fig. 2. Using the criterion of a minimum eigenvalue of unity, we retained 35 orthogonal factors that were further optimized using a varimax rotation. Together, these 35 factors explained 92.05% variability in the plasma lipidome of study participants. We next considered the possibility that the retained factors may be representative of the lipid classes. For this, we estimated the mean factor score for each factor-lipid class combination and then tested the significance of this potential association using ANOVA. Our results showed (Fig. 3) that for most of the factor-lipid class combinations, the mean factor score was near 0. This was supported by the results of ANOVA (F = 0.46, P = 0.9853), indicating that the retained factors reflected novel correlations among the lipid species that are not likely to be captured by the lipid classes.
Fig. 2.
Results of principal components analyses. The red curve associates with the left ordinate (eigenvalues), whereas the green curve associates with the right ordinate (explained variability). The abscissa represents the top 50 most significant factors in order of their ability to explain variability. All factors (n = 35) with an eigenvalue exceeding unity were retained.
Fig. 3.
Heat map representing the mean factor score for each retained factor and the lipid class. The mean factor score is represented as shown in the color index at the bottom. ANOVA P, statistical significance tested using ANOVA.
Contribution of polygenes and plasma lipidomics to variability in MS traits
We studied the interindividual variability in 14 traits related to MS (Table 3) and quantified the proportion of variability explained by the polygenic and lipidomic components. Our results indicated that traits related to glycemia, lipemia, anthropometry, and blood pressure were all significantly associated with the polygenic as well as lipidomic VCs. For the continuous traits, the polygenic contribution ranged from a minimum of 13.88% (for WHR) to a maximum of 35.26% (for SBP). In contrast, the contribution of the lipidomic component was minimum (9.06%) for SBP and maximum (30.92%) for total serum cholesterol. Not surprisingly, the strongest evidence and strength of the contribution of lipidomic VC was found for the three traits related with lipemic status: total serum cholesterol, serum TGs, and serum HDL-C levels. Interestingly, statistical significance (H0: L2 = 0; Ha: L2 > 0) at an α of 0.0036 (corrected for 14 tests using Bonferroni method) was obtained for all the traits except SBP.
TABLE 3.
Estimated polygenic and lipidomic VCs for various traits related to MS
| Trait | h2r | SE | Pa | L2 | SE | Pa |
| Fasting glucose | 0.2542 | 0.0417 | 8.44 × 10−18 | 0.2005 | 0.0263 | 3.65 × 10−10 |
| Two-hour glucose | 0.2670 | 0.0449 | 4.18 × 10−17 | 0.1814 | 0.0255 | 5.06 × 10−10 |
| Fasting insulin | 0.2434 | 0.0408 | 3.50 × 10−16 | 0.2625 | 0.0179 | 4.30 × 10−32 |
| Two-hour insulin | 0.2027 | 0.0406 | 2.65 × 10−12 | 0.2428 | 0.0211 | 1.84 × 10−20 |
| HOMA-IR | 0.2409 | 0.0399 | 9.29 × 10−17 | 0.2666 | 0.0173 | 8.18 × 10−34 |
| HOMA-β | 0.1709 | 0.0416 | 2.35 × 10−8 | 0.1765 | 0.0324 | 5.40 × 10−6 |
| Waist circumference | 0.3007 | 0.0409 | 8.27 × 10−25 | 0.2647 | 0.0162 | 1.17 × 10−40 |
| BMI | 0.3575 | 0.0439 | 1.16 × 10−30 | 0.2495 | 0.0158 | 3.85 × 10−41 |
| WHR | 0.1388 | 0.0358 | 8.38 × 10−8 | 0.2467 | 0.0252 | 2.11 × 10−15 |
| SBP | 0.3526 | 0.0538 | 2.38 × 10−19 | 0.0906 | 0.0369 | 0.0315 |
| DBP | 0.3037 | 0.0498 | 5.38 × 10−17 | 0.1537 | 0.0357 | 0.0002 |
| Serum cholesterol | 0.2049 | 0.0347 | 2.69 × 10−17 | 0.3092 | 0.0113 | 1.21 × 10−99 |
| Serum TGs | 0.2303 | 0.0383 | 3.16 × 10−17 | 0.2875 | 0.0121 | 6.99 × 10−82 |
| HDL-C | 0.3212 | 0.0429 | 3.27 × 10−26 | 0.2587 | 0.0151 | 7.92 × 10−49 |
All models are adjusted for age, age2, sex, age × sex interaction, age2 × sex interaction, and use of antidiabetic, antilipid, and antihypertensive drugs.
Significance using likelihood ratio test.
Independent contribution of the lipidomic component to MS traits
We examined the independence of the observed association in two steps. First, because the strongest association of the lipidomic component was with the routinely used indicators of lipemic status (total serum cholesterol, TGs, and HDL-C), we reasoned that the statistical association between MS traits and the plasma lipidome may have limited clinical use. To explore this, we conducted additional analyses in which we included total serum cholesterol, serum TGs, and HDL-C as additional covariates in the PL model and reran the analyses shown in Table 3. The results of these analyses are shown in Table 4. We observed (column titled “Before Adjusting for BMI”) that except for the blood pressure traits, the variability in all other traits was significantly and substantially explained by the lipidomic VC. Again, the lipidomic component significantly explained variability in the HOMA-IR and HOMA-β traits, but its most significant contribution was to the anthropometric traits capturing obesity and central obesity. Considering that obesity is a major component of MS, we repeated these analyses by additionally accounting for BMI. Our results (Table 4, column titled “After Adjusting for BMI”) showed that even after accounting for clinical covariates (age, sex, and their linear and nonlinear interactions), indicators of lipemic status (total serum cholesterol, serum TGs, and HDL-C), and obesity (BMI), the lipidomic VC continued to be an independent predictor of other MS-related traits (Bonferroni corrected P < 0.0036).
TABLE 4.
Independent contribution of the lipidomic VC to variability in traits related to type 2 diabetes, blood pressure, and obesity
| Trait | Before Adjusting for BMI |
After Adjusting for BMI |
||||
| L2 | SE | Pa | L2 | SE | Pa | |
| Fasting glucose | 0.1846 | 0.0298 | 3.1 × 10−7 | 0.1550 | 0.0353 | 0.0003 |
| Two-hour glucose | 0.1404 | 0.0255 | 8.5 × 10−5 | 0.1186 | 0.0356 | 0.0023 |
| Fasting insulin | 0.2521 | 0.0204 | 6.2 × 10−23 | 0.2181 | 0.0279 | 1.0 × 10−9 |
| Two-hour insulin | 0.2280 | 0.0243 | 1.7 × 10−14 | 0.1869 | 0.0311 | 1.1 × 10−6 |
| HOMA-IR | 0.2509 | 0.0203 | 9.0 × 10−23 | 0.2215 | 0.0270 | 1.3 × 10−10 |
| HOMA-β | 0.1518 | 0.0389 | 0.0012 | 0.1330 | 0.0448 | 0.0199 |
| Waist circumference | 0.2608 | 0.0174 | 1.0 × 10−34 | 0.1984 | 0.0390 | 3.66 × 10−6 |
| BMI | 0.2438 | 0.0169 | 3.8 × 10−35 | — | — | — |
| WHR | 0.2233 | 0.0309 | 1.2 × 10−9 | 0.1724 | 0.0438 | 0.0002 |
| SBP | 0.0000 | 0.0000 | — | 0.0000 | 0.0000 | — |
| DBP | 0.0931 | 0.0466 | 0.0726 | 0.0000 | 0.0000 | — |
All models are adjusted for age, age2, sex, age × sex interaction, age2 × sex interaction, total serum cholesterol, serum TGs, serum HDL-C, and use of antidiabetic, antilipid, and antihypertensive drugs.
Significance using likelihood ratio test.
DISCUSSION
Using a novel modification of the VC approach to analysis of complex pedigrees and the rich data from a high-risk sample of Mexican American families recruited in the SAFHS, we found that phenotypic traits reflecting glycemia, insulin resistance, central obesity, and general obesity were substantially and significantly determined by the plasma lipidomic profile (results shown in Table 4). This contribution of the plasma lipidome was independent of both the polygenic contribution, routinely used measures of lipemic status, and general obesity. Our results therefore underscore the additive value of the plasma lipidomic profile in MS.
Novelty
An important novelty of this study is the method of analysis. We used the VC approach to quantify the explained variability due to plasma lipidome (detailed in Ref. 36), but to successfully capture the variability in the plasma lipidome, we resorted to two preprocessing steps: principal components analysis and construction of the lipidomic similarity matrix. This approach had three advantages. First, the validity of using principal components was indirectly indicated by the observation that 92.05% of variability in the plasma lipidome was explained by the retained factors and that these factors were characteristically different from the lipid classes. The other advantage of using principal components was that the solution is, by design, orthogonal and therefore yields to estimation of Euclidean distances in an n-dimensional hyperspace. Second, the scaling and representation of the pair-wise Euclidean distances were useful in the construction of the λ matrix. This matrix was then readily used in the VC framework. Methodological variations in this approach that are based on weighting of factors and preminimization of correlations is also possible but would not lead to a meaningful improvement because the factor solution used in these analyses already explains most of the variability. Third, the Sequential Oligogenic Linkage Analysis Routines software is a flexible modeling environment that permits custom representation of improvised models such as the one used here, thereby facilitating the estimation of all related parameters and their statistical significance (35, 36).
Limitations
There are three limitations to the use of our analytical approach. First, VC approaches used in a cross-sectional study setting can only provide an associative estimate of the explained variability. The interpretations cannot be viewed as causal. In this vein, it should also be noted that the phrase “explained variability” as used in this paper does not connote causality but only refers to the estimated statistical contributions of one variable to the other. With regard to the potential contribution of plasma lipidomics to MS, there exists a tautological complexity such that plasma lipid species may be proximal, concomitant, or distal to the initiation of the disease (37–39). Also, because obesity is a major component of MS, it can be argued that our observations demonstrate the changes in lipidome consequent to, rather than leading to, obesity. However, the observed associations continued to hold even after adjusting for obesity. Therefore, while a confounding effect of obesity on the lipidome-MS nexus cannot be ignored, our findings indicate that lipidome was still an independent contributor to MS. Moreover, both MS and the plasma lipidomic profile can be expected to be at least partially controlled by a genetic predisposition. Nevertheless, the continued search for biomarkers of MS invokes the need to quantify the degree of variability in MS that is attributable to the diversity of plasma lipid species. Our methodological approach suited for family studies represents an important first step in that direction.
Second, the VC related to the plasma lipidome is a sum and total of all the lipid species and does not represent any single lipid class or species. Consequently, the explained variability is a complex function of all the lipid species levels as well as their correlation structure. This is both an advantage and a limitation. It is an advantage because it reduces the complexity of the lipidome and offers a first-pass screen for the potential use of lipidomic biomarkers in a specified setting. On the other hand, this approach does not permit identification of single (or a combination of) species most contributory to the observed association.
Third, a corollary to the abovementioned limitation here is that even if the proportion of trait variability explained by the plasma lipidome is nonsignificant or 0, it does not negate the possibility that one or more lipid species may be significantly associated with the trait. The preprocessing steps used in this study can mask such potential associations at the level of lipid species. For example, diacylglycerol species and ether lipid deficiencies have been shown to be significantly associated with the risk of incident or prevalent hypertension (14–16), but in this study, we found that the plasma lipidome per se was not contributory to the variability in blood pressure. We therefore recommend that use of the analytical approach outlined in this study should not preclude more detailed and lipid species-specific analyses.
Implications
Our observations have two implications. First, we demonstrated 22%, 22%, 20%, and 16% independent contribution of the lipidomic VC to variability in HOMA-IR, fasting insulin, waist circumference, and fasting glucose, respectively (Table 4). These results imply that the plasma lipidome may have an independent and additive utility in detection of insulin resistance and central obesity—conditions common in Mexican Americans (28, 40). Lipidomic studies conducted in the past (14, 41–43) have been able to detect some key associations of specific lipid species with insulin resistance, but our results indicate that the entire plasma lipidome may be associated with substantial changes in these traits. In the context of VC models as used in this study, it is important to note that these contributions of the plasma lipidome are independent of the polygenic component, indicating that the plasma lipidome may be partially tracking the environmental aspects of obesity and insulin resistance. In unison with the potentially contributory genetic pathways (44), the plasma lipidome can therefore be anticipated to act as a biomarker of obesity and insulin resistance. Further, it will also be interesting to investigate whether plasma lipidome can provide insights into future trajectories of anthropometric and other indices related to MS.
Second, we showed that the significant principal components derived from the plasma lipidome were independent of the chemically defined lipid classes (Fig. 3). This result indicates that the prevalent concentrations of lipid species in human plasma are likely a result of complex biological pathways that need to be considered rather than the more limited vision restricted to lipid classes. Indeed, future studies need to consider if biologically meaningful information can be gleaned from the correlations among all the plasma lipid species. It is interesting in this respect that only 35 principal components accounted for 92% of overall variability of the plasma lipidome, indicating that it may be possible to reduce the redundancy of the lipidome in order to better delineate the biological pathways involved in health and disease (45).
CONCLUSIONS
Using a novel analytical approach and rich data from Mexican American families recruited in the SAFHS, we have demonstrated that high-resolution plasma lipidomic studies can provide substantial and significant improvement in our understanding of interindividual variability associated with MS. Specifically, the plasma lipidome contributed to 22% variability in HOMA-IR and 16% to 22% variability in glucose, insulin, and waist circumference independent of obesity and measures of lipemic status. Future studies need to evaluate the potential role of the plasma lipidome as a biomarker of MS.
Acknowledgments
The authors are grateful to the participants of the SAFHS for their continued involvement.
Footnotes
Abbreviations:
- DBP
- diastolic blood pressure
- HDL-C
- HDL cholesterol
- HOMA
- homeostasis model of assessment
- HOMA-β
- homeostasis model of assessment-β-cell function
- HOMA-IR
- homeostasis model of assessment-insulin resistance
- MS
- metabolic syndrome
- PL
- polygenic-lipidomic
- SAFHS
- San Antonio Family Heart Study
- SBP
- systolic blood pressure
- VC
- variance component
- WHR
- waist-hip ratio
This work was supported in part by National Institutes of Health (NIH) Grants R01 DK082610 and R01 DK079169. Data collection for the San Antonio Family Heart Study was supported by NIH Grant P01 HL045522. The development of the analytical methods and software used in this study were supported by NIH Grant R37 MH059490. The AT&T Genomics Computing Center supercomputing facilities used for this work were supported in part by a gift from the AT&T Foundation and with support from the National Center for Research Resources Grant Number S10 RR029392. This investigation was conducted in facilities constructed with support from Research Facilities Improvement Program Grants C06 RR013556 and C06 RR017515 from the National Center for Research Resources of the NIH. This work was also supported by funding from the National Health and Medical Research Council of Australia; the OIS Program of the Victorian government, Australia; and Award Number 1R01DK088972-01 from the National Institute of Diabetes and Digestive and Kidney Diseases, NIH.
REFERENCES
- 1.Donovan E. L., Pettine S. M., Hickey M. S., Hamilton K. L., Miller B. F. 2013. Lipidomic analysis of human plasma reveals ether-linked lipids that are elevated in morbidly obese humans compared to lean. Diabetol. Metab. Syndr. 5: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tajima Y., Ishikawa M., Maekawa K., Murayama M., Senoo Y., Nishimaki-Mogami T., Nakanishi H., Ikeda K., Arita M., Taguchi R., et al. 2013. Lipidomic analysis of brain tissues and plasma in a mouse model expressing mutated human amyloid precursor protein/tau for Alzheimer's disease. Lipids Health Dis. 12: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mielke M. M., Haughey N. J. 2012. Could plasma sphingolipids be diagnostic or prognostic biomarkers for Alzheimer's disease? Clin. Lipidol. 7: 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishikawa M., Tajima Y., Murayama M., Senoo Y., Maekawa K., Saito Y. 2013. Plasma and serum from nonfasting men and women differ in their lipidomic profiles. Biol. Pharm. Bull. 36: 682–685. [DOI] [PubMed] [Google Scholar]
- 5.Laurila P. P., Surakka I., Sarin A. P., Yetukuri L., Hyotylainen T., Soderlund S., Naukkarinen J., Tang J., Kettunen J., Mirel D. B., et al. 2013. Genomic, transcriptomic, and lipidomic profiling highlights the role of inflammation in individuals with low high-density lipoprotein cholesterol. Arterioscler. Thromb. Vasc. Biol. 33: 847–857. [DOI] [PubMed] [Google Scholar]
- 6.Rasmiena A. A., Ng T. W., Meikle P. J. 2013. Metabolomics and ischaemic heart disease. Clin. Sci. (Lond.). 124: 289–306. [DOI] [PubMed] [Google Scholar]
- 7.Fernando H., Bhopale K. K., Kondraganti S., Kaphalia B. S., Shakeel Ansari G. A. 2011. Lipidomic changes in rat liver after long-term exposure to ethanol. Toxicol. Appl. Pharmacol. 255: 127–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kontush A., Chapman M. J. 2010. Lipidomics as a tool for the study of lipoprotein metabolism. Curr. Atheroscler. Rep. 12: 194–201. [DOI] [PubMed] [Google Scholar]
- 9.Masoodi M., Nicolaou A. 2006. Lipidomic analysis of twenty-seven prostanoids and isoprostanes by liquid chromatography/electrospray tandem mass spectrometry. Rapid Commun. Mass Spectrom. 20: 3023–3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meikle P. J., Christopher M. J. 2011. Lipidomics is providing new insight into the metabolic syndrome and its sequelae. Curr. Opin. Lipidol. 22: 210–215. [DOI] [PubMed] [Google Scholar]
- 11.Quehenberger O., Armando A. M., Brown A. H., Milne S. B., Myers D. S., Merrill A. H., Bandyopadhyay S., Jones K. N., Kelly S., Shaner R. L., et al. 2010. Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res. 51: 3299–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chua E. C., Shui G., Lee I. T., Lau P., Tan L. C., Yeo S. C., Lam B. D., Bulchand S., Summers S. A., Puvanendran K., et al. 2013. Extensive diversity in circadian regulation of plasma lipids and evidence for different circadian metabolic phenotypes in humans. Proc. Natl. Acad. Sci. U. S. A. 110: 14468–14473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Mello V. D., Lankinen M., Schwab U., Kolehmainen M., Lehto S., Seppanen-Laakso T., Oresic M., Pulkkinen L., Uusitupa M., Erkkila A. T. 2009. Link between plasma ceramides, inflammation and insulin resistance: association with serum IL-6 concentration in patients with coronary heart disease. Diabetologia. 52: 2612–2615. [DOI] [PubMed] [Google Scholar]
- 14.Graessler J., Schwudke D., Schwarz P. E., Herzog R., Shevchenko A., Bornstein S. R. 2009. Top-down lipidomics reveals ether lipid deficiency in blood plasma of hypertensive patients. PLoS ONE. 4: e6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulkarni H., Meikle P. J., Mamtani M., Weir J. M., Barlow C. K., Jowett J. B., Bellis C., Dyer T. D., Johnson M. P., Rainwater D. L., et al. 2013. Plasma lipidomic profile signature of hypertension in Mexican American families: specific role of diacylglycerols. Hypertension. 62: 621–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spijkers L. J., van den Akker R. F., Janssen B. J., Debets J. J., De Mey J. G., Stroes E. S., van den Born B. J., Wijesinghe D. S., Chalfant C. E., MacAleese L., et al. 2011. Hypertension is associated with marked alterations in sphingolipid biology: a potential role for ceramide. PLoS ONE. 6: e21817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wopereis S., Radonjic M., Rubingh C., Erk M., Smilde A., Duyvenvoorde W., Cnubben N., Kooistra T., Ommen B., Kleemann R. 2012. Identification of prognostic and diagnostic biomarkers of glucose intolerance in ApoE3Leiden mice. Physiol. Genomics. 44: 293–304. [DOI] [PubMed] [Google Scholar]
- 18.Gross R. W., Han X. 2007. Lipidomics in diabetes and the metabolic syndrome. Methods Enzymol. 433: 73–90. [DOI] [PubMed] [Google Scholar]
- 19.Puri R., Duong M., Uno K., Kataoka Y., Nicholls S. J. 2012. The emerging role of plasma lipidomics in cardiovascular drug discovery. Expert Opin. Drug Discov. 7: 63–72. [DOI] [PubMed] [Google Scholar]
- 20.Ollero M., Astarita G., Guerrera I. C., Sermet-Gaudelus I., Trudel S., Piomelli D., Edelman A. 2011. Plasma lipidomics reveals potential prognostic signatures within a cohort of cystic fibrosis patients. J. Lipid Res. 52: 1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang-Sattler R., Yu Y., Mittelstrass K., Lattka E., Altmaier E., Gieger C., Ladwig K. H., Dahmen N., Weinberger K. M., Hao P., et al. 2008. Metabolic profiling reveals distinct variations linked to nicotine consumption in humans—first results from the KORA study. PLoS ONE. 3: e3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bergheanu S. C., Reijmers T., Zwinderman A. H., Bobeldijk I., Ramaker R., Liem A. H., van der Greef J., Hankemeier T., Jukema J. W. 2008. Lipidomic approach to evaluate rosuvastatin and atorvastatin at various dosages: investigating differential effects among statins. Curr. Med. Res. Opin. 24: 2477–2487. [DOI] [PubMed] [Google Scholar]
- 23.Weir J. M., Wong G., Barlow C. K., Greeve M. A., Kowalczyk A., Almasy L., Comuzzie A. G., Mahaney M. C., Jowett J., Shaw J., et al. 2013. Plasma lipid profiling in a large population based cohort. J. Lipid Res. 54: 2898–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uhl G. R., Drgon T., Johnson C., Liu Q. R. 2009. Addiction genetics and pleiotropic effects of common haplotypes that make polygenic contributions to vulnerability to substance dependence. J. Neurogenet. 23: 272–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mercado M. M., McLenithan J. C., Silver K. D., Shuldiner A. R. 2002. Genetics of insulin resistance. Curr. Diab. Rep. 2: 83–95. [DOI] [PubMed] [Google Scholar]
- 26.Guillausseau P. J., Tielmans D., Virally-Monod M., Assayag M. 1997. Diabetes: from phenotypes to genotypes. Diabetes Metab. 23 (Suppl. 2): 14–21. [PubMed] [Google Scholar]
- 27.Mitchell B. D., Almasy L. A., Rainwater D. L., Schneider J. L., Blangero J., Stern M. P., MacCluer J. W. 1999. Diabetes and hypertension in Mexican American families: relation to cardiovascular risk. Am. J. Epidemiol. 149: 1047–1056. [DOI] [PubMed] [Google Scholar]
- 28.Razzouk L., Muntner P. 2009. Ethnic, gender, and age-related differences in patients with the metabolic syndrome. Curr. Hypertens. Rep. 11: 127–132. [DOI] [PubMed] [Google Scholar]
- 29.MacCluer J. W., Stern M. P., Almasy L., Atwood L. A., Blangero J., Comuzzie A. G., Dyke B., Haffner S. M., Henkel R. D., Hixson J. E., et al. 1999. Genetics of atherosclerosis risk factors in Mexican Americans. Nutr. Rev. 57: S59–S65. [DOI] [PubMed] [Google Scholar]
- 30.Voruganti V. S., Lopez-Alvarenga J. C., Nath S. D., Rainwater D. L., Bauer R., Cole S. A., Maccluer J. W., Blangero J., Comuzzie A. G. 2008. Genetics of variation in HOMA-IR and cardiovascular risk factors in Mexican-Americans. J. Mol. Med. (Berl.). 86: 303–311. [DOI] [PubMed] [Google Scholar]
- 31.Mamtani M., Meikle P. J., Kulkarni H., Weir J. M., Barlow C. K., Jowett J. B., Bellis C., Dyer T. D., Almasy L., Mahaney M. C., et al. 2014. Plasma dihydroceramide species associate with waist circumference in Mexican American families. Obesity (Silver Spring). 22: 950–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meikle P. J., Wong G., Barlow C. K., Weir J. M., Greeve M. A., Macintosh G. L., Almasy L., Comuzzie A. G., Mahaney M. C., Kowalczyk A., et al. 2013. Plasma lipid profiling shows similar associations with prediabetes and type 2 diabetes. PLoS ONE. 8: e74341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walder K., Segal D., Jowett J., Blangero J., Collier G. R. 2003. Obesity and diabetes gene discovery approaches. Curr. Pharm. Des. 9: 1357–1372. [DOI] [PubMed] [Google Scholar]
- 34.Bandalos D. L., Boehm-Kaufman M. R. 2009. Four common misconceptions in exploratory factor analysis. In Statistical and Methodological Myths and Urban Legends: Doctrine, Verity and Fable in the Organizational and Social Sciences. E. E. Lance and R. J. Vandenberg, editors. Taylor & Francis, New York. 61–87. [Google Scholar]
- 35.Almasy L., Blangero J. 1998. Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 62: 1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blangero J., Diego V. P., Dyer T. D., Almeida M., Peralta J., Kent J. W., Jr, Williams J. T., Almasy L., Goring H. H. 2013. A kernel of truth: statistical advances in polygenic variance component models for complex human pedigrees. Adv. Genet. 81: 1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aguilera C. M., Gil-Campos M., Canete R., Gil A. 2008. Alterations in plasma and tissue lipids associated with obesity and metabolic syndrome. Clin. Sci. (Lond.). 114: 183–193. [DOI] [PubMed] [Google Scholar]
- 38.Rader D. J. 2003. Regulation of reverse cholesterol transport and clinical implications. Am. J. Cardiol. 92 (Suppl. 1): 42–49. [DOI] [PubMed] [Google Scholar]
- 39.Matsuzawa Y., Funahashi T., Nakamura T. 1999. Molecular mechanism of metabolic syndrome X: contribution of adipocytokines adipocyte-derived bioactive substances. Ann. N. Y. Acad. Sci. 892: 146–154. [DOI] [PubMed] [Google Scholar]
- 40.Meigs J. B. 2003. Epidemiology of the insulin resistance syndrome. Curr. Diab. Rep. 3: 73–79. [DOI] [PubMed] [Google Scholar]
- 41.Zhao C., Mao J., Ai J., Shenwu M., Shi T., Zhang D., Wang X., Wang Y., Deng Y. 2013. Integrated lipidomics and transcriptomic analysis of peripheral blood reveals significantly enriched pathways in type 2 diabetes mellitus. BMC Med. Genomics. 6 (Suppl. 1): S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barber M. N., Risis S., Yang C., Meikle P. J., Staples M., Febbraio M. A., Bruce C. R. 2012. Plasma lysophosphatidylcholine levels are reduced in obesity and type 2 diabetes. PLoS ONE. 7: e41456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kotronen A., Velagapudi V. R., Yetukuri L., Westerbacka J., Bergholm R., Ekroos K., Makkonen J., Taskinen M. R., Oresic M., Yki-Jarvinen H. 2009. Serum saturated fatty acids containing triacylglycerols are better markers of insulin resistance than total serum triacylglycerol concentrations. Diabetologia. 52: 684–690. [DOI] [PubMed] [Google Scholar]
- 44.Dulloo A. G., Jacquet J., Solinas G., Montani J. P., Schutz Y. 2010. Body composition phenotypes in pathways to obesity and the metabolic syndrome. Int. J. Obes. (Lond.). 34 (Suppl. 2): S4–S17. [DOI] [PubMed] [Google Scholar]
- 45.Yetukuri L., Ekroos K., Vidal-Puig A., Oresic M. 2008. Informatics and computational strategies for the study of lipids. Mol. Biosyst. 4: 121–127. [DOI] [PubMed] [Google Scholar]