Abstract
Mass spectrometry coupled immunoprecipitation (MS-IP) studies are useful in identifying and quantitating potential binding partners of a target protein. However, they are often conducted without appropriate loading controls. Western blots are often used to analyze loading controls, yet there are limitations to their usefulness as analytical tools. One remedy for this is the use of selected reaction monitoring (SRM), where the areas under the curve (AUCs) of peptides from a protein of interest can be normalized to those from the constant regions of the immunoglobulins used for the IP. Using this normalization method, significant changes in relative peptide abundance were observed between samples when there appeared to be an unequal load based on immunoglobulin peptide abundance.
Affinity purification (AP) constitutes a category of protein enrichment strategies utilized for analysis of protein complexes.1 In order to identify and quantify proteins involved in these complexes, AP is frequently coupled with mass spectrometry (MS).1,2 One method of AP is immunoprecipitation (IP, Figure 1a), where an antibody is coupled to a solid phase (often protein A or G conjugated sepharose beads) and incubated with the biological sample of interest. The attached proteins are removed through elution or denaturation.3 The final sample for analysis contains antibody, the protein of interest, and any associated proteins. One pitfall of this strategy is that the concentration of beads can vary slightly between samples due to variable bead slurry distributions. This results in variations in the amount of antibody bound and amount of antigen and interacting proteins in the eluate. Additionally, antibody affinity for an antigen could change with mutations to the antigen. This could result in lowered recovery of the mutated antigen and be misconstrued as biologically significant. Stable isotope labeling by amino acids in cell culture (SILAC)4 is often combined with IP and ensures that samples undergo identical preparations. However, if the biological conditions of interest alter the affinity of the antigen to the antibody, sample loads would no longer be comparable. These issues indicate that a normalization method is needed in MS-IP experiments to control for discrepancies. Specifically, a constant variable between IP experiments could be used to mitigate such variations.
Figure 1.
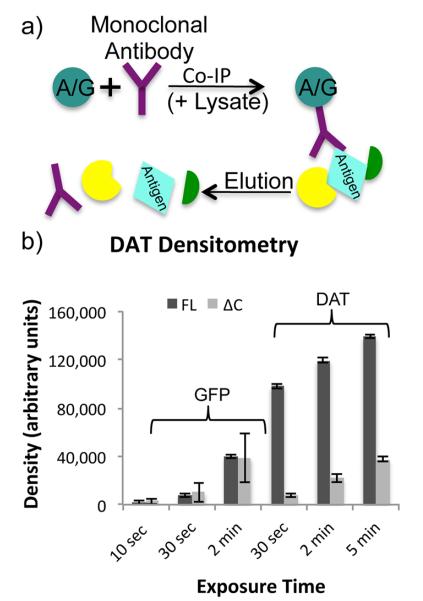
IP schematic and densitometry. (a) In a standard IP experiment, protein A or protein G coupled sepharose beads are combined with an antibody and a sample lysate. The antibody binds to both the beads and its target antigen, which may also affinity enrich a variety of interacting proteins. Proteins are eluted from the beads leaving the antibody in solution with the antigen and interacting proteins. (b) Three replicate Western blots of FL-DAT, ΔN-DAT, and ΔC-DAT αGFP IPs were performed, probing with either αDAT or αGFP primary antibody. In the αGFP blots, the ΔN-DAT was more prominent than the other lanes, while in the αDAT blots ΔN-DAT was not detected as the antibody is directed toward the N-terminus of DAT. Densitometry measurements of FL-DAT and ΔC-DAT bands at three different exposure times resulted in dramatic variations in densities.
One such variable is the amount of antibody. The immunoglobulin G (IgG) antibody class consists of two γ heavy and two light (κ or λ) chains with both variable and constant domains. Variable domain sequences differ between antibodies to form antigen-specific binding sites.5 Thus, constant regions are more reliable target regions for proteomic analyses and could be utilized for analysis of a wide variety of antibodies. Peptides from these regions have previously been utilized as internal standards for purified serum IgGs.6
The antigen of interest for this study was dopamine transporter (DAT), which is a biologically relevant 12-transmembrane domain integral membrane protein.7,8 Three dually tagged DAT constructs including full length (FL-DAT), N-terminally deleted (ΔN-DAT), and C-terminally deleted (ΔC-DAT) versions (described in the Experimental Section) with different trafficking and localization patterns were used as bait in this study.9,10 These constructs have different expression levels making it difficult to compare their binding partners without accurately comparing DAT abundance. To do this, IgG levels were used as an internal standard to which DAT levels could be normalized, allowing for the standardization of input levels based on the effectiveness of the IPs. This concept is often applied to Western blots where IgGs are used as loading controls. However, densitometry is not always an accurate representation of protein abundances in a sample, due to the limited linear dynamic range (2 orders of magnitude) of enhanced chemiluminescence (ECL), the most common method of Western blot detection.11,12 Thus, it can be inaccurate to compare these levels between samples (Figure 1b).
Rabbit and mouse antibodies targeting distinct tags on the DAT constructs were used as these are the most widely used antibody types for IPs.13 IgG and DAT levels in IP samples can be monitored via selected reaction monitoring (SRM) after selecting proteotypic peptides for quantitative analysis, and the areas under the curve (AUCs) of DAT peptides can be compared with IgG peptide AUCs for relative normalization before comparing DAT levels between samples. Normalization of peptides of interest to internal standard peptides has been shown to be a robust method that can determine relative abundance without absolute quantitation.14,15
We explored the utilization of IgGs in MS-IP studies through a relative normalization and quantitation pipeline. After identification and optimization of proteotypic peptides within IgG constant domains, and normalization of the AUC of an antigen peptide to that of an IgG peptide, we found that the relative quantitation could be significantly altered such that increased variation between IP samples resulted in significant changes between pre- and postnormalized peptide levels.
EXPERIMENTAL SECTION
Reagents and Cell Lines
Solvents were purchased from Fisher Scientific (Rockford, IL); other chemicals were from Fisher Scientific, JT Baker (Center Valley, PA), or Sigma (St. Louis, MO) unless noted.
Antibodies ordered from Abcam (Cambridge, MA) include rabbit-anti-GFP polyclonal ab290, rabbit-anti-β-actin polyclonal ab8227, rabbit-anticatalase polyclonal ab1877, and mouse-anti-GAPDH monoclonal ab9484 clone number mAbcam9484. Additional antibodies used were mouse-anti-β-actin monoclonal Sigma A5441 clone AC-15 and mouse-anti-HA.11 monoclonal Covance (Princeton, NJ) MMS-101P clone 16B12. HPLC purified peptides (>95% purity) were purchased from Thermo Scientific (Rockford, IL).
Porcine aortic endothelial (PAE) cells stably expressed dopamine transporter constructs dually tagged with yellow fluorescent protein (YFP) on the N-terminus and hemag-glutinin (HA) within the second extracellular loop. Parental cells were used along with full-length (FL), N-terminally deleted (ΔN), and C-terminally deleted (ΔC) YFP-HA-DAT expressing PAE cell lines. FL-DAT and ΔN-DAT PAE cell lines were previously described.9,10 The ΔC-DAT cell line was similarly generated, where the 30 C-terminal residues of DAT were removed, and the last two residues of the truncated protein were mutated to retain the PDZ binding domain (LAY → LKV). PAE cells were grown in Ham’s F12 medium containing 10% fetal bovine serum.
Immunoprecipitation
Cells were grown to 80-90% confluency on 245 mm square plates (Corning, NY) before two DPBS rinses and harvested using a cell scraper on ice in 2 mL of lysis buffer (20 mM Tris-HCl, pH 8, 137 mM NaCl, 10% glycerol, 1% Triton X-100, 2 mM EDTA) with protease inhibitors (aprotonin, leupeptin, and pepstatin A at 1 μg/mL and bestatin at 4 μg/mL final concentrations) added immediately prior to use. Cell pellets were broken up via pipetting. Samples were incubated 30 min with rotation at 4 °C then centrifuged 30 min at 20 817g at 4 °C to pellet cellular debris. Antibody was added to supernatant (55 μg of rabbit αGFP antibody or 30 μg of mouse αHA antibody) and samples were incubated with rotation overnight at 4 °C. Negative controls were incubated overnight without antibody. Sepharose beads (protein A conjugated for rabbit αGFP antibody samples, protein G conjugated for mouse αHA antibody samples) were added at a ratio of 100 μL of beads to 1 mL of sample, and samples were nutated 4 h at 4 °C. Samples were rinsed three times with 500 μL of lysis buffer before elution in 100 μL of 200 mM glycine, pH 2.5 for rabbit αGFP antibody samples. Mouse αHA antibody samples were denatured in 100 μL of Laemmli buffer without bromophenol blue by nutating 10 min at room temperature and boiling 5 min at 100 °C. Samples were centrifuged 15 min at 4 °C and 20 817g. Supernatants were transferred to clean tubes. In total, 50 μg of bovine serum albumin (BSA) was added to IP samples as a carrier protein for further processing steps.
MS Sample Preparation
IP samples and untreated antibodies were prepared through methanol-chloroform precipitation,16 resuspension with Rapigest, reduced with dithiothreitol, and alkylated with iodoacetamide before tryptic digestion. IP samples were cleaned using Pierce C18 tips (100 μL, Thermo Scientific, Rockford, IL) and resuspended to 0.25 mg/mL in 0.1% formic acid. For a more detailed description see the Supporting Information.
Western Blots
In total, 10 μg of each IP sample was loaded onto a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel that underwent electrophoresis. Proteins were transferred to a poly(vinylidene fluoride) (PVDF) membrane via semidry transfer. Monoclonal mouse or polyclonal rabbit primary antibodies directed to HA or GFP, respectively, and secondary antibodies from the appropriate animals conjugated with horseradish peroxidase were utilized. Bands were detected via ECL using the Pierce SuperSignal West Pico Chemiluminescent Substrate (Rockford, Il) and X-ray films and quantified using Image J software.
Nanoliquid Chromatography (NanoLC)-Mass Spectrometry
Antibody digests were analyzed using a Proxeon EASY-nLC II and an Orbitrap Elite mass spectrometer with Xcaliber software. A mass of 2 μg of each sample was loaded onto a 15 cm reversed-phase (RP) column maintained at 25 °C, using a column heater built in-house as previously described.17 Data dependent acquisition results were searched using SEQUEST,18 processed with Bullseye,19 and postprocessed using Percolator,20 and proteins were inferred using MSDaPl.21
IP digests underwent SRM analyses using a Proxeon EASY-nLC II and a TSQ Vantage Triple Stage Quadrupole mass spectrometer with Xcaliber software. In total, 1 μg of αHA IP or 2 μg of αGFP IP sample was loaded onto a 30 cm RP column maintained at 40 °C as has been previously demonstrated effective for membrane samples.17 SRM methods were created using Skyline22 for the analysis of peptides representing DAT, YFP, rabbit IgG (heavy and light chains), and mouse IgG1 (heavy and light chains). SRM data was analyzed using Skyline to measure and calculate AUC, coefficient of variance (CV), and retention time (RT). Additional LC-MS details can be found in the Supporting Information.
The upper and lower limits of detection (LOD) and quantitation (LOQ) were measured for four representative synthetic peptides, which reside in both rabbit and mouse light and heavy chains. In total, 0 mol, 10 amol, 100 amol, 1 fmol, 10 fmol, 100 fmol, and 1 pmol of each of these peptides were coinjected with both 100 fmol of bovine standard and 1 μg of whole parental PAE cell lysate tryptic digest; the four representative peptides were analyzed via SRM. The doubly charged parent ion and three to five singly charged product y-ions were monitored with a dwell time of 0.059 s (Table S-5 in the Supporting Information). Otherwise SRM methods were the same as above. Results were analyzed with Skyline.
Normalization
IgG peptide AUCs of three technical replicates were averaged for each peptide within each sample. Individual DAT and YFP peptide AUCs were each divided by the average IgG peptide AUC to create normalized AUCs. Pre- and postnormalized DAT/YFP AUCs were all divided by the average FL-YFP-HA-DAT AUC of the specific data set, such that all numbers ranged between 0 and 1 for comparison. t tests (2-tailed, type 2) were performed in Microsoft Excel comparing pre- and postnormalized AUC values for ΔN-DAT and ΔC-DAT IP samples. Four DAT/YFP peptides were normalized to each of the six IgG peptides, in both rabbit and mouse IPs.
RESULTS AND DISCUSSION
This study developed a relative normalization and quantitation pipeline for MS-IP samples through SRM analysis of IgG constant domain peptides. Tryptic digests of monoclonal and polyclonal antibodies were profiled using nanoLC-tandem mass spectrometry (MS/MS) to select for the best proteotypic IgG constant domain peptides for subsequent quantitative nanoLC-SRM assays. IPs were conducted using distinctive DAT constructs for targeted antigens. Rabbit and mouse antibodies were utilized for IPs and subsequent analysis. NanoLC-SRM was performed targeting both DAT and IgG tryptic peptides. AUCs of IgG peptides were used for the relative normalization of DAT peptides.
To assess the differences between quantitation of Western blots and MS, we conducted replicate westerns and MS with IPs from the same cell lines. Replicate Western blots of IP samples included bands corresponding to multiple forms of DAT as well as bands corresponding to IgGs (Figure 2a and Figure S-1a in the Supporting Information). However, fewer DAT bands are present in the lanes where DAT is less abundant. Particularly in the mouse αHA IP blot, it is difficult to go beyond a qualitative analysis of the samples through Western blot (Figure 2a). FL-DAT and ΔC-DAT band densities changed drastically between exposure times and with reprobing with different antibodies. The ΔN-DAT cannot be detected by the αDAT antibody but was much more abundant than the other two DAT constructs when detected with an αGFP antibody (Figure 1a and Table S-6 in the Supporting Information). Densitometry measurements of the IgGs could be used to normalize the DAT signals. However, these bands are also overexposed because IgGs are highly concentrated in order to pull down a substantial amount of DAT (Figure S-1b,c in the Supporting Information). This method did not produce a precise measurement for loading controls; the quantitative analysis of DAT levels based on densitometry was inconclusive due to the limited dynamic range of the method.
Figure 2.
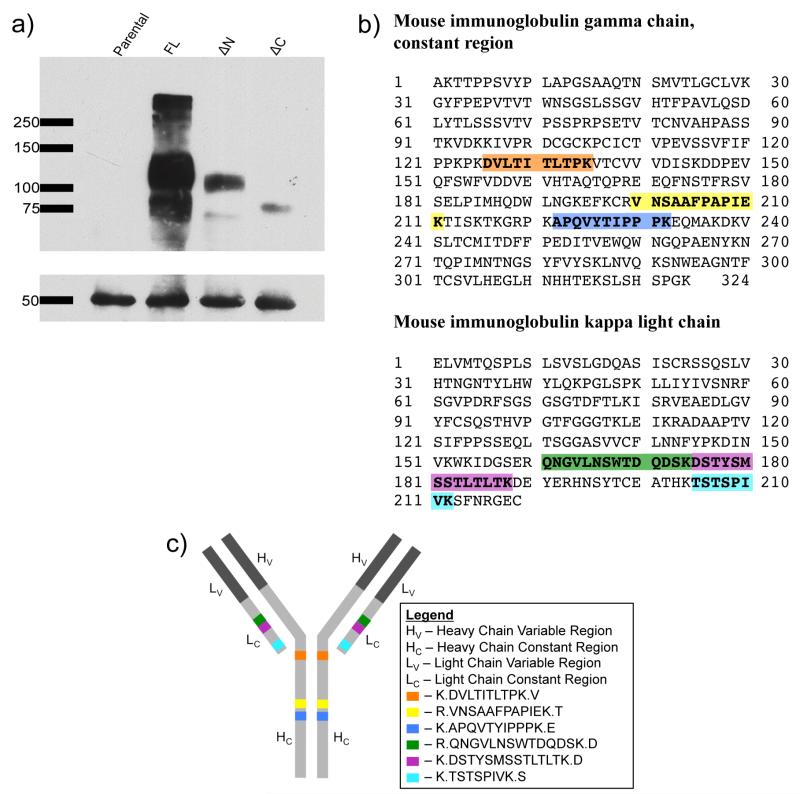
Characterization of mouse IgG: (a) Western blot showing DAT (upper box) and IgGs (lower box) in the four IP samples. The blot was probed with mouse anti-HA primary antibody followed by antimouse HRP secondary antibody that recognized both the anti-HA primary antibody attached to the DAT and that which was used in the IP. (b) Peptides are highlighted within the mouse IgG constant regions. (c) Peptides are highlighted on a cartoon representation of an antibody molecule.
To determine the representative peptides for IgG constant domains, three rabbit and two mouse antibodies were digested with trypsin and analyzed by nanoLC-MS/MS. Heavy and light chain peptides were identified across samples and technical replicates (Table S-1 in the Supporting Information). Peptides identified within this study along with peptides identified in previous MS-IP studies (data not shown) were selected for targeted analysis of IgG levels in IPed samples.
For selection of optimal proxy peptides for quantitative analysis of IgGs, 13 tryptic rabbit and 10 tryptic mouse IgG peptides were monitored using nanoLC-SRM. These peptides reside within heavy and light chain constant regions for both animals and were monitored in αGFP IP samples or αHA IP samples from parental, FL-DAT, ΔN-DAT, and ΔC-DAT PAE cells. Three technical replicates of all 8 samples and corresponding bead only control samples were produced. In both sets of IPs, six IgG peptides (Figure 2b,c and Figure S-1b,c in the Supporting Information) were reproducibly observed between biological samples and technical replicates but not in bead only control samples (Figure 3a and Figure S-1d in the Supporting Information).
Figure 3.
IgG SRM: (a) SRM analysis of six mouse IgG tryptic peptides within four IP samples. AUCs of each peptide were significantly different between samples based on ANOVA testing (p < 0.05), except for TSTSPIVK. (b, c) LOD curves of two representative mouse IgG1 peptides.
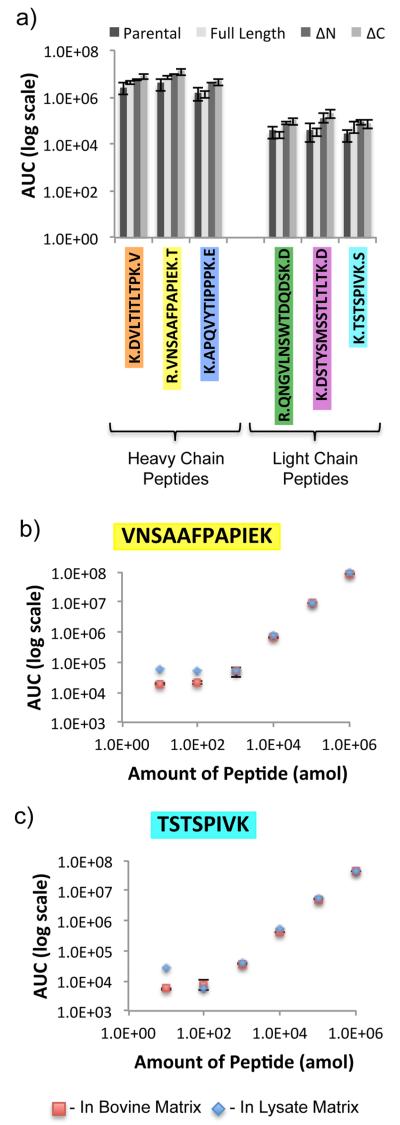
The AUC for each of these 12 IgG peptides was calculated using Skyline22 and compared between IP samples using analysis of variance (ANOVA). This analysis showed that with the rabbit antibody, two of the peptides had statistically different abundances between samples. However, with the mouse antibody, one of the monitored peptides was not present at statistically different levels between samples. The results from the rabbit αGFP IP were more consistent, as demonstrated by lower variance observed in IgG peptide abundance; whereas in the mouse αHA IP experiment, IgG abundances varied significantly.
To determine the dynamic range of detection of the peptides of interest, four representative synthetic peptides were coinjected at varying concentrations with either bovine matrix or cell lysate matrix and monitored via SRM. Three of the four peptides could be detected between 100 amol and 10 pmol in bovine matrix, while in cell lysate matrix all four peptides could be detected in the 1 fmol-10 pmol range (Figure 3b,c and Figure S-1e,f in the Supporting Information).
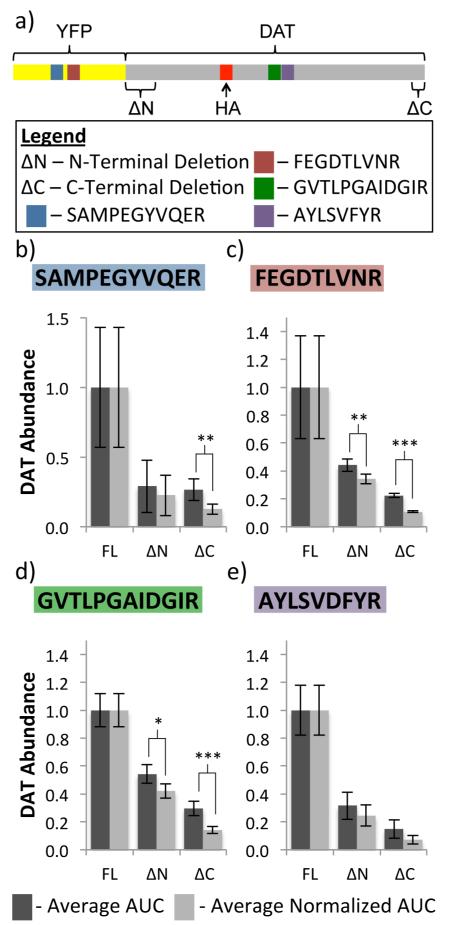
Antigen levels were also monitored via SRM. Two YFP and two DAT peptides (Figure 4a) were monitored in conjunction with IgG peptides in both αGFP and αHA IPs in all four cell lines. Normalization was performed by dividing the replicate AUCs of a peptide from the target protein (YFP-DAT) by the average AUC of an IgG peptide within the same sample, resulting in postnormalization AUC values for each YFP-DAT peptide. Pre- and postnormalization values were compared between conditions by dividing all values by the average FL-DAT AUC such that all values were between 0 and 1 (Figure 4b-e). t tests were conducted to compare the pre- and postnormalized ΔN-DAT and ΔC-DAT abundances (Tables S-7-S-10 in the Supporting Information). In rabbit αGFP IPs, the differences between the pre- and postnormalized values were less significant than those from the mouse αHA IPs. Three normalized ΔN abundance values were significantly different (p < 0.05) from the corresponding prenormalized values in the rabbit αGFP IP samples, while none of the ΔC abundance values underwent significant changes with normalization. However, in the mouse αHA IP samples, 10 of the ΔN and 18 of the ΔC abundance values resulted in significant changes after normalization (p < 0.05).
Figure 4.
Relative normalization of DAT AUCs to mouse IgG AUCs: (a) schematic of YFP and DAT peptide locations. (b-e) Comparison of pre- and postnormalization DAT abundance using one example mouse IgG heavy chain peptide (VNSAAFPAPIEK) to normalize two YFP (b,c) and two DAT (d,e) peptides. (*p < 0.10, **p < 0.05, ***p < 0.01).
The significant difference between IPs conducted with different antibodies is not surprising. There is more intersample variation in IgG abundances with the mouse antibody samples (Figure 3a and Figure S-1d in the Supporting Information). Also, there is a loose correlation between significant ANOVA analysis and significant changes between pre- and postnormalized abundance values. Peptides with significantly different abundances between samples when compared with ANOVA were more likely to result in significantly different pre- and postnormalization values (Tables S-11 and S-12 in the Supporting Information). For example, all three of the significant changes after normalization in the rabbit αGFP IP samples occurred with normalization to the LSVPTSEWQR peptide, which was shown to have significantly different levels between samples. In the mouse αHA IPs, normalization to the TSTSPIVK peptide, which did not have significantly different levels between samples, resulted in one significant normalization value, the least of any of the mouse IgG peptides. Thus, more significant changes occurred with normalization when there was a more significant difference in peptide abundance between the compared samples.
CONCLUSIONS
Appropriate loading controls are imperative in quantitative and semiquantitative experiments. For MS-IP, IgGs are an excellent loading control that has been overlooked. We propose a pipeline that uses IgG constant domain peptides for relative normalization and quantitation of an antigen of interest.
Within the proposed pipeline, after an IP is performed, IgG and antigen peptides are monitored via SRM. The resultant average DAT AUCs are divided by the average IgG AUCs to create a postnormalized AUC value. When the pre- and postnormalized AUCs are compared there can be significant differences between the two values, indicating variations in IgG levels between samples. Changes in normalized values indicate that this can result in faulty quantitation in IPs, as existing MS-IP protocols do not account for input variations. Such changes can be monitored via Western blot, but due to a limited dynamic range, more optimization is required to ensure that the relative antigen signal intensities are within the linear dynamic range of the blot. However, when comparing IgG levels by SRM on a TSQ Vantage, which has a linear dynamic range of 5 orders of magnitude,23 it is possible to compare both IgG levels between samples and IgG levels to antigen levels within the same sample, assuming the antigen is concentrated enough for detection.
This study has shown that relative normalization of antigen peptides to IgG peptides can result in significant changes in relative peptide abundance using both mouse and rabbit antibodies. If there are large inconsistencies between antigen levels in different samples, the quantitative dynamic range of ECL Western blot densitometry may not permit comparisons. Our relative normalization method allows for a confident comparison between antigens with significantly different abundances due to variations in either sample abundance or antibody affinity. We would like to continue to use this normalization pipeline in our future IP experiments for not only the antigen but for coprecipitating proteins of interest as well.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Nicholas W. Bateman for critical reading of the manuscript. This work was supported by NIH Grants U01AA016653, R01AA016171, S10RR027928, and DA014204.
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Notes
The authors declare no competing financial interest.
Supplementary methods, tables, and figures, as described in text. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Gingras A-C, Gstaiger M, Raught B, Aebersold R. Nat. Rev. Mol. Cell. Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- (2).Vermeulen M, Hubner NC, Mann M. Eur. J. Pharmacol. 2008;19:331–337. doi: 10.1016/j.copbio.2008.06.001. [DOI] [PubMed] [Google Scholar]
- (3).Bonifacino JS, Dell’Angelica EC. Curr. Protoc. Cell Biol. 2001:7.2.1–7.2.21. doi: 10.1002/0471143030.cb0702s00. [DOI] [PubMed] [Google Scholar]
- (4).Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Mol. Cell. Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- (5).Harlow E, Lane D. Antibodies: A Laboratory Manual. 1st ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1988. [Google Scholar]
- (6).Dekker LJM, Zeneyedpour L, Brouwer E, Duijn MM, Sillevis Smitt PAE, Luider TM. Anal. Bioanal. Chem. 2010;399:1081–1091. doi: 10.1007/s00216-010-4361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Kahlig KM, Galli A. Eur. J. Pharmacol. 2003;479:153–158. doi: 10.1016/j.ejphar.2003.08.065. [DOI] [PubMed] [Google Scholar]
- (8).Zahniser NR, Doolen S. Pharmacol. Ther. 2001;92:21–55. doi: 10.1016/s0163-7258(01)00158-9. [DOI] [PubMed] [Google Scholar]
- (9).Sorkina T, Miranda M, Dionne KR, Hoover BR, Zahniser NR, Sorkin AJ. Neurosci. 2006;26:8195–8205. doi: 10.1523/JNEUROSCI.1301-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Sorkina T, Richards TL, Rao A, Zahniser NR, Sorkin AJ. Neurosci. 2009;29:1361–1374. doi: 10.1523/JNEUROSCI.3250-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Schutz-Geschwender A, Zhang Y, Holt T, McDermitt D, Olive DM. Quantitative, Two-Color Western Blot Detection With; LI-COR Biosciences. Lincoln, NE; 2004. [Google Scholar]
- (12).Dickinson J, FowlerS J. In The Protein Protocols Handbook. Humana Press; Totowa, NJ: 2002. pp. 429–437. [Google Scholar]
- (13).Markham K, Bai Y, Schmitt-Ulms G. Anal. Bioanal. Chem. 2007;389:461–473. doi: 10.1007/s00216-007-1385-x. [DOI] [PubMed] [Google Scholar]
- (14).Agger SA, Marney LC, Hoofnagle AN. Clin. Chem. 2010;56:1804–1813. doi: 10.1373/clinchem.2010.152264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Sherrod SD, Myers MV, Li M, Myers JS, Carpenter KL, Maclean B, Maccoss MJ, Liebler DC, Ham A-JL. J. Proteome Res. 2012;11:3467–3479. doi: 10.1021/pr201240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Wessel D, Flügge UI. Anal. Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- (17).Blackler AR, Speers AE, Wu CC. Proteomics. 2008;8:3956–3964. doi: 10.1002/pmic.200800210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Eng JK, McCormack AL, Yates JR. J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- (19).Hsieh EJ, Hoopmann MR, Maclean B, Maccoss MJ. J. Proteome Res. 2010;9:1138–1143. doi: 10.1021/pr900816a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Käll L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Nat. Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- (21).Sharma V, Eng JK, MacCoss MJ, Riffle M. Mol. Cell. Proteomics. 2012;11:824–831. doi: 10.1074/mcp.O111.015149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Maclean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Bioinformatics. 2010;26:966. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Cell. 2009;138:795–806. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.