Abstract
Summary
The spontaneous transition of Epstein-Barr Virus (EBV) from latency to productive infection is infrequent, making its analysis in the resulting mixed cell populations difficult. We engineered cells to support this transition efficiently and developed EBV DNA variants that could be visualized and measured as fluorescent signals over multiple cell cycles. This approach revealed that EBV’s productive replication began synchronously for viral DNAs within a cell but asynchronously between cells. EBV DNA amplification was delayed until early S-phase and occurred in factories characterized by the absence of cellular DNA and histones, by a sequential redistribution of PCNA, and by localization away from the nuclear periphery. The earliest amplified DNAs lacked histones accompanying a decline in four histone chaperones. Thus, EBV transitions from being dependent on the cellular replication machinery during latency to commandeering both that machinery and nuclear structure for its own reproductive needs.
Introduction
Epstein-Barr Virus (EBV) is a human tumor virus which infects primary B-cells latently, inducing and maintaining their proliferation in cell culture and, only infrequently transitioning into its productive cycle (Sugden, 1984). EBV also infects epithelial cells to cause several carcinomas. No other herpesvirus displays its biphasic life cycle as tractably in culture, making EBV a powerful tool to reveal the steps used by a virus in commandeering its host to produce viral progeny. Much has been learned about the final stages of EBV’s productive cycle. The viral proteins needed for amplification of EBV DNA have been identified functionally (Chiu et al., 2007; Fixman et al., 1992). These EBV proteins and multiple cellular factors have been shown to localize to discrete sites in productively infected nuclei (Amon et al., 2006; Bell et al., 2000; Daikoku et al., 2005; Daikoku et al., 2006; Kudoh et al., 2009; Park et al., 2008). It has also been shown that EBV must transit from its use of its latent origin of DNA synthesis, oriP, to use two lytic origins, oriLyt L and R, to support its DNA amplification during a productive cycle (Hammerschmidt and Sugden, 1988). This transition is infrequently spontaneous, can be induced with various treatments in different cells (Chang and Liu, 2000; Chasserot-Golaz et al., 1988; Luka et al., 1979; Takada, 1984; Tovey et al., 1978; zur Hausen et al., 1978), but is usually inefficient, making it difficult not only to analyze events during EBV’s productive infection but also to distinguish between effects elicited by EBV and those induced by the treatments themselves. In addition, measurements of EBV’s productive cycle are often averaged across an asynchronous population, obscuring events that occur transiently.
We have examined the transition to and the amplification of EBV DNA during productive infection in single-cells using live-cell imaging to identify and characterize sequentially processes that are associated with the synthesis of progeny virus. To render these processes as synchronous as practical and independent of broadly active agents such as TPA and sodium butyrate, the host cells were engineered to express EBV’s immediate-early protein BZLF1 fused to the estrogen receptor ligand-binding domain (Z-ER). The translocation of Z-ER from the cytoplasm to the nucleus in cells harboring EBV latently could be induced by tamoxifen and elicited EBV’s productive cycle (Countryman and Miller, 1985; Takada et al., 1986). The productive cycle for two derivatives of EBV, Visible Amplicon and Visible EBV, following a synchronous induction occurred in either the ongoing cell cycle or in the subsequent cell cycle. Visible Amplicon contains the cis-acting viral elements required to participate in amplification and packaging (Bloss and Sugden, 1994); Visible EBV was made from the 2089 Bacmid of the B95.8 strain of EBV (Delecluse et al., 1998). Both EBV derivatives were modified to contain a polymer of lac operator binding sites (lacO) and to express the Lac repressor (LacI) fused to the tdTomato fluorescent protein to render them visible following removal of IPTG from the medium (Nanbo et al., 2007; Norby et al., 2012; Robinett et al., 1996). Following treatment with tamoxifen, host cells supported amplification of the latent, resident plasmids in up to 60% of the treated cells, which were followed hourly in live cells for 60 hr. To measure the intensities of the plasmids bound by the LacI-fluorescent fusions we developed a computer-assisted approach, termed CAPS (Computer-Assisted Plasmid Summation). CAPS uses photometry with subtraction of a background derived from an annulus of the pixels over multiple z-planes surrounding a given LacI-fluorescent signal of an EBV derivative after removal of IPTG to measure the intensities of all chosen signals. CAPS provides functions not found elsewhere and has allowed us to follow thousands of plasmids over multiple cell cycles and measure their potential overlaps with tagged proteins.
EBV DNA is synthesized using cellular machinery during its latent phase localized in replication factories that are marked by PCNA (Leonhardt et al., 2000). Induction of EBV’s productive cycle led first to dramatic alterations in nuclear morphology followed later by amplification of EBV DNA. The unlicensed synthesis of EBV’s genome in its productive cycle coincided with the maximal expression of Cdt1/eGFP revealing it began in early S-phase (Sakaue-Sawano et al., 2008). EBV’s genome synthesis occurred at distinct sites, which we term “amplification factories”, in which PCNA was redistributed. The architecture of the nucleus was reorganized so that cellular DNA and histones accumulated at its margins while the factories formed internally. Cellular histones were not detectably associated with the sites of amplifying viral DNA which correlated with marked declines in the levels of the cellular histone chaperones, CAF-1a, CAF-1b, ASF1a, and ASF1b. Nor was PCNA localized to these sites. These insights explain how EBV evades the cellular machinery that deposits histones on newly synthesized DNA (Johannsen et al., 2004). Our time-lapse analyses show that EBV transits from being a fellow-traveler dependent on cellular replication machinery during latency to commandeering both that machinery and nuclear structure for its own reproductive needs.
Results
Developing plasmids and cells to visualize EBV’s productive cycle
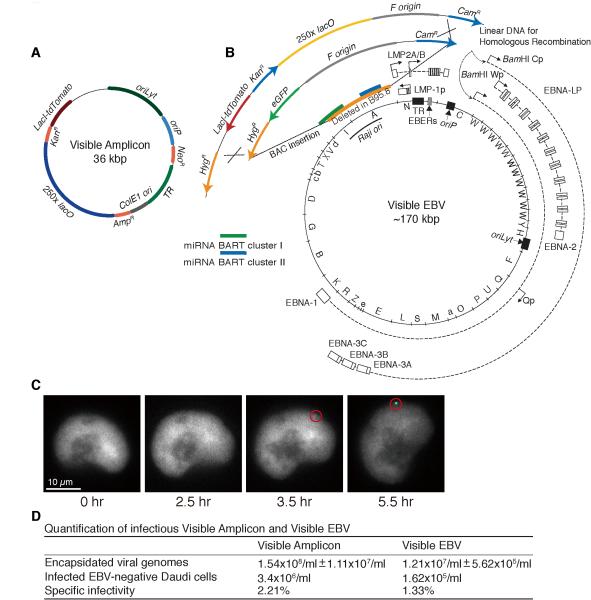
To overcome limitations inherent in studies of heterogeneous populations of cells, we have engineered two derivatives of EBV that can be analyzed visually to follow events in EBV’s productive phase. These derivatives are detected as single DNA molecules and followed in real time over days as they are amplified. One is a Visible Amplicon (Figure 1A), which contains EBV’s oriLyt, oriP, and terminal repeats. It also encodes resistance to G418, the LacI fused to the tdTomato fluorescent protein along with a nuclear localization signal, and contains 250-binding sites for this LacI-tdTomato fluorescent fusion. The other, termed Visible EBV, was constructed by introducing a DNA fragment encoding kanamycin-resistance, the LacI-tdTomato fluorescent fusion, and 250 copies of lacO sequence into an intact EBV Bacmid, 2089, to replace its eGFP gene (Figure 1B and S1A). Both the Visible Amplicon and Visible EBV, when maintained latently in mammalian cells, could be visualized as punctate fluorescent signals distributed evenly in the nuclei following removal of IPTG from the medium (Figures 1C, S1B-D, and S2D).
Figure 1.
The structures of Visible Amplicon and Visible EBV Bacmid and their characterization. (A) A DNA fragment encoding resistance to kanamycin, a fusion of LacI-tdTomato fluorescent protein, and containing 250 copies of lacO sites was inserted into an EBV-derived amplifiable plasmid, p3944. (B) That same fragment was used to substitute for the eGFP gene of EBV Bacmid 2089 by Red-mediated homologous recombination. The size of Visible EBV’s genome is predicted to be approximately 170 kbp with variable lengths of BamH-W and terminal repeats (TR) (see also Figure S1A) (Bloss and Sugden, 1994). (C) Removing IPTG from cells with Visible EBV allows binding of LacI-tdTomato fusion and its detection (red circle) in 3.5 hr. (D) The released encapsidated Visible Amplicon and Visible EBV were quantified by qPCR and by infection of EBV-negative Daudi cells. The ratio of these measurements yielded their specific infectivities. (See also Figure S1)
The Visible Amplicon contains the cis-acting elements (oriLyt, oriP, and terminal repeats) needed for EBV’s productive cycle and lacks all other EBV’s genes. It was therefore introduced into a derivative of D98/HR1 cells (iD98/HR1), which contains a variant of EBV that fully supports its productive cycle (Glaser and Rapp, 1972) and was transduced to express Z-ER. Addition of tamoxifen to these cells induced EBV’s productive cycle. The induced cells ultimately released encapsidated polymers of the Visible Amplicon (Bloss and Sugden, 1994) (Figure 1D). In parallel, Visible EBV was introduced into clones of a derivative of 293 cells (i293) transduced to express Z-ER stably. Treatment of these cells with tamoxifen also induced and supported the complete productive cycle yielding 1.2×107 Visible EBV DNA-containing particles per ml. Approximately one out of 100 DNA-containing particles was infectious (Figure 1D), an efficiency comparable to that of the B95.8 strain of EBV. The encapsidated, infectious Visible EBV was also as competent to infect and transform primary human B cells as wild-type EBV (Sugden and Mark, 1977). The infected B cells proliferated for months and displayed punctate genomes when viewed under a fluorescence microscope following removal of IPTG from the media (Figure S1D).
Developing CAPS to measure the intensities of EBV DNAs bound by fluorescent proteins
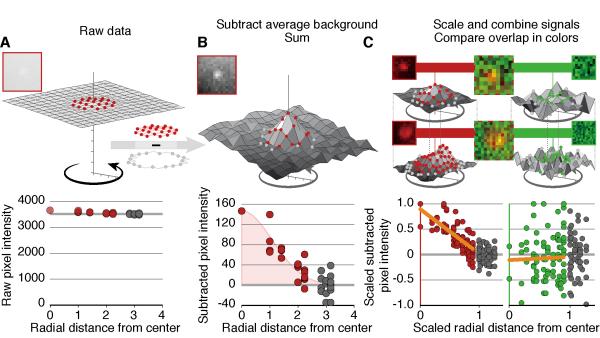
We developed CAPS to measure the intensity of fluorescent signals in microscopy images in multiple z-planes with subtraction of an annulus-background as has been applied to astronomical images (Mighell, 1999). CAPS has two libraries of code found at http://caps.grapheasy.org and instructions to help users master them. CAPS displays images of a time-series of 16-bit z-planes and allows measurements of their signals even when they are less than 5% of background (Figure 2A). It displays multiple z-planes simultaneously to aid plasmid identification. It also tracks and synchronizes measurements of signals over time, fits plasmid profiles, and allows nuclear identification. The utility of CAPS was assessed with signals derived from Visible EBV derivatives. These signal intensities revealed their near-Gaussian distribution over their radius following subtraction of the background found in an annulus of pixels surrounding the signals (Figure 2B). CAPS also sums signals and allows potential overlaps of signals from different fluorescent sources to be measured (Figure 2C), and provides a statistical test of that potential overlap. This function has allowed us to measure the possible overlap of fluorescently tagged proteins with amplifying EBV DNA.
Figure 2.
CAPS measures signal intensities. (A) Typical signals of Visible EBV in live cells contribute a peak pixel intensity of 150 (red) above a background annulus (gray) of about 3600 units per pixel. (B) CAPS averages the pixel intensities in the annulus and subtracts that average from each signal pixel and each background pixel to reveal the Gaussian distribution of Visible EBV’s signal as measured by Gaussian fitting. (C) CAPS measures the overlap of EBV bound-LacI-tdTomato signals with other florescent tags by determining signal intensities (red) as described above and then scaling both the most intense pixel in each signal to 1 and the radius of the pixels to 1 allowing them to be summed. Summed point sources show a signal that decreases as the distance from the source increases (C, lower left). To measure the overlap of another florescent tag, here green, CAPS measures intensities of the same pixels in green, measures the intensities of the pixels in the annulus in green, subtracts their average from all the pixels, sums the corrected intensities from inside the annulus, and scales them as for the LacI-tdTomato signals. An overlapping point source will have a similar profile as LacI-tdTomato signals; non-overlapping signals will not vary systematically but rather will scatter evenly independently of the distance from the source (C, lower right).
Visualization of the EBV DNAs during the productive cycle
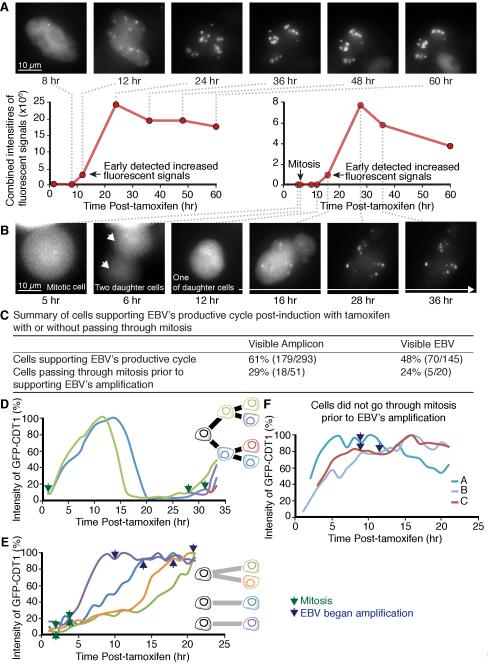
Cells carrying Visible Amplicon or Visible EBV were treated with tamoxifen to observe EBV-derived DNAs transiting from latency and progressing through the productive cycle. We used both a sensitive CCD camera and an LED light source to minimize phototoxicity (Frigault et al., 2009; Norby et al., 2012). In experiments in which two colors were followed, the same field was imaged about 10,000 times and uninduced cells still progressed through the cell cycle. These low light intensities limit resolution but allowed us to identify when we should analyze fixed cells with microscopy using more intense light to provide higher resolution. The Visible Amplicon has the advantage of containing approximately five sets of lacO sites per EBV genome length yielding a more intense signal than does Visible EBV (Figures 1A-B, 3A-B and S2A-C). However, it has the potential disadvantage of being dependent on the EBV DNA endogenous to iD98/HR1 cells which is invisible in our imaging experiments. The Visible Amplicons in individual iD98/HR1 cells were imaged following treatment with tamoxifen over a 60-hour period. Under the conditions of minimal light exposure, the fluorescent signals became intense by as early as 10-12 hr post-treatment with tamoxifen and by 22 hr showed up to a 300-fold increase in intensity as measured by CAPS (Figure 3A) (all measurements made with CAPS are from raw data of 16-bit gray scales). EBV DNA is synthesized in a licensed manner during latency so that increases in its signal intensity above two-fold indicate its productive amplification. The increased intensity of the Visible Amplicon signals were paralleled by an increased expression of lytic genes essential for the replication of EBV DNA, including BRLF1 encoding an immediate-early protein, BALF5 encoding EBV’s DNA polymerase, BMRF1 encoding a viral DNA clamp, and BcLF1 encoding a major capsid protein (Figure S1E-H). The induction of the productive cycle was efficient; amplifying signals of Visible Amplicons were detected in 60% of the cells that were treated with tamoxifen (Figure 3C). In parallel, EBV’s DNA polymerase transcript increased by more than 6,000-fold within 12 hr of the addition of tamoxifen to the iD98/HR1 cells (Figure S1F).
Figure 3.
In its productive phase EBV reorganizes the nucleus with viral DNA amplification often occurring after mitosis early in S phase. (A) iD98/HR1(Visible Amplicon) cells were treated with tamoxifen and followed over a 60-hour period. The sum of the intensities of the fluorescent signals in each nucleus for amplified Visible Amplicon was measured by CAPS. (B) Another iD98/HR1(Visible Amplicon) cell treated with tamoxifen first underwent a mitotic division and one of its daughter cells was tracked over the next 24 hours. (see also Figure S2 for examples in i293(Visible EBV) cells). (C) The fraction of cells supporting EBV’s productive cycle and going through mitosis or not is tabulated. (D-F) The time in the cell cycle at which EBV DNA first amplified was determined by measuring LacI-tdTomato signals and the intensities of introduced Cdt1/eGFP in the same cells and normalizing the maximum signal of Cdt1/eGFP for each cell cycle. (D) The profiles of these intensities in uninduced daughters shows minima at G2/M and maxima at early S. (E) Intensities of Cdt1/eGFP and LacI-tdTomato in induced iD98/HR1(Visible Amplicon) cells that passed through mitosis prior to DNA amplification show that amplification begins in early S-phase. The blue and purple plots show Cdt1/eGFP signals in separate daughter cells, the orange and green plots are signals in the daughters of one cell. (F) Tracking of DNA amplification as a function of time after tamoxifen treatment in three cells that did not pass through mitosis prior to EBV amplification. (See also Figure S2)
The amplification of visible EBV in individual i293 cells was also tracked following treatment with tamoxifen over 60 hr. The productively amplified Visible EBV genomes were seen as pronounced fluorescent signals in 48% of the i293 cells (Figure 3C). The sum of the intensities for the fluorescent signals in i293(Visible EBV) cells was determined by CAPS and increased as early as 16-20 hr post-induction and grew 20-fold by 36 hr (Figure S2A). The increased intensity of the Visible EBV signals could be detected only after removal of IPTG (Figure S2D).
The signals of Visible Amplicon and Visible EBV also increased in their diameters which were particularly evident for the amplifying Visible Amplicon; consistent with more copies of lacO sites being amplified per unit time for this replicon (Figures 1A-B, 3A-B, and S2A-C). These amplifying EBV derivatives coalesced as islands either by accretion or by fusion with adjacent sites within nuclei at later times in the productive cycle (Figures 3A-B, 4C-D, 5A-B, S2A-C, S3A-C, and S4). The architecture of these nuclei changed dramatically with their chromatin receding to their peripheries (Figures 4C and S3C). Similar changes in nuclear architecture occurred in B-cells induced to support EBV’s lytic cycle (Figure S5A-B) illustrating the generality of EBV’s appropriating cellular functions for its lytic cycle. Finally no fluorescent signals were detected in the cytoplasm after treatment with tamoxifen (Figures 3A-B, 4B, S2A-C, S3A-B, and S4A-C), indicating either that the LacI-tdTomato fusions were removed from lacO sites prior to DNA encapsidation or too few were retained to be detected.
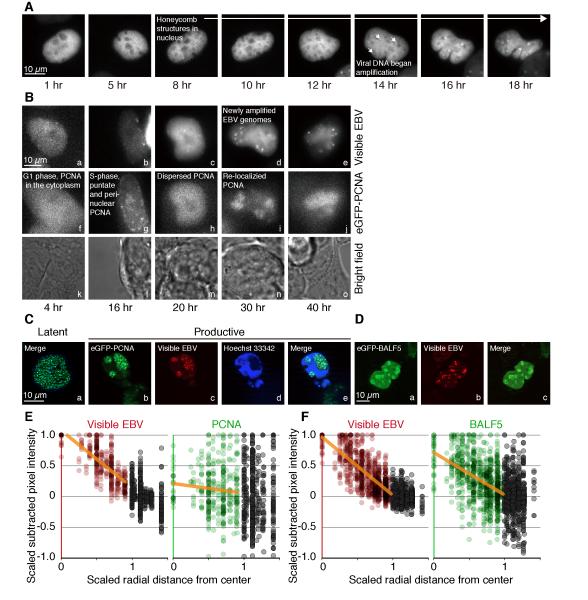
Figure 4.
Formation of EBV amplification factories is marked by PCNA. (A) In a iD98/HR1(Visible Amplicon) cell that has eGFP-PCNA, treatment with tamoxifen leads to nuclei becoming “honeycombed” hours before viral DNA is amplified by the redistribution of LacI-tdTomato fusions. (B) One i293(Visible EBV) cell expressing eGFP-PCNA was treated with tamoxifen and imaged over 40 hr. Multiple Z-planes at indicated time points are summed and illustrated. The punctate structures of ectopic eGFP-PCNA-marked replication foci were detected in i293(Visible EBV) 16 hr post-treatment with tamoxifen, subsequently were dispersed at 20 hr, and then re-localized to the general sites at which EBV DNA was being amplified (see also Figure S3 for examples in iD98/HR1(Visible amplicon) cells). (C) One i293(Visible EBV) cell that had been transduced to express eGFP-PCNA fusions was treated with tamoxifen to induce EBV’s productive cycle for 48 hr followed by staining with Hoechst 33342 prior to fixation with paraformaldehyde. It was then imaged with a Zeiss Apotome microscope and shows that the Visible EBV localized to regions of the nucleus unstained by Hoechst dye. (D) Parallel experiments to those in B were carried out with eGFP-BALF5, EBV’s DNA polymerase that mediates DNA amplification during EBV’s productive phase. (E) The signals in C of eGFP-PCNA do not overlap with those of LacI-tdTomato as determined with CAPS (see Figure 2C for an explanation). (F) The signals in D of eGFP-BALF5 do overlap with those of LacI-tdTomato as determined with CAPS. (See also Figure S3)
Figure 5.
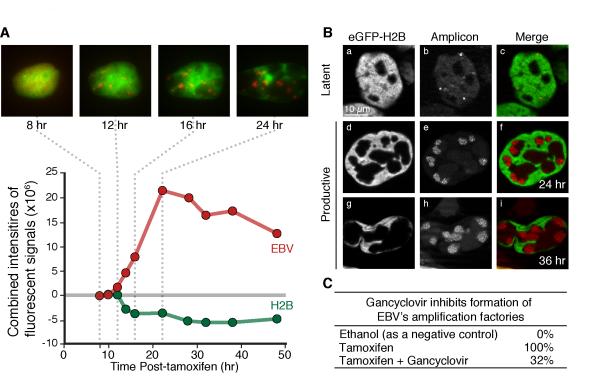
The distributions of EBV derivatives and eGFP-histone H2B fusions were tracked and measured during the productive cycle. (A) iD98/HR1(Visible Amplicon) cells transduced to express eGFP-histone H2B fusions were treated with tamoxifen and tracked with time-lapse microscopy for 50 hr. Multiple Z-planes of live-cell images at indicated time points are summed and shown. The sum of intensities of eGFP-H2B fluorescent signals (green circles) and Red fluorescent signals (red circles) for amplified Visible Amplicon in identical volumes in live cells was determined with CAPS and shows that over time eGFP-H2B signals do not increase whereas EBV DNA signals do. (Additional examples of time-lapse measurements are shown in Figure S4A.) (B) For higher resolution, similarly generated cells were examined with a Zeiss Apotome microscope following fixation with paraformaldehyde at the indicated times after tamoxifen treatment. (C) Inhibiting EBV’s DNA polymerase with gancyclovir following treatment with tamoxifen prevents formation of 68% of the amplification factories that are delineated by histone-bound chromatin. (See also Figure S4)
The time of the amplification of EBV during the cell cycle
Visible Amplicon or Visible EBV derivatives were visualized as fluorescent signals with increasing intensities during the productive cycle. Within a single cell, amplification of all plasmids became detectable during the course of 1-2 hr (Figures 3A-B, 4A-B, 5A, S2A-C, S3A-B, and S4A-C). In some cells that supported the productive cycle the EBV derivatives began to be amplified within a few hours after induction. However, in other cells amplification was detected only 30-36 hr post-induction, a time longer than the generation time of either iD98/HR1 or i293 cells (Figures 3B, S2C, and data not shown). 29% of iD98/HR1(Visible Amplicon) cells that supported the productive cycle were seen to go through mitosis prior to amplification. Amplification occurred approximately 10 hr after the cells exited mitosis. Similarly, Visible EBV signals began to be amplified detectably about 20 hr following mitosis in 24% of i293(Visible EBV) cells supporting EBV’s productive cycle (Figure 3C). The timing of amplification was determined by analyzing expression levels of a portion of Cdt1 fused to eGFP (Cdt1/eGFP) which exhibited Cdt1’s characteristic cell-cycle dependence in the same cells in which EBV DNA was amplifying (Figure 3D) (Nishitani et al., 2001; Sakaue-Sawano et al., 2008). EBV DNA began to be amplified 1-3 hours after Cdt1/eGFP reached 90% of its peak intensity corresponding to a time early in S-phase (Figure 3E-F). Once the EBV derivatives began to be amplified, those cells that had passed through mitosis and supported EBV’s productive cycle did not go through another cell division (Figures 3B, S2B-C, and S3B). Thus EBV affects passage of its host through its cell cycle, when necessary allowing it to pass through mitosis, and arresting it in early S-phase, presumably to optimize the environment for amplification of its genome. EBV also dramatically altered the morphology of its host’s nucleus. One to several hours before EBV DNA amplification could be detected in live cells the nuclei became mottled or honeycombed displaying subtle differences in the architecture of the nucleus (Figure 4A). These initial subtle changes were detected as approximately 0.5% local differences in the intensities of LacI-tdTomato and then grew more profound as the viral genomes were amplified. The Lac repressor binds non-specifically throughout the human genome so that the detected differences likely reflect differences in chromatin compaction (see below).
EBV DNA is amplified in distinctive factories
Chromosomal DNA is synthesized in replication factories that can be detected by the localization of PCNA, the DNA clamp, to them (Leonhardt et al., 2000). There are on the order of 1,000 of these factories per nucleus which form in S-phase and characteristically change their number and size during the course of DNA synthesis (ibid.). These cellular replication factories marked with eGFP-PCNA accumulated in both untreated iD98/HR1 and i293 cells in early S-phase, gave rise to punctate peri-nuclear structures in mid-S-phase, and decreased in number toward the end of S-phase (Data not shown). Unexpectedly, after treatment with tamoxifen, eGFP-PCNA-associated replication factories were detected in i293(Visible EBV) cells at the beginning of S-phase, subsequently were dispersed for approximately 2 hr, and then re-localized to the general sites at which EBV DNA was being amplified (Figure 4B). Similarly, eGFP-PCNA-associated replication factories which formed prior to treatment with tamoxifen were dispersed and re-localized generally to sites where Visible Amplicons were amplified in treated iD98/HR1 as early as 8-10 hr post-treatment (Figure S3A-B). The altered distribution of PCNA during EBV’s DNA amplification was most apparent following fixation and imaging with a Zeiss Apotome microscope which allows removal of out-of-focus light but not live-cell imaging (Figures 4C and S3C). The frequent redistribution of PCNA to these amplification factories was surprising given that EBV encodes its own DNA clamp, BMRF1, which is efficiently induced in these cells (Figure S1G) and serves as the clamp for newly amplifying EBV DNA (Makhov et al., 2004; Tsurumi et al., 1994; Tsurumi et al., 1993b). While PCNA often localized to the amplification factories, it was not detected at the sites of viral DNA synthesis (Figures 4C and 4E) (see below).
EBV’s amplification factories were prominent in cells carrying the Visible Amplicon or Visible EBV and engineered to express histones H2B, H3.1, or H3.3 each tagged with eGFP. Following treatment with tamoxifen to induce EBV’s productive cycle, the positions of the amplifying viral DNAs gradually altered. These alterations were visible in live cells (Figures 5A and S4A-C) and could be observed particularly well following fixation and imaging with a Zeiss Apotome microscope (Figures 5B and S4D-H). These morphological changes consisted of the histones and histone-bound chromosomal DNA, which was also stained with Hoechst 33342 (data not shown), moving to the margins of the nucleus and EBV’s amplification factories separating from these margins. Similar morphological changes occurred in B-cells induced to support EBV’s lytic cycle (Figure S5A-B). These morphological changes were inhibited by 68% with ganciclovir, which is activated by EBV’s protein kinase BGLF4 and inhibits the function of EBV’s DNA polymerase (Figure 5C) (Meerbach et al., 1998; Meng et al., 2010).
EBV uses two mechanisms to evade the loading of histones onto its amplifying DNA
The cell uses complex machinery to load histones onto newly synthesized DNA laying down new chromatin adjacent to the replications forks (MacAlpine and Almouzni, 2013). EBV somehow evades this machinery because histones are not found on its encapsidated DNA (Johannsen et al. 2004). To understand how EBV avoids the deposition of histones, we first measured the protein levels of the histone H2B and H3 in cells induced to support the productive cycle and found the levels did not decline (Figure 6A-B). We also measured the level of eGFP-tagged histones at the sites where EBV DNAs were being amplified. While the intensities of the EBV DNA signals rose up to 36-fold during the productive replication, the intensities of the histone signals at the identical pixel sites varied due to noise but did not increase over time (Figures 5A and S4A-C). These measurements indicate that EBV escapes the loading of histones at the time its genomes are being amplified.
Figure 6.
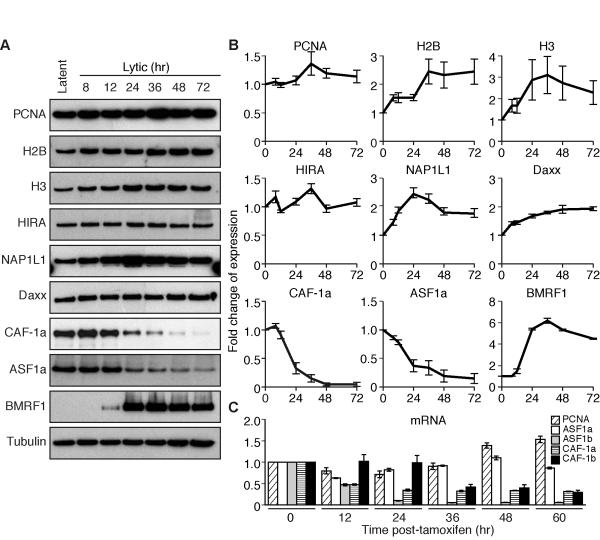
The expression of histones and their chaperones was measured during EBV’s productive cycle in iD98/HR1(Visible Amplicon) cells. (A) Gels illustrate measurements of proteins detected by immunoblotting as a function of time after treatment of cells with tamoxifen. (B) Averages of three independent experiments like those shown in A ± s.d. were measured with Image J software. The expression levels of proteins were normalized to that of α-tubulin and then compared to the expression levels in the absence of tamoxifen which was arbitrarily set to 1. (C) Levels of transcribed mRNA of histones and their chaperones after treatment with tamoxifen was quantified by qPCR. Data are the average of three independent experiments. The expression levels were normalized to the expression of mRNA for GAPDH and then compared to the expression levels in the absence of tamoxifen which was arbitrarily set to 1. (See also Figure S5 for extensions to other cell types for the expression of histones and histone chaperones during EBV’s productive cycle)
To understand this avoidance of histone deposition mechanistically, we measured the levels of constituents of the cellular machinery involved in their deposition onto DNA during EBV’s productive cycle. While the protein levels of histone chaperones HIRA, NAP1L1, and Daxx were stable throughout EBV’s productive cycle, the protein levels of the histone chaperones CAF-1a and ASF1a decreased by 80% (Figure 6A-B) and the mRNAs of CAF-1a, CAF-1b and ASF1b decreased by 80-95% (Figure 6C). These four inhibited chaperones normally contribute to deposition of histones during DNA synthesis. We asked if they were added back to cells supporting EBV’s productive cycle by the introduction of five expression vectors for their subunits, could they now deposit a tagged histone onto amplifying EBV DNA. The histone chaperones were expressed (Figure S5D), but failed to deposit the tagged histones (Figure S5C). Thus the inhibition of synthesis of histone chaperones during EBV’s lytic cycle alone does not explain the failure of histone deposition.
PCNA helps recruit histone chaperones to replicating DNA (Sharp et al., 2001; Shibahara and Stillman, 1999). We therefore assayed eGFP/PCNA for its position and found that it did not co-localize with amplifying EBV DNA (Figures 4C and 4E) while EBV’s DNA polymerase, BALF5, did (Figures 4D and 4F). Five cells induced to amplify EBV DNA and expressing eGFP/PCNA and five, similarly induced and expressing eGFP/BALF5, were assayed for their overlap of red and green signals with CAPS (one each is shown in Figures 4E-F). The correlation coefficient of the red signal of EBV DNA with the green signal of PCNA was 0.09 with a p-value of 0.29 while that of EBV DNA with BALF5 was 0.65 with a p-value of 9×10−10 (Spearman’s rank correlation test; Experimental Methods) confirming BALF5 overlapped with newly amplified EBV DNA but PCNA did not. The amplification of EBV DNA is mediated by the EBV-encoded DNA polymerase, BALF5, and the encoded DNA clamp, BMRF1 (Tsurumi et al., 1993b). We found that these two viral proteins lack the motifs known to support interactions with PCNA, [QXX(V/L/M/I)XX(F/Y)(F/Y)] or [KA(A/L/I)(A/L/Q)XX(L/V)] (Jonsson et al., 1998; Maga and Hubscher, 2003; Warbrick, 2000) consistent with PCNA’s absence from the site of EBV’s amplifying DNA. Thus during EBV’s lytic cycle, PCNA is replaced by independently acting viral machinery and not in a position to interact with the chaperones needed to deposit histones on the viral DNA. This absence along with the diminished levels of the chaperones themselves insure that EBV can amplify its DNA independently of the cellular machinery that deposits histones onto newly synthesized DNA.
Discussion
We have developed visible derivatives of EBV, an efficient way to induce their productive cycle, and photometry software to characterize EBV’s transition in real-time from latency through its genome amplification in the productive phase. The combination of these approaches has uncovered unprecedented changes in nuclear structure and cellular replication machinery that are mediated by the virus as it makes this transition.
We have used a fusion of Cdt1 to eGFP to define passage through the cell cycle in live cells (Sakaue-Sawano et al., 2008) following efficient induction of EBV’s productive cycle. About 30% of these cells went through mitosis before EBV DNAs began to be amplified. In these cells the viral DNA signals increased in intensity 10-20 hr after mitosis. The remainder of the cells supported the amplification without going through mitosis. These observations mean that a functional BZLF1 sufficient to induce EBV’s productive phase does not drive the cells into a G0/G1 arrest as has been found previously (Cayrol and Flemington, 1996). Instead, the measurements with Cdt1/eGFP show that EBV DNA began to be detectably amplified in early S-phase. The sequential measurements in live cells establish that, when necessary, EBV conditions its entry into its lytic phase by delay during a cell cycle to allow it to exploit its host’s environment in early S-phase (Kudoh et al., 2009).
The sequential remodeling of the nucleus found during EBV’s transitioning to its lytic phase was unexpected. Several hours before viral DNA was amplified in any cell, its nucleus became subtly honeycombed. These initial changes reflect small variations in concentrations of LacI-tdTomato binding non-specifically to host DNA and arise, we hypothesize, as initial stages of the recession of cellular chromatin to the nuclear margins we observed later in the same calls. The disposition of amplifying EBV plasmids during EBV’s productive cycle was particularly revealing in the presence of fluorescently labeled histones. EBV’s amplification factories were separated from the histone-bound chromosomal DNA that gradually moved to the margins of the nuclei. These amplification factories look similar to loci described for HSV-1 in infection of HeLa cells in which the volume occupied by chromosomal DNA decreased dramatically during productive infection (Monier et al., 2000).
It is not clear how EBV’s amplification factories form, but they are marked by PCNA (Daikoku et al., 2006; Sugimoto et al., 2011). We have found that EBV’s productive cycle mediates a complex, sequential redistribution of PCNA. As EBV transits into later times in its lytic phase, PCNA collects in or near viral amplification factories but is not concentrated at the sites at which EBV DNA is being amplified. Its absence at these sites reflects EBV’s DNA clamp encoded by its BMRF1 gene being required at these sites (Tsurumi et al., 1993a). In addition while the BMRF1 protein does interact with EBV’s DNA polymerase, BALF5, both BMRF1 and BALF5 lack the sequences motifs known to mediate an interaction with PCNA consistent with EBV’s genome amplification being independent of PCNA (Jonsson et al., 1998; Kiehl and Dorsky, 1995; Maga and Hubscher, 2003; Warbrick, 2000).
Another unanticipated insight into EBV’s productive cycle came from following the distribution of eGFP-labeled histones H2B, H3.1, or H3.3 relative to the amplifying viral DNA. No increase in the levels of these histones was observed to be associated with the increasing signals of amplifying EBV DNAs indicating either that they are not being loaded onto the amplified DNA or are being loaded inefficiently. We found that EBV uses two mechanisms to evade cellular machinery that deposits histones onto its newly synthesized DNA. First, we found that the levels of protein or mRNA for the cellular histone chaperones, CAF-1a, CAF-1b, ASF1a, and ASF1b declined by 72 hr to 5-20% of the levels found at the beginning of the productive cycle. These observations indicate that EBV’s productive cycle is inhibiting the synthesis of these chaperones that are involved in cell cycle-dependent deposition of histones. Second, EBV’s replacement of PCNA with its own DNA clamp means that PCNA is not positioned to recruit any remaining chaperones to sites of viral DNA amplification. We also found that EBV’s productive cycle does not affect levels of the chaperones NAP1L1, HIRA, and Daxx, which contribute to the loading of histones for transcription-coupled deposition (Lewis et al., 2010; Mosammaparast et al., 2002; Okuwaki et al., 2005; Tagami et al., 2004). Nor does EBV affect levels of PCNA, which is required for recruiting histone chaperones along with histones to the replication fork for their deposition on newly synthesized cellular DNA (Moggs et al., 2000; Shibahara and Stillman, 1999). We assume that some viral templates are bound by nucleosomes in order to be transcribed by RNA polymerase II, as has been found for HSV-1 (Kutluay and Triezenberg, 2009). Where these nucleosome-bound viral DNAs are in the nucleus is not known.
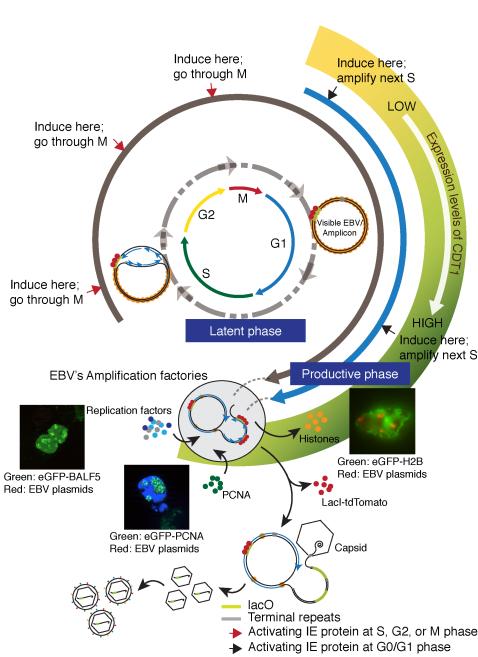
EBV supports both a latent and a lytic or productive phase in cell culture (Figure 7). Its latent origin of DNA synthesis, oriP, (Shirakata et al., 1999; Yates et al., 1984), gives way to two origins, oriLyt L and R, used by viral proteins to mediate unlicensed amplification during EBV’s productive cycle (Hammerschmidt and Sugden, 1988, 2013; Pfuller and Hammerschmidt, 1996; Tsurumi, 2001). Following EBV DNA visually has shown that its amplification is often delayed until the ensuing cell cycle insuring it occurs in early S-phase. EBV remodels nuclei to house discrete factories in which viral DNA is amplified. Late in this productive phase cellular chromatin is compacted and moves to the periphery of the nucleus. The separation of viral amplification factories from chromatin is highlighted by the redistribution of PCNA and the exclusion of histones from those factories. Thus EBV transits from a stable co-existence during latency to hijacking nuclear functions and architecture to meet its reproductive needs.
Figure 7.
Molecular events define EBV’s transition from its latent to its productive cycle. Following infection and entry of EBV’s genome into the nucleus, it circularizes through terminal repeats and is wrapped by histones as an extra-chromosomal plasmid. In the latent phase of EBV life cycle, its genome is licensed and replicates via oriP bidirectionally in S-phase as a plasmid whereas in the productive phase, amplification of EBV’s DNA is unlicensed and waits until early S-phase to begin when the expression of eGFP-Cdt1 is maximal. Once EBV’s productive cycle is activated, the viral genome is replicated efficiently via oriLyt L and R eventually as a polymer, perhaps through a rolling circle mechanism. This replication occurs in discrete factories in which the cellular clamp PCNA is redistributed and from which chromatin and histones are excluded. EBV’s productive cycle inhibits the synthesis of histone chaperones CAF-1a, CAF-1b, ASF1a, and ASF1b. EBV’s DNA clamp, BMRF1, mediates amplification insuring that PCNA does not localize to the site of amplification and cannot recruit residual chaperones to newly synthesized DNA. The replicated EBV genomes in association with polyamines are packaged into capsids.
Experimental Procedures
Generating inducible cell lines with Visible Amplicon or Visible EBV
D98/HR1 and 293 cells were transduced to express a tamoxifen-inducible BZLF1 derivative (Z-ER) and selected with 1 μg/ml of puromycin. p4012 (Visible Amplicon) was transfected into iD98/HR1 cells and p4016 (Visible EBV) was transfected into i293 cells. G418- or hygromycin-resistant clones respectively were selected. The released encapsidated DNAs of Visible Amplicon or Visible EBV from these clones were measured by qPCR to select for those supporting the complete productive cycle. Primers and probes used to quantify viral DNAs are listed in the Table S1.
Time-lapse experiments
Time-lapse experiments were performed with an image system using a Zeiss Axiovert 200M microscope as described (Norby et al., 2012). Between 60 and 86 planes were sampled at intervals of 0.35 to 0.4 μm for each z-plane each 60 to 120 min over 48-72 hr. Exposure times ranged from 50 to 200 msec for red and 5-50 msec for green signals and were chosen to allow detection of signals with minimal exposures. All images were taken with a Plan-Apochromat 63X, 1.4 N.A. oil objective and a Cascade II: 1024 EMCCD camera (Photometrics) with a 16 bit dynamic range. No images were saturated as judged by their most intense pixels being less than half of ~65,000 gray levels. Images at the indicated times following treatment with tamoxifen are displayed as the square of maximum intensity projections across the z-planes with the minimum intensity mapped to 0 and the maximum to 1. Channels were combined in ImageJ.
Quantification of intensity of fluorescence signals by CAPS
CAPS is a web-based application designed to aid in the selection and measurement of microscopy images captured on a CCD camera. Collections of images, displayed in the x-y plane, can be navigated in the z and time dimensions. CAPS measures the sum of all pixels using only raw data within the radius of the signal and subtracts the average value of the pixels in the surrounding one-pixel annulus from each pixel, thereby removing the influence of the background. The background annuli are chosen visually to encompass >90% of the signal at the most intense z-level as measured by Gaussian fitting. The value is summed for each of 1-10 z-levels spanned by the signal. CAPS is described fully at (http://caps.grapheasy.org). CAPS determines overlap by calculating the ratio of the average pixel intensity of a signal divided by the average pixel intensity of its surrounding annulus. Then, using Spearman’s rank correlation coefficient, a user measures the correlation of the ratios between one colored signal and another for multiple pairs across multiple images. No overlap between the signals in the two channels would appear uncorrelated, while overlapping signals appear correlated.
Measuring intensities of fluorescence signals in nuclei
To determine the change of intensity of fluorescent signals in nuclei over the cell cycle (for example, that of eGFP-Cdt1) we outlined the nucleus and determined the average pixel intensity within the outline. To identify the edge of the nucleus, we first defined a circle that completely surrounded the nucleus. Then, we used active contours or “snakes”, a computer vision algorithm that constricts the circle until each point along the circle has hit an edge or contour, to define the nuclear edge. This was done using the splineSnake plugin (http://bigwww.epfl.ch/jacob/software/SplineSnake/) for ImageJ and a custom macro for determining averages over time.
Supplementary Material
Highlights.
EBV sequentially reprograms the nucleus to amplify its DNA;
EBV waits until early S-phase to amplify its DNA as monitored with Cdt1/eGFP;
EBV reorganizes the nucleus to house discrete amplification factories;
EBV inhibits histone chaperones and redistributes PCNA to evade histone deposition.
Acknowledgments
This work was funded by grants from the National Cancer Institute, National Institutes of Health (grant P01 CA022443, grants R01 CA133027 and R01 CA070723). Bill Sugden is an American Cancer Society Research Professor. We thank Drs. Adams, Cardoso, Kenney and Stillman for plasmids, Danielle Westhoff Smith for isolating human primary B cells, Adityarup Chakravorty for retroviral vectors encoding eGFP-H2B and eGFP-PCNA, and Mitch Hayes for invaluable support with Zeiss software and hardware. We thank, Paul Ahlquist, Eric Johannsen, Shih-Tung Liu, Donata Oertel, Nate Sherer, and our lab colleagues for their experimental insights and suggestions. We also thank Norman Drinkwater for guiding us in the development of the Spearman’s correlation coefficient for CAPS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Document S1. five figures, one table, supplemental experimental procedures, and references
References
- Amon W, White RE, Farrell PJ. Epstein-Barr Virus origin of lytic replication mediates association of replicating episomes with promyelocytic leukaemia protein nuclear bodies and replication compartments. J Gen Virol. 2006;87:1133–1137. doi: 10.1099/vir.0.81589-0. [DOI] [PubMed] [Google Scholar]
- Bell P, Lieberman PM, Maul GG. Lytic but not latent replication of Epstein-Barr Virus is associated with PML and induces sequential release of nuclear domain 10 proteins. J Virol. 2000;74:11800–11810. doi: 10.1128/jvi.74.24.11800-11810.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloss TA, Sugden B. Optimal lengths for DNAs encapsidated by Epstein-Barr Virus. J Virol. 1994;68:8217–8222. doi: 10.1128/jvi.68.12.8217-8222.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrol C, Flemington EK. The Epstein-Barr Virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 1996;15:2748–2759. [PMC free article] [PubMed] [Google Scholar]
- Chang LK, Liu ST. Activation of the BRLF1 promoter and lytic cycle of Epstein-Barr Virus by histone acetylation. Nucleic Acids Res. 2000;28:3918–3925. doi: 10.1093/nar/28.20.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasserot-Golaz S, Schuster C, Dietrich JB, Beck G, Lawrence DA. Antagonistic action of RU38486 on the activity of transforming growth factor-beta in fibroblasts and lymphoma cells. J Steroid Biochem. 1988;30:381–385. doi: 10.1016/0022-4731(88)90127-6. [DOI] [PubMed] [Google Scholar]
- Chiu YF, Tung CP, Lee YH, Wang WH, Li C, Hung JY, Wang CY, Kawaguchi Y, Liu ST. A comprehensive library of mutations of Epstein Barr Virus. J Gen Virol. 2007;88:2463–2472. doi: 10.1099/vir.0.82881-0. [DOI] [PubMed] [Google Scholar]
- Countryman J, Miller G. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc Natl Acad Sci USA. 1985;82:4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daikoku T, Kudoh A, Fujita M, Sugaya Y, Isomura H, Shirata N, Tsurumi T. Architecture of replication compartments formed during Epstein-Barr Virus lytic replication. J Virol. 2005;79:3409–3418. doi: 10.1128/JVI.79.6.3409-3418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daikoku T, Kudoh A, Sugaya Y, Iwahori S, Shirata N, Isomura H, Tsurumi T. Postreplicative mismatch repair factors are recruited to Epstein-Barr Virus replication compartments. J Biol Chem. 2006;281:11422–11430. doi: 10.1074/jbc.M510314200. [DOI] [PubMed] [Google Scholar]
- Delecluse HJ, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. Propagation and recovery of intact, infectious Epstein-Barr Virus from prokaryotic to human cells. Proc Natl Acad Sci USA. 1998;95:8245–8250. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fixman ED, Hayward GS, Hayward SD. trans-acting requirements for replication of Epstein-Barr Virus ori-Lyt. J Virol. 1992;66:5030–5039. doi: 10.1128/jvi.66.8.5030-5039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigault MM, Lacoste J, Swift JL, Brown CM. Live-cell microscopy - tips and tools. J Cell Sci. 2009;122:753–767. doi: 10.1242/jcs.033837. [DOI] [PubMed] [Google Scholar]
- Glaser R, Rapp F. Rescue of Epstein-Barr Virus from somatic cell hybrids of Burkitt lymphoblastoid cells. J Virol. 1972;10:288–296. doi: 10.1128/jvi.10.2.288-296.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt W, Sugden B. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein-Barr Virus. Cell. 1988;55:427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt W, Sugden B. Epstein-Barr Virus DNA Replication. In: Bell S, Mechali M, D. M., editors. DNA Replication. Cold Spring Harbor Laboratory Press; NY: 2013. pp. 513–525. [Google Scholar]
- Johannsen E, Luftig M, Chase MR, Weicksel S, Cahir-McFarland E, Illanes D, Sarracino D, Kieff E. Proteins of purified Epstein-Barr Virus. Proc Natl Acad Sci USA. 2004;101:16286–16291. doi: 10.1073/pnas.0407320101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson ZO, Hindges R, Hubscher U. Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. EMBO J. 1998;17:2412–2425. doi: 10.1093/emboj/17.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehl A, Dorsky DI. Bipartite DNA-binding region of the Epstein-Barr Virus BMRF1 product essential for DNA polymerase accessory function. J Virol. 1995;69:1669–1677. doi: 10.1128/jvi.69.3.1669-1677.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudoh A, Iwahori S, Sato Y, Nakayama S, Isomura H, Murata T, Tsurumi T. Homologous recombinational repair factors are recruited and loaded onto the viral DNA genome in Epstein-Barr Virus replication compartments. J Virol. 2009;83:6641–6651. doi: 10.1128/JVI.00049-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutluay SB, Triezenberg SJ. Regulation of histone deposition on the herpes simplex virus type 1 genome during lytic infection. J Virol. 2009;83:5835–5845. doi: 10.1128/JVI.00219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt H, Rahn HP, Weinzierl P, Sporbert A, Cremer T, Zink D, Cardoso MC. Dynamics of DNA replication factories in living cells. J Cell Biol. 2000;149:271–280. doi: 10.1083/jcb.149.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci USA. 2010;107:14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luka J, Kallin B, Klein G. Induction of the Epstein-Barr Virus (EBV) cycle in latently infected cells by n-butyrate. Virology. 1979;94:228–231. doi: 10.1016/0042-6822(79)90455-0. [DOI] [PubMed] [Google Scholar]
- MacAlpine MD, Almouzni G. Chromatin and DNA replication. In: Bell S, Mechali M, D. M., editors. DNA Replication. Cold Spring Harbor Laboratory Press; NY: 2013. pp. 197–218. [Google Scholar]
- Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J Cell Sci. 2003;116:3051–3060. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- Makhov AM, Subramanian D, Holley-Guthrie E, Kenney SC, Griffith JD. The Epstein-Barr Virus polymerase accessory factor BMRF1 adopts a ring-shaped structure as visualized by electron microscopy. J Biol Chem. 2004;279:40358–40361. doi: 10.1074/jbc.M408733200. [DOI] [PubMed] [Google Scholar]
- Meerbach A, Holy A, Wutzler P, De Clercq E, Neyts J. Inhibitory effects of novel nucleoside and nucleotide analogues on Epstein-Barr Virus replication. Antivir Chem Chemother. 1998;9:275–282. doi: 10.1177/095632029800900309. [DOI] [PubMed] [Google Scholar]
- Meng Q, Hagemeier SR, Fingeroth JD, Gershburg E, Pagano JS, Kenney SC. The Epstein-Barr Virus (EBV)-encoded protein kinase, EBV-PK, but not the thymidine kinase (EBV-TK), is required for ganciclovir and acyclovir inhibition of lytic viral production. J Virol. 2010;84:4534–4542. doi: 10.1128/JVI.02487-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mighell KJ. Algorithms for CCD stellar Photometry. In: R.L.P., Mehringer DM, Roberts DA, editors. Astronomical Data Analysis Software and Systems VIII, ASP Conference Series; 1999. pp. 317–328. [Google Scholar]
- Moggs JG, Grandi P, Quivy JP, Jonsson ZO, Hubscher U, Becker PB, Almouzni G. A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol Cell Biol. 2000;20:1206–1218. doi: 10.1128/mcb.20.4.1206-1218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier K, Armas JC, Etteldorf S, Ghazal P, Sullivan KF. Annexation of the interchromosomal space during viral infection. Nat Cell Biol. 2000;2:661–665. doi: 10.1038/35023615. [DOI] [PubMed] [Google Scholar]
- Mosammaparast N, Ewart CS, Pemberton LF. A role for nucleosome assembly protein 1 in the nuclear transport of histones H2A and H2B. EMBO J. 2002;21:6527–6538. doi: 10.1093/emboj/cdf647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanbo A, Sugden A, Sugden B. The coupling of synthesis and partitioning of EBV’s plasmid replicon is revealed in live cells. EMBO J. 2007;26:4252–4262. doi: 10.1038/sj.emboj.7601853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H, Taraviras S, Lygerou Z, Nishimoto T. The human licensing factor for DNA replication Cdt1 accumulates in G1 and is destabilized after initiation of S-phase. J Biol Chem. 2001;276:44905–44911. doi: 10.1074/jbc.M105406200. [DOI] [PubMed] [Google Scholar]
- Norby K, Chiu YF, Sugden B. Monitoring plasmid replication in live mammalian cells over multiple generations by fluorescence microscopy. J Vis Exp. 2012:e4305. doi: 10.3791/4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuwaki M, Kato K, Shimahara H, Tate S, Nagata K. Assembly and disassembly of nucleosome core particles containing histone variants by human nucleosome assembly protein I. Mol Cell Biol. 2005;25:10639–10651. doi: 10.1128/MCB.25.23.10639-10651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park R, Heston L, Shedd D, Delecluse HJ, Miller G. Mutations of amino acids in the DNA-recognition domain of Epstein-Barr Virus ZEBRA protein alter its sub-nuclear localization and affect formation of replication compartments. Virology. 2008;382:145–162. doi: 10.1016/j.virol.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfuller R, Hammerschmidt W. Plasmid-like replicative intermediates of the Epstein-Barr Virus lytic origin of DNA replication. J Virol. 1996;70:3423–3431. doi: 10.1128/jvi.70.6.3423-3431.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Sharp JA, Fouts ET, Krawitz DC, Kaufman PD. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr Biol. 2001;11:463–473. doi: 10.1016/s0960-9822(01)00140-3. [DOI] [PubMed] [Google Scholar]
- Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- Shirakata M, Imadome KI, Hirai K. Requirement of replication licensing for the dyad symmetry element-dependent replication of the Epstein-Barr Virus oriP minichromosome. Virology. 1999;263:42–54. doi: 10.1006/viro.1999.9965. [DOI] [PubMed] [Google Scholar]
- Sugden B. Expression of virus-associated functions in cells transformed in vitro by Epstein-Barr Virus: Epstein-Barr Virus cell surface antigen and virus-release from transformed cells. In: Purtilo DT, editor. Immune Deficiency and Cancer : Epstein-Barr Virus and lymphoproliferative malignancies. Plenum; New York ; London: 1984. pp. 165–177. [Google Scholar]
- Sugden B, Mark W. Clonal transformation of adult human leukocytes by Epstein-Barr Virus. J Virol. 1977;23:503–508. doi: 10.1128/jvi.23.3.503-508.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto A, Kanda T, Yamashita Y, Murata T, Saito S, Kawashima D, Isomura H, Nishiyama Y, Tsurumi T. Spatiotemporally different DNA repair systems participate in Epstein-Barr Virus genome maturation. J Virol. 2011;85:6127–6135. doi: 10.1128/JVI.00258-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- Takada K. Cross-linking of cell surface immunoglobulins induces Epstein-Barr Virus in Burkitt lymphoma lines. Int J Cancer. 1984;33:27–32. doi: 10.1002/ijc.2910330106. [DOI] [PubMed] [Google Scholar]
- Takada K, Shimizu N, Sakuma S, Ono Y. Trans activation of the latent Epstein-Barr Virus (EBV) genome after transfection of the EBV DNA fragment. J Virol. 1986;57:1016–1022. doi: 10.1128/jvi.57.3.1016-1022.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovey MG, Lenoir G, Begon-Lours J. Activation of latent Epstein-Barr Virus by antibody to human IgM. Nature. 1978;276:270–272. doi: 10.1038/276270a0. [DOI] [PubMed] [Google Scholar]
- Tsurumi T. EBV replication enzymes. Curr Top Microbiol Immunol. 2001;258:65–87. doi: 10.1007/978-3-642-56515-1_5. [DOI] [PubMed] [Google Scholar]
- Tsurumi T, Daikoku T, Kurachi R, Nishiyama Y. Functional interaction between Epstein-Barr Virus DNA polymerase catalytic subunit and its accessory subunit in vitro. J Virol. 1993a;67:7648–7653. doi: 10.1128/jvi.67.12.7648-7653.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurumi T, Daikoku T, Nishiyama Y. Further characterization of the interaction between the Epstein-Barr Virus DNA polymerase catalytic subunit and its accessory subunit with regard to the 3′-to-5′ exonucleolytic activity and stability of initiation complex at primer terminus. J Virol. 1994;68:3354–3363. doi: 10.1128/jvi.68.5.3354-3363.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurumi T, Kobayashi A, Tamai K, Daikoku T, Kurachi R, Nishiyama Y. Functional expression and characterization of the Epstein-Barr Virus DNA polymerase catalytic subunit. J Virol. 1993b;67:4651–4658. doi: 10.1128/jvi.67.8.4651-4658.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warbrick E. The puzzle of PCNA’s many partners. Bioessays. 2000;22:997–1006. doi: 10.1002/1521-1878(200011)22:11<997::AID-BIES6>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Yates J, Warren N, Reisman D, Sugden B. A cis-acting element from the Epstein-Barr Viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci USA. 1984;81:3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Hausen H, O’Neill FJ, Freese UK, Hecker E. Persisting oncogenic herpesvirus induced by the tumour promotor TPA. Nature. 1978;272:373–375. doi: 10.1038/272373a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.