Abstract
Purpose.
Autosomal recessive retinitis pigmentosa (ARRP) is a genetically heterogeneous condition characterized by progressive loss of retinal photoreceptor cells. In order to gain new insights into the pathogenesis of ARRP, we evaluated the morphological, biochemical, and gene expression changes in eyes from a human donor with ARRP due to mutations in the ABCA4 gene.
Methods.
Eyes were obtained postmortem from a donor with end-stage retinitis pigmentosa. The coding sequences of the RDS, RHO, and ABCA4 genes were screened for disease-causing mutations. Morphological changes in different regions of the retina were examined histologically, and levels of lipofuscin-associated bisretinoids were measured. Gene expression was examined in retinal/choroidal tissue using microarray analysis, and all parameters were compared to those in unaffected control donors.
Results.
Genetic analysis of the donor's DNA identified two mutations in the ABCA4 gene, IVS14+1G > C and Phe1440del1 cT, each on a separate allele. Morphological evaluation revealed complete loss of the outer nuclear layer, remodeling of the inner retina, loss of retinal vasculature, and regional neovascularization. The retinal pigment epithelium and choriocapillaris exhibited regional preservation. Microarray analysis revealed loss of photoreceptor cell-associated transcripts, with preservation of multiple genes expressed specifically in inner retinal neurons.
Conclusions.
The persistence of transcripts expressed by inner retinal neurons suggests that despite significant plasticity that occurs during retinal degeneration, bipolar cells and ganglion cells remain at least partially differentiated. Findings from this study suggest that some forms of therapy currently under investigation may have benefit even in advanced retinal degeneration.
Anatomical, biochemical, and gene expression studies of donor eyes with ABCA4-associated retinitis pigmentosa revealed that photoreceptor loss precedes retinal pigment epithelium loss in some areas and that transcripts expressed by inner retinal neurons continue to be expressed for decades after complete vision loss.
Introduction
The term retinitis pigmentosa (RP) refers to a large number of syndromic and nonsyndromic heritable disorders characterized by progressive loss of vision due to degeneration of photoreceptor cells. The condition can be inherited in a dominant, recessive, X-linked, or oligogenic fashion.1,2 Mutations in dozens of genes encoding proteins with highly diverse functions expressed in photoreceptor cells, retinal pigment epithelium (RPE), or choriocapillaris have been identified that are responsible for RP, with many more remaining to be discovered (reviewed in Ref. 1).
One approach to developing an improved understanding of the pathogenesis of a rare disease is to examine biopsy or autopsy samples from the affected tissue. In the case of RP, several studies of human tissues have been performed and reveal the general characteristics of absent or severely reduced numbers of photoreceptor cells, pigment migration into the retina coincident with “bone-spicule-like pigmentation” and photoreceptor rosettes.3–6
In this study, eyes from a donor diagnosed with autosomal recessive RP (ARRP) were characterized histopathologically. We also examined the distribution of a number of histochemical markers of glial cells and other cell types and noted an increased degree of alkaline phosphatase activity in the degenerative retina that spatially colocalized with glial fibrillary acidic protein (GFAP). Sequencing of the ABCA4 gene identified both disease-causing alleles. Finally, expression microarray analysis of retinal tissue from the donor revealed the expected loss of differentiation markers of photoreceptor cells, as well as increased expression of glial markers and upregulation of a number of complement and inflammation genes. Transcripts encoded by genes expressed in inner retinal neurons were found to show continued expression. Preservation of some markers of inner retinal neurons suggests that second order neurons persist even in cases of advanced degeneration and may be able to be rescued.
Methods
Patient Description
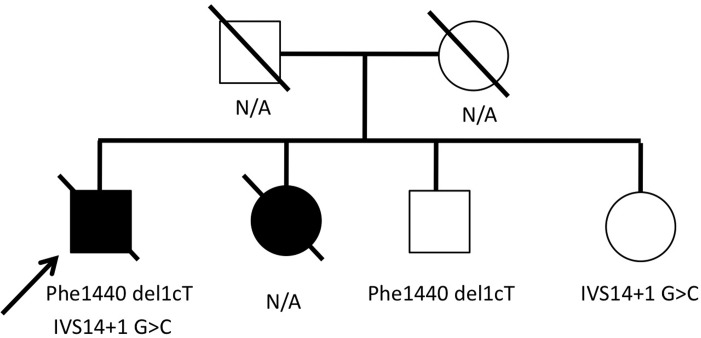
The patient was first seen in our clinic at 35 years of age (in 1961). His visual acuity was counting fingers at less than two feet OU. The fundus examination revealed an area of grayish scarring, attenuation of the retinal vasculature, and significant intraretinal pigmentation in the posterior pole, while the peripheral retina and choroid appeared to be comparatively normal. The patient's family history was remarkable for one sister with RP (Fig. 1). The electroretinogram was nonrecordable under all stimulus conditions.
Figure 1.
Genotyping of the ABCA4 gene in the proband and in two unaffected siblings. The proband and eye donor in this report harbored two ABCA4 variations, Phe1440 del1cT and IVS14+1 G > C. The proband's brother and sister each harbored only one of these mutations, demonstrating that the mutations lie on different alleles. A second sister was reportedly blind, but her DNA was not available for study.
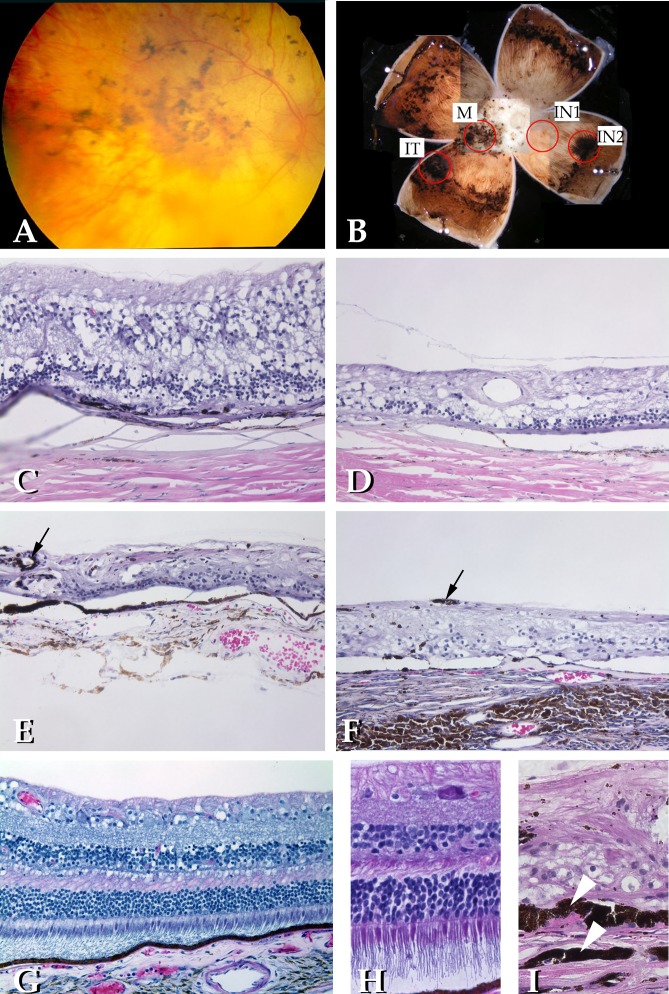
The patient's visual acuity deteriorated to hand motions at 37 years of age and to light perception at 40 years. Bone spicule-like pigmentation, chorioretinal atrophy, and arteriolar narrowing were noted at that time (Fig. 2A). At 64 years of age, the patient had no light perception. Large clumps of pigment with RPE atrophy over the posterior 75% to 80% of the fundus were observed. The patient was followed periodically over the next decade and died at age 78.
Figure 2.
Morphological evaluation of the proband's retina. (A) Fundus examination of the patient at age 40 revealed severe loss of RPE, gliosis, and pigment migration into the retina. (B) Gross view of the patient's right eye indicating the four regions sampled for microscopic analysis, including macula (M), inferotemporal pigmented mass (IT), inferonasal area of complete RPE dropout (IN1), and an inferonasal area of relatively even pigmentation (IN2). (C) Hematoxylin-eosin stain of the macular region. Extensive loss of photoreceptor cells is apparent, with small clumps of RPE still present on Bruch's membrane. Inner retinal layers are relatively preserved. (D) Appearance of the retina in the IN1 region. Loss of the outer nuclear layer, RPE, and choroid is notable. (E) IN2 region retains a largely intact layer of RPE, with some bone spicule-like pigment migration notable (arrows), especially around vascular elements in the neural retina. Photoreceptor loss in this region is complete, however. (F) Scarring was observed in the IT punch that included neovascularization and fibrosis. (G and H) Retina of an unaffected donor for comparison (original magnification in G was the same as those in C–F. (H) A higher magnification image obtained using the same conditions in I, which shows RPE hypertrophy and duplication (arrowheads) in the fibrotic lesion of the IT punch.
Genetic Analysis
The Declaration of Helsinki was followed for all experiments with human samples. Genetic testing was performed to screen the entire coding sequence of the genes encoding rhodopsin (RHO) and rds (PRPH2), and the Arg677Stop mutation in RP1. For ABCA4, seven amplimers of coding sequence were screened for mutations by using automated bidirectional DNA sequencing. Screening of the proband was discontinued after two disease-causing alleles were detected. These variants were subsequently evaluated in two of the donor's unaffected siblings to determine whether the mutations were on the same or different alleles.
Eye Processing
At the request of his family, the patient's eyes were donated for research through the Iowa Lions Eye Bank (Iowa City, IA). The right eye was fixed by immersion in 4% formaldehyde solution in 10 mM phosphate-buffered saline. From the left eye, fragments of peripheral retina-RPE/choroid tissues were collected and flash frozen in liquid nitrogen within 7 hours of death. Due to scarring between the retina and choroid, these tissues were unable to be readily separated and were therefore collected and snap frozen together. A central-to-peripheral wedge of the left eye was fixed for ∼2 hours and embedded in acrylamide solution for cryostat sectioning, as described previously.7
A 6-mm dermal biopsy punch was used to collect four regions of the fixed right eye that included the macula centered on the fovea, an inferotemporal gliotic mass admixed with pigment, an inferonasal area of apparent complete atrophy (IN1), and a more peripheral inferonasal area with pigment that appeared intact (IN2). Locations of each of these samples are indicated in Figure 2B. Fixed tissues were infiltrated in sucrose solution prior to embedment in Optimal Cutting Temperature compound (Ted Pella, Redding, CA), as described previously.8 Frozen sections were stained with hematoxylin-eosin stain or were used for histochemical analysis (described below). A control eye from a 63-year-old person with a similar postmortem preservation time was used for comparison to the RP eye.
Histological Methods
Immunohistochemical staining was performed using antibodies directed against rhodopsin (Santa Cruz Biotechnology, Santa Cruz, CA), glial fibrillar acidic protein (GFAP; Neomarkers), intercellular adhesion molecule-1 (ICAM-1; Developmental Studies Hybridoma Bank, Iowa City, IA), RPE65 (Chemicon, Temecula, CA), neurofilament heavy chain (Chemicon), and terminal complement complex (Dako). Endogenous phosphatase was detected using a nitroblue tetrazolium (NBT), 5-bromo-4-chloro-3-indolyl-phosphate (BCIP) kit (Vector Laboratories). Labeling procedures were performed as described previously.9,10
Retinoid Analysis
Paraformaldehyde-fixed human donor eyes were used for assessment of A2E levels. Disposable 6-mm trephines were used to collect peripheral punches of RPE-choroid from the left eye of the ARRP eye and from four additional, unaffected donors (ages 72, 76, 80, and 81). The region from the ARRP eye showed a complete loss of photoreceptor cells but had an intact RPE layer. Chloroform extracts of the eye punches were analyzed by normal phase on a silica column (Zorbax-Sil 5 μm, 250 mm × 4.6 mm; Agilent, Santa Clara, CA) in a high-performance liquid chromatograph (Agilent model 1100) equipped with a photodiode array detector (Agilent Technologies, Wilmington, DE). The method of extraction and analysis has been previously described.11
Microarray Analyses
A quadrant of the peripheral frozen retina-RPE-choroid complex from the left eye of this donor was used for microarray analysis. Briefly, RNA was isolated from the combined retina-RPE-choroid layers of the ARRP eye (RNeasy kit; Qiagen) and from regionally matched, pooled peripheral retina and RPE-choroid layers from an age-matched eye without retinal disease. The integrity of the isolated RNA was confirmed using a BioAnalyzer (Agilent). Reverse transcription, synthesis of complementary RNA, and hybridization to U133 plus 2.0 chips (Affymetrix, Santa Clara, CA) was performed as described in detail previously.12,13
Data were normalized using the robust microarray analysis algorithm. Only those probe sets that displayed expression values of at least 100 units in either the ARRP or the control eye were considered to be expressed and were evaluated further. Fold change ratios were determined between the ARRP eye and an unaffected control eye.
We also prospectively selected a limited number of cell type-specific genes to evaluate the status of different classes of cells in the degenerated retina. Transcripts of melanopsin, neurofilament heavy chain, and Thy1 were used to assess the integrity of ganglion cells, while transcripts of metabotropic glutamate receptor-6, calretinin, and parvalbumin were used for an assessment of bipolar and amacrine cells. Transcripts of GFAP and cellular retinaldehyde binding protein were used to evaluate astrocytes and Muller cells, while rhodopsin, ROM1, and red/green opsins were used for an assessment of photoreceptor cells. The integrity of the RPE was estimated by examining the transcripts of bestrophin-1 and RPE65, while the choriocapillaris was evaluated by quantifying transcripts of VEGFR2 and carbonic anhydrase IV.
Results
Genetic Screening
Genetic screening of RHO, PRPH2, and the RP1 Arg677Stop variation did not detect any disease-causing mutations. In contrast, DNA sequencing of ABCA4 revealed two plausible disease-causing mutations: IVS14+1G > C (c.2160+1 G > C) and a novel mutation, Phe1440del1cT (c.4318del1cT), both in the heterozygous state.
Phase was established by sequencing two unaffected siblings for the two mutations identified in the proband. The proband's unaffected brother was found to carry only the Phe1440 del1cT mutation, whereas the proband's unaffected sister was found to harbor only the IVS14+1 variant, demonstrating that both mutations lie on different alleles (Fig. 1).
Histological examination of eyes from this donor showed features consistent with other eyes with RP that have been described previously (e.g., loss of the outer nuclear layer and RPE migration into the neural retina) as summarized below.
Hematoxylin-Eosin Stain
In each of the surveyed regions, photoreceptor cell loss appeared complete. The macular region showed complete loss of the outer nuclear layer, with some evidence of inner nuclear layer and ganglion cell layer preservation (Fig. 2C). The RPE in the macula was discontinuous and in places showed migration into the outer aspect of the retina, with some cells forming a two-layered epithelium. The severely depigmented IN1 similarly showed loss of the outer nuclear layer with attenuation of the retina and choroid (Fig. 2D). Remnants of pigmentation were detected in the choroid in scattered profiles. Bruch's membrane appeared to be intact in this region. The far peripheral IN2 punch showed a relatively normal RPE layer in which the RPE was continuous and showed normal polarity, although pigment density varied regionally, with RPE cells ranging from brown to black (Fig. 2E). As in the other regions examined, however, a complete loss of outer nuclear layer and photoreceptor cells was observed. The inferotemporal region contained a large nevus with fibrovascular scar tissue between Bruch's membrane and the remnants of the neural retina. The RPE was observed in several island-like clumps at multiple layers through the scar and retina. Individual RPE cells were often hypertrophied and were present in three or more layers, in addition to accumulating around large vessels in the inner retina (Fig. 2I). The choriocapillaris was atrophic in all regions examined except for the far peripheral inferonasal punch, in which localized islands of preserved choriocapillaris were present.
Autofluorescence
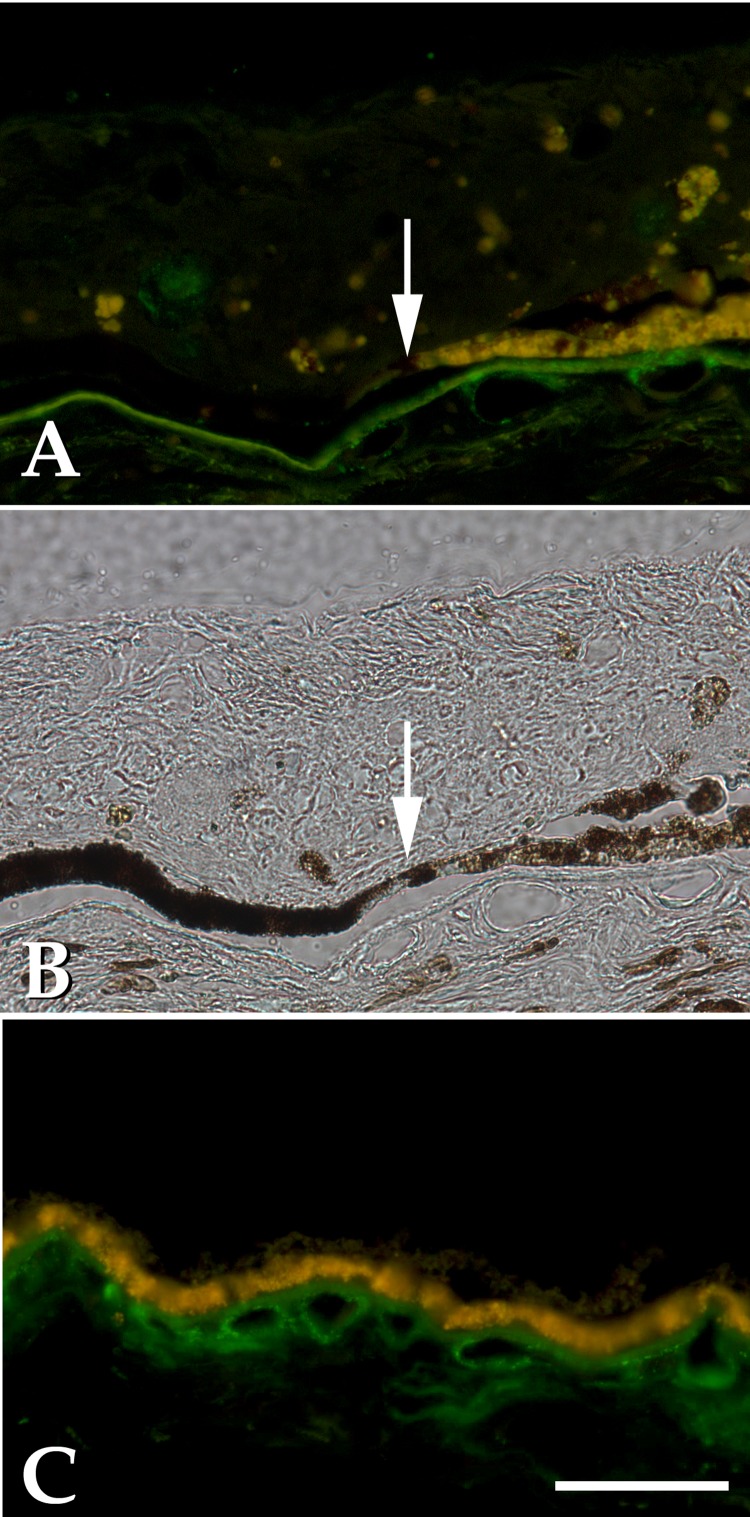
Areas of lipofuscin-laden RPE were observed in the inferonasal and inferotemporal punches. Autofluoresecent RPE cells were observed underlying retina completely devoid of photoreceptor cells (Figs. 3A and 3B). Hypofluorescent (deeply pigmented) and hyperfluorescent (normally pigmented) RPE cells were observed in this region, with a clear line of demarcation (Fig. 3, arrows). Autofluorescence characteristics of Bruch's membrane appeared normal regardless of the level of RPE pigmentation.
Figure 3.
Comparison of RPE lipofuscin (yellow-orange) and complement membrane attack complex (green) in the ARRP eye (A and B) and an age-matched control donor eye (C). Loss of RPE autofluorescence in dystrophic RPE (A and B) corresponds to loss of choroidal vasculature and decreased membrane attack complex labeling. Note the sharp demarcation between highly autofluorescent and hypofluorescent, deeply pigmented cells on either side of the arrow. Autofluorescent RPE cells were found to overlie an intact choriocapillaris, whereas nonautofluorescent RPE cells overlie an area of choriocapillaris dropout. Complement labeling in Bruch's membrane, and the choriocapillaris is observed only over the half of the field with intact capillaries and autofluorescent RPE. Merged images using FITC and tetramethyl rhodamine isothiocyanate filter sets are shown in A and C. (B) same field as A, exposed in brightfield. Bar = 50 μm.
Anti-Terminal Complement Complex C5b-9
Sections were labeled with an antibody directed against the C9 neoantigen that is exposed by complement activation. As described previously,14–17 C5b-9 labeling was observed in the macular region in Bruch's membrane and choriocapillary pillars and along the choriocapillaris (Fig. 3C). In the RP eye, the degree of C5b-9 labeling observed was generally reduced compared with that of control eyes, suggesting a role for RPE in complement activation, although the site of complement deposition in these eyes (the choriocapillaris) was generally atrophied as well. In areas of intact choriocapillaris, labeling for C5b-9 was reduced compared to that in controls but was still detectable (Fig. 3A). Figure 3 shows a field from the far peripheral inferonasal region in which RPE cells on the right half of the field are autofluorescent and overlying intact choriocapillaris, whereas the left half of the field shows hyperpigmented, nonautofluorescent RPE cells overlying an area of choriocapillaris dropout. Complement labeling in Bruch's membrane and the choriocapillaris was observed only over the half of the field with intact capillaries and autofluorescent RPE (Figs. 3A and 3C).
Anti-GFAP
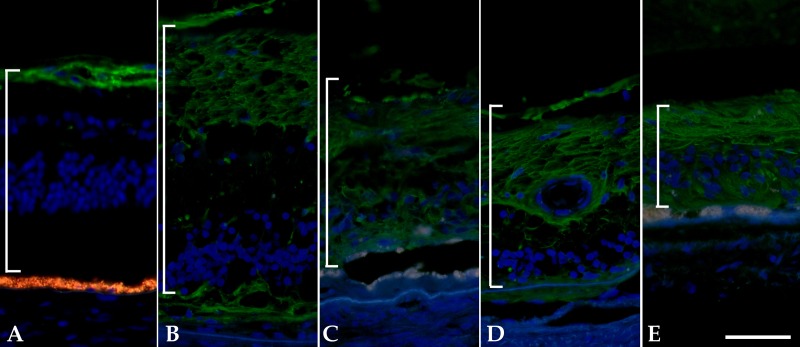
Labeling for GFAP in normal eyes was generally confined to the inner retina, along the nerve fiber layer and around large retinal vessels (Fig. 4A). In each of the regions from the RP eye, however, GFAP labeling was more intense and more widely distributed through the retina. Although the greatest intensity of labeling remained in the innermost layers, labeling was also observed throughout the remaining thickness of the retina. This was least pronounced in the macular punch where the retina was thickest.
Figure 4.
Gliosis in the ARRP eye demonstrated by labeling with anti-GFAP antibody. (A) Unaffected control eye shows labeling in the inner retina. (B–E) Labeling in RP donor. Upregulated and redistributed GFAP is present in macular (B), IT (C), IN1 (D), and IN2 (E) regions. Note the autofluorescence of the RPE still present in C and E. Brackets indicate thickness of the neural retina. Bar = 50 μm.
Alkaline Phosphatase Activity
In the right eye, alkaline phosphatase activity was not observed due to prolonged fixation. A more lightly fixed portion of the left eye was infiltrated in acrylamide and incubated with the alkaline phosphatase substrates NBT and BCIP. Whereas unaffected eyes showed reactivity of choroidal and retinal vasculature (Fig. 5A), the RP eye exhibited more widespread labeling throughout the neural retina, over approximately the same area occupied by GFAP-reactive cells (Figs. 5B and 5C). This pattern is similar to the migration columns described in animal models of retinal degeneration.18 Vascular dropout was noted in both the retina and choroid. To verify that this activity was due to alkaline phosphatase, NBT and BCIP were also prepared in neutral (pH 7.5) and acidic (pH 3.5) buffers. No labeling was observed when these solutions were used (data not shown).
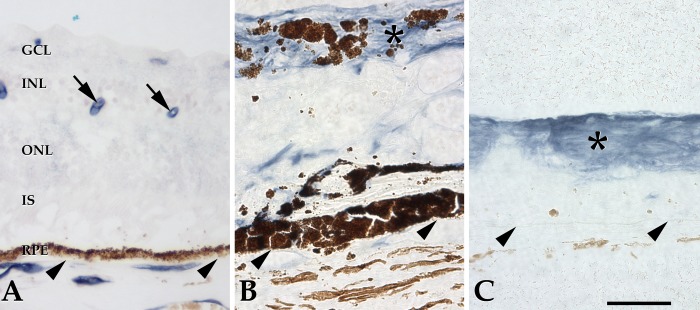
Figure 5.
Alkaline phosphatase activity in a control eye (A) and the ARRP eye (B and C). Normally alkaline phosphatase activity is restricted to retinal vessels (arrows) and choroidal endothelial cells. In the ARRP eye, elevated activity was observed throughout the inner retina (asterisks), presumably due to glial cell proliferation. Arrowheads indicate Bruch's membrane. GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer. Bar = 50 μm.
Figure 6.
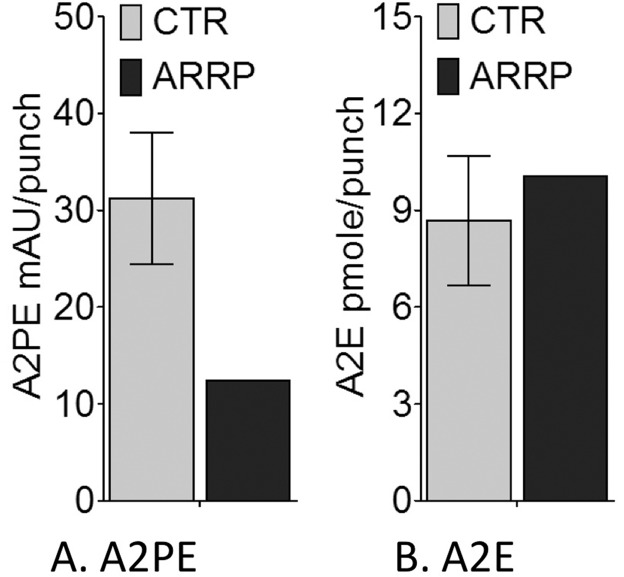
Results of high-performance liquid chromatography quantification of retinoids in ARRP and control RPE-choroid tissues. (A) A2PE expressed as the milliabsorbance unit (mAU) per punch and (B) A2E as pmol per punch. The ARRP eye showed an apparent decrease in A2PE due to loss of photoreceptor cells. A2E levels in the ARRP eye are comparable to the controls (CTR).
Peanut Agglutinin and Anti-Rhodopsin Antibody
In normal eyes, labeling of cone matrix sheaths and rod outer segments is readily detectable with peanut agglutinin (PNA) and anti-rhodopsin, respectively. In the RP eye, no evidence of immunoreactive cone or rod remnants was observed in any of the areas surveyed. In areas of retinal scarring, thin strands of PNA-binding material or cells were observed occasionally on the outer surface of the scar (data not shown).
A2E Fluorophores in ARRP
Quantification of different classes of lipofuscin fluorophores extracted from normal age-matched control eyes and the ARRP donor eye revealed that the total A2E level (defined as A2E plus iso-A2E) in the RPE underlying extremely degenerated retina of the ARRP eye was comparable to that in the normal RPE of the controls. Interestingly, whereas A2E showed similar overall levels, A2PE, a direct A2E precursor, was dramatically reduced in the ARRP eye. A2PE levels were 32% to 70% of control levels, with an average of 39.9%, reflecting significant loss of the photoreceptor cells as they are the primary source of bisretinoid formation.
Microarray Analyses
Combined neural retina and RPE/choroid layers were evaluated by microarray analysis for the ARRP donor and were compared with pooled retina/RPE/choroid layers from an unaffected donor. Due to the rarity of retinal tissue with ARRP, a rigorous statistical analysis of these data was not possible. However, Table 1 lists those genes with the largest difference in expression values, as determined by the fold change ratio (ARRP:control expression ratio). Several of these genes are represented by more than one probe set, and thus, the 100 probe sets with the most reduced expression in the RP eye correspond to 78 distinct genes. According to this analysis, the genes showing the greatest decrease in the RP eye were PDE6A, followed by CNGA1 and RHO. Notably, of the 78 genes with the most reduced expression, 28 genes are known to cause human retinal degeneration when mutated (see, e.g., RetNet; http://www.sph.uth.tmc.edu/retnet/).
Table 1.
Genes with Decreased Expression in ARRP Eye
|
Gene Symbol |
Gene Name |
ARRP/Control Expression Ratio |
| PDE6A* | Phosphodiesterase 6A, cGMP-specific, rod, alpha | 972 |
| CNGA1* | Cyclic nucleotide gated channel alpha 1 | 824 |
| RHO* | Rhodopsin (opsin 2, rod pigment) (retinitis pigmentosa 4, autosomal dominant) | 708 |
| Nonannotated | Nonannotated transcript | 708 |
| PDE6G | Phosphodiesterase 6G, cGMP-specific, rod, gamma | 693 |
| GNAT1* | Guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1 | 599 |
| IMPG1 | Interphotoreceptor matrix proteoglycan 1 | 571 |
| RDH12* | Retinol dehydrogenase 12 (all-trans/9-cis/11-cis) | 361 |
| DEFB119 | Defensin, beta 119 | 351 |
| KCNJ14 | Potassium inwardly-rectifying channel, subfamily J, member 14 | 317 |
| LOC145837 | Hypothetical protein LOC145837 | 304 |
| Nonannotated | Transcribed locus | 265 |
| Nonannotated | Transcribed locus | 252 |
| MPP4 | Membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) | 240 |
| RP1L1* | Retinitis pigmentosa 1-like 1 | 239 |
| RP1* | Retinitis pigmentosa 1 (autosomal dominant) | 228 |
| Nonannotated | Full-length insert cDNA clone YI54A07 | 220 |
| RTBDN | Retbindin | 196 |
| CNGB1* | Cyclic nucleotide gated channel beta 1 | 190 |
| PIWIL1 | Piwi-like 1 (Drosophila) | 178 |
| NRL* | Neural retina leucine zipper | 173 |
| GUCA1C | Guanylate cyclase activator 1C | 171 |
| PDC | Phosducin | 164 |
| AIPL1* | Aryl hydrocarbon receptor interacting protein-like 1 | 162 |
| SPINK4 | Serine peptidase inhibitor, Kazal type 4 | 161 |
| SAG* | S-antigen; retina and pineal gland (arrestin) | 154 |
| LOC280665 | Putative anti-CNG alpha 1 cation channel translation product | 153 |
| GNAT2* | Guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2 | 142 |
| RPGRIP1* | Retinitis pigmentosa GTPase regulator interacting protein 1 | 131 |
| NR2E3* | Nuclear receptor subfamily 2, group E, member 3 | 131 |
| Nonannotated | Full-length insert cDNA clone ZD51E06 | 130 |
| RS1* | Retinoschisis (X-linked, juvenile) 1 | 122 |
| Nonannotated | CDNA FLJ37338 fis, clone BRAMY2020466 | 117 |
| KCNV2* | Potassium channel, subfamily V, member 2 | 114 |
| LOC284422 | Similar to HSPC323 | 107 |
| PRPH2* | Peripherin 2 (retinal degeneration, slow) | 100 |
| SLC6A6 | Solute carrier family 6 (neurotransmitter transporter, taurine), member 6 | 94 |
| SLC24A1 | Solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 | 92 |
| TMEM16B | Transmembrane protein 16B | 92 |
| GUCA1B* | Guanylate cyclase activator 1B (retina) | 86 |
| PPEF2 | Protein phosphatase, EF-hand calcium binding domain 2 | 85 |
| ATP6V0D2* | ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 | 84 |
| ABCA4* | ATP-binding cassette, sub-family A (ABC1), member 4 | 82 |
| ANKRD33 | Ankyrin repeat domain 33 | 81 |
| PDE6B* | Phosphodiesterase 6B, cGMP-specific, rod, beta (congenital stationary night blindness 3, autosomal dominant) | 78 |
| GUCA1A* | Guanylate cyclase activator 1A (retina) | 77 |
| MYL4 | Myosin, light chain 4, alkali; atrial, embryonic | 76 |
| Nonannotated | Transcribed locus | 74 |
| IMPG2 | Interphotoreceptor matrix proteoglycan 2 | 73 |
| MAK* | Male germ cell-associated kinase | 73 |
| TRA@ | T-cell receptor alpha locus | 72 |
| LRRC39 | Leucine rich repeat containing 39 | 68 |
| LOC728715 /// OVOS2 | Ovostatin 2 /// similar to cDNA sequence BC048546 | 64 |
| ARR3 | Arrestin 3, retinal (X-arrestin) | 64 |
| OPN1LW* | Opsin 1 (cone pigments), medium-wave-sensitive (color blindness, deutan) | 63 |
| ZMYND19 | Zinc finger, MYND-type containing 19 | 61 |
| PTHR2 | Parathyroid hormone receptor 2 | 61 |
| RCVRN | Recoverin | 60 |
| RBP3* | Retinol binding protein 3, interstitial | 59 |
| Nonannotated | Transcribed locus | 56 |
| CASZ1 | Castor zinc finger 1 | 56 |
| CABP4* | Calcium binding protein 4 | 55 |
| ELSPBP1 | Epididymal sperm binding protein 1 | 53 |
| NT5E | 5′-Nucleotidase, ecto (CD73) | 53 |
| Nonannotated | Transcribed locus | 53 |
| RSU1 | Ras suppressor protein 1 | 53 |
| Nonannotated | CDNA FLJ90705 fis, clone PLACE1007591 | 51 |
| RAXL1* | Retina and anterior neural fold homeobox like 1 | 50 |
| Nonannotated | MRNA; cDNA DKFZp313A1040 (from clone DKFZp313A1040) | 49 |
| REEP6 | Receptor accessory protein 6 | 48 |
| PDE4DIP | Phosphodiesterase 4D interacting protein (myomegalin) | 48 |
| TBX5 | T-box 5 | 47 |
| Nonannotated | Arsenic transactivated protein 1 | 46 |
| MAP4K1 | Mitogen-activated protein kinase kinase kinase kinase 1 | 42 |
| ATP1A4 | ATPase, Na+/K+ transporting, alpha 4 polypeptide | 42 |
| PDE6C* | Phosphodiesterase 6C, cGMP-specific, cone, alpha prime | 42 |
| FUT3 | Fucosyltransferase 3 (galactoside 3(4)-l-fucosyltransferase, Lewis blood group) | 41 |
Table 1 lists genes recognized by the top 100 transcripts, showing decreased expression in the ARRP eye compared to an unaffected control eye. Note that several of the probe sets recognized genes that have not yet been annotated (probe set identifiers are appended in supplemental data, available at http://www.iovs.org/content/53/4/1883/suppl/DC1). Genes known to be associated with human retinal disease (http://www.sph.uth.tmc.edu/retnet/) indicated by an asterisk. For cases in which multiple probe sets were present for a given gene, the average ratio is shown.
Results of the reciprocal analysis of genes that are more abundantly expressed in the ARRP retina are given in Table 2. Genes showing increased expression in the RP eye included immunoglobulin superfamily genes (ICAM1, HLA-DPA1, HLA-DQB1, HLA-DOA, and ISLR) and a number of other proinflammatory genes, including complement pathway genes (CFB, C2, and complement gene homologs) and several cytokines and cytokine receptors (IL6, CXCL10, CXCL2) (Table 2).
Table 2.
Genes Showing Increased Expression in ARRP Donor Eye
|
Gene Symbol |
Gene Name |
CTL/ARRP Ratio |
| LOC728320 /// LTF | Lactotransferrin /// similar to lactotransferrin | 521 |
| IL6 | Interleukin 6 (interferon, beta 2) | 163 |
| LOC653879 | Similar to complement C3 precursor | 100 |
| SERPINA5 | Serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 | 100 |
| HAMP | Hepcidin antimicrobial peptide | 100 |
| ICAM1 | Intercellular adhesion molecule 1 (CD54), human rhinovirus receptor | 100 |
| LEFTY2 | Left-right determination factor 2 | 100 |
| ATF3 | Activating transcription factor 3 | 100 |
| CH25H | Cholesterol 25-hydroxylase | 100 |
| DIO3 | Deiodinase, iodothyronine, type III | 100 |
| AQP1 | Aquaporin 1 (Colton blood group) | 100 |
| FOXF1 | Forkhead box F1 | 100 |
| CHI3L2 | Chitinase 3-like 2 | 100 |
| MYOC | Myocilin, trabecular meshwork inducible glucocorticoid response | 100 |
| FOSL2 | FOS-like antigen 2 | 100 |
| GBP3 | Guanylate binding protein 3 | 100 |
| ANGPTL4 | Angiopoietin-like 4 | 100 |
| RFX4 | Regulatory factor X, 4 (influences HLA class II expression) | 100 |
| HLA-DPA1 | Major histocompatibility complex, class II, DP alpha 1 | 96 |
| GDF15 | Growth differentiation factor 15 | 51 |
| CFB | Complement factor B | 50 |
| FCGBP | Fc fragment of IgG binding protein | 50 |
| MCHR1 | Melanin-concentrating hormone receptor 1 | 50 |
| EFNA1 | Ephrin-A1 | 50 |
| TNFRSF1B | Tumor necrosis factor receptor superfamily, member 1B | 50 |
| Nonannotated | Transcribed locus | 50 |
| EGR2 | Early growth response 2 (Krox-20 homolog, Drosophila) | 50 |
| EPO | Erythropoietin | 50 |
| PMP2 | Peripheral myelin protein 2 | 50 |
| CXCL10 | Chemokine (C-X-C motif) ligand 10 | 50 |
| NNMT | Nicotinamide N-methyltransferase | 44 |
| HIG2 | Hypoxia-inducible protein 2 | 42 |
| MRAP | Melanocortin 2 receptor accessory protein | 42 |
| F5 | Coagulation factor V (proaccelerin, labile factor) | 39 |
| MAFF | v-Maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian) | 38 |
| PTGS1 | Prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) | 36 |
| TNC | Tenascin C (hexabrachion) | 33 |
| CD14 | CD14 molecule | 33 |
| HBEGF | Heparin-binding EGF-like growth factor | 33 |
| PGF | Placental growth factor, vascular endothelial growth factor-related protein | 33 |
| C21orf37 | Chromosome 21 open reading frame 37 | 33 |
| Nonannotated | Transcribed locus, moderately similar to XP_001,162,191.1 complement component 4A isoform 2 (Pan troglodytes) | 33 |
| GPRC5A | G protein-coupled receptor, family C, group 5, member A | 33 |
| RNASE2 | Ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) | 33 |
| SERPINE1 | Serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 | 33 |
| CD300A | CD300a molecule | 33 |
| Nonannotated | Transcribed locus | 33 |
| HSPA6 | Heat shock protein, 70-kDa protein 6 (HSP70B') | 33 |
| ISLR | Ig superfamily containing leucine-rich repeat | 33 |
| C2 | Complement component 2 | 33 |
| hCG_1,998,957 /// HLA-DR(locus) | Major histocompatibility complex, class II (locus) | 33 |
| FAM20A | Family with sequence similarity 20, member A | 33 |
| PAX2 | Paired box 2 | 33 |
| INHA | Inhibin, alpha | 33 |
| RSPO4 | R-spondin family, member 4 | 33 |
| Nonannotated | Transcribed locus | 33 |
| GADD45B | Growth arrest and DNA-damage-inducible, beta | 33 |
| HLA-DQB1 | Major histocompatibility complex, class II, DQ beta 1 | 29 |
| SOCS3 | Suppressor of cytokine signaling 3 | 25 |
| TGFBI | Transforming growth factor, beta-induced, 68 kDa | 25 |
| GBP2 | Guanylate binding protein 2, interferon-inducible | 25 |
| HP | Haptoglobin | 25 |
| ISG15 | ISG15 ubiquitin-like modifier | 25 |
| HLA-DOA | Major histocompatibility complex, class II, DO alpha | 25 |
| BAALC | Brain and acute leukemia, cytoplasmic | 25 |
| ABCC3 | ATP-binding cassette, sub-family C (CFTR/MRP), member 3 | 25 |
| CXCL2 | Chemokine (C-X-C motif) ligand 2 | 25 |
| MGC45438 | Hypothetical protein MGC45438 | 25 |
| DCT | Dopachrome tautomerase (dopachrome delta-isomerase, tyrosine-related protein 2) | 25 |
| TEAD4 | TEA domain family member 4 | 25 |
| SCARA3 | Scavenger receptor class A, member 3 | 25 |
| OAS1 | 2′,5′-oligoadenylate synthetase 1, 40/46 kDa | 25 |
| ATP6V1C2 | ATPase, H+ transporting, lysosomal 42 kDa, V1 subunit C2 | 25 |
| FCGR2B /// FCGR2C | Fc fragment of IgG, low affinity IIb, receptor (CD32) /// Fc fragment of IgG, low affinity IIc, receptor for (CD32) | 25 |
| CA12 | Carbonic anhydrase XII | 25 |
| EGR1 | Early growth response 1 | 25 |
| Nonannotated | CDNA clone IMAGE:6,025,865 | 25 |
| Nonannotated | 25 | |
| PAX8 | Paired box 8 | 25 |
| SPON2 | Spondin 2, extracellular matrix protein | 25 |
| CLDN1 | Claudin 1 | 25 |
Table 2 lists genes recognized by the top 100 transcripts showing increased expression in the ARRP eye compared to an unaffected control eye. Several of the differentially expressed genes are involved in immune mediated processes. Several of the probe sets recognize genes that have not yet been annotated (probe set identifiers are appended in Supplemental Data available at http://www.iovs.org/content/53/4/1883/suppl/DC1). For cases in which multiple probe sets were present for a given gene, the average ratio is shown.
We also examined a list of prospectively selected genes associated with different retinal cell types for expression changes in the ARRP eye (Table 3). Consistent with our immunohistochemical findings, the expression of GFAP mRNA was increased as well. Interestingly, most of the assessed inner retinal neuron genes showed persistent expression, despite the structural alterations and plasticity observed in this eye. For example, genes encoding neurofilament heavy chain, Thy1, parvalbumin, and metabotropic glutamate receptor 6 showed continued expression despite severe retinal degeneration. As might be expected, genes expressed primarily by photoreceptor cells showed a profound decrease in expression.
Table 3.
Relative expression of selected genes expressed in specific retinal cell types
|
Gene name |
Protein |
ARRP expression |
Control expression |
| OPN4 | Melanopsin | 78, P | 11, A |
| NEFH | Neurofilament heavy chain | 816, P | 535, P |
| THY1 | THY1/CD90 antigen | 993, P | 568, P |
| PVALB | Parvalbumin | 2108, P | 2202, P |
| GRM6 | Metabotropic glutamate receptor-6 | 2114, P | 2000, P |
| CALB2 | Calretinin | 1426, P | 845, P |
| RLBP1 | Cellular retinaldehyde binding protein | 3791, P | 5330, P |
| GFAP | Glial fibrillary acidic protein | 4217, P | 777, P |
| RHO | Rhodopsin | 33, P | 8039, P |
| ROM1 | Rod outer segment protein 1 | 130, P | 3449, P |
| OPN1LW and OPN1MW | Long wavelength (red cone) opsin | 53, P | 3335, P |
| RPE65 | RPE-Specific Protein, 65-KDa | 434, P | 3324, P |
| BEST1 | Bestrophin-1 | 563, P | 1803, P |
| CA4 | Carbonic anhydrase 4 | 8, P | 8, P |
| KDR | Vascular endothelial growth factor receptor 2 | 887, P | 551, P |
Table 3 lists relative expression of a small number of cell type-specific genes in the ARRP donor eye and a control eye. Preservation of several inner retina neuron specific transcripts is observed.
Discussion
The ABCA4 gene encodes a 2273-amino acid transmembrane protein of the photoreceptor outer segment disk, whose role is to couple the hydrolysis of ATP to the transport of a visual cycle intermediate, N-retinylidene-phosphatidylethanolamine, from the inner to the outer leaflet of the disk membrane.19 Mutations in ABCA4 that reduce or abolish this flippase activity result in the formation of toxic bisretinoids that can either injure the photoreceptors directly or accumulate in the underlying RPE, causing dysfunction or loss of that tissue and secondary injury to the photoreceptors. The close physical relationship between the photoreceptors and the RPE and the fact that retinoids pass through both cell types during the course of the visual cycle explain in large measure the wide range of clinical findings that are observed in patients with ABCA4 dysfunction. The mildest disease-causing genotypes cause a disorder known as Stargardt disease; moderate genotypes cause selective injury to cone photoreceptors that are manifest clinically as cone or cone-rod dystrophy; and the most severe genotypes cause direct injury to both rod and cone photoreceptors, which appears clinically as ARRP20–23 (recently reviewed in Refs. 24 and 25).
In this article, we report the first clinicopathologic study of the eyes of an individual affected with molecularly confirmed ABCA4-related ARRP. In most previous histopathological studies of retinitis pigmentosa, eye donors were believed to have an autosomal dominant form of the disease.3–6,26–28 In this study, the donor's pedigree structure (Fig. 1) coupled with the finding that each of his two unaffected siblings shared one of his two disease-causing mutations in ABCA4 provide strong evidence for the diagnosis of ABCA4-associated ARRP in this patient. Specifically, the donor was found to carry two possible disease-causing ABCA4 variations: IVS14+1G > C, which he shared with his unaffected sister, and Phe1440del1cT, which he shared with his unaffected brother. Bioinformatic analysis suggests that the IVS14+1G > C mutation abolishes the donor splice site of intron 14.29,30 This mutation has been previously reported in Stargardt disease31 but, to our knowledge, has not been observed in RP. The Phe1440del1cT frame shift mutation causes a stop codon 16 bases downstream of the deletion. This mutation would be expected to encode a protein lacking the last five transmembrane domains and the NBD2 domain responsible for proper localization to the outer segment disk.32
One of the major motivations for studying human donor tissue from individuals affected with rare diseases is to gain sufficient insight into the pathogenesis of these diseases that more effective treatments can be developed. There are at least three treatment-related facts that this donor tissue allowed us to investigate: (1) the order in which the RPE and photoreceptors succumbed in the face of ABCA4 dysfunction; (2) the longevity of the inner retinal neurons following the loss of the photoreceptor cells; and (3) the list of human genes abundantly expressed in the outer retina, which could themselves be candidates for involvement in other forms of heritable photoreceptor disease.
The order in which the RPE and photoreceptors are lost in ABCA4-related retinal diseases is relevant to the design of both gene- and cell-based therapies for these conditions. If, for example, the RPE cells are always lost first and photoreceptor cells die only as a consequence of this RPE loss, then one might envision conferring some RPE resistance to bisretinoid accumulation using gene transfer of a modifying gene to these cells. Transfer of a normal ABCA4 gene to the RPE would not be expected to be helpful as this gene is not normally expressed in this cell type and would have no obvious physiologic role there. One could also consider replacing the injured RPE with stem cell-derived RPE cells,33,34 but that strategy alone might not be a permanent solution for a patient because the source of the toxic bisretinoids, the photoreceptors, would not be corrected with such a transplant. The situation is even more challenging if bisretinoid accumulation is directly toxic to the photoreceptor cells and if photoreceptors die in significant numbers before the loss of the RPE. In this case, treatments directed toward the RPE would only be helpful if coupled to a rescue or replacement of photoreceptor cells.
Our histological studies in this case revealed complete loss of the photoreceptors, which was consistent with the patient's clinical history of a complete inability to perceive light for decades. Our finding that some areas of the RPE were preserved beneath a completely atrophic outer nuclear layer supports the notion that photoreceptor loss precedes loss of the RPE in at least some cases of ABCA4-associated ARRP.
Immunohistochemical evaluation of the retina from this donor also revealed complete loss of photoreceptor-specific markers and an increase of GFAP immunoreactivity throughout the retina. Gliosis with increased expression of GFAP has been previously observed in association with RP.35 We also observed upregulation of endogenous alkaline phosphatase activity. Alkaline phosphatase is a membrane protein that is a useful marker for vascular endothelial cells and is normally localized to viable retinal and choroidal vessels (Fig. 4A).36,37 In our case, distribution of AP reactivity was observed throughout the retina, suggesting that AP upregulation may also be a marker of gliosis.
In addition to performing morphological studies, we also analyzed the retinoid levels in the donor tissue and compared them to those in unaffected control eyes. Quantification of A2E from the most intact region of the eye revealed that bisretinoids can persist for decades in eyes that are completely blind and have no morphological evidence of functional photoreceptor cells (Fig. 3). In fact, the total A2E level in the ABCA4-associated RP eye was comparable to those in age-matched controls with intact photoreceptor cells. However, A2PE, an immediate A2E precursor, was dramatically reduced in the RP eye compared to those in controls (39.9% of the average control value), consistent with the loss of the photoreceptor cells.
We also compared gene expression in the ABCA4-associated RP retina to that of an unaffected control. Given the advanced state of disease, differences in expression between the RP retina and the control retina are more reflective of differences in the viable cell populations of these specimens than they are of transcriptional differences within comparable cell types. Despite this limitation, several insights may be obtained from these data. First, many of the transcripts with decreased abundance in the RP retina correspond to genes that are known to cause inherited photoreceptor disease when they are mutated in humans. This is not terribly surprising because it has been known for decades that there is correspondence between genes with high photoreceptor expression and retinal disease.38 Thus, the degree of decreased expression of a given gene in this RP eye is an indication that the gene is highly expressed in the outer retina in a normal eye. In fact, there is a good correlation between these human microarray data and similar data obtained in mice deficient for Bbs4 (see supplemental tables in Ref. 12), suggesting that cellular similarities exist in the late stages of RP regardless of species and specific genotype. The practical utility of this observation is that the list of genes with decreased mRNA abundance in the ABCA4-associated human RP retina and the Bbs4-associated murine RP retina includes RP-causing genes whose role in this disease have yet to be identified. This is of great interest because a large number of ARRP genes remain to be discovered,1 and methods for evaluating such genes in moderate to large patient populations are rapidly improving.
Another group of genes with elevated expression levels in the RP retina encode proteins involved in immunological processes, such as ICAM1, MHC antigens,39 and complement genes. With respect to complement genes, it is intriguing that upregulation of the complement system has been described in various animal models of retinal degeneration40,41 and, overall, supports a role for the immune system in either exacerbating damage or removing cellular debris from the injured retina.42 These findings are also consistent with experiments that suggest a protective role for immune suppression in the degenerating retina.43
Of greatest clinical relevance is our observation that sets of genes that are representative of multiple classes of inner retinal neurons were still expressed in the retina even after decades of no light perception (Table 3). The persistence of transcripts for bipolar, amacrine, and ganglion cells suggests that despite the extensive plasticity of the degenerating neural retina,18,44 there is persistence of some inner retinal neurons for many years after vision is lost. This preservation, at least at the molecular level, provides support for the possibility of successful intervention with stem cell-based photoreceptor replacement,45 optogenetic sensitization of inner retinal neurons,46 or prosthetic electrical stimulation of the inner retina, even very late in the clinical course of the disease.
Supplementary Material
Acknowledgments
The authors thank the Iowa Lions Eye Bank for support of our research and Elizabeth Faidley for technical assistance.
Disclosure: R.F. Mullins, None; M.H. Kuehn, None; R.A. Radu, None; G.S. Enriquez, None; J.S. East, None; E.I. Schindler, None; G.H. Travis, None; E.M. Stone, None
Supported by the Howard Hughes Medical Institute (EMS); NEI Grants EY-017451 (RFM), EY-016822 (EMS), and EY-019485 (MHK); the Macula Vision Research Foundation (RFM); the Iowa Biosciences Advantage, funded by National Institutes of Health National Institute of General Medical Sciences Grant 58939; and the Hansjoerg Kolder Professorship in Best Disease Research (RFM).
References
- 1. Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368:1795–1809. [DOI] [PubMed] [Google Scholar]
- 2. Daiger SP, Sullivan LS, Bowne SJ, et al. Targeted high-throughput DNA sequencing for gene discovery in retinitis pigmentosa. Adv Exp Med Biol. 664:325–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kolb H, Gouras P. Electron microscopic observations of human retinitis pigmentosa, dominantly inherited. Invest Ophthalmol. 1974;13:487–498. [PubMed] [Google Scholar]
- 4. Tulvatana W, Adamian M, Berson EL, Dryja TP. Photoreceptor rosettes in autosomal dominant retinitis pigmentosa with reduced penetrance. Arch Ophthalmol. 1999;117:399–402. [DOI] [PubMed] [Google Scholar]
- 5. To K, Adamian M, Dryja TP, Berson EL. Histopathologic study of variation in severity of retinitis pigmentosa due to the dominant rhodopsin mutation Pro23His. Am J Ophthalmol. 2002;134:290–293. [DOI] [PubMed] [Google Scholar]
- 6. To K, Adamian M, Dryja TP, Berson EL. Retinal histopathology of an autopsy eye with advanced retinitis pigmentosa in a family with rhodopsin Glu181Lys. Am J Ophthalmol. 2000;130:790–792. [DOI] [PubMed] [Google Scholar]
- 7. Johnson L, Blanks J. Application of acrylamide as an embedding medium in studies in lectin and antibody binding in the vertebrate retina. Curr Eye Res. 1984;3:969–974. [DOI] [PubMed] [Google Scholar]
- 8. Barthel L, Raymond P. Improved method for obtaining 3-microns cryosections for immunocytochemistry. J Histochem Cytochem. 1990;38:1383–1388. [DOI] [PubMed] [Google Scholar]
- 9. Mullins R, Grassi M, Skeie J. Glycoconjugates of choroidal neovascular membranes in age-related macular degeneration. Mol Vis. 2005;11:509–517. [PubMed] [Google Scholar]
- 10. Mullins R, Skeie J, Malone E, Kuehn M. Macular and peripheral distribution of ICAM-1 in the human choriocapillaris and retina. Mol Vis. 2006;12:224–235. [PubMed] [Google Scholar]
- 11. Radu RA, Hu J, Yuan Q, et al. Complement system dysregulation and inflammation in the retinal pigment epithelium of a mouse model for Stargardt macular degeneration. J Biol Chem. 2011;286:18593–18601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Swiderski R, Nishimura D, Mullins R, et al. Gene expression analysis of photoreceptor cell loss in bbs4-knockout mice reveals an early stress gene response and photoreceptor cell damage. Invest Ophthalmol Vis Sci. 2007;48:3329–3340. [DOI] [PubMed] [Google Scholar]
- 13. Lively GD, Jiang B, Hedberg-Buenz A, et al. Genetic dependence of central corneal thickness among inbred strains of mice. Invest Ophthalmol Vis Sci. 51:160–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gerl VB, Bohl J, Pitz S, Stoffelns B, Pfeiffer N, Bhakdi S. Extensive deposits of complement C3d and C5b-9 in the choriocapillaris of eyes of patients with diabetic retinopathy. Invest Ophthalmol Vis Sci. 2002;43:1104–1108. [PubMed] [Google Scholar]
- 15. Hageman GS, Anderson DH, Johnson LV, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A. 2005;102:7227–7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Seth A, Cui J, To E, Kwee M, Matsubara J. Complement-associated deposits in the human retina. Invest Ophthalmol Vis Sci. 2008;49:743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Skeie JM, Fingert JH, Russell SR, Stone EM, Mullins RF. Complement component C5a activates ICAM-1 expression on human choroidal endothelial cells. Invest Ophthalmol Vis Sci. 2010;51:5336–5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jones BW, Marc RE. Retinal remodeling during retinal degeneration. Exp Eye Res. 2005;81:123–137. [DOI] [PubMed] [Google Scholar]
- 19. Weng J, Mata N, Azarian S, Tzekov R, Birch D, Travis G. Insights into the function of rim protein in photoreceptors and etiology of Stargardt's disease from the phenotype in abcr knockout mice. Cell. 1999;98:13–23. [DOI] [PubMed] [Google Scholar]
- 20. Allikmets R, Singh N, Sun H, et al. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat Genet. 1997;15:236–246. [DOI] [PubMed] [Google Scholar]
- 21. Martinez-Mir A, Paloma E, Allikmets R, et al. Retinitis pigmentosa caused by a homozygous mutation in the Stargardt disease gene ABCR. Nat Genet. 1998;18:11–12. [DOI] [PubMed] [Google Scholar]
- 22. Rozet J, Gerber S, Ghazi I, et al. Mutations of the retinal specific ATP binding transporter gene (ABCR) in a single family segregating both autosomal recessive retinitis pigmentosa RP19 and Stargardt disease: evidence of clinical heterogeneity at this locus. J Med Genet. 1999;36:447–451. [PMC free article] [PubMed] [Google Scholar]
- 23. Fishman GA, Stone EM, Eliason DA, Taylor CM, Lindeman M, Derlacki DJ. ABCA4 gene sequence variations in patients with autosomal recessive cone-rod dystrophy. Arch Ophthalmol. 2003;121:851–855. [DOI] [PubMed] [Google Scholar]
- 24. Schindler EI, Nylen EL, Ko AC, et al. Deducing the pathogenic contribution of recessive ABCA4 alleles in an outbred population. Hum Mol Genet. 2010;19:3693–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sheffield VC, Stone EM. Genomics and the eye. N Engl J Med. 2011;364:1932–1942. [DOI] [PubMed] [Google Scholar]
- 26. Rodrigues M, Wiggert B, Hackett J, Lee L, Fletcher R, Chader G. Dominantly inherited retinitis pigmentosa. Ultrastructure and biochemical analysis. Ophthalmology. 1985;92:1165–1172. [DOI] [PubMed] [Google Scholar]
- 27. Meyer K, Heckenlively J, Spitznas M, Foos R. Dominant retinitis pigmentosa. A clinicopathologic correlation. Ophthalmology. 1982;89:1414–1424. [DOI] [PubMed] [Google Scholar]
- 28. Vinores SA, Kuchle M, Derevjanik NL, et al. Blood-retinal barrier breakdown in retinitis pigmentosa: light and electron microscopic immunolocalization. Histol Histopathol. 1995;10:913–923. [PubMed] [Google Scholar]
- 29. Rogan P, Schneider T. Using information content and base frequencies to distinguish mutations from genetic polymorphisms in splice junction recognition sites. Hum Mutat. 1995;6:74–76. [DOI] [PubMed] [Google Scholar]
- 30. Rogan P, Faux B, Schneider T. Information analysis of human splice site mutations. Hum Mutat. 1998;12:153–171. [DOI] [PubMed] [Google Scholar]
- 31. Lewis R, Shroyer N, Singh N, et al. Genotype/Phenotype analysis of a photoreceptor-specific ATP-binding cassette transporter gene, ABCR, in Stargardt disease. Am J Hum Genet. 1999;64:422–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhong M, Molday LL, Molday RS. Role of the C terminus of the photoreceptor ABCA4 transporter in protein folding, function, and retinal degenerative diseases. J Biol Chem. 2009;284:3640–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Buchholz DE, Hikita ST, Rowland TJ, et al. Derivation of functional retinal pigmented epithelium from induced pluripotent stem cells. Stem Cells. 2009;27:2427–2434. [DOI] [PubMed] [Google Scholar]
- 34. Carr AJ, Vugler AA, Hikita ST, et al. Protective effects of human iPS-derived retinal pigment epithelium cell transplantation in the retinal dystrophic rat. PLoS One. 2009;4:e8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rodrigues MM, Bardenstein D, Wiggert B, Lee L, Fletcher RT, Chader G. Retinitis pigmentosa with segmental massive retinal gliosis. An immunohistochemical, biochemical, and ultrastructural study. Ophthalmology. 1987;94:180–186. [DOI] [PubMed] [Google Scholar]
- 36. McLeod D, Lutty G. High-resolution histologic analysis of the human choroidal vasculature. Invest Ophthalmol Vis Sci. 1994;35:3799–3811. [PubMed] [Google Scholar]
- 37. McLeod DS, Grebe R, Bhutto I, Merges C, Baba T, Lutty GA. Relationship between RPE and choriocapillaris in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2009;50:4982–4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dryja TP, McGee TL, Reichel E, et al. A point mutation of the rhodopsin gene in one form of retinitis pigmentosa. Nature. 1990;343:364–366. [DOI] [PubMed] [Google Scholar]
- 39. Detrick B, Rodrigues M, Chan CC, Tso MO, Hooks JJ. Expression of HLA-DR antigen on retinal pigment epithelial cells in retinitis pigmentosa. Am J Ophthalmol. 1986;101:584–590. [DOI] [PubMed] [Google Scholar]
- 40. Rohrer B, Guo Y, Kunchithapautham K, Gilkeson GS. Eliminating complement factor D reduces photoreceptor susceptibility to light-induced damage. Invest Ophthalmol Vis Sci. 2007;48:5282–5289. [DOI] [PubMed] [Google Scholar]
- 41. Kuehn MH, Kim CY, Ostojic J, et al. Retinal synthesis and deposition of complement components induced by ocular hypertension. Exp Eye Res. 2006;83:620–628. [DOI] [PubMed] [Google Scholar]
- 42. Kuehn MH, Kim CY, Jiang B, Dumitrescu AV, Kwon YH. Disruption of the complement cascade delays retinal ganglion cell death following retinal ischemia-reperfusion. Exp Eye Res. 2008;87:89–95. [DOI] [PubMed] [Google Scholar]
- 43. Glybina IV, Kennedy A, Ashton P, Abrams GW, Iezzi R. Photoreceptor neuroprotection in RCS rats via low-dose intravitreal sustained-delivery of fluocinolone acetonide. Invest Ophthalmol Vis Sci. 2009;50:4847–4857. [DOI] [PubMed] [Google Scholar]
- 44. Marc RE, Jones BW, Watt CB, Strettoi E. Neural remodeling in retinal degeneration. Prog Retin Eye Res. 2003;22:607–655. [DOI] [PubMed] [Google Scholar]
- 45. Tucker BA, Park IH, Qi SD, et al. Transplantation of adult mouse iPS cell-derived photoreceptor precursors restores retinal structure and function in degenerative mice. PLoS One. 2011;6:e18992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Doroudchi MM, Greenberg KP, Liu J, et al. Virally delivered channelrhodopsin-2 safely and effectively restores visual function in multiple mouse models of blindness. Mol Ther. 2011;19:1220–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.