Abstract
Background
The epidermal growth factor receptor (EGFR) is differently expressed in breast cancer, and its presence may favor cancer progression. We hypothesized that two EGFR functional polymorphisms, a (CA)n repeat in intron 1, and a single nucleotide polymorphism, R497K, may affect EGFR expression and breast cancer clinical profile.
Methods
The study population consisted of 508 Brazilian women with unilateral breast cancer, and no distant metastases. Patients were genotyped for the (CA)n and R497K polymorphisms, and the associations between (CA)n polymorphism and EGFR transcript levels (n = 129), or between either polymorphism and histopathological features (n = 505) were evaluated. The REMARK criteria of tumor marker evaluation were followed.
Results
(CA)n lengths ranged from 14 to 24 repeats, comprehending 11 alleles and 37 genotypes. The most frequent allele was (CA)16 (0.43; 95% CI = 0.40–0.46), which was set as the cut-off length to define the Short allele. Variant (CA)n genotypes had no significant effect in tumoral EGFR mRNA levels, but patients with two (CA)n Long alleles showed lower chances of being negative for progesterone receptor (ORadjusted = 0.42; 95% CI = 0.19–0.91). The evaluation of R497K polymorphism indicated a frequency of 0.21 (95% CI = 0.19 – 0.24) for the variant (Lys) allele. Patients with variant R497K genotypes presented lower proportion of worse lymph node status (pN2 or pN3) when compared to the reference genotype Arg/Arg (ORadjusted = 0.32; 95% CI = 0.17–0.59), which resulted in lower tumor staging (ORadjusted = 0.34; 95% CI = 0.19-0.63), and lower estimated recurrence risk (OR = 0.50; 95% CI = 0.30-0.81). The combined presence of both EGFR polymorphisms (Lys allele of R497K and Long/Long (CA)n) resulted in lower TNM status (ORadjusted = 0.22; 95% CI = 0.07-0.75) and lower ERR (OR = 0.25; 95% CI = 0.09-0.71). When tumors were stratified according to biological classification, the favorable effects of variant EGFR polymorphisms were preserved for luminal A tumors, but not for other subtypes.
Conclusions
The data suggest that the presence of the variant forms of EGFR polymorphisms may lead to better prognosis in breast cancer, especially in patients with luminal A tumors.
Keywords: Breast cancer, EGFR, Gene polymorphisms, Gene expression, Prognostic estimates
Background
Breast cancer is the most frequent type of cancer in women both in the developed and the developing world [1]. It is a very heterogeneous disease with regards to its molecular profile [2], and clinical course, which presents great interpatient variability. Although conventional histopathological characteristics remain the most important prognostic determinants of survival [3], there is a continuous search for new biomarkers or stage models that could help predicting clinical evolution [4], or improving therapy selection. In this regard, genetic variations in carcinogenesis-related processes are natural candidates for exploring new prognostic factors or potential targets for specific therapies [5,6].
The epidermal growth factor receptor (EGFR) is a transmembrane tyrosine kinase (TK) receptor of the ErbB family, whose activation leads to mitogenic signaling [7]. EGFR is frequently overexpressed in many tumors, including breast cancer, and its activation contributes to unrestricted proliferation, advanced stages of disease, resistance to conventional treatments, and poor prognosis [8]. Despite the recognition that EGFR overexpression in breast tumors may affect disease progression [8], the responses of anti-EGFR therapies in breast cancer are not fully satisfactory [9], and the reasons for this clinical variability are not fully understood.
The EGFR gene, located at 7p12.3-p.1, contains multiple polymorphisms [10], two of which are recognized for their functional effects: a dinucleotide (CA)n repeat sequence polymorphism in intron 1 (rs72554020) affects gene transcription [11], and appears to modulate EGFR expression in breast tumors [12], and a single nucleotide change (G → A) in exon 13 leads to an Arginine (Arg) → Lysine (Lys) substitution in codon 497 (rs11543848), resulting in attenuated TK activity, with consequent reductions in ligand binding, growth stimulation, and induction of proto-oncogenes myc, fos, and jun[13].
In the present work, we aimed to describe the frequency of these two EGFR polymorphisms among Brazilian breast cancer patients, and to evaluate their impact on breast cancer prognosis, exploring the effects of (CA)n polymorphism on EGFR transcript levels, and the associations of both polymorphisms with histopathological features and prognostic estimates.
Materials and methods
Subjects and study design
The study population consisted of a prospective cohort of Brazilian women with first diagnosis of unilateral breast cancer and no distant metastases, admitted at the Brazilian National Cancer Institute (INCA) during the period from February 2009 to April 2011, and who were assigned for tumor resection as their first therapeutic approach. The recruitment occurred before surgery, but the inclusion was only completed after diagnosis confirmation by histopathological evaluation of the resected tumor. The study protocol was approved by the Ethics Committee of the Brazilian National Cancer Institute (INCA #129/08), and all patients gave written consent to participate. The REMARK guidelines (REporting recommendations for tumor MARKer prognostic studies) were followed [14].
Histopathological characterization
The histopathological evaluation of resected tumors was performed following institutional routine procedures, and all individual data were obtained from electronic medical records. The histopathological characterization was based on the TNM classification by the American Joint Committee on Cancer [15] and on the Elston Ellis histological grading system [16].
The data on hormone receptors, i.e. Estrogen Receptor (ER), and Progesterone Receptor (PR), and on the Human Epidermal growth factor Receptor 2 (HER2) status were used for biological classification of the tumors, as proposed by Huober et al. [17]. The Estimated Recurrence Risk (ERR) was inferred by a combination of all histopathological features, as proposed by the Early Breast Cancer Trialists’ Collaborative Group [18], with the following categories: “Low Risk”, characterized by the presence of [age ≥ 35 years, N0 (absence of tumor cells in lymph nodes), G1 (histological grade 1), T1 (tumor size lower than 2 cm), (ER+ or PR+), HER2-], and absence of peritumoral vascular invasion; “Intermediate Risk”, characterized by N0 in the presence of [age < 35 years, or T ≥ 2, or G ≥ 2, or (ER- and PR-), or HER2+], or by N1 (presence of tumor cells in 1 to 3 lymph nodes) in the presence of [HER2-, and (ER + or PR+)]; and “High Risk”, characterized by N1 in the presence of [HER2+, or (ER- and PR-)], or by N ≥ 2 (presence of tumor cells in more than 3 lymph nodes).
Genotyping analyses
Peripheral blood samples (3 mL) were collected from the subjects, and DNA was extracted using the Blood Genomic Prep Mini Spin Kit (GE Heathcare, Buckinghamshire, UK), following the procedures recommended by the manufacturer. The genotyping analyses were performed using PCR-RFLP for the SNP R497K (rs11543848) or by capillary electrophoresis for the (CA)n repeat polymorphism in intron 1 (rs72554020). The PCR amplifications were performed with the following primers (Life Thechnology, Carlsbad, CA, USA): 5′-AGGTCTGCCATGCCTTGT-3′ (sense) and 5′-CAACGCAAGGGGATTAAAGA-3′ (antisense) for R497K; or 5′-TTCTCCTCAAAACCCGGAGAC-3′ labeled with 6-FAM™ (sense) and 5′-GTCACGAAGCCAGACTCGCT-3′ for (CA)n repeat (antisense). The R497K PCR products (5 μL) were digested with 5U of BstN1 restriction enzyme (New England BioLabs, Northbrook, IL, USA) at 60°C for 3 hours, and the digestion products were resolved on 2% agarose gel and stained with ethidium bromide for visualization under UV light. The digestion of the homozygous G alleles (Arginine) produced two fragments (100 bp and 56 bp), whereas the homozygous A alleles (Lysine) remained intact (156 bp). The method was validated by direct sequencing of four samples of each genotype.
The (CA)n repeat PCR products (0.5 μl) were denatured at 95°C for 3 min in the presence of 0.5 μl of the GeneScan™ 400HD ROX molecular weight standard (Applied Biosystems, Foster City, CA, USA) and 9.0 μl of Hi-Di™ Formamide (Life Thechnology, Carlsbad, CA, USA), refrigerated to 4°C for 2 min, and then submitted to separation by capillary electrophoresis in ABI Prism® 3130 Genetic Analyzer, using POP7™ polymer (Applied Biosystems, Foster City, CA, USA). The analyses were performed using the GeneMapper® Software v.3.7 (Applied Biosystems, Foster City, CA, USA). The PCR products identified as homozygous, i.e. those presenting a single retention time at the capillary electrophoresis, were submitted to direct sequencing, using the BigDye® Terminator Kit (Applied Biosystems, Foster City, CA, USA), in order to establish a correspondence between each retention time and the respective number of CA repeats (or allele length).
Quantification of EGFR mRNA
Fresh specimens of breast tumors were dissected by clinical pathologists after tumor resection, frozen in liquid N2, and stored at the Brazilian National Bank of Tumors (BNT-INCA). Frozen sections of breast specimens (with approximately 2 mm) were used for RNA isolation, which was performed using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA), following the manufacturer’s instructions. The RNA samples were stored in RNAse-free distilled water at -80°C, and the corresponding cDNA was synthesized using 2 μg of RNA, with High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA), according to the manufacturer’s instructions.
The relative quantification of EGFR transcripts was performed using quantitative real-time RT-PCR (TaqMan) assays, in an ABI PRISM 7500 Sequence Detector System (Applied Biosystems, Foster City, CA, USA). Each reaction contained: cDNA templates (approximately 40 ng), 10 μl of reaction mix containing 5 μl Taqman® Gene Expression Master Mix, and Taqman® probes, which were as follows: EGFR Hs01076078_m1 (with FAM), PPIA 4326316E (with VIC) (Applied Biosystems, Foster City, CA, USA). The thermal cycling conditions comprised an initial denaturation step at 95°C for 10 min, followed by 40 cycles of 95°C denaturation for 15 sec, and annealing at 60°C for 1 min. The experiments were carried out in 96-well plates, including a nontemplate control, and a reference control, consisting of cDNA obtained from a commercial Human Mammary Gland (HMG) total RNA (Clontech Laboratories, Mountain View, CA, USA). The relative quantification of EGFR mRNA was calculated as the average 2-ΔΔCt, where ΔΔCT = ΔCTEGFR - ΔCTHMG, and ΔCTEGFR = CtEGFR - CtPPIA, and ΔCTHMG = CtHMG - CtPPIA. All data were generated in triplicates and expressed as median +/- SD with the 25–75 percentiles.
Statistical analyses
A descriptive study of the cohort was conducted, presenting measures of central tendency and dispersion for continuous variables, or relative frequencies for each categorical variable. Allelic and genotypic frequencies were derived by gene counting. The histopathological features were dichotomized for better and worse prognostic values, and their associations with EGFR genotypes were evaluated by the Chi-square or Fisher’s exact tests. In the cases of significant associations between EGFR genotypes and independent histopathological variables, the odds ratios (OR) and their respective 95% confidence intervals (95% CI) were tested for linear-by-linear associations, with calculation of trend significances (Ptrend), and definition of phenotypic inheritance models. The odds ratios between EGFR phenotypic groups and histopathological categorical features were adjusted for all other independent clinical variables (ORadjusted) using multiple regression analyses. The comparison of the relative quantities of EGFR mRNA as a function of histopathological features or EGFR genotypes was performed with the GraphPad Prism 5.0 software (GraphPad Software, La Jolla, CA, USA), using the non-parametric Mann–Whitney U-test for comparison of two groups, or the Kruskal-Wallis test for comparison of multiple groups. All other statistical analyses were conducted using SPSS 13.0 for Windows (SPSS Inc., Chicago, Illinois). The threshold for significance was set at P < 0.05.
Results
Characterization of the cohort
A total of 576 patients were recruited when admitted for surgery, and 528 had the diagnosis and inclusion criteria confirmed after pathological evaluation of their resected tumor. Blood samples were available for 511 cases, and 508 of them had good DNA quality for genotyping assays. Table 1 presents the main clinical and histopathological characteristics, as well as the genotypic distribution of EGFR polymorphisms for the 508 patients evaluated. The median age was 59 years old, ranging from 27 to 92.
Table 1.
Histopathological characteristics, tumor classification and EGFR genotypes in Brazilian breast cancer patients
| Independent variables | n | % | Tumor classifications | n | % |
|---|---|---|---|---|---|
| Histological type |
|
|
|
|
|
| Ductal invasive |
440 |
8606 |
0 |
35 |
7.1 |
| Ductal in situ |
34 |
6.7 |
IA |
157 |
31.7 |
| Lobular in situ |
1 |
0.2 |
IB |
7 |
1.4 |
| Lobular invasive |
29 |
5.7 |
II A |
148 |
29.9 |
| Mixed |
4 |
0.8 |
II B |
70 |
14.1 |
| |
|
|
III A |
52 |
10.5 |
| Tumor grade |
|
|
III B |
1 |
0.2 |
| Grade 1 |
60 |
12.6 |
III C |
25 |
5.1 |
| Grade 2 |
193 |
40.5 |
missing |
13 |
|
| Grade 3 |
223 |
46.8 |
|
|
|
| Missing |
32 |
|
Biological classifiation |
|
|
| |
|
|
luminal A |
293 |
64.8 |
| Tumor size |
|
|
luminal B |
89 |
5.8 |
| pTis |
35 |
7.1 |
HER2 like |
26 |
5.8 |
| pT1 |
226 |
45.7 |
triple negative |
44 |
9.7 |
| pT2 |
220 |
44.4 |
missing |
56 |
|
| pT3 |
13 |
2.6 |
|
|
|
| pT4 |
1 |
0.2 |
ERR categories |
|
|
| Missing |
13 |
|
low |
20 |
4.2 |
| |
|
|
intermediate |
357 |
75.5 |
| Lymph node status |
|
|
high |
96 |
20.3 |
| pN0 |
312 |
61.4 |
missing |
35 |
|
| pN1 |
120 |
23.6 |
|
|
|
| pN2 |
50 |
9.8 |
|
|
|
| pN3 |
26 |
5.1 |
|
|
|
| Estrogen receptor |
|
|
EGFR Genotypes |
n
|
% |
| Negative |
76 |
15.0 |
R497K |
|
|
| Positive |
414 |
84.5 |
Arg/arg |
308 |
61.0 |
| Missing |
18 |
|
Arg/Lys |
176 |
34.9 |
| |
|
|
Lys/Lys |
21 |
4.2 |
| Progesterone receptor |
|
|
missing |
3 |
|
| Negative |
134 |
26.4 |
|
|
|
| Positive |
355 |
72.6 |
|
|
|
| Missing |
19 |
|
|
|
|
| |
|
|
(CA)n |
|
|
| HER2 |
|
|
Short/Short |
108 |
22.6 |
| Negative |
378 |
74.4 |
Short/Long |
230 |
48.2 |
| Indeterminate |
15 |
3.2 |
Long/Long |
139 |
29.1 |
| Positive |
73 |
15.7 |
missing |
31 |
|
| Missing | 42 |
Data are expressed as the number of patients and their respective percentage in each category. (CA)n Short: n≤ 16; (CA)n Long: n > 16. Abbreviations: ERR Estimated Recurrence Risk, HER2 Human Epidermal growth factor Receptor 2.
Characterization of EGFR polymorphisms
The genotyping of the R497K polymorphism was obtained for 505 patients, whereas the characterization of the number of (CA)n repeats by electrophoresis was conclusive in 477 cases (Table 1). The frequency of the variant R497K allele (Lys) was 0.21 (95% CI = 0.19-0.24).
The evaluation of the (CA)n lengths indicated a range of 14 to 24 repeats (Figure 1), comprehending 11 alleles and 37 genotypes. The most frequent allele was (CA)16 (0.43; 95% CI = 0.40–0.46), which was taken as the cut-off length to group Short alleles. All the other variant alleles, with more than 16 (CA) repeats were considered as Long alleles. Thus, the genotypic distribution used for further analyses was: Short/Short (reference homozygous genotype), Short/Long (heterozygous) and Long/Long (variant homozygous genotype).
Figure 1.
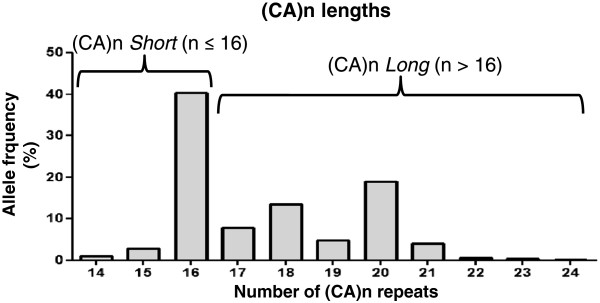
Evaluation of the (CA)n lengths in the Brazilian cohort of breast cancer patients: (CA)n ≤ 16 were grouped as Short alleles, whereas (CA)n > 16 were grouped as Long alleles.
Characterization of EGFR mRNA expression in breast tumors, and evaluation of the influence of (CA)n genotypes and of histopathological characteristics
The EGFR mRNA expression levels were evaluated in fresh-frozen tumor samples from 129 patients. Table 2 shows the main clinical and histopathological characteristics, as well as the genotypic distribution of EGFR polymorphisms in this subcohort. The data are presented in comparison with those described for the general population (Table 1). The results indicate that the subcohort whose tumors were used for expression analyses is similar to the general population, except for tumor size, and nodal status, which tend to be higher in the former. This difference is caused by the institutional biobank policy, which restricts collection of tumors with less than 1 cm for non-diagnostic purposes.
Table 2.
Characterization of a subcohort used for tumoral RNA analyses, and comparison with the complete cohort
| |
Complete cohort |
Tumor subcohort |
|
||
|---|---|---|---|---|---|
|
Individual variables |
Total |
n = 508 |
Total |
n = 129 |
P |
| n | % | n | % | ||
| Age |
|
|
|
|
|
| > 35 years-old |
496 |
97.6 |
125 |
96.9 |
0.63 |
| ≤ 35 years-old |
12 |
2.4 |
4 |
3.1 |
|
| Family history |
|
|
|
|
|
| No |
400 |
87.7 |
85 |
85.9 |
0.61 |
| Yes |
56 |
12.3 |
14 |
14.1 |
|
| Histological type |
|
|
|
|
|
| in situ |
35 |
7.0 |
2 |
1.6 |
0.018 |
| Invasive |
462 |
93.0 |
127 |
98.4 |
|
| Tumor grade |
|
|
|
|
|
| Grade 1 |
60 |
12.6 |
10 |
7.8 |
0.14 |
| Grade 2 or 3 |
416 |
87.4 |
117 |
90.7 |
|
| Tumor size |
|
|
|
|
|
| pT ≤ 2 cm |
226 |
49.1 |
42 |
33.1 |
0.001 |
| pT > 2 cm |
234 |
50.9 |
85 |
66.9 |
|
| Lymph node status |
|
|
|
|
|
| pN0 or pN1 |
432 |
85.0 |
99 |
76.7 |
0.024 |
| pN2 or pN3 |
76 |
15.0 |
30 |
23.3 |
|
| ER/PR status |
|
|
|
|
|
| Negative (both) |
76 |
15.5 |
27 |
20.9 |
0.14 |
| Positive (either) |
414 |
84.5 |
102 |
79.1 |
|
| HER2 status |
|
|
|
|
|
| Negative |
378 |
83.8 |
102 |
81.0 |
0.45 |
| Positive |
73 |
16.2 |
24 |
19.0 |
|
| Biological classification |
|
|
|
|
|
| Non triple negative |
440 |
90.9 |
112 |
86.8 |
0.17 |
| Triple negative |
44 |
9.1 |
17 |
13.2 |
|
|
EGFR genotypes |
|
|
|
|
|
|
R497K |
|
|
|
|
|
| Arg/Arg |
308 |
61.0 |
82 |
64.1 |
0.380 |
| Arg/Lys |
176 |
34.9 |
38 |
29.7 |
|
| Lys/Lys |
21 |
4.2 |
8 |
6.3 |
|
|
(CA)n |
|
|
|
|
|
| Short/Short |
108 |
22.6 |
30 |
23.6 |
0.358 |
| Short/Long |
230 |
48.2 |
68 |
53.5 |
|
| Long/Long | 139 | 29.1 | 29 | 22.8 | |
Abbreviations: ER Estrogen receptor, PR Progesterone receptor, HER2 Human Epidermal groth factor Receptor 2. Statistically significant differences are presented in bold characters.
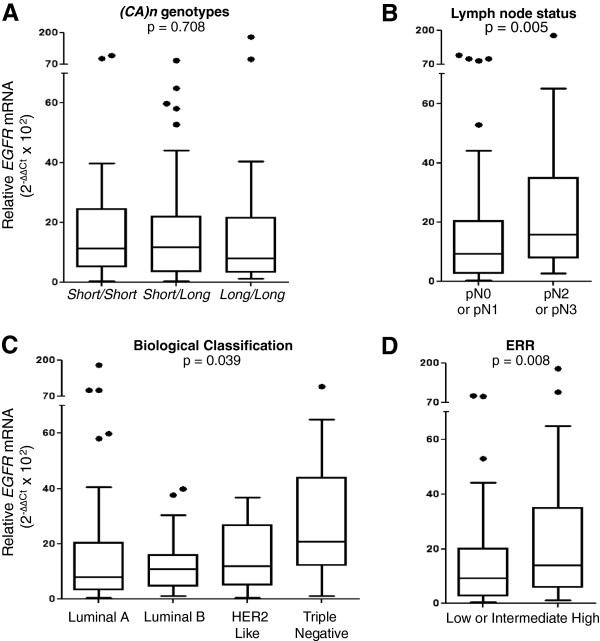
The relationship between EGFR mRNA expression levels and (CA)n genotypes or prognostic categories of breast tumors were explored (Figure 2). The results indicate no differences related to (CA)n genotypes (Figure 2A), whereas the lymph node status (Figure 2B) and the biological subclassification (Figure 2C) showed significant influences. The EGFR mRNA expression levels were significantly higher for patients with worse lymph node status, as well as for triple-negative tumors when compared to all other subgroup classifications (p = 0.003). As a consequence of these two associations, patients with higher ERR presented higher EGFR mRNA expression levels (Figure 2D).
Figure 2.
Expression levels of EGFR mRNA in breast tumors according to EGFR genotypes and histopathological prognostic estimates: (A)(CA)n genotypes, presented as Short/Short, (n = 30) Short/Long (n = 68) or Long/Long (n = 29); (B) Lymph node status, presented as pN0 or pN1 (n = 99) or pN2 or pN3 (n = 30); (C) Biological classification of tumors, presented as Luminal A (n = 69), Luminal B (n = 30), HER2 like (n = 10) or triple negative (n = 17); (D) Estimated Recurrence Risk (ERR), divided in “Low or intermediate” (n = 50) and “High” (n = 50).
Association between EGFR genotypes and prognostic variables
Table 3 presents the distribution of R497K and (CA)n genotypes according to prognostic categories. The distribution of R497K genotypes was statistically different as a function of the lymph node status, whereas the distribution of (CA)n genotypes was statistically different as a function of the PR status.
Table 3.
Distribution of the R497K and (CA)n genotypes according to prognostic variables
|
Prognostic variables |
|
R497K
|
|
(CA)n
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
Arg/Arg
|
Arg/Lys
|
Lys/Lys
|
|
Short/short
|
Short/long
|
Long/long
|
|
|||
|
Total |
n = 308 |
n = 176 |
n = 21 |
Total |
n = 108 |
n = 230 |
n = 139 |
|
||
| n | % | % | % | P | n | % | % | % | P | |
| Age |
|
|
|
|
|
|
|
|
|
|
| > 35 years-old |
493 |
61.7 |
34.1 |
4.3 |
0.06 |
466 |
22.7 |
48.5 |
28.8 |
0.48 |
| ≤ 35 years-old |
112 |
33.3 |
66.7 |
0.0 |
|
11 |
18.2 |
36.4 |
45.5 |
|
| Familial history |
|
|
|
|
|
|
|
|
|
|
| No |
400 |
60.0 |
36.0 |
4.0 |
0.99 |
376 |
21,5 |
49,7 |
28,7 |
0.38 |
| Yes |
56 |
60.7 |
35.7 |
3.6 |
|
53 |
26,4 |
39,6 |
34.0 |
|
| Histological type |
|
|
|
|
|
|
|
|
|
|
| in situ |
35 |
62.9 |
34.3 |
2.9 |
0.91 |
33 |
15.2 |
48.5 |
36.4 |
0.44 |
| Invasive |
459 |
61.0 |
34.6 |
4.4 |
|
434 |
23.3 |
48.6 |
28.1 |
|
| Tumor grade |
|
|
|
|
|
|
|
|
|
|
| Grade 1 |
60 |
56.7 |
40.0 |
3.3 |
0.64 |
58 |
25.9 |
46.6 |
27.6 |
0.74 |
| Grade 2 or 3 |
413 |
61.3 |
34.1 |
4.6 |
|
389 |
21.3 |
48.8 |
29.8 |
|
| Tumor size |
|
|
|
|
|
|
|
|
|
|
| pT ≤ 2 cm |
224 |
59.4 |
36.6 |
4.0 |
0.65 |
214 |
22.9 |
45.3 |
31.8 |
0.24 |
| pT > 2 cm |
233 |
62.7 |
32.6 |
4.7 |
|
218 |
23.4 |
51.8 |
29.8 |
|
| Lymph node status |
|
|
|
|
|
|
|
|
|
|
| pN0 or pN1 |
429 |
58.0 |
37.3 |
4.7 |
0.005 |
405 |
22.7 |
47.2 |
30.1 |
0.47 |
| pN2 or pN3 |
76 |
77.6 |
21.1 |
1.3 |
|
72 |
22.2 |
54.2 |
23.6 |
|
| ER status |
|
|
|
|
|
|
|
|
|
|
| Negative |
76 |
68.4 |
27.6 |
3.9 |
0.38 |
72 |
26.4 |
51.4 |
22.2 |
0.29 |
| Positive |
411 |
60.1 |
35.5 |
4.4 |
|
387 |
21.2 |
47.8 |
31.0 |
|
| PR status |
|
|
|
|
|
|
|
|
|
|
| Negative |
133 |
63.2 |
32.3 |
4.5 |
0.87 |
128 |
28.1 |
50.0 |
21.9 |
0.031 |
| Positive |
353 |
60.9 |
34.8 |
4.2 |
|
330 |
19.4 |
47.9 |
32.7 |
|
| HER2 status |
|
|
|
|
|
|
|
|
|
|
| Negative |
376 |
59.3 |
35.6 |
5.1 |
0.13 |
356 |
21.9 |
50.0 |
28.1 |
0.30 |
| Positive |
72 |
65.3 |
34.7 |
0.0 |
|
67 |
20.9 |
41.8 |
37.3 |
|
| Biological classification |
|
|
|
|
|
|
|
|
|
|
| Luminal A |
291 |
58.8 |
36.4 |
4.8 |
0.68 |
275 |
20.0 |
49.8 |
30.2 |
0.35 |
| Luminal B |
88 |
60.2 |
36.4 |
3.4 |
83 |
21.7 |
43.4 |
34.9 |
||
| HER2 like |
26 |
73.1 |
26.9 |
0.0 |
24 |
37.5 |
41.7 |
20.8 |
||
| Triple negative | 44 | 63.6 | 29.5 | 6.8 | 42 | 23.8 | 54.8 | 21.4 | ||
Data are expressed as the number of patients and respective percentages in each category. (CA)n Short: n ≤ 16; (CA)n Long: n > 16. Abbreviations: ER Estrogen Receptor, PR Progesterone Receptor, HEr2 Human Epidermal groth factor Receptor 2. Statistically significant differences are presented in bold characters.
The association between R497K genotypes and lymph node status, or between (CA)n genotypes and the PR status is further explored in Figure 3. Figure 3A shows that patients with the heterozygous genotype Arg/Lys presented lower proportion of the worse lymph node status (pN2 or pN3), when compared to the reference homozygous genotype Arg/Arg (OR = 0.42; 95% CI = 0.23–0.76), whereas among patients with the homozygous variant genotype Lys/Lys (n = 21), there was only 1 case of pN2 or pN3 (OR = 0.21; 95% CI = 0.028–1.60). These results indicate that the magnitude of the association between R497K polymorphism and lymph node status depends on the number of variant Lys alleles (Ptrend = 0.001). Similarly, Figure 3B shows the impact of the number of variant (CA)n alleles on the proportion of negative PR status. The results indicate an apparently progressive effect of the number of long (CA)n alleles (Ptrend = 0.008). Thus, patients with the Short/Long genotype showed a slightly lower proportion of negative PR status when compared to the reference Short/Short genotype (OR = 0.72; 95% CI = 0.44–1.19), and a significant protective effect was observed for the variant Long/Long genotype (OR = 0.46; 95% CI = 0.26–0.83).
Figure 3.
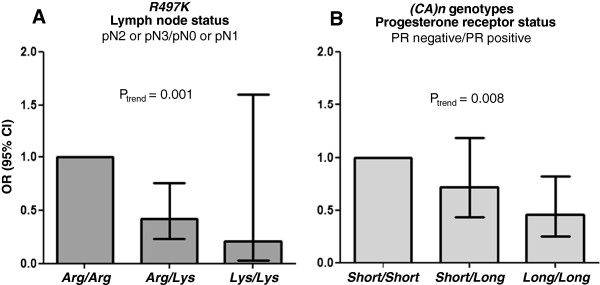
Significant influences of EGFR polymorphisms on histopathological features of brast cancer: (A) Proportion of lymph node status (pN2 or pN3/pN0 or pN1) according to R497K genotypic groups; (B) proportion of progesterone receptor status (PR negative/PR positive) according to (CA)n genotypic groups.
Interaction between EGFR polymorphisms
The above trend analyses suggested an inheritance model of codominance for the association between R497K polymorphism and lymph node status and of recessiveness for (CA)n Long allele and PR status. Thus, the genotypes Arg/Lys and Lys/Lys were grouped for evaluation of their impact on lymph node status, whereas the (CA)n Long/Long genotype was evaluated in comparison with the combined Short/Short and Short/Long genotypes for its effect on the PR status. The two EGFR polymorphisms were also evaluated in a combined analysis in order to investigate a possible interaction between them on the distribution of breast cancer prognostic features (Table 4).
Table 4.
Impact of EGFR polymorphisms on histopathological features and prognostic estimates of breast cancer
| |
R497K
variant genotypes |
(CA)n
variant genotypes |
EGFR
combined variant genotypes |
|||||
|---|---|---|---|---|---|---|---|---|
|
Lymph node status |
TNM status |
ERR |
PR status |
TNM status |
ERR |
TNM status |
ERR |
|
|
Tumor stratification |
N2/N3 |
≥ IIIA |
High |
negative |
≥ IIIA |
High |
≥ IIIA |
High |
|
OR adjusted |
OR adjusted |
OR |
OR adjusted |
OR adjusted |
OR |
OR adjusted |
OR |
|
|
(IC95%) |
(IC95%) |
(IC95%) |
(IC95%) |
(IC95%) |
(IC95%) |
(IC95%) |
(IC95%) |
|
| P | P | P | P | P | P | P | P | |
| Complete cohort |
0.32 |
0.34 |
0.50 |
0.42 |
0.74 |
0.74 |
0.22 |
0.25 |
| |
(0.17-0.59) |
(0.19-0.63) |
(0.30-0.81) |
(0.19-0.91) |
(0.40-1.32) |
(0.43-1.25) |
(0.07-0.75) |
(0.09-0.71) |
|
< 0.001 |
0.001 |
0.005 |
0.030 |
0.53 |
0.26 |
0.018 |
0.005 |
|
| Luminal A |
0.34 |
0.38 |
0.35 |
NA |
0.62 |
0.60 |
0.32 |
0.24 |
| |
(0.15-0.75) |
(0.18-0.80) |
(0.17-0.75) |
|
(0.29-1.36) |
(0.27-1.32) |
(0.092-1.12) |
(0.06-1.03) |
|
0.007 |
0.011 |
0.005 |
|
0.22 |
0.20 |
0.075 |
0.038 |
|
| Luminal B |
0.057 |
0.060 |
0.58 |
0.080 |
1.29 |
1.29 |
NC |
0.61 |
| |
(0.006-0.53) |
(0.007-0.53) |
(0.21-1.60) |
(0.012-0.54) |
(0.35-4.72) |
(0.48-3.48) |
|
(0.12-3.10) |
|
0.012 |
0.012 |
0.29 |
< 0.001 |
0.26 |
0.62 |
|
0.54 |
|
| HER2-like |
0.18 |
0.22 |
0.27 |
NA |
1.14 |
2.91 |
NC |
NC |
| |
(0.02-2.02) |
(0.02-2.28) |
(0.05-1.64) |
|
(0.15-8.59) |
(0.27-31.21) |
|
|
| 0.18 |
0.22 |
0.14 |
|
0.90 |
0.36 |
|
|
|
| Triple negative |
5.2 |
5.1 |
2.0 |
NA |
NC |
NC |
NC |
NC |
| |
(0.48-55.27) |
(0.48-54.03) |
(0.48-8.40) |
|
|
|
|
|
| 0.17 | 0.18 | 0.349 | ||||||
R497K variant genotypes include Arg/Lys and Lys/Lys. The genotype Arg/Arg was taken as reference for OR calculation; (CA)n variant genotypes include two Long alleles in combination (Long/Long), where the Long allele corresponds to (CA)n lengths with more than 16 repeats. The genotypes Short/Short and Short/Long were considered together as reference for OR calculation. EGFR combined variants include patients with genotypes Arg/Lys or Lys/Lys for R497K and Long/Long for (CA)n. The reference genotypes were R497K Arg/Arg and (CA)n Short/Short or Short/Long. The OR calculation for lymph node status was adjusted for age, hystological type, tumor grade, tumor size, and for ER, PR and HER2 status. The OR calculation for PR status was adjusted for age, hystological type, tumor grade, tumor size, lymph node status and for ER and HER2 status. The OR calculation for TNM status was adjusted for age, hystological type, tumor grade and for ER, PR and HER2 status. Statistically significant differences are presented in bold characters. Abbreviations: TNM Tumor staging according to tumor size, lymph node status and distant metastases, ERR Estimated Recurrence Risk, PR Progesterone Receptor, HER2 Human Epidermal growth factor Recetor 2, NA not applicable, NC not calculated (the number of samples was not sufficient for OR calculation).
The results indicate a significantly protective effect of the Lys allele on the proportion of the worse lymph node status after adjustment for other independent individual prognostic variables. As a consequence, patients carrying the Lys allele showed lower TNM status and lower ERR. With regards to the (CA)n polymorphism, the association between (CA)n Long/Long genotype and PR negative status also remained significant after adjustment for other independent individual prognostic variables (ORadjusted = 0.42; 95% CI = 0.19-0.91), but did not affect TNM status or the ERR. When the two EGFR polymorphisms are present, there is lower TNM status (ORadjusted = 0.22; 95% CI = 0.07-0.75) and lower ERR (OR = 0.25; 95% CI = 0.09-0.71).
The stratification of breast tumors according to their biological classification indicates that the association between combined variant EGFR polymorphisms and better lymph node status occurs for tumors classified as luminal A, but not for the other biological subtypes.
Discussion
The distribution of the two EGFR functional polymorphisms in the Brazilian population was not known before the current study. Our data indicate a frequency of 0.21 (95% CI = 0.19 – 0.24) for the 497 K (Lys allele), and of 0.43 (95% CI = 0.40 – 0.46) for the (CA)16. These results are similar to the frequencies reported for Europeans and North-Americans (including African-Americans), either for R497K polymorphisms [19,20] or (CA)n[12,21]. Asian populations, however, appear to have higher frequencies of the Lys allele [22,23], and different patterns of (CA)n alleles [12,21,24,25].
One difficulty of evaluating the effects of (CA)n polymorphism in gene transcriptional activity in vivo is the vast distribution of the number of (CA) repeats, with various possible heterozygous genotypes, and no clear model on how the two alleles interact for the final cell phenotype. Amador et al. [26] considered the sum of CA repeats of both alleles and showed an inverse correlation between this combined length and the levels of EGFR mRNA in head and neck cancer cell lines. Buerger et al. [12], studying breast tumors, considered the length of the smaller allele, and showed a non-significant tendency for lower EGFR protein expression with increasing allele length. Accordingly, Buerger et al. [27] showed that breast tumors from Japanese patients, who present high frequencies of (CA)20 and other long alleles, had lower amounts of EGFR protein than tumors from German patients, who have a predominance of (CA)16 and other short alleles. Other authors, however, found no correlation between the length of the (CA)n region and the relative quantification of EGFR mRNA [28] or EGFR protein expression [29].
Our data confirm the great dispersion of (CA) lengths and indicate great variability on the expression of EGFR mRNA, with no apparent inverse correlation between the number of (CA) repeats, considering either the smaller allele or the combined length within each genotype (data not shown). In order to investigate a possible effect of somatic mutations on the tumoral (CA)n genotype, we evaluated a set of 40 tumor samples. The number of CA repeats was preserved in relation to genomic DNA in all cases (data not shown). Although we did not extend such analyses to all patients, it appears that mutational events, such as loss of heterozigosity, are not affecting the EGFR locus of breast tumors. Nevertheless, an accurate characterization of the impact of EGFR polymorphisms on the gene transcriptional activity in vivo would ideally include quantification of gene amplification in the tumors [27]. In addition, there are two other EGFR polymorphisms (-216G/T or rs712829 and -191C/A or rs712830), located in the promoter region, which might have functional impact on EGFR transcriptional activity [30]. Finally, epigenetic variations may also interfere with EGFR expression [31].
The evaluation of the impact of EGFR polymorphisms on histopathological and molecular characteristics of breast cancer indicated significant association between R497K variant genotypes and better lymph node status, corroborating the findings of Kallel et al. [32], and between Long/Long (CA)n genotypes and positive PR status. These two associations seem protective in relation to breast cancer evolution, since a greater number of affected lymph nodes increases the risk of systemic metastasis [33], and the lack of PR expression increases the risk of disease progression, especially in post-menopausal women [34].
With regards to the molecular mechanisms underlying lymph node metastases, EGFR appears to activate integrins [35] and metaloproteinases [36], favoring cell differentiation towards an invasive phenotype. The association between the variant allele (Lys) and better lymph node status appear to corroborate the notion of reduced signaling with the variant EGFR isoform [13], leading to lower invasiveness, which reinforce the role of EGFR in breast cancer pathogenesis.
The interaction between the EGFR activity and the PR status might occur via a cross-talk mechanism between steroid and growth factor receptors [37], resulting in activation of the PIK3-Akt-mTOR pathway, which appears to negatively modulate the transcriptional activity of the PR [38]. This negative modulation of ER-mediated functions in breast cancer via EGFR signaling may underlie the mechanism of resistance to hormone therapy observed in tumors with high EGFR expression [39]. Taken together, the association between EGFR polymorphisms and lymph node metastases and negative PR status appear to corroborate the role of EGFR in breast cancer pathogenesis.
The combined presence of Long/Long (CA)n genotypes and Lys R497K alleles appears to favor better prognostic estimates in breast cancer. Other studies involving different types of cancer also point to an interaction between the two EGFR polymorphisms, with a combined protective effect in relation to disease progression. Zhang et al. [40], evaluating pelvic recurrence in patients with rectal cancer treated with chemoradiation, showed that the highest risk for local recurrence was seen in patients with the reference genotypes, i.e., both 497 Arg alleles and <20 CA repeats. Bandrés et al. [41], studying head and neck cancer, showed that patients with at least one 497 Arg allele and both (CA)n repeats ≤ 16 presented higher risk of death. Press et al. [42], studying metastatic colon cancer, found that men with the Arg/Arg genotype and two short alleles (< 20 CA repeats) had shorter overall survival than men with the Lys/Lys or Arg/Lys variant genotypes and any long allele (≥ 20 CA repeats).
The stratification of breast tumors according to their biological subtypes suggests that the apparently protective effects of EGFR polymorphisms are characteristic of luminal A tumors. This apparently selective effect of EGFR polymorphisms might be due to the lower genomic instability of luminal A tumors in relation to other subtypes, which present more aggressive phenotypes due to superposed molecular alterations [43]. Nevertheless, the small number of non-luminal A tumors limits the statistical power of the analyses, and the confidence of this assumption. In addition, the apparently favorable associations of EGFR polymorphisms with prognostic features at diagnosis cannot be considered as actually predictive of disease progression or therapy response,
Conclusions
In conclusion, the current results indicate a potential benefit of EGFR polymorphisms as independent prognostic factors, especially in early-stage luminal A tumors, as they might contribute to identify patients at higher risk of progression. We propose that EGFR genotyping should be further evaluated for their prognostic value in prospective studies of breast cancer survival.
Ethical standards
The study was conducted following the international precepts of ethics in research and of good clinical practice. The authors complied with the Brazilian regulation of clinical research. The protocol was approved by the Ethics Committee of the Brazilian National Cancer Institute (INCA #129/08), and all patients gave written consent to participate.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MSL recruited patients, collected clinical information, set the genotyping and expression assays, characterized genotypes and haplotypes, performed statistical analyses, generated tables and figures, and drafted the manuscript. LCG recruited patients, collected clinical information, and helped with genotyping assays. DNP set and performed the expression assays, and helped revising the manuscript. JSF-V collected and revised histopathological data, and helped revising the manuscript. VI-do-B recruited patients, collected clinical information, and helped with the statistical analyses. SK conceived the epidemiological design of the cohort. RSMN coordinated the genotyping of (CA)n polymorphism. MAC coordinated the expression assays, collaborated with data interpretation and revised the manuscript. RV-J conceived, designed, and coordinated the study, analyzed the data, wrote and revised the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Marcelo Sobral Leite, Email: m.leite@nki.nl.
Letícia Carlos Giacomin, Email: lelcg@hotmail.com.
Diogo Nascimento Piranda, Email: diogopiranda@gmail.com.
Juliana Simões Festa-Vasconcellos, Email: juliana.s.festa@gmail.com.
Vanessa Indio-do-Brasil, Email: vicosta@inca.gov.br.
Sérgio Koifman, Email: koifman@ensp.fiocruz.br.
Rodrigo Soares de Moura-Neto, Email: rsmouraneto@gmail.com.
Marcelo Alex de Carvalho, Email: marcelo.carvalho@ifrj.edu.br.
Rosane Vianna-Jorge, Email: farmaco@inca.gov.br.
Acknowledgements
The authors thank Dr. Guilherme Suarez-Kurtz for the use of laboratory facilities, and the personnel from the Breast Cancer Hospital (HC3-INCA) and from the National Bank of Tumors in the Brazilian National Cancer Institute (BNT-INCA), for logistic support in sample and data collection.
Financial support
This study was supported by grants from Conselho Nacional de Pesquisa e Desenvolvimento (CNPq474522/2010-5), from Fundação Carlos Chagas Filho de Amparo à Pesquisa no Rio de Janeiro (FAPERJ E-26/110356/2010), from Fundação para o Desenvolvimento Científico e Tecnológico em Saúde (FIOTEC/FIOCRUZ; Projeto Inova ENSP), and from INCT para Controle do Câncer (CNPq 573806/2008-0; FAPERJ E26/170.026/2008). MSL, DNP, JSFV, and VI-B received scholarships from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), MSL received a scholarship from Ministério da Saúde – INCA, and LCG received a scholarship from CNPq.
References
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- Sørlie T, Perou CM, Tibshiranie R, Aasf T, Geislerg S, Johnsenb H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lønning PE, Børresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soerjomataram I, Louwman MWJ, Ribot JG, Roukema JA, Coebergh JWW. An overview of prognostic factors for long-term survivors of breast cancer. Breast Cancer Res Treat. 2008;107:309–330. doi: 10.1007/s10549-007-9556-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danova M, Delfanti S, Manzoni M, Mariucci S. Tissue and soluble biomarkers in breast cancer and their applications: ready to use? J Natl Cancer Inst Monogr. 2011;2011:75–78. doi: 10.1093/jncimonographs/lgr023. [DOI] [PubMed] [Google Scholar]
- Ruiz C, Tolnay M, Bubendorf L. Application of personalized medicine to solid tumors: opportunities and challenges. Swiss Med Wkly. 2012;142:w13587. doi: 10.4414/smw.2012.13587. [DOI] [PubMed] [Google Scholar]
- Chung CC, Chanock SJ. Current status of genome-wide association studies in cancer. Hum Genet. 2011;130:59–78. doi: 10.1007/s00439-011-1030-9. [DOI] [PubMed] [Google Scholar]
- Brandt B, Meyer-Staeckling S, Schmidt H, Agelopoulos K, Buerger H. Mechanisms of egfr gene transcription modulation: relationship to cancer risk and therapy response. Clin Cancer Res. 2006;12:7252–7260. doi: 10.1158/1078-0432.CCR-06-0626. [DOI] [PubMed] [Google Scholar]
- Klijn JG, Look MP, Portengen H, Alexieva-Figusch J, van Putten WL, Foekens JA. The prognostic value of epidermal growth factor receptor (EGF-R) in primary breast cancer: results of a 10-year follow-up study. Breast Cancer Res Treat. 1994;29:73–83. doi: 10.1007/BF00666183. [DOI] [PubMed] [Google Scholar]
- Flynn JF, Wong C, Wu JM. Anti-EGFR therapy: mechanism and advances in clinical efficacy in breast cancer. J Oncol. 2009. p. 16. [DOI] [PMC free article] [PubMed]
- Liu G, Gurubhagavatula S, Zhou W, Wang Z, Yeap BY, Asomaning K, Su L, Heist R, Lynch TJ, Christiani DC. Epidermal growth factor receptor polymorphisms and clinical outcomes in non-small-cell lung cancer patients treated with gefitinib. Pharmacogenomics J. 2008;8:129–138. doi: 10.1038/sj.tpj.6500444. [DOI] [PubMed] [Google Scholar]
- Gebhardt F, Zänker KS, Brandt B. Modulation of epidermal growth factor receptor gene transcription by a polymorphic dinucleotide repeat in intron 1. J Biol Chem. 1999;274:13176–13180. doi: 10.1074/jbc.274.19.13176. [DOI] [PubMed] [Google Scholar]
- Buerger H, Gebhardt F, Schmidt H, Beckmann A, Hutmacher K, Simon R, Lelle R, Boecker W, Brandt B. Length and loss of heterozygosity of an intron 1 polymorphic sequence of egfr is related to cytogenetic alterations and epithelial growth factor receptor expression. Cancer Res. 2000;60:854–857. [PubMed] [Google Scholar]
- Moriai T, Kobrin MS, Hope C, Speck L, Korc M. A variant epidermal growth factor receptor exhibits altered type alpha transforming growth factor binding and transmembrane signaling. Proc Natl Acad Sci. 1994;91:10217–10221. doi: 10.1073/pnas.91.21.10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting Recommendations for Tumor Marker Prognostic Studies. J Clin Oncol. 2005;23(36):9067–9072. doi: 10.1200/JCO.2004.01.0454. [DOI] [PubMed] [Google Scholar]
- Lester S, Bose S, Chen Y, Connolly J, de Baca M, Fitzgibbons P, Hayes DF, Kleer C, O'Malley FP, Page DL, Smith BL, Tan LK, Weaver DL, Winer E. Protocol for the examination of specimens from patients with invasive carcinoma of the breast. Arch Pathol Lab Med. 2009;133:1515–1538. doi: 10.5858/133.10.1515. [DOI] [PubMed] [Google Scholar]
- Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- Huober J, von Minckwitz G, Denkert C, Tesch H, Weiss E, Zahm DM, Belau A, Khandan F, Hauschild M, Thomssen C, Högel B, Darb-Esfahani S, Mehta K, Loibl S. Effect of neoadjuvant anthracycline-taxane-based chemotherapy in different biological breast cancer phenotypes: overall results from the GeparTrio study. Breast Cancer Res Treat. 2010;124:133–140. doi: 10.1007/s10549-010-1103-9. [DOI] [PubMed] [Google Scholar]
- Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- Gonçalves A, Esteyries S, Taylor-Smedra B, Lagarde A, Ayadi M, Monges G, Bertucci F, Esterni B, Delpero JR, Turrini O, Lelong B, Viens P, Borg JP, Birnbaum D, Olschwang S, Viret F. A polymorphism of EGFR extracellular domain is associated with progression free-survival in metastatic colorectal cancer patients receiving cetuximab-based treatment. BMC Cancer. 2008;8:169. doi: 10.1186/1471-2407-8-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKibbin T, Zhao W, Tagen M, Daw NC, Furman WL, McGregor LM, Geyer JR, Allen JW, Stewart CF. Epidermal growth factor receptor polymorphisms and risk for toxicity in paediatric patients treated with gefitinib. Eur J Cancer. 2010;46:2045–2051. doi: 10.1016/j.ejca.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Innocenti F, Chen P, Das S, Cook EH, Ratain MJ. Interethnic difference in the allelic distribution of human epidermal growth factor receptor intron 1 polymorphism. Clin Cancer Res. 2003;9:1009–1012. [PubMed] [Google Scholar]
- Nomura M, Shigematsu H, Li L, Suzuki M, Takahashi T, Estess P, Siegelman M, Feng Z, Kato H, Marchetti A, Shay JW, Spitz MR, Wistuba II, Minna JD, Gazdar AF. Polymorphisms, mutations, and amplification of the EGFR gene in non-small cell lung cancers. PLoS Med. 2007;4:e125. doi: 10.1371/journal.pmed.0040125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Okuda K, Shimizu S, Takada M, Kawahara M, Kitahara N, Okumura M, Matsumura A, Iuchi K, Kawaguchi T, Kubo A, Kawano O, Yukiue H, Yano M, Fujii Y. EGFR R497K polymorphism is a favorable prognostic factor for advanced lung cancer. J Cancer Res Clin Oncol. 2009;135:313–318. doi: 10.1007/s00432-008-0464-5. [DOI] [PubMed] [Google Scholar]
- Huang C-L, Yang C-H, Yeh K-H, Hu F-C, Chen K-Y, Shih J-Y. EGFR intron 1 dinucleotide repeat polymorphism is associated with the occurrence of skin rash with gefitinib treatment. Lung Cancer. 2009;64:346–351. doi: 10.1016/j.lungcan.2008.09.009. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Cheung Y, Jada S, Lim W, Kuo W, Gray J, Lee AS, Chowbay B. EGFR Intron 1 polymorphism in Asian populations and its correlation with EGFR gene expression and amplification in breast tumor tissues. Cancer Biol Ther. 2006;5:1445–1449. doi: 10.4161/cbt.5.11.3457. [DOI] [PubMed] [Google Scholar]
- Amador ML, Oppenheimer D, Perea S, Maitra A, Cusatis G, Cusati G, Iacobuzio-Donahue C, Baker SD, Ashfaq R, Takimoto C, Forastiere A, Hidalgo M. An epidermal growth factor receptor intron 1 polymorphism mediates response to epidermal growth factor receptor inhibitors. Cancer Res. 2004;64:9139–9143. doi: 10.1158/0008-5472.CAN-04-1036. [DOI] [PubMed] [Google Scholar]
- Buerger H, Packeisen J, Boecker A, Tidow N, Kersting C, Bielawski K, Isola J, Yatabe Y, Nakachi K, Boecker W, Brandt B. Allelic length of a CA dinucleotide repeat in the egfr gene correlates with the frequency of amplifications of this sequence–first results of an inter-ethnic breast cancer study. J Pathol. 2004;203:545–550. doi: 10.1002/path.1542. [DOI] [PubMed] [Google Scholar]
- Buisine M-P, Wacrenier A, Mariette C, Leteurtre E, Escande F, Aissi S, Ketele A, Leclercq A, Porchet N, Lesuffleur T. Frequent mutations of the CA simple sequence repeat in intron 1 of EGFR in mismatch repair-deficient colorectal cancers. World J Gastroenterol. 2008;14:1053–1059. doi: 10.3748/wjg.14.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Grimaldi M-C, Pereira S, Magné N, Formento J-L, Francoual M, Fontana X, Demard F, Dassonville O, Poissonnet G, Santini J, Bensadoun RJ, Szepetowski P, Milano G. Analysis of the dinucleotide repeat polymorphism in the epidermal growth factor receptor (EGFR) gene in head and neck cancer patients. Ann Oncol. 2005;16:934–941. doi: 10.1093/annonc/mdi189. [DOI] [PubMed] [Google Scholar]
- Liu W, Innocenti F, Wu MH, Desai AA, Dolan ME, Cook EH, Ratain MJ. A functional common polymorphism in a Sp1 recognition site of the epidermal growth factor receptor gene promoter. Cancer Res. 2005;65:46–53. [PubMed] [Google Scholar]
- Gömöri E, Pál J, Kovács B, Dóczi T. Concurrent hypermethylation of DNMT1, MGMT and EGFR genes in progression of gliomas. Diagn Pathol. 2012;7:8. doi: 10.1186/1746-1596-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallel I, Rebai M, Khabir A, Farid NR, Rebaï A. Genetic polymorphisms in the EGFR (R521K) and estrogen receptor (T594T) genes, EGFR and ErbB-2 protein expression, and breast cancer risk in Tunisia. J Biomed Biotechnol. 2009;2009:753683. doi: 10.1155/2009/753683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagpar AB, Camp RL, Rimm DL. Lymph node ratio should be considered for incorporation into staging for breast cancer. Ann Surg Oncol. 2011;18:3143–3148. doi: 10.1245/s10434-011-2012-9. [DOI] [PubMed] [Google Scholar]
- Balleine RL, Earl MJ, Greenberg ML, Clarke CL. Absence of progesterone receptor associated with secondary breast cancer in postmenopausal women. Br J Cancer. 1999;79:1564–1571. doi: 10.1038/sj.bjc.6690249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricono JM, Huang M, Barnes LA, Lau SK, Weis SM, David D, Schlaepfer DD, Hanks SK, Cheresh DA. Specific cross-talk between epidermal growth factor receptor and integrin alphavbeta5 promotes carcinoma cell invasion and metastasis. Cancer Res. 2010;69:1383–1391. doi: 10.1158/0008-5472.CAN-08-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson L, Moss N, Stack M. EGF-receptor regulation of matrix metalloproteinases in epithelial ovarian carcinoma. Future Oncol. 2009;5:323–338. doi: 10.2217/fon.09.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox EM, Andrade J, Shupnik MA. Novel actions of estrogen to promote proliferation: integration of cytoplasmic and nuclear pathways. Steroids. 2009;74:622–627. doi: 10.1016/j.steroids.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakkar JP, Mehta DG. A review of an unfavorable subset of breast cancer: estrogen receptor positive progesterone receptor negative. Oncologist. 2011;16:276–285. doi: 10.1634/theoncologist.2010-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpino G, Weiss H, Lee AV, Schiff R, De Placido S, Osborne CK, Elledge RM. Estrogen receptor-positive, progesterone receptor-negative breast cancer: association with growth factor receptor expression and tamoxifen resistance. J Natl Cancer Inst. 2005;97:1254–1261. doi: 10.1093/jnci/dji249. [DOI] [PubMed] [Google Scholar]
- Zhang W, Park DJ, Lu B, Yang DY, Gordon M, Groshen S, Yun J, Press OA, Vallböhmer D, Rhodes K, Lenz HJ. Epidermal growth factor receptor gene polymorphisms predict pelvic recurrence in patients with rectal cancer treated with chemoradiation. Clin Cancer Res. 2005;11:600–605. [PubMed] [Google Scholar]
- Bandrés E, Barricarte R, Cantero C, Honorato B, Malumbres R, Zárate R, Alcalde J, García-Foncillas J. Epidermal growth factor receptor (EGFR) polymorphisms and survival in head and neck cancer patients. Oral Oncol. 2007;43:713–719. doi: 10.1016/j.oraloncology.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Press OA, Zhang W, Gordon MA, Yang D, Lurje G, Iqbal S, El-Khoueiry A, Lenz HJ. Gender-related survival differences associated with EGFR polymorphisms in metastatic colon cancer. Cancer Res. 2008;68:3037–3042. doi: 10.1158/0008-5472.CAN-07-2718. [DOI] [PubMed] [Google Scholar]
- Kwei KA, Kung Y, Salari K, Holcomb IN, Pollack JR. Genomic instability in breast cancer: Pathogenesis and clinical implications. Mol Oncol. 2010;4:255–266. doi: 10.1016/j.molonc.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]