Abstract
Being a well-characterized pathway, JAK-STAT signaling serves as a valuable paradigm for studying the architecture of gene regulatory networks. The discovery of untranslated or non-coding RNAs, namely microRNAs and long non-coding RNAs, provides an opportunity to elucidate their roles in such networks. In principle, these regulatory RNAs can act as downstream effectors of the JAK-STAT pathway and/or affect signaling by regulating the expression of JAK-STAT components. Examples of interactions between signaling pathways and non-coding RNAs have already emerged in basic cell biology and human diseases such as cancer, and can potentially guide the identification of novel biomarkers or drug targets for medicine.
Keywords: microRNA, long non-coding RNA, gene expression program, posttranscriptional regulation, epigenetics, cellular differentiation
Introduction
Signaling through the JAK-STAT pathway can lead to cell proliferation, survival, and differentiation (reviewed in refs. 1 and 2). By coordinating these basic cellular functions, JAK-STAT mediates many important biological phenomena such as hematopoiesis, immune development and function, mammary gland development, and lactation.3-5 Accordingly, germline mutations in the pathway can result in human disease, particularly of the immune system.6 Somatic gain-of-function alterations in JAK-STAT signaling can lead to uncontrolled cell proliferation that drives cancer growth and myeloproliferative diseases.7,8 For example, in acute lymphoblastic leukemia, JAK2 is hyperactivated,9 and mutations in suppressor of cytokine signaling 1 (SOCS1) are seen in cases of Hodgkin lymphoma.10 Therefore, it is understandable why studying the details of this pleiotropic signaling pathway is an important area of research.
Schematically, canonical JAK-STAT signaling is often depicted as a simple linear pathway, consisting of three sequential elements: (1) a cytokine or growth factor receptor, (2) Janus kinase (JAK), and (3) signal transducer and activator of transcription (STAT). The JAK family of tyrosine kinases contains four members: JAK1, JAK2, JAK3, and tyrosine kinase 2 (TYK2). All are expressed in most cells except for JAK3, which appears to be restricted to hematopoietic cells and is required for common gamma-chain (γc) cytokine receptor signaling.11 Mutations in JAK3 or γc result in severe combined immunodeficiency.12-14
Upon cytokine binding, JAK proteins interacting with the cytoplasmic tails of cytokine receptors are induced to trans-phosphorylate each other.14,15 Activated JAK proteins bind and phosphorylate STAT transcription factors latent in the cytoplasm.16 Phosphorylated STATs form homodimers and sometimes heterodimers, translocate to the nucleus, and bind consensus DNA sequences to regulate transcription.
In total, there are seven STAT proteins in humans: STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B, and STAT6. Particular cytokines lead to activation of certain JAKs and STATs.1 Each STAT has a slightly different DNA binding motif, allowing them to regulate a distinct subset of genes.1 Autosomal dominant mutations of STAT1 or STAT3 result in primary immunodeficiency called chronic mucocutaneous candidiasis or Job syndrome, respectively.17,18
JAK-STAT signaling is transient and regulated by negative feedback loops. Four common mechanisms for this are currently recognized, and include:
1) Internalization and degradation of the receptors by the lysosome and proteasome pathways,19
2) Phosphatase (e.g., SHP-1) recruitment to the receptor, leading to dephosphorylation of JAKs,20
3) Induction of inhibitors such as the suppressors of cytokine signaling (SOCS), SOCS1–7,21 and
4) Sumoylation of STATs by protein inhibitor of activated STAT (PIAS) family members, PIAS1–4.22
These inhibitory mechanisms are usually activated by JAK-STAT signaling, forming a negative feedback loop that ensures JAK-STAT signaling is transient, preventing the uncontrollable cell growth frequently seen in cancer and myeloproliferative diseases.
Recently, an expanded search for new players in JAK-STAT signaling has identified several non-coding RNA species. These findings have important implications for our understanding of JAK-STAT signaling, which will be discussed in this review. To date, non-coding RNAs are categorized somewhat arbitrarily by size. Short non-coding RNAs (<200 base pairs) include microRNAs, tRNAs, small nucleolar RNAs, and several others. Of this group, only microRNAs have been found to associate with JAK-STAT, whereas the others largely perform housekeeping functions. Long non-coding RNAs (>200 base pairs) include long intervening non-coding RNAs (lincRNA), pseudogenes, circular RNAs, anti-sense RNAs, and enhancer RNAs.
Overview of MicroRNA Pathway
MicroRNAs are evolutionarily conserved, small, untranslated RNAs of ~21 nucleotides in length that silence gene expression posttranscriptionally (reviewed by Ambros23). To date, more than 1000 miRNA genes have been cataloged in the human genome.24 Altogether, miRNAs are predicted to regulate approximately 60% of protein-coding transcripts,25 and represent an important regulatory layer of gene expression. The vast majority of miRNA genes are transcribed by RNA polymerase II, and as such their expression can be regulated by transcription factors and chromatin regulators, similar to conventional protein-coding genes.26,27 Historically, searches for STAT targets have focused on protein-coding genes; however, it is likely that STATs will regulate the expression of non-coding RNAs with yet unappreciated results (Fig. 1A).
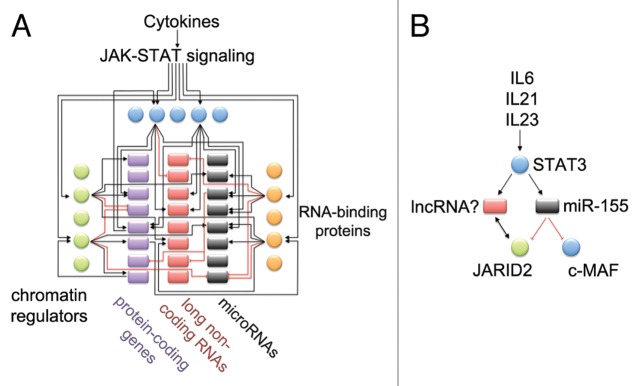
Figure 1. How are JAK-STAT signaling networks wired? (A) MicroRNAs, lncRNAs, and RNA-binding proteins need to be considered in building predictive models of regulatory circuits that control gene expression programs. Extracellular signals are conveyed from the cell surface to the nucleus using signaling pathways such as JAK-STAT. In the nucleus, transcription factors, such as STAT proteins, bind to specific DNA sequence motifs; however, accessibility of binding sites is determined by chromatin regulators. Some chromatin regulators also interact with long non-coding RNAs, and this interaction can modify their function. Once transcription factors bind to DNA, often at promoter or enhancer sites, they can induce or inhibit the expression of many genes, sometimes even triggering cell differentiation. However, this gene expression program can be fine-tuned further, through posttranscriptional control of mRNA levels. The Argonaute family of RNA-binding proteins, which are guided by miRNAs, bind to cognate mRNA transcripts, and can silence the expression of target mRNAs. (B) As a specific example of a JAK-STAT regulatory circuit, the cytokines IL-6, IL-21, and IL-23 can activate STAT3 in CD4+ T helper cells. MiR-155 expression is induced by STAT3. Mir-155 can silence the expression of JARID2, a component of the chromatin modifying PRC2 complex. LncRNAs are also able to interact with JARID2, possibly influencing its function. We predict that STAT3 will regulate the expression of lncRNAs.
Nascent parental miRNA transcripts are not destined for translation by the ribosome but are instead processed by a series of enzymatic cleavages to generate a functional mature miRNA. Typically, the larger parental transcript, called a primary miRNA (pri-miRNA), undergoes 7-methylguanosine (m7G) capping at the 5′ end and polyadenylation at the 3′ end to prevent exonucleolytic degradation, and in some cases can undergo splicing.28 The pri-miRNA is processed in the nucleus by the Drosha-DGCR8 complex (called Microprocessor), which recognizes the junction between single stranded and double stranded regions of stem-loop RNA structures, and cleaves ~11 bp downstream of the junction. The excised stem-loop is called a precursor miRNA (pre-miRNA) if it harbors a mature miRNA sequence within the stem.29 The pre-miRNA is exported out of the nucleus by exportin-5. Once in the cytoplasm, Dicer further processes the pre-miRNA into a double stranded duplex ~21 base pairs long. One strand from this duplex, known as the guide strand, is loaded into Argonaute proteins within the miRNA-induced silencing complex (miRISC). In this complex, the guide strand acts as a template to allow miRISC to recognize mRNA targets in a sequence-specific manner. The “seed region” of the guide strand, bases 2–8 at the 5′ end, are evolutionarily conserved and believed to be the most important for recognizing mRNA targets.30
Once the miRISC complex interacts with a target, it can promote deadenylation of the mRNA and/or inhibit it from being translated, effectively leading to posttranscriptional silencing of gene expression. Most studies of miRNA-mRNA interactions have focused on the 3′ UTR of mRNAs. It is believed that the 3′ UTR is generally accessible to the RISC complex even when an mRNA is being actively translated by ribosomes. However, recent transcriptome-wide analyses of miRNA-mRNA interactions have revealed that miRNA binding sites can also be found elsewhere in the mRNA31 and in non-coding RNAs.32 Although much is known about miRNAs and their mechanism of action, it remains a great challenge to demonstrate the functionality of miRNA binding sites in vivo, since the ultimate validation would be mutation of these binding sites in their endogenous context.
Transcriptional Regulation of miRNA Genes by JAK-STAT
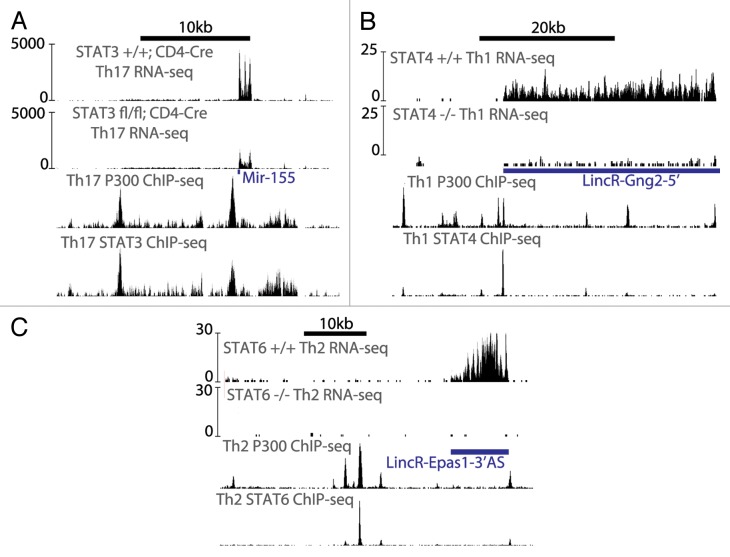
While the bulk of research has focused on finding protein-coding genes that are transcriptionally activated or repressed by JAK-STAT, a comprehensive understanding of regulatory networks must also identify non-coding RNAs whose expression is regulated by the JAK-STAT pathway. As would be predicted, STATs can directly transactivate the expression of non-coding RNA genes, which act as downstream players in the JAK-STAT pathway. One of the first miRNAs shown to be induced by JAK-STAT, miR-21, resembles an oncogene in function.33 It is expressed in multiple myeloma cells and head and neck squamous cell carcinoma, and can contribute to cell proliferation, as well as resistance to apoptosis and chemotherapy.33,34 Another microRNA that acts as an oncogene, miR-155, is induced by STAT3, and there are at least two peaks of STAT3 enrichment at the Mir155 locus that coincides with P300 binding (Fig. 2A).35
Figure 2. Transcriptional regulation of non-coding RNAs in JAK-STAT network. In T cells, STAT proteins activate the expression of microRNAs and lincRNAs. (A) In Th17 cells, optimal expression of primary miR-155 transcript requires STAT3 (data from GSE40918; Ciofani et al.78). (B) In Th1 cells, STAT4 directly binds to the LincR-Gng2-5′ locus. In STAT4-deficient CD4+ T cells cultured under Th1 conditions, very little expression of LincR-Gng2-5′ is seen. (Data from GSE48138 and GSE22105; Hu et al. and Wei et al.70,79) (C) Similarly, LincR-Epas1-3′AS is regulated by STAT6 in Th2 cells. (Data from GSE48138 and GSE22105; Hu et al. and Wei et al.70,79).
MiR-29a and miR-29b-1 are also induced by JAK-STAT, however in contrast to miR-21, miR-29 may be a tumor suppressor. MiR-29a and miR-29b-1 are expressed when melanoma cells are exposed to the antitumor cytokine IFNγ. IFNγ activates STAT1, which binds to five gamma interferon activation site (GAS) elements in the promoter of the miR-29a~29b-1 cluster (GAS elements contain a DNA sequence motif that is recognized by STAT1).36
These examples show that JAK-STAT can directly induce miRNA expression via the binding of STAT proteins to miRNA promoters and enhancers, and it is likely that this is a common mechanism. It would be important to analyze this on a genome-wide level, to identify more miRNAs regulated by JAK-STAT under different situations.
Posttranscriptional Silencing of JAK-STAT Components
JAK-STAT has proven to be a robust, fundamental signaling pathway for many eukaryotic organisms, having orthologs in species distant from humans (such as C. elegans and D. melanogaster). It has become apparent that JAK-STAT signaling cannot function properly with just the core pathway components, but is in fact much more complex. The PIAS, SOCS, and protein tyrosine phosphatase (PTPs) families regulate the JAK-STAT pathway at various steps (reviewed by Shuai et al.37). It is now becoming appreciated that miRNAs can provide an additional layer of control for JAK-STAT signaling (see Table 1). Although still a new area of research, there have been major findings in cell biology as well as health and disease. These non-coding RNAs need to be systematically investigated in future studies of JAK-STAT as well as during the search for treatments.
Table 1. Human JAK-STAT components predicted to be targets of broadly conserved miRNAs.
| Target | 3′ UTR length | miRNAs | Publication |
| JAK1 | 1.3 kb | miR-17/20/93/106 | |
| JAK2 | 1.4 kb | miR-135a | Wu et al.80 |
| JAK3 | 2.0 kb | ||
| TYK2 | 0.3 kb | ||
| STAT1 | 1.7 kb | miR-145,a miR-221/222a | Gregerson et al.,81 Lu et al.82 |
| STAT2 | 1.8 kb | ||
| STAT3 | 2.5 kb | let-7,a miR-17/20/93/106, miR-124, miR-125/351 | Koukos et al.,83 Wang et al.,84 |
| STAT4 | 0.3 kb | ||
| STAT5A | 1.3 kb | ||
| STAT5B | 2.5 kb | miR-23 | |
| STAT6 | 1.2 kb | miR-135 | |
| SOCS1 | 0.4 kb | miR-19, miR-30/384 | |
| SOCS2 | 1.0 kb | ||
| SOCS3 | 1.6 kb | miR-19, miR-30/384, miR-148/152, miR-218 | |
| SOCS4 | 5.1 kb | let-7/98, miR-9a | Zhuang et al.85 |
| SOCS5 | 2.6 kb | miR-124 | |
| SOCS6 | 3.9 kb | miR-15/16/195/322, miR-17/20/93/106, miR-25/32/92/363/367, miR-27, miR-30/384, miR-128, miR-130/301, miR-137, miR-142 | |
| SOCS7 | 6.1 kb | let-7/98, miR-17/20/93/106, miR-26, miR-29, miR-96, miR-218 | |
| PIAS1 | 0.2 kb | ||
| PIAS2 | 0.3 kb | ||
| PIAS3 | 0.9 kb | miR-9, miR-18,amiR-21a | Wu et al.,86 Xiong et al.34 |
| PIAS4 | 0.2 kb | miR-29 |
Sites with higher probability of preferential conservation are reported from TargetScan Release 6.2 (published interactions shown in bold).25 Due to space constraints only miR-1 up to miR-400 are included, and sites with lower probability of conservation are not shown. aHuman miRNA-target relationships reported in the literature but not predicted by TargetScan to have higher probability of preferential conservation (also shown in bold). For brevity, published mouse miRNA-target relationships are not listed.
Recently, posttranscriptional regulation has emerged as a new mechanism of regulating JAK-STAT components, with effects on strength and temporal aspects of signaling. Effects mediated via miRNAs, once better understood, may be useful to guide drug development. A number of examples have been uncovered in the immune system, where cytokine signaling, and thus the JAK-STAT pathway, is used to communicate between cells and coordinate an appropriate immune response. If this system could be manipulated, it might be possible to instruct immune cells to perform specific functions.
Intersection of miR-155 and JAK-STAT in CD4+ T Cells
JAK-STAT signaling in CD4+ T cells leads to the activation and differentiation of T helper cells, such as the Th1, Th2, and Th17 lineages, as well as regulatory T (Treg) cells.38 The lineage that develops following activation of naïve CD4+ T cells is determined by cytokines in the microenvironment. For example, cytokines that activate STAT3 in naïve CD4+ T cells lead to the development of Th17 cells characterized by the expression of the RORγt transcription factor and IL17 cytokine. Recently, we found that miR-155 is a direct transcriptional target of STAT3 in Th17 cells (Fig. 2A).35 Without miR-155, Th17 cells do not effectively produce IL-17.39 However, it is not understood how miR-155 facilitates Th17 functionality.
It is known that miR-155 plays an important role in the differentiation of both T and B lymphocytes, central components of the adaptive immune system. MiR-155 knockout mice are immunodeficient, and have problems with both B and T cell responses.40,41 Consequently, they have trouble developing appropriate immune memory, and do not respond well to vaccinations or infections. CD8+ T cell function is dependent on miR-155, and viral clearance is impaired in miR-155-deficient CD8+ T cells.42 Interestingly, the optimal homeostasis of Treg cells depends on miR-155 targeting SOCS1, a negative regulator of JAK-STAT signaling.43,44
SOCS proteins form a negative feedback loop for JAK-STAT signaling: once STAT proteins translocate to the nucleus, they activate expression of SOCS proteins, which then repress the activity of JAK proteins.45 This suggests that miR-155, which is itself induced during T cell activation, is important for maintaining low levels of SOCS1 in recently activated CD4+ T cells, and in this manner it contributes to the robustness of JAK-STAT signaling by dulling a negative feedback loop.
The situation is likely more complex, however, as miRNAs do not act as on-off switches for gene expression but rather as fine-tuners.46 miRNAs silence gene expression from 1.2- to 4-fold, yet since they mediate many major biological processes, miRNAs may have evolved to control key regulatory proteins that are sensitive to this range of modulation, as well as buffer undesirable transcriptional noise. In the immune system, where each response must be correctly tailored to combat individual infections, diverse mechanisms of control are advantageous. If the immune response is too small or too slow, the pathogen will prevail, and the infection could lead to death of the host organism. On the other hand, high levels of inflammation can be detrimental to the host, leading to local tissue damage, or even worse, fatal systemic problems such as a cytokine storm.
MicroRNAs and JAK-STAT in Disease
Dysregulation of miR-155 expression has been linked to cancer and, based on mouse studies, possibly immune disease as well.39,47,48 Recently, the role of miR-155 has been studied in two mouse models of inflammatory disease: experimental autoimmune encephalitis (EAE) and experimental autoimmune uveitis (EAU). EAE pathogenesis is characterized by inflammatory foci in the brain and spinal cord, mediated by Th17 cells, that is reminiscent of multiple sclerosis in humans.49 Blocking or eliminating miR-155 effectively reduced pathogenic Th17 cells, as well as symptoms of the EAE disease in mice.39,50,51 As higher levels of miR-155 are seen in patients with MS,52 this line of research could lead to a new treatment of MS by blocking miR-155. In a similar fashion, miR-155 appears to be pathogenic in EAU.35 Another miRNA, miR-301a, was also shown to participate in EAE pathology by silencing PIAS3, a protein that inhibits STAT3 by sumoylating the transactivation domain.53 These examples demonstrate the promising medical potential of studying the roles of non-coding RNA within the JAK-STAT regulatory network.
Overview of Long Non-Coding RNAs
Long non-coding RNAs (lncRNA) are defined as transcripts of more than 200 base pairs in length that do not contain discernable open reading frames. LncRNAs have been further classified based on their size and genomic relationship to protein-coding genes, for example, by the GENCODE consortium, which has produced the most comprehensive database of human lncRNAs.54
However, this classification scheme is based on currently available annotation data only. Another group of non-coding RNA expressed from certain enhancers, known as enhancer associated RNA (eRNA), has been proposed based on their putative function.55-57 These transcripts range in size from 50 to 2000 bp, so some actually would be categorized as small non-coding RNA, while others would be considered lncRNA. It is possible that as we uncover the functions of lncRNAs, different families will be recognized, and a functional classification scheme may be favored.
A handful of important lncRNAs have been appreciated for many years, however it was not until recently that the existence of thousands of lncRNAs was recognized. Some of the early examples of lncRNAs are Terc and XIST. Terc, discovered 1995, serves as the RNA template used for telomere elongation.58 XIST was discovered in 1991, and controls X chromosome inactivation in eutherians.59,60
Discovery and Functions of Long Non-Coding RNA
After the human genome project and the ENCODE pilot project, widespread transcription was seen from regions of the genome that did not contain genes. Due to low conservation of these transcripts, it was thought that most were merely transcriptional noise. Later, with improvements in genome-wide tiling microarrays, and eventually RNA-sequencing (RNA-seq), it became clear that thousands of lncRNAs exist, and these are highly regulated in cells. These lncRNAs are polyadenylated, spliced, and show highly tissue-specific patterns of expression.54,61,62
Although the function for most of these lncRNAs is still not known (or if all are even functional), a few have been shown to associate with chromatin modifying enzymes, acting as tethers or guides to help change the pattern of gene expression.63 Thus, there are emerging clues which suggest that lncRNAs act as important mediators of gene expression. The lincRNA TINCR coordinates differentiation of skin cells by regulating the expression of hundreds of important genes in this process.64 It works by binding the Staufen1 protein, forming a complex that regulates the stability of mRNAs such as KRT80.
HOTAIR and XIST are two other lincRNAs with important effects on gene expression.59,60,63,65,66 They associate with polycomb repressor complex 2 (PRC2), leading to transcriptional silencing of HoxD genes, in the case of HOTAIR, and an entire female X chromosome, in the case of XIST. Recently, lncRNAs were found to interact directly with Jarid2, an accessory component of PRC2, and play a role in chromatin targeting.67 When these lncRNAs interact with PRC2, they guide the complexes to target locations on the genome, where PRC2 is able to catalyze H3K27me3 histone marks, which silences chromatin and gene expression.68,69 Thus, even though PRC2 is ubiquitous, in concert with lncRNAs it may be able to target specific genomic regions in different cell types.
Long Non-Coding RNAs are Targets of JAK-STAT during T Helper Cell Differentiation
Recently, JAK-STAT signaling has been shown to regulate the expression of hundreds of long intergenic non-coding RNAs (lincRNAs) when naïve CD4+ T cells differentiate into either Th1 or Th2 lineages.70 Expression of 90 lincRNAs was reduced in STAT4-deleted cells grown in Th1 differentiation conditions, and expression of 56 lincRNAs were reduced in STAT6-deleted cells grown under Th2 differentiation conditions. Consistent with these observations, STAT4 and STAT6 bound to the genomic vicinity of lincRNAs preferentially expressed in Th1 or Th2 cells, respectively (examples in Fig. 2B and C).70
These lincRNAs are candidates for factors that help drive cell differentiation toward the Th1 and Th2 lineages. STAT proteins initiate a gene expression program for cell differentiation by activating or repressing transcription of key genes that mediate changes in cell identity, including surface receptors, cell cycle genes, and of particular importance, transcription factors such as TBX21, GATA3, and RORC (expressed in Th1, Th2, and Th17 cells, respectively). Future work will reveal which lincRNAs, if any, also play important roles in T helper cell differentiation. NeST, an enhancer-like lncRNA, has been found to recruit WDR5 and promote histone H3 lysine 4 (H3K4) trimethylation at the IFNG locus.71 NeST is expressed in Th1 cells but not Th2 cells,71 and its expression is dependent on STAT4 and TBX21.72
With these examples in mind, investigating lincRNAs that regulate gene expression should be promoted in JAK-STAT research, or any other signaling pathway for that matter. LincRNAs likely work alongside other genes induced by signaling pathways to help orchestrate the complex changes in gene expression and chromatin remodeling that leads to cell differentiation. The highly cell-specific expression of lincRNAs is a key feature that implicates them in this role. LincRNAs could, for example, help guide ubiquitously expressed chromatin remodeling factors or transcription factors to locations in the genome important for cell differentiation, using their cell-specific expression as a mechanism to mediate the variation in binding patterns of these factors seen in different tissues.
The rapid production of lincRNAs relative to proteins, as well as prompt control of their breakdown by cells, are features of this transcript species that make them useful during cell differentiation. Most lncRNAs are processed immediately after transcription, and polyadenylated,62 similar to mRNAs, however, they do not require the extra translation and posttranslational modification steps that are necessary for the production and activity of proteins, and thus can be produced much more rapidly. This could be valuable when rapid cell division and differentiation is needed.
Furthermore, RNA degradation is generally a quicker and more efficient process than protein degradation. RNA can be degraded by ribonucleases such as the exosome complex using a hydrolytic reaction. Although lincRNA half-lives can range from less than 2 h to more than 16 h, on average, they have shorter half-lives than mRNAs.73 As an extreme example, some long non-coding RNAs named promoter associated pervasive transcripts (PROMPTs) are degraded almost immediately after they are transcribed, and can only be detected if the RNA degradation machinery is disrupted.74 Cells likely take advantage of the rapid production and turnover of lincRNAs to mediate cellular differentiation and other processes; in a similar sense, lincRNAs may also contribute to the functional pleiotropy of JAK-STAT by acting as cell specific regulators of gene expression.
Non-Coding RNA-Based Treatments
The JAK-STAT pathway has been targeted for the treatment of rheumatoid arthritis, psoriasis, and inflammatory bowel disease (for an excellent review, see ref. 75). To date, 13 JAK inhibitors (Jakinibs) have been approved for clinical use for rheumatoid arthritis and other diseases, and many others are in development or in clinical trials. Importantly, Jakinibs have been shown to be efficacious in patients that did not respond to biological drugs, demonstrating that targeting this pathway is a sensible approach for treating many immune diseases. Unfortunately, patients who received Jakinibs have reported increased risk of serious infection,76 and Jakinibs are generally not used as first line therapy in most autoimmune diseases. Thus it would be beneficial to find alternative methods of selectively inhibiting JAK-STAT signaling, until an approach is found with therapeutic efficacy and minimal side effects.
Studies in animal models reveal that miRNAs could serve as attractive drug targets for the treatment of autoimmune disease. Blocking miR-155, for example, has potential for the treatment of multiple sclerosis and uveitis, as it can prevent EAE and EAU pathogenesis in mice.35,51 As some miRNAs and lncRNAs are more tissue specific than other JAK-STAT components that have been therapeutically investigated, fewer side effects would be predicted. Although clinical trials are evaluating the potential of miR-122 blockers to treat hepatitis C viral infections,77 the technology for delivering this type of drug generally requires further improvement. However, it will be more straightforward to rationally design drugs against miRNAs, as therapies would be designed based on a linear nucleic acid sequence, which could drastically cut down on the cost of drug screening and accelerate drug development.
Conclusions
Future studies will hopefully provide a deeper understanding to these findings. Additionally, as tools and methods improve to predict, identify, and validate miRNA targets, it will be easier to demonstrate novel interactions with the JAK-STAT pathway. This could lead to a better understanding of diseases and new treatments. Knowledge of lncRNAs has not yet caught up to our understanding of miRNAs; however, recent studies have provided a strong foundation to propel the field forward, through the creation of high quality expression catalogs and transcript models. Given the high number of known lncRNAs, it is very probable that some will be found to interact with the JAK-STAT signaling pathway, and enact several different mechanisms, such as transcriptional, posttranscriptional, or translational control. Additionally, as expression of lncRNAs is much more tissue specific than genes,62 they may serve as important biomarkers.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank C. Kanellopoulou and E. Metzakopian for critically reading this manuscript; A. Bradley for support. S.W. is a MSTP student and NIH-Cambridge Scholar. The Integrative Immunobiology Unit is supported by the Intramural Research Program of the NIAID, NIH.
Footnotes
Previously published online: www.landesbioscience.com/journals/jak-stat/article/28055
References
- 1.Leonard WJ, O’Shea JJ. Jaks and STATs: biological implications. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 2.Schindler C, Darnell JE., Jr. Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–51. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 3.Hou J, Schindler U, Henzel WJ, Wong SC, McKnight SL. Identification and purification of human Stat proteins activated in response to interleukin-2. Immunity. 1995;2:321–9. doi: 10.1016/1074-7613(95)90140-X. [DOI] [PubMed] [Google Scholar]
- 4.Watson CJ, Burdon TG. Prolactin signal transduction mechanisms in the mammary gland: the role of the Jak/Stat pathway. Rev Reprod. 1996;1:1–5. doi: 10.1530/ror.0.0010001. [DOI] [PubMed] [Google Scholar]
- 5.Liu X, Robinson GW, Wagner KU, Garrett L, Wynshaw-Boris A, Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–86. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 6.Casanova JL, Holland SM, Notarangelo LD. Inborn errors of human JAKs and STATs. Immunity. 2012;36:515–28. doi: 10.1016/j.immuni.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen E, Staudt LM, Green AR. Janus kinase deregulation in leukemia and lymphoma. Immunity. 2012;36:529–41. doi: 10.1016/j.immuni.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, Vassiliou GS, Bench AJ, Boyd EM, Curtin N, et al. Cancer Genome Project Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–61. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 9.Lacronique V, Boureux A, Valle VD, Poirel H, Quang CT, Mauchauffé M, Berthou C, Lessard M, Berger R, Ghysdael J, et al. A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science. 1997;278:1309–12. doi: 10.1126/science.278.5341.1309. [DOI] [PubMed] [Google Scholar]
- 10.Weniger MA, Melzner I, Menz CK, Wegener S, Bucur AJ, Dorsch K, Mattfeldt T, Barth TF, Möller P. Mutations of the tumor suppressor gene SOCS-1 in classical Hodgkin lymphoma are frequent and associated with nuclear phospho-STAT5 accumulation. Oncogene. 2006;25:2679–84. doi: 10.1038/sj.onc.1209151. [DOI] [PubMed] [Google Scholar]
- 11.Chen M, Cheng A, Chen YQ, Hymel A, Hanson EP, Kimmel L, Minami Y, Taniguchi T, Changelian PS, O’Shea JJ. The amino terminus of JAK3 is necessary and sufficient for binding to the common gamma chain and confers the ability to transmit interleukin 2-mediated signals. Proc Natl Acad Sci U S A. 1997;94:6910–5. doi: 10.1073/pnas.94.13.6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macchi P, Villa A, Giliani S, Sacco MG, Frattini A, Porta F, Ugazio AG, Johnston JA, Candotti F, O’Shea JJ, et al. Mutations of Jak-3 gene in patients with autosomal severe combined immune deficiency (SCID) Nature. 1995;377:65–8. doi: 10.1038/377065a0. [DOI] [PubMed] [Google Scholar]
- 13.Russell SM, Tayebi N, Nakajima H, Riedy MC, Roberts JL, Aman MJ, Migone TS, Noguchi M, Markert ML, Buckley RH, et al. Mutation of Jak3 in a patient with SCID: essential role of Jak3 in lymphoid development. Science. 1995;270:797–800. doi: 10.1126/science.270.5237.797. [DOI] [PubMed] [Google Scholar]
- 14.Noguchi M, Yi H, Rosenblatt HM, Filipovich AH, Adelstein S, Modi WS, McBride OW, Leonard WJ. Interleukin-2 receptor gamma chain mutation results in X-linked severe combined immunodeficiency in humans. Cell. 1993;73:147–57. doi: 10.1016/0092-8674(93)90167-O. [DOI] [PubMed] [Google Scholar]
- 15.Remy I, Wilson IA, Michnick SW. Erythropoietin receptor activation by a ligand-induced conformation change. Science. 1999;283:990–3. doi: 10.1126/science.283.5404.990. [DOI] [PubMed] [Google Scholar]
- 16.Ihle JN. The Janus protein tyrosine kinase family and its role in cytokine signaling. Adv Immunol. 1995;60:1–35. doi: 10.1016/S0065-2776(08)60582-9. [DOI] [PubMed] [Google Scholar]
- 17.van de Veerdonk FL, Plantinga TS, Hoischen A, Smeekens SP, Joosten LA, Gilissen C, Arts P, Rosentul DC, Carmichael AJ, Smits-van der Graaf CA, et al. STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. N Engl J Med. 2011;365:54–61. doi: 10.1056/NEJMoa1100102. [DOI] [PubMed] [Google Scholar]
- 18.Holland SM, DeLeo FR, Elloumi HZ, Hsu AP, Uzel G, Brodsky N, Freeman AF, Demidowich A, Davis J, Turner ML, et al. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med. 2007;357:1608–19. doi: 10.1056/NEJMoa073687. [DOI] [PubMed] [Google Scholar]
- 19.Royer Y, Staerk J, Costuleanu M, Courtoy PJ, Constantinescu SN. Janus kinases affect thrombopoietin receptor cell surface localization and stability. J Biol Chem. 2005;280:27251–61. doi: 10.1074/jbc.M501376200. [DOI] [PubMed] [Google Scholar]
- 20.Klingmüller U, Lorenz U, Cantley LC, Neel BG, Lodish HF. Specific recruitment of SH-PTP1 to the erythropoietin receptor causes inactivation of JAK2 and termination of proliferative signals. Cell. 1995;80:729–38. doi: 10.1016/0092-8674(95)90351-8. [DOI] [PubMed] [Google Scholar]
- 21.van de Geijn GJ, Gits J, Aarts LH, Heijmans-Antonissen C, Touw IP. G-CSF receptor truncations found in SCN/AML relieve SOCS3-controlled inhibition of STAT5 but leave suppression of STAT3 intact. Blood. 2004;104:667–74. doi: 10.1182/blood-2003-08-2913. [DOI] [PubMed] [Google Scholar]
- 22.Palvimo JJ. PIAS proteins as regulators of small ubiquitin-related modifier (SUMO) modifications and transcription. Biochem Soc Trans. 2007;35:1405–8. doi: 10.1042/BST0351405. [DOI] [PubMed] [Google Scholar]
- 23.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 24.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–83. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karp DR, Long EO. Identification of HLA-DR1 beta chain residues critical for binding staphylococcal enterotoxins A and E. J Exp Med. 1992;175:415–24. doi: 10.1084/jem.175.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–40. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 30.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 31.Loeb GB, Khan AA, Canner D, Hiatt JB, Shendure J, Darnell RB, Leslie CS, Rudensky AY. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol Cell. 2012;48:760–70. doi: 10.1016/j.molcel.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–52. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bourguignon LY, Earle C, Wong G, Spevak CC, Krueger K. Stem cell marker (Nanog) and Stat-3 signaling promote MicroRNA-21 expression and chemoresistance in hyaluronan/CD44-activated head and neck squamous cell carcinoma cells. Oncogene. 2012;31:149–60. doi: 10.1038/onc.2011.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiong Q, Zhong Q, Zhang J, Yang M, Li C, Zheng P, Bi LJ, Ge F. Identification of novel miR-21 target proteins in multiple myeloma cells by quantitative proteomics. J Proteome Res. 2012;11:2078–90. doi: 10.1021/pr201079y. [DOI] [PubMed] [Google Scholar]
- 35.Escobar T, Yu CR, Muljo SA, Egwuagu CE. STAT3 activates miR-155 in Th17 cells and acts in concert to promote experimental autoimmune uveitis. Invest Ophthalmol Vis Sci. 2013;54:4017–25. doi: 10.1167/iovs.13-11937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitt MJ, Philippidou D, Reinsbach SE, Margue C, Wienecke-Baldacchino A, Nashan D, Behrmann I, Kreis S. Interferon-γ-induced activation of Signal Transducer and Activator of Transcription 1 (STAT1) up-regulates the tumor suppressing microRNA-29 family in melanoma cells. Cell Commun Signal. 2012;10:41. doi: 10.1186/1478-811X-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shuai K, Liu B. Regulation of JAK-STAT signalling in the immune system. Nat Rev Immunol. 2003;3:900–11. doi: 10.1038/nri1226. [DOI] [PubMed] [Google Scholar]
- 38.O’Shea JJ, Lahesmaa R, Vahedi G, Laurence A, Kanno Y. Genomic views of STAT function in CD4+ T helper cell differentiation. Nat Rev Immunol. 2011;11:239–50. doi: 10.1038/nri2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Connell RM, Kahn D, Gibson WS, Round JL, Scholz RL, Chaudhuri AA, Kahn ME, Rao DS, Baltimore D. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–19. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–11. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–8. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 42.Gracias DT, Stelekati E, Hope JL, Boesteanu AC, Doering TA, Norton J, Mueller YM, Fraietta JA, Wherry EJ, Turner M, et al. The microRNA miR-155 controls CD8(+) T cell responses by regulating interferon signaling. Nat Immunol. 2013;14:593–602. doi: 10.1038/ni.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kohlhaas S, Garden OA, Scudamore C, Turner M, Okkenhaug K, Vigorito E. Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells. J Immunol. 2009;182:2578–82. doi: 10.4049/jimmunol.0803162. [DOI] [PubMed] [Google Scholar]
- 44.Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K, Loeb GB, Lee H, Yoshimura A, Rajewsky K, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alexander WS. Suppressors of cytokine signalling (SOCS) in the immune system. Nat Rev Immunol. 2002;2:410–6. doi: 10.1038/nri818. [DOI] [PubMed] [Google Scholar]
- 46.Sevignani C, Calin GA, Siracusa LD, Croce CM. Mammalian microRNAs: a small world for fine-tuning gene expression. Mamm Genome. 2006;17:189–202. doi: 10.1007/s00335-005-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S, Kroesen BJ, van den Berg A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–9. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 48.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci U S A. 2005;102:3627–32. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gold R, Linington C, Lassmann H. Understanding pathogenesis and therapy of multiple sclerosis via animal models: 70 years of merits and culprits in experimental autoimmune encephalomyelitis research. Brain. 2006;129:1953–71. doi: 10.1093/brain/awl075. [DOI] [PubMed] [Google Scholar]
- 50.Hu R, Huffaker TB, Kagele DA, Runtsch MC, Bake E, Chaudhuri AA, Round JL, O’Connell RM. MicroRNA-155 confers encephalogenic potential to Th17 cells by promoting effector gene expression. J Immunol. 2013;190:5972–80. doi: 10.4049/jimmunol.1300351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murugaiyan G, Beynon V, Mittal A, Joller N, Weiner HL. Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. J Immunol. 2011;187:2213–21. doi: 10.4049/jimmunol.1003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paraboschi EM, Soldà G, Gemmati D, Orioli E, Zeri G, Benedetti MD, Salviati A, Barizzone N, Leone M, Duga S, et al. Genetic association and altered gene expression of mir-155 in multiple sclerosis patients. Int J Mol Sci. 2011;12:8695–712. doi: 10.3390/ijms12128695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mycko MP, Cichalewska M, Machlanska A, Cwiklinska H, Mariasiewicz M, Selmaj KW. MicroRNA-301a regulation of a T-helper 17 immune response controls autoimmune demyelination. Proc Natl Acad Sci U S A. 2012;109:E1248–57. doi: 10.1073/pnas.1114325109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–7. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mousavi K, Zare H, Dell’orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, Hager GL, Sartorelli V. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell. 2013;51:606–17. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, Adams RR, Chang E, Allsopp RC, Yu J, et al. The RNA component of human telomerase. Science. 1995;269:1236–41. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 59.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 60.Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–26. doi: 10.1016/0092-8674(92)90519-I. [DOI] [PubMed] [Google Scholar]
- 61.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–10. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–5. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32:1616–25. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaneko S, Bonasio R, Saldana-Meyer R, Yoshida T, Son J, Nishino K, Umezawa A, Reinberg D. Interactions between JARID2 and Noncoding RNAs Regulate PRC2 Recruitment to Chromatin. Mol Cell. 2014;53:290–300. doi: 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK, Kesner BA, Maier VK, Kingston RE, Lee JT. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013;504:465–9. doi: 10.1038/nature12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu G, Tang Q, Sharma S, Yu F, Escobar TM, Muljo SA, Zhu J, Zhao K. Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat Immunol. 2013;14:1190–8. doi: 10.1038/ni.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell. 2013;152:743–54. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM. Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J Immunol. 2012;189:2084–8. doi: 10.4049/jimmunol.1200774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Clark MB, Johnston RL, Inostroza-Ponta M, Fox AH, Fortini E, Moscato P, Dinger ME, Mattick JS. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012;22:885–98. doi: 10.1101/gr.131037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–4. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 75.O’Shea JJ, Kontzias A, Yamaoka K, Tanaka Y, Laurence A. Janus kinase inhibitors in autoimmune diseases. Ann Rheum Dis. 2013;72(Suppl 2):ii111–5. doi: 10.1136/annrheumdis-2012-202576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fleischmann R, Kremer J, Cush J, Schulze-Koops H, Connell CA, Bradley JD, Gruben D, Wallenstein GV, Zwillich SH, Kanik KS, ORAL Solo Investigators Placebo-controlled trial of tofacitinib monotherapy in rheumatoid arthritis. N Engl J Med. 2012;367:495–507. doi: 10.1056/NEJMoa1109071. [DOI] [PubMed] [Google Scholar]
- 77.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–94. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 78.Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkurst CN, Muratet M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wei L, Vahedi G, Sun HW, Watford WT, Takatori H, Ramos HL, Takahashi H, Liang J, Gutierrez-Cruz G, Zang C, et al. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity. 2010;32:840–51. doi: 10.1016/j.immuni.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu H, Huang M, Cao P, Wang T, Shu Y, Liu P. MiR-135a targets JAK2 and inhibits gastric cancer cell proliferation. Cancer Biol Ther. 2012;13:281–8. doi: 10.4161/cbt.18943. [DOI] [PubMed] [Google Scholar]
- 81.Gregersen LH, Jacobsen AB, Frankel LB, Wen J, Krogh A, Lund AH. MicroRNA-145 targets YES and STAT1 in colon cancer cells. PLoS One. 2010;5:e8836. doi: 10.1371/journal.pone.0008836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu MC, Lai NS, Chen HC, Yu HC, Huang KY, Tung CH, Huang HB, Yu CL. Decreased microRNA(miR)-145 and increased miR-224 expression in T cells from patients with systemic lupus erythematosus involved in lupus immunopathogenesis. Clin Exp Immunol. 2013;171:91–9. doi: 10.1111/j.1365-2249.2012.04676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Koukos G, Polytarchou C, Kaplan JL, Morley-Fletcher A, Gras-Miralles B, Kokkotou E, Baril-Dore M, Pothoulakis C, Winter HS, Iliopoulos D. MicroRNA-124 regulates STAT3 expression and is down-regulated in colon tissues of pediatric patients with ulcerative colitis. Gastroenterology. 2013;145:842–52, e2. doi: 10.1053/j.gastro.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang Y, Lu Y, Toh ST, Sung WK, Tan P, Chow P, Chung AY, Jooi LL, Lee CG. Lethal-7 is down-regulated by the hepatitis B virus x protein and targets signal transducer and activator of transcription 3. J Hepatol. 2010;53:57–66. doi: 10.1016/j.jhep.2009.12.043. [DOI] [PubMed] [Google Scholar]
- 85.Zhuang G, Wu X, Jiang Z, Kasman I, Yao J, Guan Y, Oeh J, Modrusan Z, Bais C, Sampath D, et al. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. 2012;31:3513–23. doi: 10.1038/emboj.2012.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu W, Takanashi M, Borjigin N, Ohno SI, Fujita K, Hoshino S, Osaka Y, Tsuchida A, Kuroda M. MicroRNA-18a modulates STAT3 activity through negative regulation of PIAS3 during gastric adenocarcinogenesis. Br J Cancer. 2013;108:653–61. doi: 10.1038/bjc.2012.587. [DOI] [PMC free article] [PubMed] [Google Scholar]