Abstract
Background
Trichomonas gallinae is a protozoan parasite causing trichomonosis in many species of domestic poultry and birds world-wide. microRNAs (miRNAs) are a class of small non-coding RNAs that play key roles in gene regulation. However, no miRNAs have been characterized from T. gallinae.
Methods
Here, we investigated the global miRNA profile of this parasite by high throughput sequencing technology, bioinformatics platform analysis and quantitative RT-PCR.
Results
Three miRNA candidates, with typical precursor stem-loop structures, were identified from 11.13 million raw sequencing reads. Three miRNAs, Tga-miR-1, Tga-miR-2 and Tga-miR-3 had no homologues in publically available miRNA databases, suggesting that they may be T. gallinae-specific. Tga-miR-2 and Tga-miR-3 occupied only one location each on the reference genome, while Tga-miR-1 was found at 3 locations.
Conclusions
The results of the present study provided a sound basis for the further understanding of gene regulation in this parasite of animal health significance, with the potential to inform the development of novel control reagents and strategies and also inform a more in-depth understanding of the evolution of miRNAs.
Keywords: Trichomonas gallinae, MicroRNA (miRNA), Quantitative RT-PCR, Gene regulation, Richomonosis
Background
Trichomonas gallinae is a flagellated protozoon, which causes avian trichomonosis in many species of domestic poultry and birds, including chickens, pigeons, and parrots. It mainly lives in the anterior digestive tract and causes granulomatous lesions or canker in the host [1]. Different strains of T. gallinae vary in their pathogenicity, resulting in a range of clinical outcomes from asymptomatic to lethal infections [2,3]. Certain highly pathogenic strains can affect other organs of birds and cause necrotic foci [4] and peptidases secreted by the parasite were recently reported to have cytopathogenic effects on chicken liver cells [5]. The clinical signs of trichomonosis normally involve anorexia, vomiting, ruffled feathers, weight loss and, in some severe cases, death as a result of starvation [6,7]. Major outbreaks of the parasite cause epidemic mortality, especially breeding populations [8-10]. T. gallinae therefore has important welfare and commercial implications for the poultry industry as well as game bird and pigeon breeding and rearing [11]. Trichomonosis has also now been highlighted as a major threat to some endangered wild bird populations, such as the Pink Pigeon in Mauritius [12].
MicroRNAs (miRNAs) are a set of small non-coding RNAs that are now considered as a key mechanism of gene regulation and are essential for the complex life cycles of different parasites [13-16], regulating gene expression at the post-transcriptional level and resulting in post-transcriptional repression [17]. MiRNAs are conserved in metazoans and have been reported in diverse organisms from viruses to mammals [18]. However, despite the veterinary and commercial importance of T. gallinae there have been no published studies to date on their miRNAs.
Here we investigated the global miRNA expression profile of T. gallinae using a combined platform of next-generation sequencing technology, bioinformatic analysis and real-time quantitative PCR. Due to the similarities between the Trichomonas spp., miRNA profile research in T. gallinae will shed light on gene regulation studies in other species such as T. vaginalis, T. buccalis and T. hominis, which are medically important parasites in humans.
Methods
Ethics statement
The pigeons from which T. gallinae were collected, were handled in accordance with good animal practices required by the Animal Ethics Procedures and Guidelines of the People's Republic of China. The present study was approved by the Animal Ethics Committee of Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences (Approval No. LVRIAEC2011-007).
Parasites
T. gallinae was isolated from pigeons and cultured as described previously with modifications, as follows [19]. T. gallinae was collected from the oral cavity of a pigeon with a cotton swab and cultivated in vitro in Diamond’s medium supplemented with 10% calf serum and antibiotics (50 IU gentamicin–streptomycin). Cultures were incubated at 36°C for 24 h. The dense cultures were then washed with 0.9% saline for 3 times, and then flash frozen in liquid nitrogen and stored at -80°C. The identity of the cultured parasites was confirmed by sequencing of the ITS of rDNA following PCR amplification with oligonucleotide primers as follows: NC5: 5′-GTAGGTGAACCTGCGGAAGGATCATT-3′; NC2: 5′-TTAGTTTCTTTTCCTCC GCT-3′ (data not shown).
Total RNA and small RNA isolation
Total RNA of T. gallinae was prepared with Trizol reagent according to the manufacturer’s protocol (Invitrogen Co. Ltd). Small RNA was prepared as described previously [20]. Briefly, small RNAs of 20–35 bases in length were isolated from 10 μg total RNA using a 15% TBE-Urea polyacrylamide gel. After adding the 5’ and 3’ adaptors (Illumina Co. Ltd), the fragments were reverse transcribed and then purified with 6% TBE PAGE gel. All gels and kits were purchased from Invitrogen Co. Ltd.
High-throughput sequencing and computational analysis
Samples were sequenced using a Solexa (Illumina) sequencer. Adaptors and low quality reads were removed from the raw dataset during the base-calling stage. Non-coding RNAs, including rRNA, tRNA, snRNA and snoRNA, were removed by mapping with the Rfam database (http://rfam.sanger.ac.uk/) using BLAST software [21]. Repetitive sequences were removed by searching against the RepeatMasker (http://www.repeatmasker.org) database. Because no publically available genome is currently accessible for T. gallinae, the genome of the closely related and well researched species, T. vaginalis, from the EuPathDB database (http://eupathdb.org/eupathdb/) was used as a reference genome with reads mapped using SOAP [22]. Mfold was used for the prediction of miRNA candidates [23]. The identified miRNA candidates were then searched against the Sanger miRBase to identify known miRNAs. Potential targets of known miRNAs were predicted with RNAhybrid software [24]. To reduce false-positive results, two extra parameters were applied to the analyzed results: 1) the △△G was set as lower than -25 kcal/mol; 2) P-value was set as ≤ 0.01. The Gene Ontology (GO, http://www.geneontology.org/) database was used for the functional analysis of predicted targets.
Analysis of novel miRNA expression
Novel miRNAs were analyzed using a modified stem-loop real-time RT-PCR (ABI PRISM® 7300 Sequence Detection System) as described previously [25]. Synthetic lin-4 was used as the endogenous control [26]. The SYBR Green PCR Master Mix was purchased from the TOYOBO Co. Ltd. The amplification cycle conditions were as follows: 95°C for 5 min, followed by 30 cycles of 95°C for 15 s, 65°C for 15 s, and 72°C for 32 s. All reactions were carried out in triplicate. The quantification of each miRNA relative to the lin-4 was calculated using the equation: N = 2-ΔCt, ΔCt = CtmiRNA-Ctlin4[27].
Results
Profile characteristics of short RNAs
High throughput sequencing yielded 11.13 million raw reads from the total RNA of T. gallinae among which 10.76 million had high quality with no adaptors or polyA regions. Repeat analysis revealed 1,891 sequences as the repeat type LTR:1, 6 sequences as DNA/Maverick:1 and one sequence as DNA/Maverick:0. Therefore, repeat sequences occupied a very small percentage of the high quality reads. rRNA was found to be responsible for a relatively high percentage of the reads (35.71%); and tRNA accounted for 1.84% of the total. Other non-coding RNAs, including snRNA and snoRNA, represented only 0.03% of the reads.
miRNA profile analysis
When mapped onto the T. vaginalis genome, 4.47 million reads were successfully mapped. However, of these, only 3 miRNA candidates with precursors having stem-loop structures met the criteria we imposed for miRNA selection (Table 1). These three miRNAs, Tga-miR-1, Tga-miR-2 and Tga-miR-3 had no homologues in the miRBase database. Tga-miR-1 was identified at 3 locations on the reference genome, (scaffolds gi|121819238, gi|121897016 and gi|121907615), while Tga-miR-2 and Tga-miR-3 occupied only one location each on the reference genome. The precursor and stem-loop structure of Tga-miR-1 are shown in Figure 1.
Table 1.
miRNA profiles in Trichomonas gallinae
| Name | Location | Arms | Mfe | Count | Sequence |
|---|---|---|---|---|---|
| Tga-miR-1 |
gi|121819238|:570:634:- |
5p |
-25.8 |
6 |
TGGTGCTACAGTCGGCTCT |
| Tga-miR-2 |
gi|121883425|:16027:16113:+ |
3p |
-21.5 |
19 |
GAAGAATTGTTGGAAGAA |
| Tga-miR-3 | gi|121907444|:136045:136145:- | 5p | -21.2 | 4 | AGAAGGTCTCCTGCCCAC |
Figure 1.
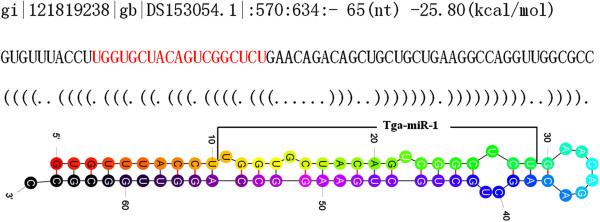
The stem-loop structure of Tga-miR-1. The top line indicates gene location, length of precursor, and the energy of the stem-loop structure. The mature miRNA in the precursor is shown in red text and marked in the stem-loop structure.
Target prediction and function analysis
A total of 60,816 annotated transcripts of T. vaginalis were downloaded from the EuPathDB database (http://eupathdb.org/eupathdb/) and used for target prediction of the 3 miRNA candidates. Targets were successfully predicted for the 3 miRNAs with total transcript target numbers being 86 (Tga-miR-1), 2 (Tga-miR-2) and 20 (Tga-miR-3), and with the best matched targets as TVAG_212500, TVAG_158870 and TVAG_068410 for each of the miRNAs respectively. The 3 best matched targets all originated from T. vaginalis G3. TVAG_212500 and TVAG_158870 encode hypothetical proteins, while TVAG_068410 encodes a legumain-like cysteine peptidase.
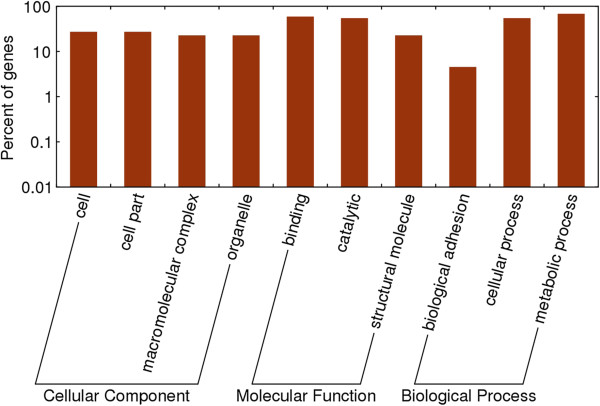
For all the 108 targets, 77 (71.3%) were hypothetical proteins, 4 targets were pe-pgrs proteins and 2 targets, (TVAG_480240, and TVAG_165450), were CAMK family protein kinases. In addition, one transcription activator (TVAG_078540, a “metallothionein-i gene transcription activator”), and one kinase receptor (TVAG_283180, a “leukocyte tyrosine kinase receptor precursor-related protein”) were also found as targets of the 3 miRNAs. Enrichment analysis showed that the molecular function of the targets were biased towards binding and catalytic function (Figure 2).
Figure 2.
Enrichment analysis of the predicted targets of Trichomonas gallinae miRNAs.
miRNAs quantification
Each of the 3 miRNAs could be successfully amplified using qRT-PCR. The relative expression level of Tga-miR-1 (1 ± 0.16) and Tga-miR-2 (1.05 ± 0.12) were similar to the endogenous control. Tga-miR-3 had a significantly higher relative expression level (20.61 ± 1.3) than the other two miRNAs investigated.
Discussion and conclusions
Avian trichomonosis, caused by the protozoan T. gallinae, has been considered as a major disease for numerous avian species. Control by chemotherapy is the main approach to control the disease at present. However, drug resistance coupled with high costs of developing new drugs is still a bottleneck for treatment and prevention of the disease. Therefore, new approaches including immunological, biotechnological and genetic methods are more desirable alternatives [28]. MiRNAs perform a variety of pivotal functions within cells, including regulation of growth, metabolism, development and cell differentiation. They are now considered as biomarkers of parasite invasion, key regulators of gene expression at the post-transcriptional level and potential new tools for disease diagnostics and control [15,29-32].
We herein investigated the global miRNAs of the protozoan T. gallinae and 3 miRNA candidates were identified. The number of miRNAs of T. gallinae was similar to the miRNAs described by Lin et al. [33], who revealed 9 novel miRNAs from T. vaginalis, a closely-related parasite. The three novel miRNAs from T. gallinae described herein did not possess homologues in miRNAs from T. vaginalis, and therefore, the three miRNA candidates identified in the present study may be T. gallinae-specific.
Target prediction revealed more than one hundred targets for the 3 miRNAs candidates. However, 71.3% of these targets encoded hypothetical proteins indicating the lack of published confirmatory data for gene prediction and function in Trichomonas genomics, in spite of the identification of ~60,000 putative genes in the 170 MB genome of T. vaginalis[34]. We identified some miRNA targets as kinases, transcription activators, and receptors, which have fundamental functions in the growth and metabolism of the parasite.
The results of the present study have therefore provided a sound basis for the further understanding of gene regulation in this species, with the potential to inform the development of novel control reagents and strategies and also inform a more in-depth understanding of the evolution of miRNAs.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
XQZ and MJX conceived and designed the study, and critically revised the manuscript. MJX, SBQ and JHF performed the experiments, analyzed the data and drafted the manuscript. AJN, and CCS helped in study design, study implementation and manuscript revision. All authors read and approved the final manuscript.
Contributor Information
Min-Jun Xu, Email: xuminjun@caas.cn.
Shen-Ben Qiu, Email: shenbenqiu@126.com.
Alasdair J Nisbet, Email: alasdair.nisbet@moredun.ac.uk.
Jing-Hua Fu, Email: fujinghua@scau.edu.cn.
Chang-Chun Shao, Email: 7330968@163.com.
Xing-Quan Zhu, Email: xingquanzhu1@hotmail.com.
Acknowledgements
This work was supported in part by the International Science & Technology Cooperation Program of China (Grant No. 2013DFA31840), the Science Fund for Creative Research Groups of Gansu Province (Grant No. 1210RJIA006) and the Scientific and Technological Program of Guangdong Province, China (Grant No. 2009B020307005).
References
- Daly JJ. The maltose metabolism of Trichomonas gallinae (Rivolta, 1878). II. Metabolic studies. J Parasitol. 1971;57:370–374. doi: 10.2307/3278045. [DOI] [PubMed] [Google Scholar]
- Amin A, Bilic I, Berger E, Hess M. Trichomonas gallinae, in comparison to Tetratrichomonas gallinarum, induces distinctive cytopathogenic effects in tissue cultures. Vet Parasitol. 2012;186:196–206. doi: 10.1016/j.vetpar.2011.11.037. [DOI] [PubMed] [Google Scholar]
- Stabler RM. Trichomonas gallinae: a review. Exp Parasitol. 1954;3:368–402. doi: 10.1016/0014-4894(54)90035-1. [DOI] [PubMed] [Google Scholar]
- Zimre-Grabensteiner E, Arshad N, Amin A, Hess M. Genetically different clonal isolates of Trichomonas gallinae, obtained from the same bird, can vary in their drug susceptibility, an in vitro evidence. Parasitol Int. 2011;60:213–215. doi: 10.1016/j.parint.2011.02.003. [DOI] [PubMed] [Google Scholar]
- Amin A, Nobauer K, Patzl M, Berger E, Hess M, Bilic I. Cysteine peptidases, secreted by Trichomonas gallinae, are involved in the cytopathogenic effects on a permanent chicken liver cell culture. PLoS ONE. 2012;7:e37417. doi: 10.1371/journal.pone.0037417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecco R, Preis IS, Vilela DA, Luppi MM, Malta MC, Beckstead RB, Stimmelmayer R, Gerhold RW. Molecular confirmation of Trichomonas gallinae and other parabasalids from Brazil using the 5.8S and ITS-1 rRNA regions. Vet Parasitol. 2012;190:36–42. doi: 10.1016/j.vetpar.2012.05.029. [DOI] [PubMed] [Google Scholar]
- Sansano-Maestre J, Garijo-Toledo MM, Gomez-Munoz MT. Prevalence and genotyping of Trichomonas gallinae in pigeons and birds of prey. Avian Pathol. 2009;38:201–207. doi: 10.1080/03079450902912135. [DOI] [PubMed] [Google Scholar]
- Lawson B, Cunningham AA, Chantrey J, Hughes LA, John SK, Bunbury N, Bell DJ, Tyler KM. A clonal strain of Trichomonas gallinae is the aetiologic agent of an emerging avian epidemic disease. Infect Genet Evol. 2011;11:1638–1645. doi: 10.1016/j.meegid.2011.06.007. [DOI] [PubMed] [Google Scholar]
- Robinson RA, Lawson B, Toms MP, Peck KM, Kirkwood JK, Chantrey J, Clatworthy IR, Evans AD, Hughes LA, Hutchinson OC, John SK, Pennycott TW, Perkins MW, Rowley PS, Simpson VR, Tyler KM, Cunningham AA. Emerging infectious disease leads to rapid population declines of common British birds. PLoS ONE. 2010;5:e12215. doi: 10.1371/journal.pone.0012215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neimanis AS, Handeland K, Isomursu M, Agren E, Mattsson R, Hamnes IS, Bergsjø B, Hirvelä-Koski V. First report of epizootic trichomoniasis in wild finches (family Fringillidae) in southern Fennoscandia. Avian Dis. 2010;54:136–141. doi: 10.1637/8952-060509-Case.1. [DOI] [PubMed] [Google Scholar]
- McKeon T, Dunsmore J, Raidal SR. Trichomonas gallinae in budgerigars and columbid birds in Perth, Western Australia. Aust Vet J. 1997;75:652–655. doi: 10.1111/j.1751-0813.1997.tb15363.x. [DOI] [PubMed] [Google Scholar]
- Bunbury N, Jones CG, Greenwood AG, Bell DJ. Trichomonas gallinae in Mauritian columbids: implications for an endangered endemic. J Wildl Dis. 2007;43:399–407. doi: 10.7589/0090-3558-43.3.399. [DOI] [PubMed] [Google Scholar]
- Wang Z, Xue X, Sun J, Luo R, Xu X, Jiang Y, Zhang Q, Pan W. An “in-depth” description of the small non-coding RNA population of Schistosoma japonicum schistosomulum. PLoS Negl Trop Dis. 2010;4:e596. doi: 10.1371/journal.pntd.0000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu MJ, Wang CR, Huang SY, Fu JH, Zhou DH, Chang QC, Zheng X, Zhu XQ. Identification and characterization of microRNAs in the pancreatic fluke Eurytrema pancreaticum. Parasit Vectors. 2013;6:25. doi: 10.1186/1756-3305-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao GH, Xu MJ, Zhu XQ. Identification and characterization of microRNAs in Baylisascaris schroederi of the giant panda. Parasit Vectors. 2013;6:216. doi: 10.1186/1756-3305-6-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Liu X, Jia B, Lu H, Peng S, Piao X, Hou N, Cai P, Yin J, Jiang N, Chen Q. A comparative study of small RNAs in Toxoplasma gondii of distinct genotypes. Parasit Vectors. 2012;5:186. doi: 10.1186/1756-3305-5-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blahna MT, Hata A. Smad-mediated regulation of microRNA biosynthesis. FEBS Lett. 2012;586:1906–1912. doi: 10.1016/j.febslet.2012.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang Q, Pan X. MicroRNAs and their regulatory roles in animals and plants. J Cell Physiol. 2007;210:279–289. doi: 10.1002/jcp.20869. [DOI] [PubMed] [Google Scholar]
- Reinmann K, Muller N, Kuhnert P, Campero CM, Leitsch D, Hess M, Henning K, Fort M, Müller J, Gottstein B, Frey CF. Tritrichomonas foetus isolates from cats and cattle show minor genetic differences in unrelated loci ITS-2 and EF-1alpha. Vet Parasitol. 2012;185:138–144. doi: 10.1016/j.vetpar.2011.09.032. [DOI] [PubMed] [Google Scholar]
- Xu MJ, Liu Q, Nisbet AJ, Cai XQ, Yan C, Lin RQ, Yuan ZG, Song HQ, He XH, Zhu XQ. Identification and characterization of microRNAs in Clonorchis sinensis of human health significance. BMC Genomics. 2010;11:521. doi: 10.1186/1471-2164-11-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount DW. Using the Basic Local Alignment Search Tool (BLAST) CSH Protoc. 2007. pdb.top17 http://www.ncbi.nlm.nih.gov/pubmed/21357135. [DOI] [PubMed]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- Zuker MM. fold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger J, Rehmsmeier M. NAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;R34:W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed N, Bentwich Z, Hod M, Goren Y, Chajut A. Serum microRNAs are promising novel biomarkers. PLoS ONE. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luo F, Li GQ, Su RQ, Liang G, Chen ZH, Hicham W. Cloning and sequencing of adhesion protein gene of Trichomonas gallinae from pigeon. Vet Parasitol. 2010;168:125–129. doi: 10.1016/j.vetpar.2009.10.017. [DOI] [PubMed] [Google Scholar]
- Du T, Zamore PD. Beginning to understand microRNA function. Cell Res. 2007;17:661–663. doi: 10.1038/cr.2007.67. [DOI] [PubMed] [Google Scholar]
- Zhang B, Farwell MA. microRNAs: a new emerging class of players for disease diagnostics and gene therapy. J Cell Mol Med. 2008;12:3–21. doi: 10.1111/j.1582-4934.2007.00196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Sai X, Chen C, Zhang Y, Xu X, Zhang D, Pan W. Host serum miR-223 is a potential new biomarker for Schistosoma japonicum infection and the response to chemotherapy. Parasit Vectors. 2013;6:272–279. doi: 10.1186/1756-3305-6-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu MJ, Zhou DH, Nisbet AJ, Huang SY, Fan YF, Zhu XQ. Characterization of mouse brain microRNAs after infection with cyst-forming Toxoplasma gondii. Parasit Vectors. 2013;6:154–160. doi: 10.1186/1756-3305-6-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WC, Li SC, Lin WC, Shin JW, Hu SN, Yu XM, Huang TY, Chen SC, Chen HC, Chen SJ, Huang PJ, Gan RR, Chiu CH, Tang P. Identification of microRNA in the protist Trichomonas vaginalis. Genomics. 2009;93:487–493. doi: 10.1016/j.ygeno.2009.01.004. [DOI] [PubMed] [Google Scholar]
- Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, Zhao Q, Wortman JR, Bidwell SL, Alsmark UC, Besteiro S, Sicheritz-Ponten T, Noel CJ, Dacks JB, Foster PG, Simillion C, Van de Peer Y, Miranda-Saavedra D, Barton GJ, Westrop GD, Müller S, Dessi D, Fiori PL, Ren Q, Paulsen I, Zhang H, Bastida-Corcuera FD, Simoes-Barbosa A, Brown MT, Hayes RD, Mukherjee M. et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]