Abstract
The newly discovered transactivation function of ErbB4 receptor tyrosine kinase is believed to be mediated by virtue of the ability of its proteolytically-cleaved intracellular domain (ICD) to physically associate with YAP2 transcriptional regulator. In an effort to unearth the molecular basis of YAP2-ErbB4 interaction, we have conducted a detailed biophysical analysis of the binding of WW domains of YAP2 to PPXY motifs located within the ICD of ErbB4. Our data show that the WW1 domain of YAP2 binds to PPXY motifs within the ICD in a differential manner and that this behavior is by and large replicated by the WW2 domain. Remarkably, while both WW domains absolutely require the integrity of the PPXY consensus sequence, non-consensus residues within and flanking this motif do not appear to be critical for binding. In spite of this shared mode of binding, the WW domains of YAP2 display distinct conformational dynamics in complex with PPXY motifs derived from ErbB4. Collectively, our study lends new insights into the molecular basis of a key protein-protein interaction involved in a diverse array of cellular processes.
Keywords: WW-ligand thermodynamics, PPXY Motifs, Structural analysis, Conformational dynamics
INTRODUCTION
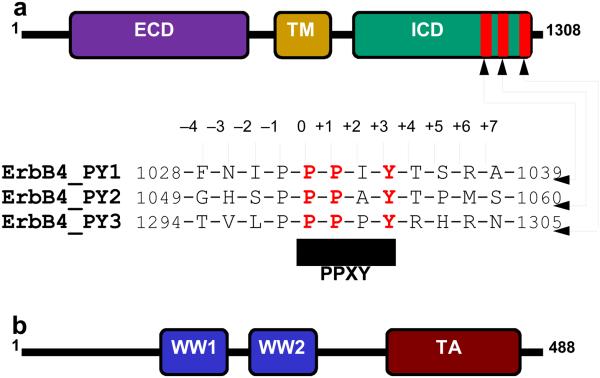
A key member of the receptor tyrosine kinase (RTK) family, ErbB4 is comprised of a central single-helical transmembrane (TM) domain flanked between an N-terminal extracellular domain (ECD) and a C-terminal intracellular domain (ICD) (Figure 1a). Upon stimulation with its extracellular ligand heregulin or in response to TPA (12-O-tetradecanoylphorbol-13-acetate)-induced activation of protein kinase C, the ErbB4 receptor tyrosine kinase undergoes intracellular proteolytic cleavage by γ-secretase [1, 2]. This coupled event culminates in the release of ICD and marks the initiation of ErbB4 intracellular signaling. Notably, the ICD of ErbB4 harbors putative PPXY motifs (designated PY1, PY2 and PY3)—the chemical baits that serve as recognition sites for the recruitment of WW-containing proteins such as YAP (YES-associated protein) transcriptional regulator [3, 4], WWOX (WW-containing oxidoreductase) tumor suppressor [5], and ITCH ubiquitin ligase [6]. The physical association between YAP and ICD facilitates translocation of the latter to the nucleus [3], where it is believed to regulate the transcription of hitherto unidentified target genes involved in key cellular processes including embryonic development [7]. While YAP acts as transcriptional co-activator of ErbB4, interaction with WWOX not only results in the cytoplasmic sequestration of ICD but also suppresses its transcriptional co-activation by YAP [5]. On the other hand, binding to ITCH promotes polyubiquitination and degradation of ErbB4, thereby regulating its stability and the availability of ICD for subsequent transcriptional regulation in the nucleus [6]. The concerted action of WWOX and ITCH thus antagonizes the co-activation function of YAP by virtue of their ability to bind to the ICD of ErbB4 in a competitive manner.
Figure 1.
Modular organization of human ErbB4 and YAP2 proteins. (a) ErbB4 contains the canonical ECD-TM-ICD receptor tyrosine kinase modular cassette, where the central single-helical transmembrane (TM) domain is flanked between an N-terminal extracellular domain (ECD) and a C-terminal intracellular domain (ICD). The three PPXY motifs (designated PY1, PY2 and PY3) within the ICD are located at the extreme C-terminus. Note that the amino acid sequence of 12-mer peptides containing the PPXY motifs and flanking residues are provided. The numerals indicate the nomenclature used in this study to distinguish residues within and flanking the PPXY motifs relative to the first consensus proline, which is arbitrarily assigned zero. (b) YAP2 is comprised of a tandem copy of WW domains, designated WW1 and WW2, located N-terminal to the trans-activation (TA) domain.
Originally identified as a binding partner of YES tyrosine kinase [8], YAP is comprised of two major isoforms termed YAP1 and YAP2, also referred to as YAP1-1 and YAP1-2 on the basis of new nomenclature [9]. While YAP2 contains a tandem copy of WW domains (termed WW1 and WW2) located N-terminal to the transactivation (TA) domain (Figure 1b), WW2 domain is deleted in YAP1 through RNA splicing [10]. In addition to its key role in mediating the transactivation function of ErbB4 receptor tyrosine kinase [3, 4], YAP also serves as a transcriptional regulator of a multitude of cellular factors including p73, RUNX, TEAD, LATS1, ErbB4 and, in particular, plays a key role in mediating the Hippo signaling pathway [11-18]—involved in regulating the size of organs and in the suppression of tumors through inhibiting cellular proliferation and promoting apoptosis. Consistent with these observations, YAP-knockout in mice results in embryonic lethality [19]. Most importantly, the YAP-ICD interaction is mediated by the canonical binding of WW domains of YAP to PPXY motifs located within the ICD of ErbB4 (Figures 1a and 1b). It should be noted here that ICD of ErbB4 is a much more potent co-activator of YAP2 than YAP1 [3]. This finding most likely argues in favor of a multivalent interaction between the tandem WW domains of YAP2 and PPXY motifs within ICD. In an effort to uncover the molecular basis of YAP-ErbB4 interaction, we report herein a detailed biophysical analysis of the binding of WW domains of YAP2 to PPXY motifs located within the ICD of ErbB4. Briefly, our data show that the WW1 domain of YAP2 binds to PPXY motifs within the ICD in a differential manner and that this behavior is by and large replicated by the WW2 domain. Remarkably, while both WW domains absolutely require the integrity of the PPXY consensus sequence, non-consensus residues within and flanking this motif do not appear to be critical for binding. In spite of this shared mode of binding, the WW domains of YAP2 display distinct conformational dynamics in complex with PPXY motifs derived from ErbB4.
MATERIALS and METHODS
Protein preparation
WW1 domain (residues 171-205) and WW2 domain (residues 230-264) of human YAP2 were cloned into pET30 bacterial expression vectors with an N-terminal His-tag using Novagen ligation-independent cloning (LIC) as described earlier [20]. Recombinant proteins were subsequently expressed in Escherichia coli BL21*(DE3) bacterial strain (Invitrogen) and purified on a Ni-NTA affinity column using standard procedures [20]. Further treatment on a Hiload Superdex 200 size-exclusion chromatography (SEC) column coupled in-line with GE Akta FPLC system led to purification of WW domains to apparent homogeneity as judged by SDS-PAGE analysis. Final yield was typically between 50-100mg protein of apparent homogeneity per liter of bacterial culture. Protein concentration was determined spectrophotometrically on the basis of extinction coefficients calculated for each protein construct using the online software ProtParam at ExPasy Server [21].
Peptide synthesis
12-mer wildtype and mutant peptides spanning various PPXY motifs within the ICD of human ErbB4 were commercially obtained from GenScript Corporation. The wildtype sequence of these peptides is shown in Figure 1a. The peptide concentrations were measured gravimetrically.
Isothermal titration calorimetry
Isothermal titration calorimetry (ITC) experiments were performed on a Microcal VP-ITC instrument and data were acquired and processed using the integrated Microcal ORIGIN software. All measurements were repeated at least three times. Briefly, WW domains of YAP2 and ErbB4 peptides were dialyzed in 50mM Sodium phosphate, 100mM NaCl, 1mM EDTA and 5mM β-mercaptoethanol at pH 7.0. The experiments were initiated by injecting 25 x 10μl aliquots of 4 mM of each peptide from the syringe into the calorimetric cell containing 1.46ml of 40-60 μM of each WW domain at 25°C. The change in thermal power as a function of each injection was automatically recorded using the ORIGIN software and the raw data were further processed to yield binding isotherms of heat release per injection as a function of molar ratio of each peptide to WW domain construct. The heats of mixing and dilution were subtracted from the heat of binding per injection by carrying out a control experiment in which the same buffer in the calorimetric cell was titrated against each peptide in an identical manner. To extract the binding constant (Kd) and binding enthalpy (ΔH), the ITC isotherms were iteratively fit to a one-site model by non-linear least squares regression analysis using the integrated ORIGIN software as described earlier [22, 20]. The free energy change (ΔG) upon peptide binding was calculated from the relationship:
| (1) |
where R is the universal molar gas constant (1.99 cal/K/mol) and T is the absolute temperature. The entropic contribution (TΔS) to the free energy of binding was calculated from the relationship:
| (2) |
where ΔH and ΔG are as defined above.
Circular dichroism
Far-UV circular dichroism (CD) measurements were conducted on a Jasco J-815 spectropolarimeter thermostatically controlled at 25°C. Briefly, ErbB4 peptides were dialyzed in 10mM Sodium phosphate at pH 7.0 and experiments were conducted on 100μM sample of each peptide. Data were collected using a quartz cuvette with a 2-mm pathlength in the 185-255 nm wavelength range and with a slit bandwidth of 2 nm at a scan rate of 10 nm/min. All data were normalized against reference spectra to remove the contribution of buffer. Each data set represents an average of four scans acquired at 0.1 nm intervals. Data were converted to mean ellipticity, [θ], as a function of wavelength (λ) of electromagnetic radiation using the following equation:
| (3) |
where Δε is the observed ellipticity in mdeg, c is the peptide concentration in μM, and l is the cuvette pathlength in cm.
Molecular modeling
Structural models of WW1 and WW2 domains of YAP2 in complex with a peptide containing the PY3 motif located within the ICD of ErbB4 (ErbB4_PY3) were built using the MODELLER software based on homology modeling [23]. In each case, four NMR structures of WW domains of YAP bound to peptides containing the PPXY motif were used in a multi-template alignment fashion (PDBIDs 2LAW, 2LTV and 2LTW and 1JMQ). A total of 100 atomic models were calculated and the structure with the lowest energy, as judged by the MODELLER Objective Function, was selected for further analysis. The atomic models were rendered using RIBBONS [24].
Molecular dynamics
Molecular dynamics (MD) simulations were performed with the GROMACS (version 4.54) software [25] using the integrated AMBER99SB-ILDN force field [26]. Briefly, the structural models of WW1 and WW2 domains of YAP2 in complex with ErbB4_PY3 peptide were each centered in a cubic box and explicitly hydrated with a water layer that extended 10Å (box size) from the protein surface along each orthogonal direction using the extended simple point charge (SPC/E) water model [27, 28]. The ionic strength of solution was set to 100mM with NaCl and the hydrated structures were energy-minimized with the steepest descent algorithm prior to equilibration under the NPT ensemble conditions, wherein the number of atoms (N), pressure (P) and temperature (T) within the system were kept constant. The Particle-Mesh Ewald (PME) method [29] was employed to compute long-range electrostatic interactions with a spherical cut-off of 10Å and a grid space of 1.6Å with a fourth order interpolation. The Linear Constraint Solver (LINCS) algorithm was used to restrain bond lengths [30]. All MD simulations were performed under periodic boundary conditions (PBC) at 310K using the leap-frog integrator with a time step of 2fs. For the final MD production runs, data were collected every ns over a time scale of 1μs.
RESULTS and DISCUSSION
WW domains of YAP2 bind to PPXY motifs in ErbB4 in a differential manner
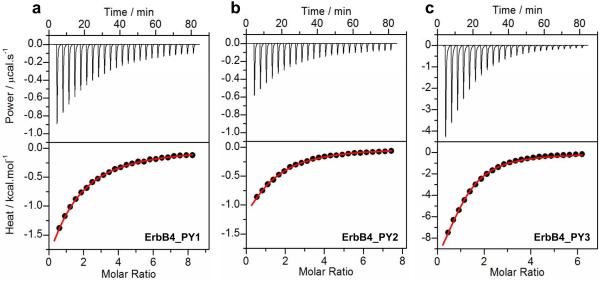
To understand the molecular basis of YAP2-ErbB4 interaction, we analyzed the binding of WW domains of YAP2 to various ErbB4 peptides containing PPXY motif using ITC (Figure 2 and Table 1). Our analysis shows that both WW1 and WW2 domains recognize all three PPXY motifs in a physiologically-relevant manner with affinities in the order of tens of micromolar. However, both WW domains appear to preferentially bind to ErbB4_PY3 peptide as evidenced by an affinity that is two-fold greater relative to that observed toward the ErbB4_PY1 and ErbB4_PY2 peptides. Such differences in binding are further reflected in the underlying thermodynamics. Thus, while the binding of both WW domains of Yap2 to ErbB4_PY3 peptide is driven by favorable enthalpic forces accompanied by unfavorable entropic changes, the role of enthalpy becomes increasingly less important in mediating binding to ErbB4_PY1 and ErbB4_PY2 peptides, wherein entropic penalty is either completely eliminated or also contributes favorably to binding. These observations suggest the differential role of intermolecular forces such as hydrogen bonding, ion pairing and van der Waals contacts in driving the binding of YAP2 to specific PPXY motifs in ErbB4.
Figure 2.
Representative ITC isotherms for the binding of WW1 domain of YAP2 to ErbB4_PY1 (a), ErbB4_PY2 (b) and ErbB4_PY3 (c) peptides. The upper panels show the raw ITC data expressed as change in thermal power with respect to time over the period of titration. In the lower panels, change in molar heat is expressed as a function of molar ratio of corresponding peptide to WW1 domain. The red solid lines in the lower panels show the fit of data to a one-site binding model using the integrated ORIGIN software as described earlier [22, 20].
In order to uncover the origin of the differential binding of WW domains of YAP2 to various PPXY motifs within ErbB4 (Table 1), we next carried out alanine scan on the ErbB4_PY3 peptide and measured the binding of each mutant peptide to WW domains as described above (Tables 2 and 3). Intriguingly, our data reveal that alanine substitution of non-consensus residues within and flanking the PPXY motif within the ErbB4_PY3 peptide does not dramatically affect the binding of WW domains. This finding suggests that non-consensus residues within and flanking the PPXY motifs are not critical for driving the YAP2-ErbB4 interaction but they may be important for stabilizing the conformation of PPXY peptides. It should however be noted that a number of positions appear to play an important but not critical role. Thus, alanine substitution of P-1 (PY3_A-1) and P+2 (PY3_A+2) results in the reduction of binding affinity of both WW domains to ErbB4_PY3 peptide by two-fold. This implies that both WW domains have a preference for a proline at the -1 and +2 positions. While the -1 position is occupied by a proline in all three ErbB4 peptides, the +2 position in ErbB4_PY1 and ErbB4_PY2 peptides is replaced by non-proline residues. Thus, the binding of WW domains of YAP2 to ErbB4_PY3 peptide with higher affinity relative to ErbB4_PY1 and ErbB4_PY2 peptides could in part be due to the lack a proline at the +2 position in the latter peptides.
Table 1.
Thermodynamic parameters for the binding of WW1 and WW2 domains of YAP2 to PPXY peptides derived from the ICD of ErbB4
| Peptide | Sequence | Kd / μM | ΔH / kcal.mol−1 | TΔS / kcal.mol−1 | ΔG / kcal.mol−1 |
|---|---|---|---|---|---|
| WW1 Domain | |||||
| ErbB4_PY1 |

|
113 ± 18 | −6.14 ± 0.31 | −0.74 ± 0.40 | −5.40 ± 0.09 |
| ErbB4_PY2 |
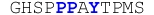
|
84 ± 11 | −2.94 ± 0.17 | +2.63 ± 0.25 | −5.57 ± 0.08 |
| ErbB4_PY3 |

|
31 ± 3 | −13.35 ± 0.49 | −7.19 ± 0.55 | −6.16 ± 0.05 |
| WW2 Domain | |||||
| ErbB4_PY1 |

|
100 ± 5 | −3.90 ± 0.52 | +1.57 ± 0.55 | −5.47 ± 0.03 |
| ErbB4_PY2 |
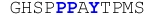
|
116 ± 17 | −4.23 ± 0.45 | +1.15 ± 0.53 | −5.38 ± 0.08 |
| ErbB4_PY3 |

|
56 ± 4 | −7.72 ± 0.54 | −1.90 ± 0.58 | −5.81 ± 0.04 |
Note that the consensus residues within the PPXY motif of each peptide are colored blue for clarity. Binding stoichiometries generally agreed to within ±10%. Errors were calculated from at least three independent measurements. All errors are given to one standard deviation.
Table 2.
Thermodynamic parameters for the binding of WW1 domain of YAP2 to wildtype (PY3_WT) and single alanine mutants of ErbB4_PY3 peptide
| Peptide | Sequence | Kd / μM | ΔH / kcal.mol−1 | TΔS / kcal.mol−1 | ΔG / kcal.mol−1 |
|---|---|---|---|---|---|
| PY3_WT |
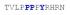
|
31 ± 3 | −13.35 ± 0.49 | −7.19 ± 0.55 | −6.16 ± 0.05 |
| PY3_A-3 |
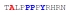
|
46 ± 7 | −16.95 ± 0.64 | −11.02 ± 0.73 | −5.93 ± 0.09 |
| PY3_A-2 |
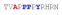
|
45 ± 10 | −16.05 ± 0.64 | −10.11 ± 0.78 | −5.94 ± 0.13 |
| PY3_A-1 |
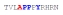
|
70 ± 14 | −15.90 ± 0.71 | −10.22 ± 0.83 | −5.68 ± 0.12 |
| PY3_A0 |
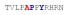
|
NBD | NBD | NBD | NBD |
| PY3_A+1 |
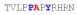
|
NBD | NBD | NBD | NBD |
| PY3_A+2 |

|
63 ± 8 | −12.65 ± 0.49 | −6.91 ± 0.57 | −5.74 ± 0.07 |
| PY3_A+3 |

|
NBD | NBD | NBD | NBD |
| PY3_A+4 |
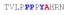
|
53 ± 12 | −9.80 ± 0.28 | −3.95 ± 0.42 | −5.85 ± 0.14 |
| PY3_A+5 |
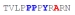
|
15 ± 4 | −14.55 ± 0.35 | −7.95 ± 0.52 | −6.60 ± 0.17 |
| PY3_A+6 |
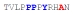
|
36 ± 4 | −10.35 ± 0.35 | −4.27 ± 0.29 | −6.08 ± 0.06 |
Note that the alanine substitutions within the ErbB4_PY3 peptide are colored red and underlined for clarity. Binding stoichiometries generally agreed to within ±10%. Errors were calculated from at least three independent measurements. All errors are given to one standard deviation. NBD indicates no binding observed.
Table 3.
Thermodynamic parameters for the binding of WW2 domain of YAP2 to wildtype (PY3_WT) and single alanine mutants of ErbB4_PY3 peptide
| Peptide | Sequence | Kd / μM | ΔH / kcal.mol−1 | TΔS / kcal.mol−1 | ΔG / kcal.mol−1 |
|---|---|---|---|---|---|
| PY3_WT |
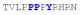
|
56 ± 4 | −7.72 ± 0.54 | −1.90 ± 0.58 | −5.81 ± 0.04 |
| PY3_A-3 |
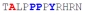
|
62 ± 17 | −10.30 ± 0.28 | −4.54 ± 0.45 | −5.76 ± 0.16 |
| PY3_A-2 |
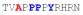
|
83 ± 17 | −13.05 ± 0.21 | −7.47 ± 0.09 | −5.58 ± 0.12 |
| PY3_A-1 |
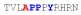
|
110 ± 22 | −10.70 ± 0.28 | −5.29 ± 0.40 | −5.41 ± 0.12 |
| PY3_A0 |
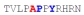
|
NBD | NBD | NBD | NBD |
| PY3_A+1 |

|
NBD | NBD | NBD | NBD |
| PY3_A+2 |

|
126 ± 16 | −10.08 ± 0.32 | −4.74 ± 0.40 | −5.33 ± 0.08 |
| PY3_A+3 |

|
NBD | NBD | NBD | NBD |
| PY3_A+4 |
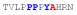
|
96 ± 10 | −8.61 ± 0.07 | −3.12 ± 0.01 | −5.49 ± 0.06 |
| PY3_A+5 |
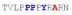
|
46 ± 7 | −14.55 ± 0.35 | −8.62 ± 0.26 | −5.93 ± 0.09 |
| PY3_A+6 |
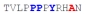
|
85 ± 16 | −9.06 ± 0.11 | −3.49 ± 0.01 | −5.57 ± 0.11 |
Note that the alanine substitutions within the ErbB4_PY3 peptide are colored red and underlined for clarity. Binding stoichiometries generally agreed to within ±10%. Errors were calculated from at least three independent measurements. All errors are given to one standard deviation. NBD indicates no binding observed.
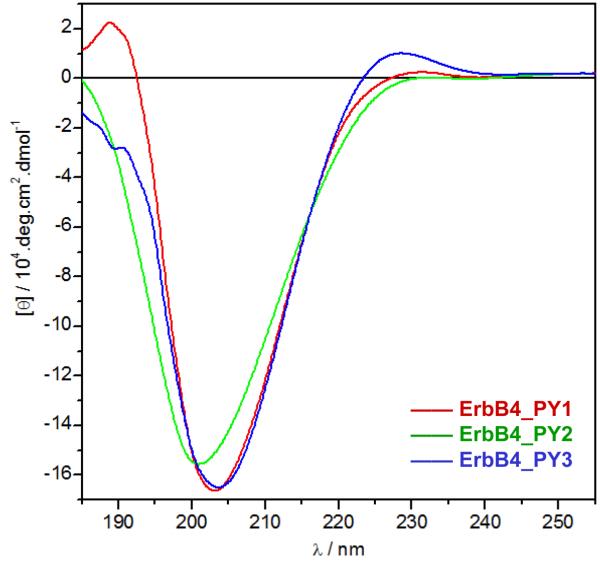
In particular, the importance of a proline at the +2 position may be accounted for by the virtue of its ability to buttress the polyproline II (PPII) helical conformation of ErbB4_PY3 peptide required for its optimal binding to WW domains [31-34]. This notion is indeed further corroborated by our far-UV CD analysis (Figure 3). Thus, while the far-UV spectra of all three ErbB4 peptides are characterized by the presence of a large negative band centered around 205nm, they also exhibit a shoulder at 225nm of varying intensity. These spectral features are well-documented for proline-rich peptides harboring random coil and PPII-helical conformations in equilibrium exchange, with the intensity of the 225-nm shoulder rapidly increasing with increasing degree of PPII-helical content [35, 36]. Importantly, the fact that the 225-nm shoulder appears to be much more pronounced in ErbB4_PY3 peptide is indicative of its higher PPII-helical content relative to the other two ErbB4 peptides. This could in part account for the preferential binding of WW domains of YAP2 to ErbB4_PY3 peptides. Accordingly, the binding of ErbB4_PY3 peptide to WW domains should be expected to encounter smaller entropic penalty compared to the other two peptides. Yet, our thermodynamic analysis suggests otherwise (Table 1). We believe that such discrepancy is probably reflective of the fact that the overall change in thermodynamic parameters is based on many other physical factors in addition the conformation of an individual binding partner. Thus, it is likely that the stronger binding of ErbB4_PY3 peptide to WW domains results in the complexes becoming more constrained so as to override the entropic advantage conferred by the PPII-helical conformation of the peptide. Equally importantly, binding of ErbB4_PY3 peptide to WW domains may also result in the greater entrapment of waters at interfacial surfaces and this would also be expected to reduce the favorable contribution of solvent release to the overall change in entropic penalty.
Figure 3.
Far-UV CD spectra of ErbB4_PY1 (red), ErbB4_PY2 (green) and ErbB4_PY3 (blue) peptides. Note that the mean ellipticity, [θ], was calculated using Eq [3].
On the other hand, while alanine substitution of H+5 (PY3_A+5) augments binding affinity of WW1 domain to ErbB4_PY3 peptide by two-fold, a similar effect is not observed for the WW2 domain. This salient observation argues that the WW1 domain most likely prefers a non-bulky residue at the +5 position. In short, our thermodynamic analysis shows that the WW domains of YAP2 bind to ErbB4 peptides in a differential manner with varying contribution of non-consensus residues within and flanking the PPXY motifs to the overall free energy of binding.
Structural models provide physical basis for the binding of WW domains of YAP2 to PPXY motifs within ErbB4
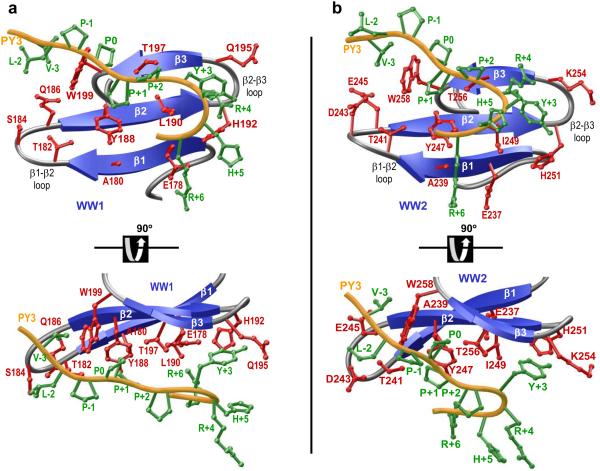
Our data presented above suggest that both WW domains of YAP2 bind to ErbB4 peptides in a manner that is more or less indistinguishable from each other. In order to understand the physical basis of such shared mode of binding, we modeled atomic structures of WW1 and WW2 domains in complex with ErbB4_PY3 peptide (Figure 4). Notably, these modeled structures were built on the basis of four known NMR structures of WW domains of YAP bound to PPXY-containing peptides in a multi-template alignment fashion [34, 37, 38]. Thus, the accuracy of these models can be relied upon with a high degree of confidence. Our models show that the PPXY peptide roughly adopts the PPII-helical conformation and binds within the hydrophobic groove on the concave face of the triple-stranded β-sheet fold of the WW domains in a canonical manner [31-34, 37, 38]. Notably, the C-terminus of the peptide undergoes a sharp 180°-bend so as to fold back onto the WW domains, a feature that somewhat mimics the formation of β-hairpin conformation observed in the binding of the template Smad peptides to YAP WW domains [37, 38]. More importantly, in agreement with our thermodynamic data presented above, only the consensus residues within the PPXY motif appear to be engaged in key intermolecular van der Waals contacts and hydrogen bonding with specific residues lining the hydrophobic groove of the WW domains (Figures 4a and 4b). In particular, the engagement of ErbB4_PY3 peptide in such intermolecular contacts with the WW1 domain is almost indistinguishable from that observed with the WW2 domain (Figures 4a and 4b). Thus, while the pyrrolidine moiety of P0, the first proline within the PPXY motif, stacks against the indole sidechain of W199 in WW1 domain, this contact is fully conserved due to the presence of W258 in the structurally-equivalent position within WW2 domain. In a similar manner, while the pyrrolidine moiety of P+1 is sandwiched by the sidechains of Y188/T197 in WW1 domain, the presence of Y247/T256 at the structurally-equivalent positions in WW2 domains extends the fidelity of this contact. Finally, while the L190/H192/Q195 trio in WW1 domain escorts the phenyl moiety of Y+3, this role is fulfilled by the relatively well-conserved structurally-equivalent I249/H251/K254 trio in WW2 domain. It is important to note here that, although within close proximity, the Hη atom of Y+3 within the ErbB4_PY3 peptide does not appear to engage in hydrogen bonding contacts with Nδ1 atom of H192 and H251 located within WW1 and WW2 domains, respectively. However, it is more likely that the Hη atom of Y+3 may hydrogen bond to the backbone O atom of H192/H251.
Figure 4.
Structural models of WW1 (a) and WW2 (b) domains of YAP2 in complex with ErbB4_PY3 peptide containing the PPXY motif. The β-strands in the WW domains are shown in blue with loops depicted in gray and the peptide is colored yellow. Note that two orientations related by a 90°-rotation about the horizontal axis are depicted for the inquisitive eye. The sidechain moieties of all residues, including the PPXY motif (which corresponds to P0, P+1 and Y+3), within the bound peptide are shown in green. For the WW domains, the sidechain moieties colored in red denote all residues pointing toward the peptide on the concave side.
While non-consensus residues within and flanking the PPXY motif do not appear to engage in any discernable contacts with residues lining the concave binding groove of the WW domains, three flanking residues draw particular attention. Firstly, V-3 within the PY3 peptide appears to be within van der Waals distance of the indole moiety of W199 in WW1 domain—a feat that is also replicated via W258 in WW2 domain. Secondly, L-2 within the PY3 peptide lies within van der Waals distance of Q186 within WW1 domain but this potential contact does not appear to be reproduced in the case of the WW2 domain. This distinguishing feature may in part result from the fact that the neutral Q186 within the WW1 domain is substituted by the acidic E245 at the structurally-equivalent position in WW2 domain. Thirdly, R+6 within the PY3 peptide seemingly lies within ion pairing and/or hydrogen bonding distance of E178 in WW1 domain. While the presence of E237 at the structurally-equivalent position ensures conservation of a glutamate within the WW2 domain, R+6 points away rather than toward E237. Given that these potential contacts made by the above-mentioned flanking residues largely reside on the periphery of WW domains, they are likely to represent transient rather than stable WW-peptide interactions in agreement with our thermodynamic data. Nevertheless, the varying contribution of flanking residues to the stabilization of PY3 peptide is likely to account for its differential binding to WW1 and WW2 domains (Table 1).
In sum, our structural models provide the physical basis for the binding of WW domains of YAP2 to ErbB4_PY3 peptide, and by extension to other ErbB4 peptides, with very similar affinities. Although non-consensus residues within and flanking the PPXY motif make no discernable contacts with any residues within the WW domains, they may be important for stabilizing the PPII-helical conformation of the peptide, while others may be destabilizing either through their engagement in unfavorable contacts or by simply compromising the peptide conformation that best fits the hydrophobic groove within the WW domains. Such a scenario could account for the differential binding of each WW domain to ErbB4 peptides as noted in Table 1.
WW domains of YAP2 display distinct conformational dynamics in complex with PPXY motifs within ErbB4
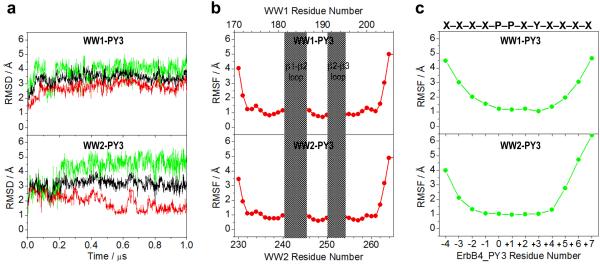
To further understand the molecular basis of the binding of WW domains of YAP2 to PPXY motifs within ErbB4, we next conducted MD simulations on the structural models of WW domains of YAP2 bound to ErbB4_PY3 peptide over a μs timescale (Figure 5). It is noteworthy that these modeled structures were built on the basis of four known NMR structures of WW domains of YAP bound to PPXY-containing peptides in a multi-template alignment fashion [34, 37, 38]. Thus, the accuracy of these starting models used in our MD simulations can be relied upon with a high degree of confidence and the associated dynamics should be reflective of the WW-peptide interactions at atomic level. Moreover, the choice of the starting structure in MD simulations is also important. Toward this goal, a total of 100 atomic models were calculated and the structure with the lowest energy, as judged by the MODELLER Objective Function, was selected for MD analysis. As shown in Figure 5a, the MD trajectories reveal that both WW domains in complex with ErbB4_PY3 peptide reach structural equilibrium with a root mean square deviation (RMSD) for the backbone atoms of just over 3Å. This implies that both complexes are relatively flexible and that their overall dynamic behavior is comparable with no significant differences in agreement with our thermodynamic and structural data presented above. In order to further compare their dynamic behavior, we next deconvoluted the overall RMSD of each WW-peptide complex into its constituent components, namely the WW domain and the ErbB4_PY3 peptide (Figure 5a). Such analysis shows that while both WW1 domain and peptide rapidly reach structural equilibrium and exhibit remarkably similar dynamics to the overall WW1-peptide complex, the WW2 domain and its peptide counterpart follow significantly different trajectories compared to the WW2-peptide complex. Thus, while the peptide in association with WW2 domain appears to be somewhat destabilized relative to the WW2-peptide complex, this is largely offset by an equal but opposite behavior of WW2 domain. In particular, the WW2 domain undergoes slow dynamic relaxation upon binding to the peptide and becomes substantially more stabilized with an equilibrium RMSD value hovering just above 1Å. This argues that while both WW1 and peptide contribute more or less equally to the instability of WW1-peptide complex, the flexibility of WW2-peptide complex is largely due to the unstable peptide. These observations clearly suggest that while the overall dynamics of both WW-peptide complexes are comparable at equilibrium, the WW2 domain undergoes substantial relaxation to attain a thermodynamically more stable conformation upon ligand binding.
Figure 5.
Conformational dynamics as probed through MD simulations conducted on WW1 and WW2 domains of YAP2 in complex with ErbB4_PY3 peptide containing the PPXY motif. (a) RMSD of backbone atoms (N, Cα and C) within each simulated structure relative to the initial modeled structure of WW1 (top panel) and WW2 (bottom panel) domains in complex with ErbB4_PY3 peptide as a function of simulation time. Note that the overall RMSD for each WW-peptide complex (black) is deconvoluted into the WW domain alone (red) and the peptide alone (green). (b) RMSF of backbone atoms (N, Cα and C) averaged over the entire course of corresponding MD trajectory of WW1 (top panel) and WW2 (bottom panel) domains in complex with ErbB4_PY3 peptide as a function of residue number within each WW domain. The shaded vertical rectangular boxes denote residues located within the β1-β2 and β2-β3 loops. (c) RMSF of backbone atoms (N, Cα and C) averaged over the entire course of corresponding MD trajectory of WW1 (top panel) and WW2 (bottom panel) domains in complex with ErbB4_PY3 peptide as a function of residue number within the peptide (see Figure 1a for nomenclature). The PPXY motif and the flanking residues are overlayed for reference.
An alternative means to assess mobility and stability of macromolecular complexes is through an assessment of the root mean square fluctuation (RMSF) of specific atoms over the course of MD simulation. Such analysis for the backbone atoms of each residue within both the WW domains and the bound ErbB4_PY3 peptide is provided (Figures 5b and 5c). As expected, residues encompassing the N- and C-termini within both WW domains appear to be relatively destabilized. However, residues within the core region of WW2 domain are somewhat more stabilized relative to their counterparts within the WW1 domain, a feature particularly notable for β1-β2 loop. Thus, the molecular origin of the higher stability of WW2 domain relative to WW1 domain, as noted above, lies in the ability of core resides to adopt a more stable conformation as opposed to the stability of the termini. Unsurprisingly, the RMSF analysis also paints notable differences between the motional behavior of ErbB4_PY3 peptide in complex with WW1 and WW2 domains (Figure 5c), in agreement with the corresponding RMSD analysis presented above. Thus, while residues corresponding to the PPXY motif and those located C-terminal to this consensus sequence display similar fluctuations within both complexes, the C-terminal residues appear to be much more mobile in complex with the WW2 domain relative to WW1 domain. Taken together, our MD analysis lends new insights into how the motional properties of WW-peptide complexes fine tune their thermodynamic stability and binding.
WW1 and WW2 domains of YAP2 in complex with PPXY motifs within ErbB4 display remarkable similarities with respect to the stability of intermolecular contacts
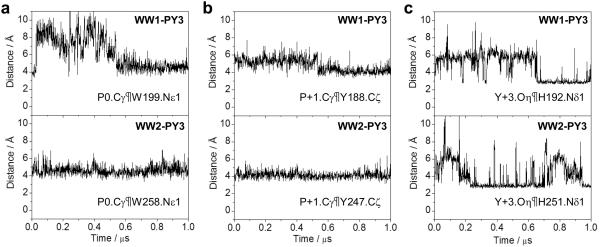
Prompted by their differential dynamics, we next assessed and compared the stability of intermolecular contacts between consensus residues within the PPXY motifs of ErbB4 and their counterparts within WW domains of YAP2 on the basis of our MD simulations (Figure 6). In particular, our structural models presented above suggest that P0, P+1 and Y+3 within the PPXY motif may engage in van der Waals and hydrogen bonding contacts with specific residues located within WW1 and WW2 domains (Figure 4). Consistent with this observation, the intermolecular interaction between Cγ pyrrolidine carbon of P0 within the PPXY motif and Nε1 indole nitrogen of W258 located within WW2 domain appears to be very stable with an equilibrium distance of around 5Å (Figure 6a). In contrast, this structurally analogous interaction between Cγ pyrrolidine carbon of P0 within the PPXY motif and Nε1 indole nitrogen of W199 within WW1 domain undergoes substantial dynamic fluctuations with RMSD reaching as high as 10Å prior to stabilization. A remarkably similar trend is also observed for the intermolecular interaction between Cγ pyrrolidine carbon of P+1 within the PPXY motif and Cζ phenolic carbons of Y188 and Y247 located respectively within WW1 and WW2 domains (Figure 6b). On the other hand, the intermolecular interaction between Oη phenolic oxygen of Y+3 within the PPXY motif and Nδ1 imidazole nitrogens of H192 and H251 located respectively within WW1 and WW2 domains is observed to be highly unstable in both cases (Figure 6c), albeit with subtle differences. This finding implies that the hydrogen bonding contact between Hη atom of Y+3 with Nδ1 atom of H192 and H251 is highly compromised and that such instability is likely to confer a level of flexibility upon both WW complexes.
Figure 6.
Intermolecular distances, as probed through MD simulations, between consensus residues within the PPXY motif of ErbB4_PY3 peptide and residues lining the binding groove within WW1 and WW2 domains of YAP2. (a) Distance between Cγ pyrrolidine carbon of P0 within the PPXY motif and Nε1 indole nitrogens of W199 and W258 located respectively within WW1 (top panel) and WW2 (bottom panel) domains. (b) Distance between Cγ pyrrolidine carbon of P+1 within the PPXY motif and Cζ phenolic carbons of Y188 and Y247 located respectively within WW1 (top panel) and WW2 (bottom panel) domains. (c) Distance between Oη phenolic oxygen of Y+3 within the PPXY motif and Nδ1 imidazole nitrogens of H192 and H251 located respectively within WW1 (top panel) and WW2 (bottom panel) domains.
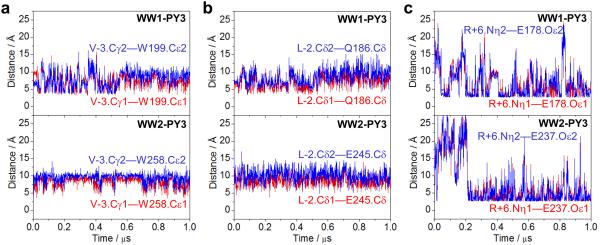
While our thermodynamic analysis reveals that non-consensus residues within and flanking the PPXY motif do not appear to be critical for the binding of WW domains (Tables 2 and 3), it is nonetheless conceivable that these residues may engage in transient interactions. Notably, our structural models presented above suggest that non-consensus residues such as V-3, L-2 and R+6 may play an accessory role in buttressing the WW-peptide complexes (Figure 4). To test this hypothesis further, we also assessed the stability of intermolecular contacts between non-consensus residues within PPXY motif and WW domains (Figure 7). As shown in Figure 7a, the potential van der Waals contact between Cγ1/Cγ2 methyl carbons of V-3 within the PPXY motif and Cδ1/Cδ2 indole carbons of W199 and W258 located respectively within WW1 and WW2 domains is marked by intermolecular distance fluctuating between 5-10Å. A similar trend is also observed for the der Waals contact between Cδ1/Cδ2 methyl carbons of L-2 within the PPXY motif and Cδ carbons of Q186 and E245 located respectively within WW1 and WW2 domains. On the other hand, the potential hydrogen bonding contact between Nη1/Nη2 guanidine nitrogens of R+6 within the PPXY motif and Oε1/Oε2 carbonyl oxygens of E178 and E237 located respectively within WW1 and WW2 domains appears to be highly unstable at the start of the trajectory but slowly achieves some level of stability at equilibrium. Collectively, these observations imply that the intermolecular interactions between non-consensus residues flanking the PPXY motif and WW domains are at best transient in nature in remarkable agreement with our thermodynamic and structural data.
Figure 7.
Intermolecular distances, as probed through MD simulations, between residues flanking the PPXY motif of ErbB4_PY3 peptide and residues lining the binding groove within WW1 and WW2 domains of YAP2. (a) Distance between Cγ1/Cγ2 methyl carbons of V-3 within the PPXY motif and Cδ1/Cδ2 indole carbons of W199 and W258 located respectively within WW1 (top panel) and WW2 (bottom panel) domains. (b) Distance between Cδ1/Cδ2 methyl carbons of L-2 within the PPXY motif and Cδ carbons of Q186 and E245 located respectively within WW1 (top panel) and WW2 (bottom panel) domains. (c) Distance between Nη1/Nη2 guanidine nitrogens of R+6 within the PPXY motif and Oε1/Oε2 carbonyl oxygens of E178 and E237 located respectively within WW1 (top panel) and WW2 (bottom panel) domains.
CONCLUSIONS
While the role of YAP as a transcriptional co-activator of ErbB4 is well-documented [3, 4], little is understood about the molecular basis of this protein-protein interaction. Given their antagonistic roles in a diverse array of cellular signaling, with ErbB4 promoting oncogenic pathways [39-46], while YAP mediating the Hippo tumor suppressor pathway [11-18], the importance of understanding the YAP-ErbB4 interaction at molecular level could not be over-emphasized. Toward this goal, our biophysical analysis presented herein provides new insights into YAP2-ErbB4 interaction. In particular, our study shows that all three PPXY motifs located within the ICD of ErbB4 act as bona fide binding sites for the WW domains of YAP2. However, both WW domains display a slight preference for the C-terminal PY3 motif. Nonetheless, this finding strongly argues that these multiple docking sites within the ICD may positively cooperate with the tandem WW domains to mediate the YAP2-ErbB4 interaction in a multivalent manner. Such a tempting scenario would clearly synergize this key protein-protein interaction and thereby could result in an affinity much higher than that reported here for individual pairs of WW domains and PPXY motifs within the context of cellular environment. Our future efforts will be directed at unraveling the role of multivalent interactions in driving the assembly of YAP2-ErbB4 signaling complex.
Notably, the affinities observed here for the binding of WW domains of YAP2 to ErbB4 are in line with the canonical binding of WW domains to their cognate PPXY motifs, which typically lie in the tens of micromolar regime [34, 47-49, 38]. On the other hand, the fact that non-consensus residues within and flanking the PPXY motifs located within ErbB4 do not appear to be critical for the binding of WW domains of YAP2 seems to be an exception rather than a rule in the context of WW-PPXY interactions. Recent work from our lab has shown that WW domains of YAP optimally bind to the PPXYXG motif within WBP2 signaling adaptor [20], thereby implicating a key role of a non-bulky and flexible glycine residue at the +5 position located outside the PPXY motif. On the other hand, the WW1 domain of WWOX recognizes the PXPPXYY motif within WBP2 with optimal affinity [50], implying that flanking proline and tyrosine respectively at the -2 and +4 positions buttress the WWOX-WBP2 interaction. It is noteworthy that the TVV motif spanning the last three C-terminal residues and located immediately C-terminal to PY3 motif in ErbB4 represents a docking site for PDZ-containing partners of ErbB4 at neuronal synapses [51]. While it has not been possible to explore how the binding of PDZ domains to ErbB4 would affect the binding of WW domains of YAP2 and vice versa in this study, we suspect that they in all probability would bind in a competitive and mutually-exclusive manner due to steric hindrance as a result of the close proximity of TVV and PY3 motifs. In light of the knowledge that several PDZ-containing adaptors have been shown to regulate shuttling of proteins from the cytoplasm to the cell nucleus [52], it would indeed be interesting to conduct detailed analysis on the extent of competition between WW domains of YAP2 and PDZ proteins for binding to ErbB4 and how such competition affects cellular homeostasis.
Importantly, our structural and dynamic analysis of WW domains in complex with PPXY motifs lays the framework for understanding YAP2-ErbB4 interaction at atomic level with potential for the design of novel therapies, particularly in light of the increasing role of ErbB4 in the development of human cancer [39]. In this regard, two promising approaches have recently emerged in terms of regulating YAP-ligand complexes. Firstly, in silico predictions have suggested two small compounds with potential to disrupt WW–peptide complexes of YAP though they have not been verified experimentally. These include digitoxin and endohedral metallofullerenol [53-55]. While digitoxin is a cardiac glycoside used in the treatment of congestive heart failure and cardiac arrhythmia [56], endohedral metallofullerenol has been shown to inhibit tumor growth [57]. Since these are small molecules with hydrophobic character they spontaneously cross cell membrane and thus harbor great potential for the development of anti-cancer therapies. Importantly, our work bears implications on the optimization of these small compounds or development thereof of similar compounds specifically targeting the YAP2-ErbB4 interaction in cancer cells. Secondly, exogenous expression of WW1 domain of YAP2 has been shown to inhibit GAG-PPXY mediated budding of Rous sarcoma virus [58]. Moreover, a growing body of data suggest that the new adenovirus-based vectors with PPXY-containing dodecahedron proteins may provide an efficient system for delivering WW domains to cells at high efficiency [59, 60]. With these new tools at our disposal, it may also become feasible to deliver engineered WW domains to cancer cells so as to inhibit the YAP2-ErbB4 interaction.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health Grant R01-GM083897 and funds from the USylvester Braman Family Breast Cancer Institute (to AF), and by Breast Cancer Coalition grants (RFA #50709 & RFA #60707) from the Department of Health of Pennsylvania (to MS). CBM is a recipient of a postdoctoral fellowship from the National Institutes of Health (Award# T32-CA119929).
ABBREVIATIONS
- ErbB4
Erythroblastic (Erb) leukemia viral oncogene homolog B4
- ITC
Isothermal titration calorimetry
- ITCH
Ubiquitin ligase itchy homolog
- LIC
Ligation-independent cloning
- MM
Molecular modeling
- PPII
Polyproline type II (helix)
- RTK
Receptor tyrosine kinase
- SEC
Size-exclusion chromatography
- SH3
Src homology 3
- SLS
Static light scattering
- TPA
12-O-tetradecanoylphorbol-13-acetate
- YAP
YES-associated protein
- WWOX
WW-containing oxidoreductase
REFERENCES
- 1.Ni CY, Murphy MP, Golde TE, Carpenter G. gamma-Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science. 2001;294:2179–2181. doi: 10.1126/science.1065412. [DOI] [PubMed] [Google Scholar]
- 2.Lee HJ, Jung KM, Huang YZ, Bennett LB, Lee JS, Mei L, Kim TW. Presenilin-dependent gamma-secretase-like intramembrane cleavage of ErbB4. J. Biol. Chem. 2002;277:6318–6323. doi: 10.1074/jbc.M110371200. [DOI] [PubMed] [Google Scholar]
- 3.Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J. Biol. Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- 4.Omerovic J, Puggioni EM, Napoletano S, Visco V, Fraioli R, Frati L, Gulino A, Alimandi M. Ligand-regulated association of ErbB-4 to the transcriptional co-activator YAP65 controls transcription at the nuclear level. Exp. Cell. Res. 2004;294:469–479. doi: 10.1016/j.yexcr.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Aqeilan RI, Donati V, Palamarchuk A, Trapasso F, Kaou M, Pekarsky Y, Sudol M, Croce CM. WW domain-containing proteins, WWOX and YAP, compete for interaction with ErbB-4 and modulate its transcriptional function. Cancer Res. 2005;65:6764–6772. doi: 10.1158/0008-5472.CAN-05-1150. [DOI] [PubMed] [Google Scholar]
- 6.Omerovic J, Santangelo L, Puggioni EM, Marrocco J, Dall'Armi C, Palumbo C, Belleudi F, Di Marcotullio L, Frati L, Torrisi MR, Cesareni G, Gulino A, Alimandi M. The E3 ligase Aip4/Itch ubiquitinates and targets ErbB-4 for degradation. FASEB J. 2007;21:2849–2862. doi: 10.1096/fj.06-7925com. [DOI] [PubMed] [Google Scholar]
- 7.Hoeing K, Zscheppang K, Mujahid S, Murray S, Volpe MV, Dammann CE, Nielsen HC. Presenilin-1 processing of ErbB4 in fetal type II cells is necessary for control of fetal lung maturation. Biochim. Biophys. Acta. 2011;1813:480–491. doi: 10.1016/j.bbamcr.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sudol M. Yes-associated protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the Yes proto-oncogene product. Oncogene. 1994;9:2145–2152. [PubMed] [Google Scholar]
- 9.Gaffney CJ, Oka T, Mazack V, Hilman D, Gat U, Muramatsu T, Inazawa J, Golden A, Carey DJ, Farooq A, Tromp G, Sudol M. Identification, basic characterization and evolutionary analysis of differentially spliced mRNA isoforms of human YAP1 gene. Gene. 2012;509:215–222. doi: 10.1016/j.gene.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sudol M, Bork P, Einbond A, Kastury K, Druck T, Negrini M, Huebner K, Lehman D. Characterization of the mammalian YAP (Yes-associated protein) gene and its role in defining a novel protein module, the WW domain. J. Biol. Chem. 1995;270:14733–14741. doi: 10.1074/jbc.270.24.14733. [DOI] [PubMed] [Google Scholar]
- 11.Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, Zheng P, Ye K, Chinnaiyan A, Halder G, Lai ZC, Guan KL. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hao Y, Chun A, Cheung K, Rashidi B, Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J. Biol. Chem. 2008;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- 14.Zhao B, Ye X, Yu J, Li L, Li W, Li S, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, Guan KL. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertini E, Oka T, Sudol M, Strano S, Blandino G. YAP: at the crossroad between transformation and tumor suppression. Cell Cycle. 2009;8:49–57. doi: 10.4161/cc.8.1.7259. [DOI] [PubMed] [Google Scholar]
- 16.Sudol M. Newcomers to the WW Domain-Mediated Network of the Hippo Tumor Suppressor Pathway. Genes Cancer. 2010;1:1115–1118. doi: 10.1177/1947601911401911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sudol M, Harvey KF. Modularity in the Hippo signaling pathway. Trends Biochem. Sci. 2010;35:627–633. doi: 10.1016/j.tibs.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 18.Salah Z, Aqeilan RI. WW domain interactions regulate the Hippo tumor suppressor pathway. Cell Death Dis. 2011;2:e172. doi: 10.1038/cddis.2011.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morin-Kensicki EM, Boone BN, Howell M, Stonebraker JR, Teed J, Alb JG, Magnuson TR, O'Neal W, Milgram SL. Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol. Cell Biol. 2006;26:77–87. doi: 10.1128/MCB.26.1.77-87.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDonald CB, McIntosh SK, Mikles DC, Bhat V, Deegan BJ, Seldeen KL, Saeed AM, Buffa L, Sudol M, Nawaz Z, Farooq A. Biophysical Analysis of Binding of WW Domains of the YAP2 Transcriptional Regulator to PPXY Motifs within WBP1 and WBP2 Adaptors. Biochemistry. 2011;50:9616–9627. doi: 10.1021/bi201286p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein Identification and Analysis Tools on the ExPASy Server. In: Walker JM, editor. The Proteomics Protocols Handbook. Humana Press; Totowa, New Jersey, USA: 2005. pp. 571–607. [Google Scholar]
- 22.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 23.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative Protein Structure Modeling of Genes and Genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 24.Carson M. Ribbons 2.0. J. Appl. Crystallogr. 1991;24:958–961. [Google Scholar]
- 25.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 26.Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toukan K, Rahman A. Molecular-dynamics study of atomic motions in water. Physical Review B. 1985;31:2643–2648. doi: 10.1103/physrevb.31.2643. [DOI] [PubMed] [Google Scholar]
- 28.Berendsen HJC, Grigera JR, Straatsma TP. The Missing Term in Effective Pair Potentials. J. Phys. Chem. 1987;91:6269–6271. [Google Scholar]
- 29.Darden TA, York D, Pedersen L. Particle mesh Ewald: An N.log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 30.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 31.Macias MJ, Hyvonen M, Baraldi E, Schultz J, Sudol M, Saraste M, Oschkinat H. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature. 1996;382:646–649. doi: 10.1038/382646a0. [DOI] [PubMed] [Google Scholar]
- 32.Huang X, Poy F, Zhang R, Joachimiak A, Sudol M, Eck MJ. Structure of a WW domain containing fragment of dystrophin in complex with beta-dystroglycan. Nat. Struct. Biol. 2000;7:634–638. doi: 10.1038/77923. [DOI] [PubMed] [Google Scholar]
- 33.Kanelis V, Rotin D, Forman-Kay JD. Solution structure of a Nedd4 WW domain-ENaC peptide complex. Nat. Struct. Biol. 2001;8:407–412. doi: 10.1038/87562. [DOI] [PubMed] [Google Scholar]
- 34.Pires JR, Taha-Nejad F, Toepert F, Ast T, Hoffmuller U, Schneider-Mergener J, Kuhne R, Macias MJ, Oschkinat H. Solution structures of the YAP65 WW domain and the variant L30 K in complex with the peptides GTPPPPYTVG, N-(n-octyl)-GPPPY and PLPPY and the application of peptide libraries reveal a minimal binding epitope. J. Mol. Biol. 2001;314:1147–1156. doi: 10.1006/jmbi.2000.5199. [DOI] [PubMed] [Google Scholar]
- 35.Rabanal F, Ludevid MD, Pons M, Giralt E. CD of proline-rich polypeptides: application to the study of the repetitive domain of maize glutelin-2. Biopolymers. 1993;33:1019–1028. doi: 10.1002/bip.360330704. [DOI] [PubMed] [Google Scholar]
- 36.Woody RW. Circular dichroism spectrum of peptides in the poly(Pro)II conformation. J. Am. Chem. Soc. 2009;131:8234–8245. doi: 10.1021/ja901218m. [DOI] [PubMed] [Google Scholar]
- 37.Aragon E, Goerner N, Zaromytidou AI, Xi Q, Escobedo A, Massague J, Macias MJ. A Smad action turnover switch operated by WW domain readers of a phosphoserine code. Genes Dev. 2011;25:1275–1288. doi: 10.1101/gad.2060811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aragon E, Goerner N, Xi Q, Gomes T, Gao S, Massague J, Macias MJ. Structural basis for the versatile interactions of Smad7 with regulator WW domains in TGF-beta Pathways. Structure. 2012;20:1726–1736. doi: 10.1016/j.str.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roskoski R., Jr. The ErbB/HER receptor protein-tyrosine kinases and cancer. Biochem. Biophys. Res. Commun. 2004;319:1–11. doi: 10.1016/j.bbrc.2004.04.150. [DOI] [PubMed] [Google Scholar]
- 40.Burgess AW. EGFR family: structure physiology signalling and therapeutic targets. Growth Factors. 2008;26:263–274. doi: 10.1080/08977190802312844. [DOI] [PubMed] [Google Scholar]
- 41.Sundvall M, Iljin K, Kilpinen S, Sara H, Kallioniemi OP, Elenius K. Role of ErbB4 in breast cancer. J. Mammary Gland Biol. Neoplasia. 2008;13:259–268. doi: 10.1007/s10911-008-9079-3. [DOI] [PubMed] [Google Scholar]
- 42.Sundvall M, Veikkolainen V, Kurppa K, Salah Z, Tvorogov D, van Zoelen EJ, Aqeilan R, Elenius K. Cell death or survival promoted by alternative isoforms of ErbB4. Mol. Biol. Cell. 2010;21:4275–4286. doi: 10.1091/mbc.E10-04-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veikkolainen V, Vaparanta K, Halkilahti K, Iljin K, Sundvall M, Elenius K. Function of ERBB4 is determined by alternative splicing. Cell Cycle. 2011;10:2647–2657. doi: 10.4161/cc.10.16.17194. [DOI] [PubMed] [Google Scholar]
- 44.Hollmen M, Liu P, Kurppa K, Wildiers H, Reinvall I, Vandorpe T, Smeets A, Deraedt K, Vahlberg T, Joensuu H, Leahy DJ, Schoffski P, Elenius K. Proteolytic processing of ErbB4 in breast cancer. PLoS One. 2012;7:e39413. doi: 10.1371/journal.pone.0039413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wadugu B, Kuhn B. The role of neuregulin/ErbB2/ErbB4 signaling in the heart with special focus on effects on cardiomyocyte proliferation. Am. J. Physiol. Heart Circ. Physiol. 2012;302:H2139–2147. doi: 10.1152/ajpheart.00063.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paatero I, Lassus H, Junttila TT, Kaskinen M, Butzow R, Elenius K. CYT-1 isoform of ErbB4 is an independent prognostic factor in serous ovarian cancer and selectively promotes ovarian cancer cell growth in vitro. Gynecol. Oncol. 2013;129:179–187. doi: 10.1016/j.ygyno.2012.12.044. [DOI] [PubMed] [Google Scholar]
- 47.Macias MJ, Wiesner S, Sudol M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 2002;513:30–37. doi: 10.1016/s0014-5793(01)03290-2. [DOI] [PubMed] [Google Scholar]
- 48.Morales B, Ramirez-Espain X, Shaw AZ, Martin-Malpartida P, Yraola F, Sanchez-Tillo E, Farrera C, Celada A, Royo M, Macias MJ. NMR structural studies of the ItchWW3 domain reveal that phosphorylation at T30 inhibits the interaction with PPxY-containing ligands. Structure. 2007;15:473–483. doi: 10.1016/j.str.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 49.Webb C, Upadhyay A, Giuntini F, Eggleston I, Furutani-Seiki M, Ishima R, Bagby S. Structural features and ligand binding properties of tandem WW domains from YAP and TAZ, nuclear effectors of the Hippo pathway. Biochemistry. 2011;50:3300–3309. doi: 10.1021/bi2001888. [DOI] [PubMed] [Google Scholar]
- 50.McDonald CB, Buffa L, Bar-Mag T, Salah Z, Bhat V, Mikles DC, Deegan BJ, Seldeen KL, Malhotra A, Sudol M, Aqeilan RI, Nawaz Z, Farooq A. Biophysical basis of the binding of WWOX tumor suppressor to WBP1 and WBP2 adaptors. J. Mol. Biol. 2012;422:58–74. doi: 10.1016/j.jmb.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia RA, Vasudevan K, Buonanno A. The neuregulin receptor ErbB-4 interacts with PDZ-containing proteins at neuronal synapses. Proc. Natl. Acad. Sci. 2000;97:3596–3601. doi: 10.1073/pnas.070042497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oka T, Remue E, Meerschaert K, Vanloo B, Boucherie C, Gfeller D, Bader GD, Sidhu SS, Vandekerckhove J, Gettemans J, Sudol M. Functional complexes between YAP2 and ZO-2 are PDZ domain-dependent, and regulate YAP2 nuclear localization and signalling. Biochem. J. 2010;432:461–472. doi: 10.1042/BJ20100870. [DOI] [PubMed] [Google Scholar]
- 53.Casey FP, Pihan E, Shields DC. Discovery of small molecule inhibitors of protein-protein interactions using combined ligand and target score normalization. J. Chem. Inf. Model. 2009;49:2708–2717. doi: 10.1021/ci900294x. [DOI] [PubMed] [Google Scholar]
- 54.Kang SG, Huynh T, Zhou R. Non-destructive inhibition of metallofullerenol Gd@C(82)(OH)(22) on WW domain: implication on signal transduction pathway. Sci. Rep. 2012;2:957. doi: 10.1038/srep00957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sudol M, Shields DC, Farooq A. Structures of YAP protein domains reveal promising targets for development of new cancer drugs. Semin. Cell. Dev. Biol. 2012;23:827–833. doi: 10.1016/j.semcdb.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Belz GG, Breithaupt-Grogler K, Osowski U. Treatment of congestive heart failure--current status of use of digitoxin. Eur. J. Clin. Invest. 2001;31(Suppl 2):10–17. doi: 10.1046/j.1365-2362.2001.0310s2010.x. [DOI] [PubMed] [Google Scholar]
- 57.Kang SG, Zhou G, Yang P, Liu Y, Sun B, Huynh T, Meng H, Zhao L, Xing G, Chen C, Zhao Y, Zhou R. Molecular mechanism of pancreatic tumor metastasis inhibition by Gd@C82(OH)22 and its implication for de novo design of nanomedicine. Proc. Natl. Acad. Sci. 2012;109:15431–15436. doi: 10.1073/pnas.1204600109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patnaik A, Wills JW. In vivo interference of Rous sarcoma virus budding by cis expression of a WW domain. J. Virol. 2002;76:2789–2795. doi: 10.1128/JVI.76.6.2789-2795.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garcel A, Gout E, Timmins J, Chroboczek J, Fender P. Protein transduction into human cells by adenovirus dodecahedron using WW domains as universal adaptors. J. Gene. Med. 2006;8:524–531. doi: 10.1002/jgm.862. [DOI] [PubMed] [Google Scholar]
- 60.Villegas-Mendez A, Fender P, Garin MI, Rothe R, Liguori L, Marques B, Lenormand JL. Functional characterisation of the WW minimal domain for delivering therapeutic proteins by adenovirus dodecahedron. PLoS One. 2012;7:e45416. doi: 10.1371/journal.pone.0045416. [DOI] [PMC free article] [PubMed] [Google Scholar]