Abstract
Type 1 diabetes (T1D) shows ~ 40% concordance rate in monozygotic twins (MZ) suggesting a role for environmental factors and/or epigenetic modifications in the etiology of the disease. The aim of our study was to dissect the contribution of epigenetic factors, particularly, DNA methylation (DNAm), to the incomplete penetrance of T1D. We performed DNAm profiling in lymphocyte cell lines from 3 monozygotic (MZ) twin pairs discordant for T1D and 6 MZ twin pairs concordant for the disease using HumanMethylation27 BeadChip. This assay assesses the methylation state of 27,578 CpG sites, mostly located within proximal promoter regions. We identified 88 CpG sites displaying significant methylation changes in all T1D-discordant MZ twin pairs. Functional annotation of the genes with distinct CpG methylation profiles in T1D samples showed differential DNAm of immune response and defense response pathways between affected and unaffected twins. Integration of DNAm data with GWAS data mapped several known T1D associated genes, HLA, INS, IL-2RB, CD226, which showed significant differences in DNAm between affected and unaffected of twins. Our findings suggest that abnormalities of DNA methylation patterns, known to regulate gene transcription, may be involved in the pathogenesis of T1D.
Key Terms: type 1 diabetes, DNA methylation, monozygotic twins, transcriptional regulation
1. INTRODUCTION
Type 1 diabetes (T1D) is one of the commonest autoimmune diseases and has a prevalence in the US of ~0.4% [1]. T1D is characterized by formation of islet specific T-cells and autoantibodies and inflammatory destruction of the beta cells [2], leading to hyperglycemia, and micro- and macro-vascular complications [3]. Genetic susceptibility plays a major role in the etiology of T1D and several T1D genes have been identified; most notably the MHC locus which confers ~50% of the heritable risk for T1D [4]. However, non-genetic factors must play a key role in the etiology of T1D since the average concordance rate of T1D among MZ twins varies between 13–67.7% with an average of 46.2% [5]. Moreover, the incidence of T1D has been doubling every 20 years in the past several decades, a trend that cannot be explained by genetic factors and that suggests strong environmental effects [6]. Therefore, it is likely that non-genetic factors interact with susceptibility genes to trigger the development of T1D. Recently, epigenetic regulation has been proposed as a key mechanism by which environmental influences interact with genetic factors to trigger T1D [7]. However, little is known about the contribution of epigenetic mechanisms to the etiology of T1D.
Along with histone modifications and noncoding RNAs, DNA methylation represents a crucial epigenetic mechanism for regulating gene transcription, and is involved in the transcriptional regulation of a wide variety of genes. DNA is methylated by the addition of methyl groups to cytosine residues usually when they are located before guanine residues. We hypothesized that abnormal DNA methylation may play a role in the etiology of T1D. One of the most powerful methods to analyze the contribution of abnormal DNA methylation to complex diseases is by comparisons among discordant MZ twins [8]. Therefore, we performed a genome-wide DNA methylation analysis in lymphocyte DNA from MZ twins discordant and concordant for T1D. We report here identification of methyaltion changes specific to T1D.
2. MATERIALS AND METHODS
2.1 Patients
The study was approved by the Mount Sinai Institutional Review Board. Global DNAm was assessed in B cell lines from twin pairs discordant and concordant for T1D. The DNA from EBV immortalized B cell lines were obtained from NDRI. All subjects had confirmed diagnosis of type 1 diabetes based on the ADA guidelines [9]. All T1D patients were members of the NDRI T1D family registry and their DNA samples were provided to us deidentified. We studied 9 monozygotic (MZ) twin pairs (n=18 T1D patients): 7 female MZ twin pairs (n = 14), 3 discordant and 4 concordant for T1D; and 2 male MZ twin pairs (n=4), both concordant for T1D. The age at diagnosis was between 1 to 14 years old (median diagnosis age for discordant twins was 7 years old, and for concordant twins 10 years old). Median duration of T1D was 11 years and the range was 1 to 29 years (Table 1).
Table 1.
Age of T1D onset and gender of twin pairs analyzed.
| ID | Age of the onset (years) | Gender | |
|---|---|---|---|
| Twin 1 | Twin 2 | ||
| CT1 | 1 | 1 | F |
| CT2 | 14 | 13 | M |
| CT3 | 10 | 12 | F |
| CT4 | 1.6 | 1.5 | F |
| CT5 | 10 | 11 | M |
| CT6 | 12 | 10 | F |
| DT1 | Un-affected | 7 | F |
| DT2 | Un-affected | 11 | F |
| DT3 | Un-affected | 4 | F |
2.2 Confirmation of monozygotic twinning
To confirm twinning we used the Applied Biosystems Indentifiler assay (Life Technologies, Grand Island, NY).
2.3 DNA methylation and Bioinformatic Data analysis
DNA samples were subjected to methylation analysis using Illumina 27K methyl BeadChip. After raw intensity data were extracted from chip and quantile-normalized, methylation level for each probe was calculated in GenomeStudio (Illumina). Probes with a low signal (detection p>0.05) across all samples were excluded from analysis.
To identify differentially methylation levels in discordant twins, paired LIMMA test was performed between affected and unaffected pairs and p values were corrected for multiple testing by FDR. Top 1000 differentially methylated sites between each affected vs. unaffected discordant twin pair were selected by the fold change with a minimum of 25% methylation level for hyper-methylated sites and their corresponding genes were subjected to the Gene Ontology (GO)(http://www.geneontology.org/) enrichment analysis. The median methylation level of all sites was 6% and 30% of the sites had a methylation level greater than 25%. The distribution of differential methylation sites related to TSS within or without CpG islands were calculated and assessed for deviation from that of all the sites on the chip using Fisher-exact test. The differential methylation sites and enriched GO categories were compared among individual twin pairs to identify the common sites or GO function categories.
To identify differentially methylated sites in concordant affected twins compared to unaffected twins, we applied two approaches: (1) LIMMA test was performed comparing averaged methylation levels of 6 concordant affected twin pairs versus 3 unaffected individuals; (2) Using methylation level of unaffected individuals as the reference, the common differential sites in each concordant twin pair from top 1000 were identified as described above and subjected to GO analysis. Finally, common differential sites from both analyses of discordant twins and concordant twins were identified and visualized in heatmap in TMEV (http://www.tm4.org/mev/).
2.4 Direct sequencing of bisulfite treated DNA
Methylation status of the CpG sites in the Kv2.2 and CCDC3 promoters was determined by cloning and bisulfate sequencing. After bisulfite treatment, DNA was amplified by PCR using primers: Kv2.2: (F) 5′-GGGGTAGGAGATAGGTAGAGGATA-3′ (R) 5′-AACCCTAAATCCAAAACTCTCTAA-3′; CCDC3: (F) 5′-GAAATGTAGAGA AAATGTAAAA-3′ (R) 5′-CCTCCACTCGAAAAACAACTAA-3′. PCR products were cloned into pCR-TOPO plasmid using TOPO TA Cloning Kit (Life Techologies, Grand Island, NY). DNA was sequenced using ABI 3130 Genetic Analyzer. Five to eight clones for each sample were sequenced and their methylation status determined based on protection from conversion of cytosine to uracil residues.
3. RESULTS
3.1 Identification of unique T1D-associated DNA methylation profiles
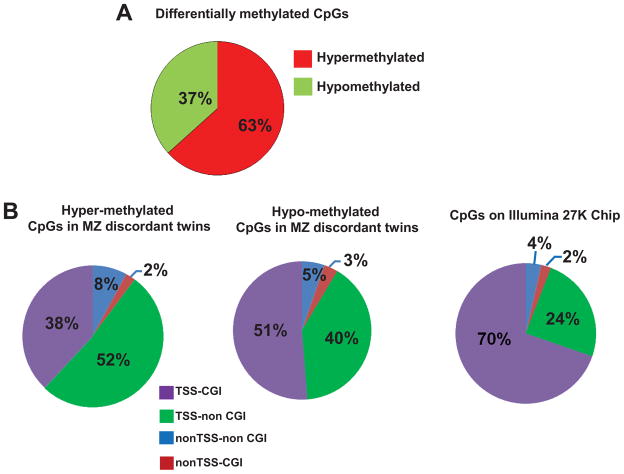
Using the LIMMA paired test we identified 191 hyper- and 115 hypo-methylated sites from at least 2 out of 3 twin pairs including 55 hyper- and 33 hypo- methylated sites in all 3 twin pairs (p = 2.24x10−14 and 3.64x10−8, respectively) (Figures 1A). GO analysis identified immune defense genes and genes involved in cell signaling (Table 2).
Figure 1.
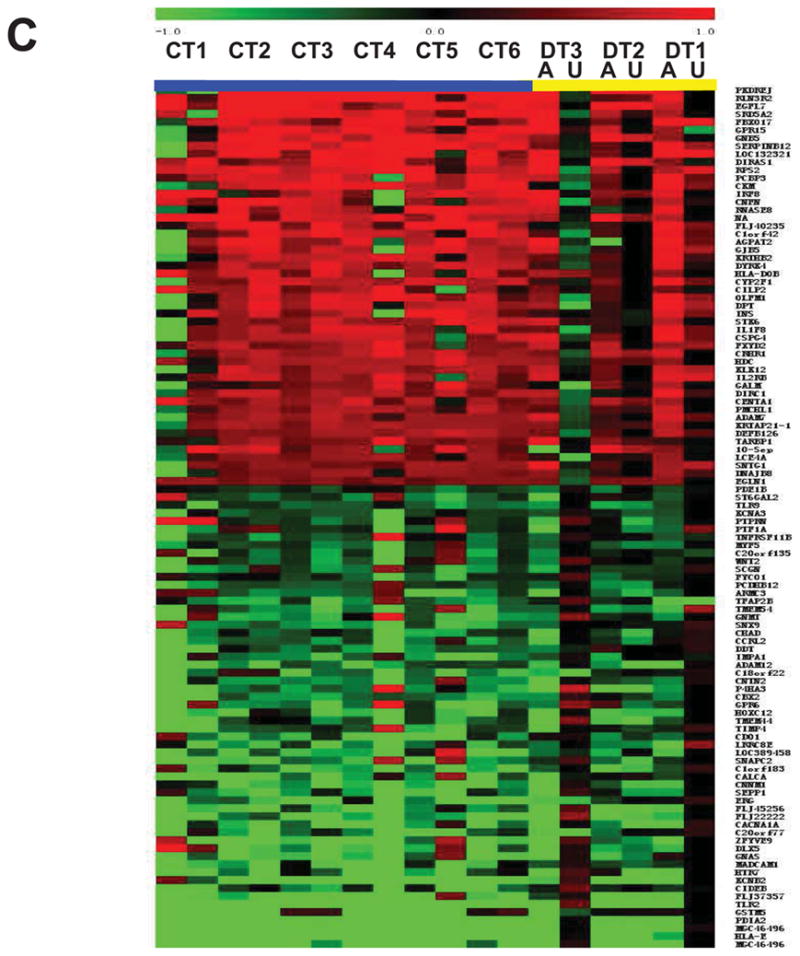
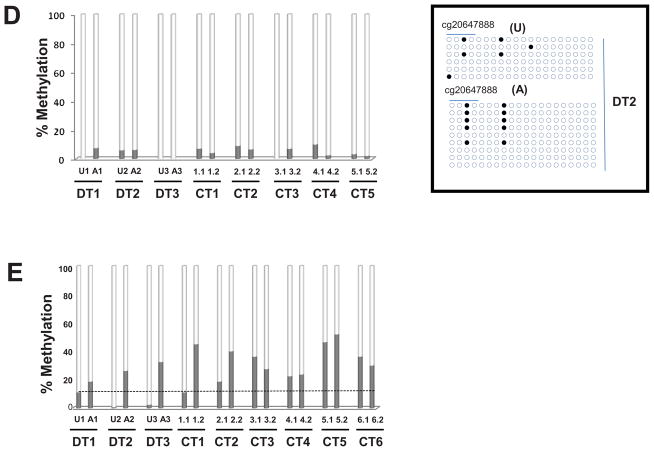
(A) Pie chart showing the 88 CpG sites displaying significant methylation changes in all T1D-discordant MZ twin pairs. More CpG sites were hyper-methylated (55 sites) compared with hypo-methylated (33 sites); (B) Distribution of hyper- and hypo-methylated CpG islands (CGI) across four annotation sets relative to the TSS (± 1 kb from TSS) in discordant twins. TSS-CGI = CGIs present 1Kb up- and down-stream the TSS; TSS-non CGI = no CGIs present 1Kb up- and down-stream the TSS; nonTSS-nonCGI = regions outside TSS with no CGIs; nonTSS-CGI = CGIs present outside TSS ± 1Kb; (C) Heatmap of differentially methylated CpG loci showing different methylation patterns in affected vs. unaffected individuals for both discordant (yellow line) and concordant (blue line) MZ twin pairs; Median methylation level of unaffected twins was used as baseline. 1.0 and −1.0 represents 2-fold increase or decrease methylation respectively compared to the baseline. A, affected twin; U, unaffected twin; CT, concordant MZ twin pairs; DT, discordant MZ twin pairs; (D) DNA methylation levels in the CCDC3 gene: 20 CpGs from promoter CGI were analyzed in 8 twin pairs. All samples showed reduced DNA methylation; the levels of methylation in the cg20647888 array probe did not differ from the overall level of the entire CGI. Insert: an example of bisulfite sequencing of multiple clones from the DT2 twin pair. Schematic representation of the methylation status of each CpG. Black and white circles indicate methylated and unmethyalted CpGs, respectively. Horizontal line indicates methylation sites at the 4 CpGs present on cg20647888. (E) Confirmation of the DNAm levels in the Kv2.2 gene in all twin pairs by direct sequencing of bisulfite modified DNA. 10 CpGs form Kv2.2 CGI were analyzed in all twin pairs. With exception of one CT pair (CT1) all affected individuals had increased CpG methylation within Kv2.2 promoter; U = unaffected twin, A = affected twin.
Table 2.
Top biological pathways attributed by Gene ontology annotations associated with differentially methylated CpGs in discordant twin pairs.
| Hypermethylated CpGs | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| GO ID | Go attribute | #genes within GO attribute | Pair 1 | Pair 2 | Pair 3 | Common genes | |||
| # Genes | p-value | # Genes | p-value | # Genes | p-value | ||||
| GO:0006952 | defense response | 683 | 60 | 9.39E-07 | 51 | 2.5E-04 | 65 | 4.46E-08 | APOA4, C4BPA, CLEC7A, SFTPD |
| GO:0006955 | immune response | 754 | 61 | 7.18E-05 | 59 | 3.35E-04 | 70 | 3.84E-08 | APOA4, C4BPA, CD1E, CLEC7A, CYSLTR2, SFTPD |
| GO:0048878 | chemical homeostasis | 538 | 49 | 2.16E-05 | 51 | 9.83E-06 | 47 | 3.71E-05 | APOA4, CLDN11, RYR1, SFTPD |
| Hypomethylated CpGs | |||||||||
| GO:0007267 | cell-cell signaling | 620 | 60 | 3.17E-08 | 51 | 4.55E-05 | 56 | 1.04E-06 | APITD1, CALCA, FKBP1B, GATA4, MTNR1B, SEMA3B, SLC12A5 |
To identify differentially methylated CpGs within CpG islands (CGIs) we plotted the T1D-associated Methylation Variable Positions (MVPs) at Transcription Start Sites (TSS) of genes (± 1 Kb) and sorted by the presence of CGIs. Interestingly, both hyper- and hypo-methylated T1D-associated MVPs were more prevalent in the non CGI regions (60% and 45%, respectively) compared to CpGs represented on the chip (28%) (enrichment p-value < 1x10−10, Figure 1B).
Comparing concordant affected twin pairs to unaffected twins, we identified 110 hyper- and 132 hypo-methylated sites in at least 4 out of 6 affected twin pairs. Of these, 2 hyper- and 10 hypo-methylated were common in all 6 twin concordant affected pairs. GO terms included acute inflammatory response, immune response, and defense responses for hyper-methylated sites and cell adhesion and signaling for hypo-methylated sites (p<0.05) (Supplementary Table 1). Finally we combined the differentially methylated sites of affected concordant and discordant twins and found 109 common sites (51 hyper- and 58 hypo-methylated), including sites for important T1D genes (eg. HLA, INS, see below) (Figure 1C).
3.2 Confirmation of T1D-associated DNA methylation profiles by cloning and bisulfite sequencing
To confirm the accuracy of the methylation arrays we performed bisulfite DNA sequencing of two representative genes, the coiled-coil domain containing 3 (CCDC3) and Kv2.2 genes. We cloned and bisulfite sequenced the fragments corresponding to the oligonucleotide probes of the methylation arrays. This approach allowed us to test if information provided by the array probes, which cover ~ 50 nucleotides containing ~ 1–8 CpGs, can be extrapolated to the entire CGI within a gene promoter. The CCDC3 gene was found to be uniformly hypomethylated in all twins by the arrays. Analysis of 20 CpGs in 4–8 clones from all twins’ DNA showed no differences between the degree of methylation of the GpG represented on the probe and the neighboring CpGs, confirming that the array results can be extrapolated to neighboring CpG’s (Figure 1D).
Next we analyzed the methylation levels of 10 CpGs within the promoter of the Kv2.2 gene which was found to be differentially methylated by the arrays in all T1D-affected twins compared to unaffected twins. Kv2.2 encodes for a voltage-gated potassium channel with a role in hormone secretion [10;11]. Cloning and bisulfite sequencing confirmed the array analysis (Figure 1E).
3.3 Pathway analysis of differentially methylated CpGs in T1D
The most significant pathways identified for hypermethylated CpG sites common for all 3 T1D discordant twin pairs were defense response and immune response pathways (Table 1). Only one GO functional category, cell-signaling pathway, was found to be significantly hypomethylated in the affected twins in all T1D discordant pairs.
3.4 Integration of DNA methylation data with genome wide association (GWAS) data
We identified 6 genes, known to be associated with T1D from GWAS studies that showed differential CpG methylation between T1D affected and non-affected twins. Of these, HLA-E was the only T1D-associated gene showing hypomethylated CpG sites in affected twins; the differential methylation was in the promoter CGI. Three genes, HLA-DOB, INS and IL-2RB, showed hypermethylated MVPs in the majority of affected discordant and concordant twins (at least 2 out of 3 of discordant twin pairs and at least 4 out of 6 concordant twin pairs), and two T1D genes, HLA-DQA2 and CD226, showed hypermethylated CpG sites in affected discordant twins.
4. DISCUSSION
Genome wide association studies (GWAS) have identified > 40 T1D loci [5;12]. How these polymorphisms interact with environmental factors to trigger T1D is unknown, but recent evidence suggests that epigenetic mechanisms play a key role [7]. To study the contribution of epigenetics, in particular DNA methylation, to the etiology of T1D we analyzed global methylation profiles in B-cells DNA from MZ twin pairs concordant and discordant for the disease. DNA methylation profiles showed more similarity among T1D affected twins than to their unaffected MZ twins, suggesting that epigenetic modifications are critical to the etiology of the disease, and contributed to the discordance in discordant MZ twins.
So far, several studies have examined the role of DNA methylation in the development of autoimmune diseases. For example, in rheumatoid arthritis DNA methylation has been shown to regulate expression of the CXCL12 gene in synovial fibroblasts [13]. In addition, methylation status at the promoters of IL6 [14] and death receptor 3 genes [15] was associated with disease. Analysis of DNA methylation in MZ twins discordant for systemic lupus erythematosus (SLE) found widespread changes in the DNA methylation status of genes with immune functions in SLE twins compared with their healthy co-twins [16]. Only a few studies have addressed the role of DNA methylation in the pathogenesis of T1D. Rakyan et al, showed that T1D-associated methylation variable positions (T1D-MVPs) are associated with the presence of islet autoantibodies and that these modifications are detectable before the diagnosis of disease [17]. A recent study on global DNA methylation in one set of MZ quadruplets discordant for T1D showed MVPs in diabetes-associated genes (INS-IGF2, SH2B3, MEG3 and ORMDL3) in the affected twins [18]. Genome-wide DNA methylation for T1D-associated nephropathy identified 19 potential CpG sites associated with risk for the disease [19]. Our results showing association of DNA methylation patterns with T1D in discordant MZ twins are consistent with these studies, and support a major role for differential methylation patterns in the etiology of T1D.
In order to analyze the role of genetic-epigenetic interactions in the etiology of T1D we integrated genome-wide methylation profiles with T1D GWAS data to look for overlapping gene-loci. Intriguingly, we mapped differentially methylated CpG sites at 6 major T1D susceptibility genes. One of these was the MHC locus, known to confer the strongest risk for T1D [5]; HLA-DOB and HLA-DQA2 were hypermethylated in T1D affected twins and HLA-E was hypomethylated. Rakyan et al also reported T1D-associated differential methylation at the MHC II locus [17]. These data suggest that differences in DNA methylation at multiple genes in the MHC region may contribute to the etiology of T1D. Another major T1D gene, the insulin (INS) gene, showed differential methylation, within intron 1 (+558 bp from TSS, cg13993218), consistent with another study that reported methylation specific patterns at CpG sites upstream of the INS gene TSS in T1D patients [20]. Interestingly, differential MVPs at the INS gene were shown to be influenced by the genotype of rs689, a SNP associated with T1D and located +22 bp from the TSS [20]. A potential limitation of our study is related to the imbalance between female and male twin pairs we analyzed (7 pairs of female and 2 pairs of male twins). Indeed, a minor male excess in children diagnosed with T1D under age 14 years has been reported in populations of high incidence (European origin) [21;22]. To test if gender bias could affect our results we performed LIMMA analysis by comparing DNAm profiles of females vs. male twins. This analysis did not show any significant DNAm changes related with gender. Another limitation of our study is the small number of twin pairs analyzed. Therefore, our data need confirmation in larger cohorts. In conclusion we used global DNA methylation analysis to identify Methylation Variable Positions (MVPs) that are associated with T1D in MZ twins discordant for the disease. We identified DNA methylation patterns at several T1D-associated genes and immunoregulatory pathways that were distinct in affected compared to unaffected MZ twins. If replicated our findings can provide a foundation for development of epigenetic markers in T1D.
Supplementary Material
Biological pathways, determined by Gene ontology annotations, that are associated with differentially methylated CpGs in concordant twin pairs.
Highlights.
we analyzed global DNA methylation in MZ twins discordant and concordant for T1D
we found an overall increase in DNAm in affected twins
increased DNAm was found in genes of defense and immune response pathways
we identified DNAm changes in MHC genes and other T1D susceptibility genes
DNAm modifications may be involved in the etiology of T1D
Acknowledgments
We thank the National Disease Research Interchange (NDRI, Philadelphia, PA) for providing us with the DNA samples of the diabetes twins. This work was supported in part by grants DK061659, DK067555 & DK073681 from NIDDK (to YT). In addition this material is based upon work supported in part by the Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, and by the VA Biomedical Laboratory Research and Development Merit Award 1I01BX002031 (to YT).
Footnotes
Conflict of interest:
There are no conflicts of interest to disclose associated with this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.LaPorte RE, Fishbein HA, Drash AL, Kuller LH, Schneider BB, Orchard TJ, Wagner DK. The Pittsburgh Insulin-dependent diabetes mellitus (IDDM) registry. The incidence of Insulin-dependent diabetes mellitus in Allegheny County, Pennsylvania (1965–1976) Diabetes. 1981;30:279–284. doi: 10.2337/diab.30.4.279. [DOI] [PubMed] [Google Scholar]
- 2.Falorni A, Kockum CB, Sanjeevi CB, Lernmark A. The pathogenesis of insulin-dependent diabetes mellitus. Baillière’s Clin Endocrinol Metab. 1995;9:25–46. doi: 10.1016/s0950-351x(95)80803-5. [DOI] [PubMed] [Google Scholar]
- 3.The Diabetes Control and Complications Trial Research Group. The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. The Diabetes Control and Complications Trial Research Group. N Engl J Med. 1993;329:977–986. doi: 10.1056/NEJM199309303291401. [DOI] [PubMed] [Google Scholar]
- 4.Jahromi MM, Eisenbarth GS. Genetic determinants of type 1 diabetes across populations. Ann N Y Acad Sci. 2006;1079:289–299. doi: 10.1196/annals.1375.044. [DOI] [PubMed] [Google Scholar]
- 5.Huber A, Menconi F, Corathers S, Jacobson EM, Tomer Y. Joint genetic susceptibility to type 1 diabetes and autoimmune thyroiditis: from epidemiology to mechanisms. Endocr Rev. 2008;29:697–725. doi: 10.1210/er.2008-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hasham A, Tomer Y. The recent rise in the frequency of type 1 diabetes: who pulled the trigger? J Autoimmun. 2011;37:1–2. doi: 10.1016/j.jaut.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macfarlane AJ, Strom A, Scott FW. Epigenetics: deciphering how environmental factors may modify autoimmune type 1 diabetes. Mamm Genome. 2009;20:624–632. doi: 10.1007/s00335-009-9213-6. [DOI] [PubMed] [Google Scholar]
- 8.Bell JT, Spector TD. A twin approach to unraveling epigenetics. Trends Genet. 2011;27:116–125. doi: 10.1016/j.tig.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Report of the Expert Committee on the Diagnosis and Classification of Diabetes Mellitus. Diabetes Care. 1997;20:1183–1197. doi: 10.2337/diacare.20.7.1183. [DOI] [PubMed] [Google Scholar]
- 10.Li XN, Herrington J, Petrov A, Ge L, Eiermann G, Xiong Y, Jensen MV, Hohmeier HE, Newgard CB, Garcia ML, Wagner M, Zhang BB, Thornberry NA, Howard AD, Kaczorowski GJ, Zhou YP. The role of voltage-gated potassium channels Kv2.1 and Kv2. 2 in the regulation of insulin and somatostatin release from pancreatic islets. J Pharmacol Exp Ther. 2013;344:407–416. doi: 10.1124/jpet.112.199083. [DOI] [PubMed] [Google Scholar]
- 11.Jensen MV, Haldeman JM, Zhang H, Lu D, Huising MO, Vale WW, Hohmeier HE, Rosenberg P, Newgard CB. Control of Voltage-gated Potassium Channel Kv2.2 Expression by Pyruvate-Isocitrate Cycling Regulates Glucose-stimulated Insulin Secretion. J Biol Chem. 2013;288:23128–23140. doi: 10.1074/jbc.M113.491654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polychronakos C, Li Q. Understanding type 1 diabetes through genetics: advances and prospects. Nat Rev Genet. 2011;12:781–792. doi: 10.1038/nrg3069. [DOI] [PubMed] [Google Scholar]
- 13.Karouzakis E, Gay RE, Michel BA, Gay S, Neidhart M. DNA hypomethylation in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum. 2009;60:3613–3622. doi: 10.1002/art.25018. [DOI] [PubMed] [Google Scholar]
- 14.Nile CJ, Read RC, Akil M, Duff GW, Wilson AG. Methylation status of a single CpG site in the IL6 promoter is related to IL6 messenger RNA levels and rheumatoid arthritis. Arthritis Rheum. 2008;58:2686–2693. doi: 10.1002/art.23758. [DOI] [PubMed] [Google Scholar]
- 15.Takami N, Osawa K, Miura Y, Komai K, Taniguchi M, Shiraishi M, et al. Hypermethylated promoter region of DR3, the death receptor 3 gene, in rheumatoid arthritis synovial cells. Arthritis Rheum. 2006;54:779–787. doi: 10.1002/art.21637. [DOI] [PubMed] [Google Scholar]
- 16.Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20:170–179. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rakyan VK, Beyan H, Down TA, Hawa MI, Maslau S, Aden D, et al. Identification of type 1 diabetes-associated DNA methylation variable positions that precede disease diagnosis. PLoS Genet. 2011;7:e1002300. doi: 10.1371/journal.pgen.1002300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Disanto G, Vcelakova J, Pakpoor J, Elangovan RI, Sumnik Z, Ulmannova T, et al. DNA methylation in monozygotic quadruplets affected by type 1 diabetes. Diabetologia. 2013;56:2093–2095. doi: 10.1007/s00125-013-2972-3. [DOI] [PubMed] [Google Scholar]
- 19.Bell CG, Teschendorff AE, Rakyan VK, Maxwell AP, Beck S, Savage DA. Genome-wide DNA methylation analysis for diabetic nephropathy in type 1 diabetes mellitus. BMC Med Genomics. 2010;3:33. doi: 10.1186/1755-8794-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fradin D, Le FS, Mille C, Naoui N, Groves C, Zelenika D, et al. Association of the CpG methylation pattern of the proximal insulin gene promoter with type 1 diabetes. PLoS One. 2012;7:e36278. doi: 10.1371/journal.pone.0036278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gale EA, Gillespie KM. Diabetes and gender. Diabetologia. 2001;44:3–15. doi: 10.1007/s001250051573. [DOI] [PubMed] [Google Scholar]
- 22.Karvonen M, Pitkaniemi M, Pitkaniemi J, Kohtamaki K, Tajima N, Tuomilehto J. Sex difference in the incidence of insulin-dependent diabetes mellitus: an analysis of the recent epidemiological data. World Health Organization DIAMOND Project Group Diabetes. Metab Rev. 1997;13:275–291. doi: 10.1002/(sici)1099-0895(199712)13:4<275::aid-dmr197>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Biological pathways, determined by Gene ontology annotations, that are associated with differentially methylated CpGs in concordant twin pairs.