Abstract
Chlamydia trachomatis infection is the most common sexually transmitted bacterial infection in the United States and a significant health burden worldwide. Protection from Chlamydia infection in the genital mucosa is dependent on interferon-gamma (IFN-γ) derived from CD4+ Th1 cells. These CD4+ T cells must home successfully to the genital tract to exert their effector function and decrease C. trachomatis burden. Although adhesion receptors expressed by CD4+ T cells in the genital tract have been characterized, the integrin receptor required for Chlamydia-specific CD4+ T cell-mediated protection has not been explored. Here we demonstrate that C. trachomatis infection of the upper genital tract results in recruitment of Chlamydia-specific CD4+ T cells robustly expressing the integrin α4β1. Interfering with α4β1, but not α4β7, function resulted in defective CD4+ T cell trafficking to the uterus and high bacterial load. We conclude that integrin α4β1 is necessary for CD4+ T cell-mediated protection against C. trachomatis infection in the genital mucosa. By identifying homing molecules required for successful CD4+ T cell trafficking to C. trachomatis infected tissues, we will be better equipped to design vaccines that elicit sterilizing, long lasting immunity without inducing immune pathologies in the upper genital tract.
INTRODUCTION
Chlamydia trachomatis is the most common cause of bacterial sexually transmitted infection in the United States and the leading cause of preventable blindness worldwide (1). C. trachomatis is an obligate intracellular pathogen that infects conjunctival and genital tract epithelial cells. In the upper genital tract, complications from C. trachomatis infection include pelvic inflammatory disease, ectopic pregnancy and infertility (2, 3). The high frequency of infection, low incidence of acquired immunity and lack of an effective vaccine make C. trachomatis a continuing public health concern.
Protection of the genital mucosa from C. trachomatis is dependent on the production of IFN-γ (4). IFN-γ protects through the upregulation of IDO, NOS and IRGs that interfere with various aspects of the pathogen’s developmental cycle and reduce growth (5–8). Mice that are deficient in IFN-γ production have delayed resolution of infection in the genital mucosa (9). CD4+ T cells must produce IFN-γ in order to mediate protection, as transfer of Chlamydia-specific CD4+ T cells only protect naïve mice against challenge when IFN-γ is produced by those T cells. It is also critical that antigen-specific, IFN-γ-secreting CD4+ T cells efficiently traffic to the genital mucosa in response to Chlamydia infection in order to drive protective immunity (10, 11).
Homing receptors mediate the migration of immune cells towards specific signals in order to exit the circulation and enter target tissues (12). Integrins are a family of adhesion receptors consisting of α and β heteroduplexes that direct signaling from both outside and inside of the cell membrane (13). The role of certain integrin members on leukocytes has been studied extensively. For example, LFA-1 has been shown to play a crucial function in the arrest of leukocytes in the blood vessels at the site of inflammation (14, 15). Other integrin heterodimers, namely α4β1 and α4β7, provide tissue-specificity to T cells when homing to different areas of the body. Descriptions of how lymphocytes traffic to the gastrointestinal tract and central nervous system (CNS) have been reported. Lymphocyte recruitment to the gastrointestinal tract is largely mediated by the chemokine receptor CCR9 and the integrin receptor α4β7 (16). On the other hand, integrin α4β1 regulates trafficking to the CNS. In these models, interfering with α4β1 and α4β7 profoundly impairs immune cell recruitment to the respective tissues (17, 18). In fact, integrin-specific antibodies are used clinically to block immune cell infiltration and provide relief from autoimmune diseases such as ulcerative colitis and multiple sclerosis (19, 20). Unfortunately, our understanding of how CD4+ T cells traffic to the genital mucosa has been limited, including what combination of adhesion receptors is required for successful migration.
In this study, we interrogated the importance of α4β1 and α4β7 integrin heterodimers in promoting Chlamydia-specific CD4+ T cell recruitment to the genital mucosa and protecting mice from C. trachomatis infection. We show that integrin α4β1 is dramatically increased on the surface of both polyclonal and Chlamydia-specific CD4+ T cells in the uterus following infection. We find that blocking or deleting integrin α4β1, but not α4β7, on pathogen-specific CD4+ T cells results in the impairment of trafficking to the uterus and a decrease in the protective capacity of CD4+ T cells. We conclude that integrin α4β1 is necessary for CD4+ T cell-mediated protection against C. trachomatis. Identifying the receptors required for CD4+ T cell trafficking to the genital tract in response to C. trachomatis is important in designing a vaccine that elicits sterilizing, long lasting immunity against the pathogen while limiting the extent of tissue pathology.
MATERIALS AND METHODS
Mice
C57BL/6, B6.PL-Thy1a (CD90.1 congenic), C57BL/6, B6.SJL-Ptprca Pep3/BoyJ (CD45.1 congenic), B6.Cg-Tg548Jxm/J (Lck-CRE), C57BL/6-Itgb7tm1Cgn/J (Itgb7−/−), and B6;129-Itgb1tm1Efu/J (Itgb1flox/flox) mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and NR1 mice were previously described (21). B6;129-Itgb1tm1Efu/J (Itgb1flox/flox) mice were crossed with C57BL/6 for greater than 10 generations. C. trachomatis-specific integrin-deficient mice were generated by breeding NR1 mice to either Lck-CRE and Itgb1flox/flox or Itgb7−/− mice. All mice were maintained in facilities managed by the Harvard Medical School Center for Animal Resources and Comparative Medicine. To normalize for the murine estrous cycle, mice were treated subcutaneously with 2.5 mg of medroxyprogesterone 7 days prior to infection. Harvard’s institutional animal care and use committee approved all the experiments described. A minimum of 4 mice per group was used in each experiment.
Growth, isolation and detection of C. trachomatis
C. trachomatis serovar L2 (434/Bu) was propagated using McCoy cell monolayers grown in Eagle’s MEM (Invitrogen, Grand Island, NY) plus 10% FCS, 1.5 g/l sodium bicarbonate, 0.1 M nonessential amino acids, and 1 mM sodium pyruvate. Infected McCoy cells were detached from plates using sterile glass beads and then sonicated to disrupt C. trachomatis inclusions. Density gradient centrifugation was used to purify elementary bodies(22). Aliquots were stored at −80°C in a medium containing 250 mM sucrose, 10 mM sodium phosphate, and 5 mM L-glutamic acid.
Transfer of NR1 cells, infection of mice, and tissue preparation
C. trachomatis-specific CD4+ T cells were isolated from the lymph nodes and spleens of naïve donor NR1 mice. Recipient mice received 106 NR1 cells and were infected the following day with 106 C. trachomatis inclusion forming units (IFU) in 10 µl of sucrose-phosphate-glutamate media. We used the NSET device (ParaTechs) to bypass the cervix and directly infect the uterine horns (10). The uterus was harvested and disaggregated by digestion with 1 mg/ml of type XI collagenase (Sigma, St. Louis, MO) and 50 Kunitz/ml of DNase (Sigma) for 30 min at 37°C. Single cell suspensions from tissues were obtained by mechanical disaggregation prior to staining. Suspensions of splenocytes were treated with a hypotonic buffer to lyse red blood cells prior to use.
Flow cytometry
Single cell suspensions were stained immediately for activation markers or stimulated for 5 hours with 100 ng/ml PMA (Alexis Biochemical) and 1 µg/ml ionomycin (Calbiochem) in brefeldin A (BD Biosciences) for intracellular cytokine staining. Cells were treated with anti-FcγR (BioXCell) before staining with combinations of the following antibodies: anti-β1 Pacific Blue, anti-β7 FITC, anti-TCRvα2 allophycocyanin, anti-CD90.1 peridinin chlorophyll protein, anti-CD45.2 phycoerythrin (PE), anti-CD90.2 FITC, anti-IFN-γ PE, anti-TNF-α PE-cy7, anti-CD25 PE, anti-CD44 PE or Pacific Blue, anti-CD62L FITC (Biolegend), anti-CD3ε allophycocyanin, anti-α4 PE (BD Biosciences), anti-CD4 Qdot605 and a LIVE/DEAD dead cell stain kit (Invitrogen). The efficacy of all antibodies used in this study was confirmed extensively in vitro and compared to isotype control antibodies. For cytokine staining, cells were permeabilized using a Cytofix/Cytoperm Plus Kit following manufacturer’s instructions (BD Biosciences). Cell number was determined with AccuCheck Counting Beads (Invitrogen). Flow cytometry data were collected on a modified FACSCalibur (Cytek Development) or an LSRII (BD Biosciences) and analyzed using FlowJo (Tree Star).
Th1 polarization and protection against C. trachomatis
CD4+ T cells were harvested from the lymph nodes and spleens of naïve NR1 mice and enriched with a mouse CD4 negative isolation kit (Invitrogen) following the manufacturer’s protocol. CD4+ T cells were cultured in media consisting of RPMI 1640 (Invitrogen), 10% FCS, L-glutamine, HEPES, 50 µM 2-ME, 50 U/ml penicillin, and 50 mg/ml streptomycin. NR1 cells were activated by co-culture with irradiated or mitomycin-treated splenocytes pulsed with 5 µM of Cta1133–152 peptide at a stimulator:T cell ratio of 4:1. Th1 polarization was achieved by supplying cultures with 10 ng/ml of IL-12 (Peprotech, Rocky Hill, NJ) and 10 µg/ml of anti-IL-4 antibody (Biolegend). After 5 days of stimulation, NR1 Th1 cells were transferred i.v. into naïve recipient mice. In integrin antibody blocking experiments, NR1 cells were treated with 100 ug of anti-α4, anti-α4β7 or isotype antibody (BioXCell) for 1 hour at room temperature prior to transfer into recipients. The next day, mice were challenged with 5×106 C. trachomatis IFUs and the upper genital tract was analyzed for burden 5 days after infection. Mice were treated i.p. with 200 ug of the respective antibody at 1 and 3 days post-infection for integrin blocking experiments.
Competitive homing
For this assay an equivalent number of integrin wildtype NR1 cells (CD45.2/CD90.1) and congenic wildtype, Lck-CRE/Itgb1flox/flox, or Itgb7−/− NR1 cells (CD45.2/CD90.2) were combined and transferred into congenically mismatched CD45.1 hosts. The following day, mice were transcervically infected with 5×106 C. trachomatis IFUs. Tissues were harvested and analyzed 7 days after infection.
Quantitative PCR
Bacterial burden was evaluated by quantifying C. trachomatis 16S DNA relative to mouse GADPH DNA(21). The uterus was homogenized and DNA was extracted using the DNeasy blood and tissue kit (Qiagen). DNA was analyzed using C. trachomatis- and mouse-specific primer pairs and dual-labeled probes. Threshold values were detected by an ABI Prism 7000 sequence system. The ratio of C. trachomatis to host DNA was obtained using a standard curve.
Statistical analysis
Statistical significance between groups was determined using an unpaired two-tailed t test and depicted within figures as * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001.
RESULTS
C. trachomatis infection leads to robust integrin α4β1 surface expression on bulk CD4+ T cells in the uterus
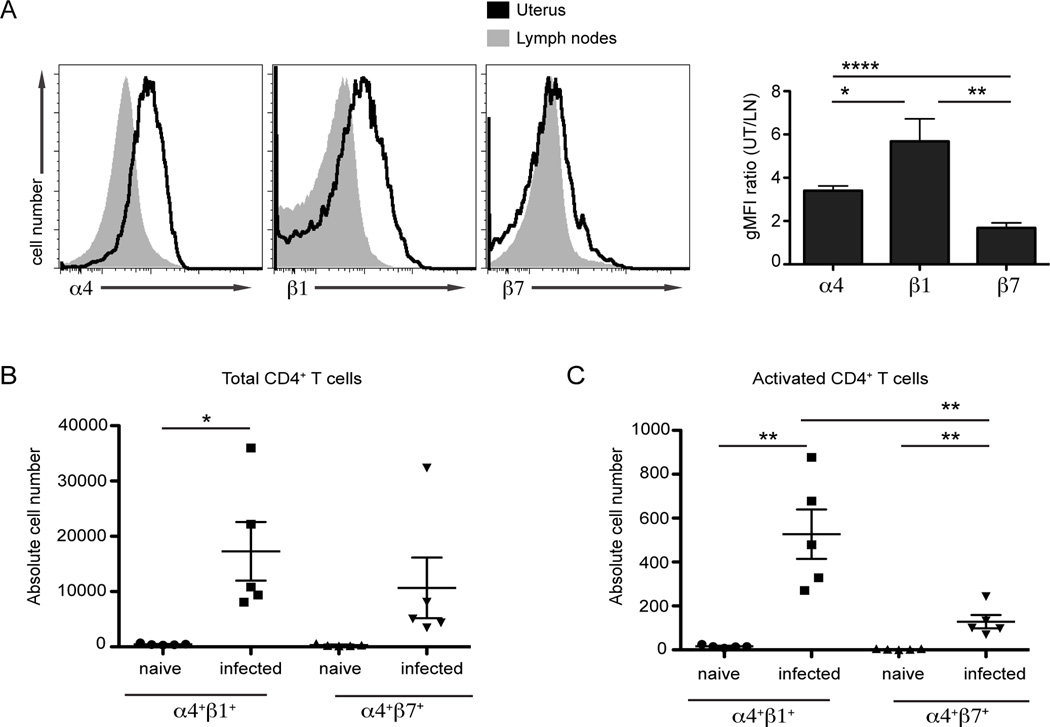
Previous reports differ regarding the levels of α4β1 and α4β7 expression on T cells in the uterus during Chlamydia infection (23–25). As a first step to resolve these discrepancies, we examined the surface expression of α4, β1, and β7 on CD4+ T cells responding to C. trachomatis infection in the genital mucosa. To test differences in integrin expression, mice were infected transcervically with C. trachomatis (10). Seven days later, the uterus and draining (iliac) lymph nodes were isolated and examined for the surface expression of integrins on endogenous CD4+ T cells by flow cytometry. We found that α4 and β1 were dramatically upregulated on the surface of CD4+ T cells in the uterus relative to those present in the draining lymph nodes of infected mice (Fig 1A). In contrast, the surface expression of β7 was only modestly increased on CD4+ T cells in the genital mucosa compared to α4 and β1. We next quantified the absolute number of α4+β1+ - and α4+β7+ -expressing CD4+ T cells in the genital tract (Fig 1B). Very few CD4+ T cells were found in the uterus during steady state in naïve mice. These extremely low numbers precluded conclusive interpretations about integrin staining differences in naïve mice. Following infection, the absolute number of α4+β1+ CD4+ T cells in the upper genital tract significantly increased whereas the number of α4+β7+ cells did not. We next compared the number of activated α4+β1+ or α4+β7+ CD4+ T cells responding to the genital mucosa by gating for populations expressing high levels of CD44. Interestingly, both the number of activated α4+β1+ and α4+β7+ CD4+ T cells were significantly increased in infected animals relative to naïve controls (Fig 1C). Nonetheless, there was a more robust recruitment of activated α4+β1+ CD4+ T cells to infected uteri compared to α4+β7+ CD4+ T cells. Our results show that while both α4+β1+ and α4+β7+ CD4+ T cells are found in the infected genital mucosa, β1 is more highly expressed. These observations on endogenous T cells suggest that α4β1 is the primary integrin driving CD4+ T cell recruitment to the genital mucosa in response to C. trachomatis infection.
FIGURE 1.
C. trachomatis infection leads to robust integrin α4β1 surface expression on polyclonal CD4+ T cells in the genital tract. C57BL/6 mice were transcervically infected with 106 IFU of C. trachomatis. Seven days following infection the indicated tissues were harvested and prepared for flow cytometry. After gating on live cells that were CD3+CD4+ the surface expression of α4, β1 and β7 was quantified. (A) The integrin surface expression was analyzed by comparing the geometric mean fluorescence intensity (gMFI) ratio of CD4+ T cells localized in the genital mucosa to those in the draining lymph node. (B) The absolute number of α4+β1+ and α4+β7+ CD4+ T cells were quantified in the genital tract of naïve or mice infected with C. trachomatis for seven days. (C) The absolute number of activated α4+β1+ and α4+β7+ CD4+ T cells was quantified in the genital tract of naïve or infected mice; activation was determined by CD44+ staining. Shown are representative results from one of two independent experiments. * p < 0.05, ** p < 0.01, and **** p < 0.0001.
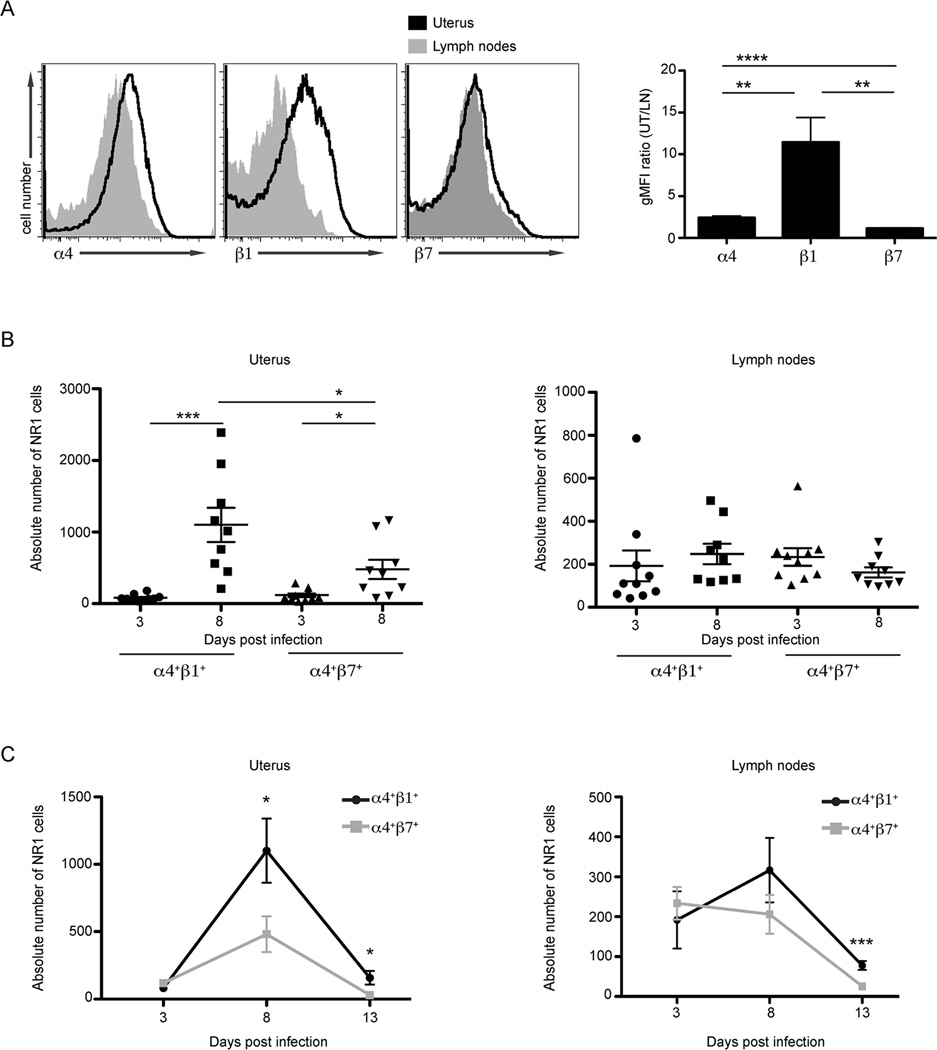
Infection leads to increased α4β1 surface expression on C. trachomatis-specific CD4+ T cells in the uterus
During Chlamydia infection, antigen-specific T cells are primed and recruited specifically to the genital tract. However, inflammatory cytokines can also activate bystander T cells at the site of infection independently of antigen specificity (26). Previous work characterizing integrin receptors required for T cell recruitment to the genital mucosa focused exclusively on memory CD4+ T cell lines (23), polyclonal CD4+ T cells (24), or bulk CD4+ T cells (25) but never examined naïve C. trachomatis-specific CD4+ T cells. However, only by using naïve antigen-specific T cells is it possible to properly model primary infection. To directly interrogate the integrin profile of Chlamydia-specific CD4+ T cells, we took advantage of C. trachomatis-specific TCR transgenic CD4+ T cells that are locked into specificity for the Chlamydia antigen Cta1 (referred to subsequently as NR1 cells, 21). Based on the results described in the section above, we hypothesized that NR1 cells would display significantly increased surface β1, rather than β7, upon trafficking to infected uteri. To test this prediction, we transferred 106 NR1 cells intravenously into congenic naïve animals that were then infected transcervically the following day with C. trachomatis. The integrin profile of naïve NR1 cells did not have major differences prior to transfer (data not shown). In agreement with our observations with polyclonal CD4+ T cells (Fig 1), 8 days after infection there was over a 10-fold increase in the surface expression of integrin β1 on NR1 cells in the uterus compared to NR1 cells in the draining lymph nodes (Fig 2A). To better understand the dynamics of integrin expression we monitored the surface expression of integrins on NR1 cells in the genital tract and draining lymph nodes at three time points following C. trachomatis infection. Three, 8, and 13 days after infection were chosen to coincide with T cell activation, peak of infiltration to the site of infection, and contraction, respectively (Fig 2B and C). There was a surge of both α4+β1+ and α4+β7+ NR1 cells in the genital tract 8 days after infection compared to what was observed with these populations 3 days after infection (Fig 2B). Even though both NR1 populations increased during infection, the magnitude of infiltration of the α4+β1+ NR1 cells to the uterus was far more robust (***p<0.001) than the α4+β7+ population (*p<0.05) (Fig 2B, left). Surface integrin profiles of NR1 cells in the draining lymph nodes were statistically indistinguishable between these two time points (Fig 2B, right). Thirteen days after infection the decline of NR1 cells (Fig 2C) corresponded to the resolution of C. trachomatis infection in the genital mucosa (10). Even at this late time point, α4+β1+ NR1 cells were found significantly more frequently than α4+β7+ NR1 cells in the genital tract. These observations suggest that α4+β1+ NR1 cells are either recruited or retained in the uterus for a longer period of time than other T cell populations. We confirmed that the limited β7 expression observed in Figures 1 and 2 was not due to poor antibody staining. Robust β7 staining was detected on NR1 cells activated in vitro in the presence of retinoic acid, a metabolite previously shown to induce α4β7 expression on T cells (data not shown). Taken together, these findings demonstrate that both polyclonal and Chlamydia-specific CD4+ T cells, preferentially express α4 and β1 on their surface in the murine genital mucosa in response to C. trachomatis infection.
FIGURE 2.
C. trachomatis infection leads to robust α4β1 surface expression on C. trachomatis-specific CD4+ T cells responding to the genital tract. One million CD90.1+ NR1 cells were transferred intravenously into CD90.2+ recipient mice. The following day mice were infected transcervically with 106 IFU of C. trachomatis. The uterus and draining lymph nodes were harvested at the indicated time points following infection and prepared for flow cytometry. For integrin surface staining analysis, cells were pre-gated as live Vα2+CD4+CD90.1+ cells and then examined for the surface expression of α4, β1, and β7. (A) Eight days after C. trachomatis infection, the integrin surface expression on NR1 cells was analyzed by comparing the gMFI ratio of NR1 cells in uterus to those localized in the draining lymph nodes. (B) The absolute numbers of α4+β1+ and α4+β7+ NR1 cells was quantified 3 and 8 days after transcervical infection in the uterus (left) and the draining lymph nodes (right). (C) Quantification of the trafficking kinetics of α4+β1+ and α4+β7+ NR1 cells in the uterus (left) and draining lymph nodes (right) at the indicated time points. Shown are representative results from one of three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001.
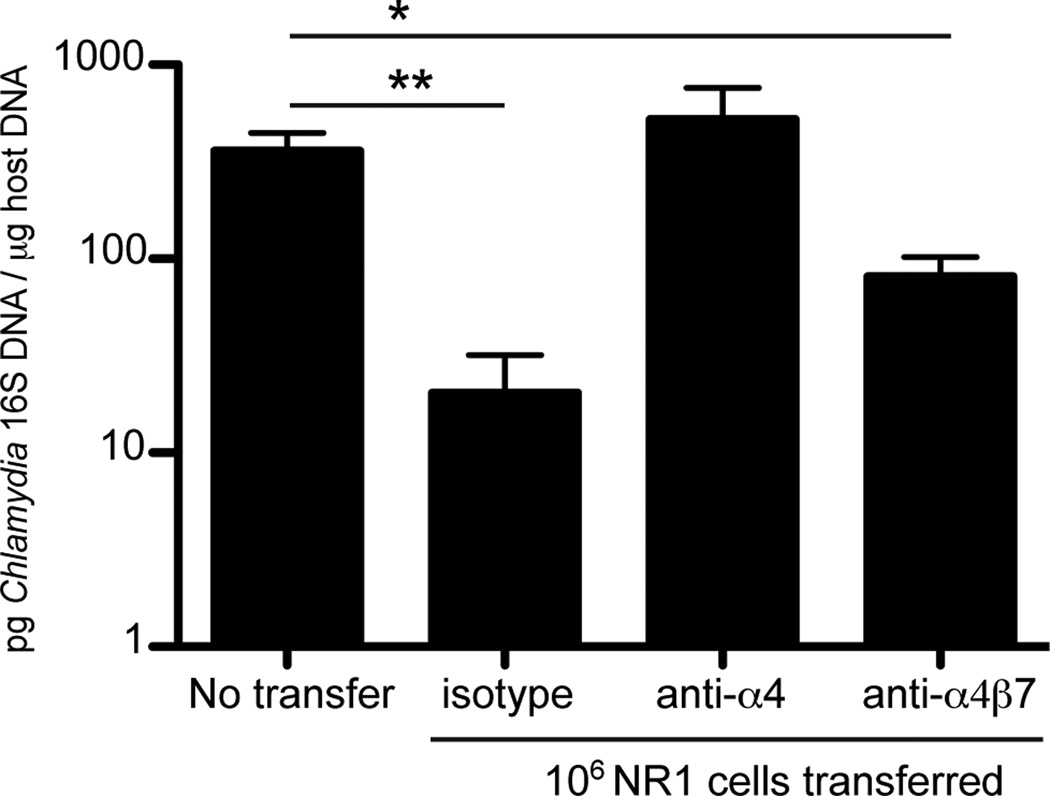
Blocking integrin α4 but not α4β7 increases C. trachomatis burden
Since we identified a strong upregulation of α4β1 on the surface of CD4+ T cells responding to C. trachomatis infection, we next assessed the functional role of β1 and β7 in promoting protective immunity. Our group has previously shown that transfer of in vitro-activated Th1-skewed NR1 cells into naïve mice confers significant protection from C. trachomatis infection compared to mice that receive no T cells (10). We hypothesized that if specific integrins are required for recruitment to the genital mucosa, blocking these integrins on activated NR1 cells may alter their protective capacity. To test this and determine the relative contributions of β1 and β7 to CD4+ T cell-mediated protection, we selectively blocked integrin receptors using antibodies (Fig 3). NR1 cells from naïve mice were harvested and polarized in vitro to a Th1 phenotype. IFN-γ production by the NR1 cells was assayed to confirm their Th1 phenotype by flow cytometry prior to transfer (data not shown). One million NR1 Th1 cells were pre-treated with antibody that blocked α4 (blocks both α4β1 and α4β7), α4β7, or an isotype control and then transferred intravenously into host mice that were then transcervically infected with C. trachomatis the following day. To ensure robust blockade of integrins, each group of mice was also treated with antibody 1 and 3 days following infection. Five days after infection, C. trachomatis burden in the uterus was measured using quantitative PCR (qPCR). Mice that were treated with blocking antibody against α4 had a higher C. trachomatis burden in the genital tract than isotype control treated animals, similar to bacterial levels found in mice that received no NR1 cells at all. On the other hand, mice treated with α4β7 blocking antibody had significantly lower C. trachomatis levels than the no transfer group and similar bacterial burden to the isotype antibody treated mice. Despite the lack of an antibody that specifically blocks α4β1, we can infer the importance of α4β1 indirectly. Because blocking α4 with antibody prevents both α4β1 and α4β7 signaling, the differences observed in protection between α4 and α4β7 antibody treatment groups point towards a function of α4β1. Since the integrin α4 chain dimerizes with either β1or β7(27), the results shown in Figure 3 indirectly confirm that only α4β1 is required for CD4+ T cell-mediated protection against C. trachomatis in the genital mucosa.
FIGURE 3.
Antibody blockade of α4 but not α4β7 exacerbates C. trachomatis burden in the genital mucosa. NR1 cells were skewed in vitro to a Th1 phenotype for 5 days. NR1 cells were pretreated with the indicated antibodies and then transferred intravenously into naïve recipients. The following day, mice were infected transcervically with 5×106 IFU of C. trachomatis. Groups were injected with the respective integrin or isotype control antibody 1 and 3 days following infection. Five days after infection, the genital tract was isolated and genomic DNA was purified. The levels of Chlamydia 16S DNA relative to the levels of host GADPH were quantified using qPCR. Shown are representative results from one of two independent experiments. * p < 0.05, and ** p < 0.01.
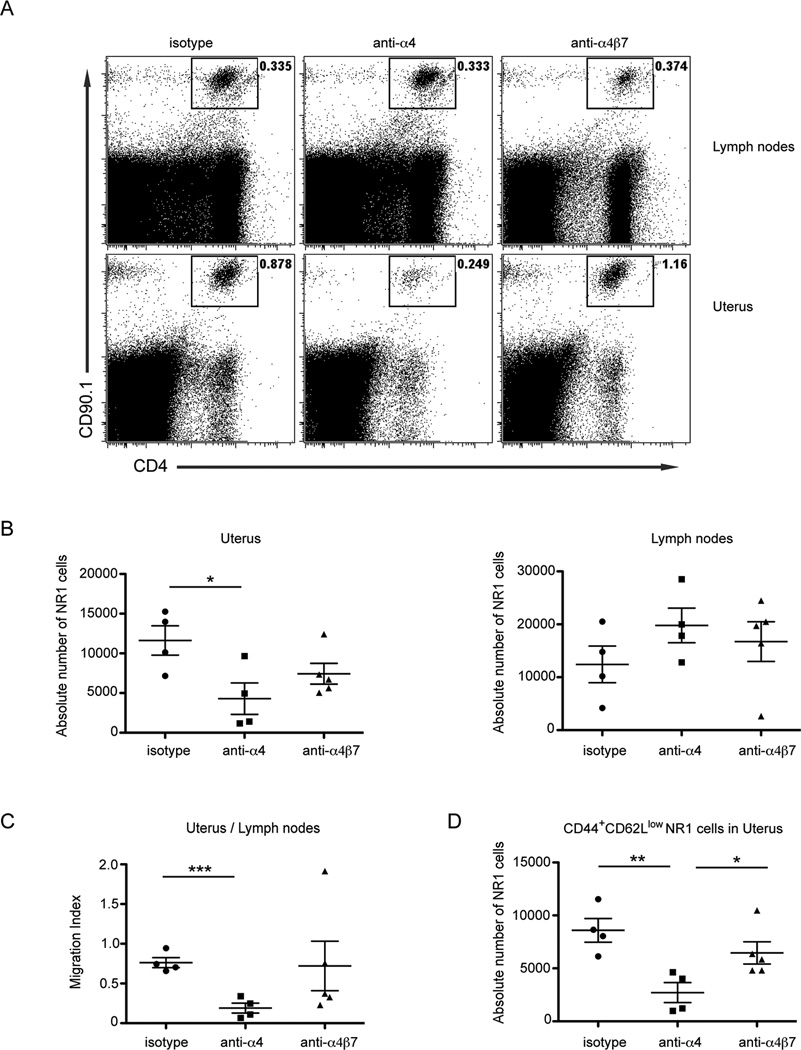
A reduction in CD4+ T cells is responsible for the higher C. trachomatis burden in anti-α4 antibody-treated mice
Although blocking α4 was sufficient to prevent CD4+ T cell-mediated protection in the genital mucosa, we had not determined the mechanism responsible for higher burden. We predicted that the loss of protection seen in mice treated with α4-blocking antibody was due to diminished recruitment of NR1 cells to the infected genital mucosa. To test this possibility, we monitored the trafficking of NR1 cells to the infected genital tract after integrin blockade. NR1 cells were skewed to the Th1 phenotype in vitro, and then transferred into congenically mismatched host mice. The next day mice were infected transcervically with C. trachomatis. Prior to transfer, NR1 cells were pretreated with individual integrin blocking antibodies or an isotype control. Mice were also treated with the same blocking antibody or isotype control 1 and 3 days after infection. Five days after infection, we examined the number of NR1 cells present in the genital tract and draining lymph nodes by flow cytometry. We noted that the number of NR1 cells in the genital mucosa was significantly diminished following treatment with the α4 blocking antibody relative to the isotype treated control mice (Fig 4A and B). In contrast, α4β7 antibody treatment did not impact NR1 cell recruitment to the uterus, as absolute numbers were similar between isotype- and α4β7-treated groups. The absolute numbers of NR1 cells present in the draining lymph nodes were not significantly different between the groups suggesting no general defect in trafficking of NR1 cells following antibody treatment. We also calculated a migration index (the ratio of live NR1 cells in the uteri to draining lymph nodes within the same animal) to normalize for mouse-to-mouse variability (Fig 4C). We found that the migration index was profoundly decreased in mice treated with α4 blocking antibody compared to isotype-treated mice. These results show that blocking α4 prevents efficient CD4+ T cell trafficking from the draining lymph nodes to the uteri following C. trachomatis infection. The migration index of mice treated with α4β7 antibody was not statistically different from isotype control treated mice demonstrating that α4β7 plays a limited role in NR1 cell trafficking to the genital mucosa in response to C. trachomatis infection.
FIGURE 4.
Blockade of α4 but not α4β7 impairs C. trachomatis-specific CD4+ T cell trafficking to the genital tract following infection. NR1 cells were skewed in vitro to a Th1 phenotype for 5 days. NR1 cells were pretreated with the indicated antibodies and then 106 CD90.1+ NR1 cells were transferred intravenously into CD90.2+ host mice. The following day mice were infected transcervically with 5×106 IFU of C. trachomatis. Groups were injected with the respective integrin or control antibody 1 and 3 days following infection. The uterus and draining lymph nodes were harvested 5 days following infection and prepared for flow cytometry. For quantification of NR1 cell trafficking, we gated on live Vα2+CD4+CD90.1+ cells. (A) Representative flow cytometry plots indicate the percentage of NR1 cells following the indicated antibody treatment in the draining lymph nodes (top) and uterus (bottom) (B) The absolute number of NR1 cells were quantified in the uterus (left) and draining lymph nodes (right). (C) A migration index for each antibody treatment was calculated by comparing the absolute number of live NR1 cells in the uterus directly to the number of NR1 cells in draining lymph nodes within each animal. A lower migration index ratio indicates decreased NR1 cell recruitment to the uterus. (D) The absolute number of effector NR1 cells was determined by examining the CD44+CD62Llow gated population in the genital mucosa. Shown are representative results from one of two independent experiments. * p < 0.05, ** p < 0.01, and *** p < 0.001.
We next examined how antibody treatment directly alters the recruitment of effector CD4+ T cell populations in the genital mucosa. NR1 cells were stained for CD44 and CD62L in order to evaluate the presence of effector CD4+ T cells at the site of the infection. The absolute number of CD44+CD62Llow NR1 cells in the uterus was significantly decreased only in the α4 antibody-treated group compared to isotype-treated mice (Fig 4D). Moreover, anti-α4β7 antibody treatment did not significantly decrease the absolute number of effector NR1 cells in the genital tract relative to the isotype-treated group. We confirmed the efficiency of α4β7 antibody blockade in vitro (data not shown). These results show that disrupting the integrin α4β1 but not α4β7 on NR1 cells is sufficient to eliminate CD4+ T cell-mediated protection provided following C. trachomatis infection in the genital tract.
Integrin β1-deficient C. trachomatis-specific CD4+ T cells are unable to protect the uterus
To complement the antibody-blocking experiments showing that α4β1 is required for Chlamydia-specific CD4+ T cells to home to and protect the genital mucosa, we generated TCR transgenic mice in which the T cells are deficient in either integrin β1 or β7. Because loss of β1 results in embryonic lethality (28), we used a CRE-Flox system to generate NR1 cells conditionally deficient in β1. NR1 transgenic mice were crossed to Lck-CRE and Itgb1flox/flox animals such that only the lymphocytes were deficient in β1. We also crossed NR1 transgenic mice with Itgb7−/− mice to generate Chlamydia-specific CD4+ T cells deficient in β7. We first confirmed that integrin surface expression was significantly altered for each knockout T cell genotype (Supp. Fig 1A and B). Interestingly, loss of β1 led to a concomitant increase of surface β7 on NR1 cells similar to a previous report showing that α4β7 heterodimers form more readily in the absence of the β1 chain (29). The loss of β7 also led to an increase of the percentage of β1+ NR1 cells after in vitro activation. LFA-1 was robustly upregulated on NR1 cells from the three different genotypes. However, no compensatory LFA-1 expression was observed between integrin deficient T cells. In addition, NR1 cells do not express CD103 upon in vitro activation (data not shown). We next confirmed that NR1 cells deficient in individual integrins proliferated normally. Integrin-deficient NR1 cells were harvested from the knockout mice and polarized in vitro for 5 days to a Th1 phenotype. We found no significant difference between groups in the total number of recovered NR1 cells 5 days following activation, demonstrating that all the genotypes are viable (Fig 5A). We next examined the activation and cytokine production for each group. For all genotypes examined, NR1 cells were robustly activated as determined by staining for the activation markers CD25 and CD44 (Fig 5B). When we assayed cytokine profiles using intracellular cytokine staining, we found similar levels of IFN-γ and TNF-α in all groups, demonstrating that loss of integrin β chains does not negatively impact Th1 differentiation (Fig 5C). Therefore, the absence of either β1 or β7 does not interfere with expansion, activation, and Th1 cytokine production of NR1 cells in vitro.
FIGURE 5.
Chlamydia-specific CD4+ T cells deficient in integrin β1 are unable to protect mice from C. trachomatis infection. The indicated NR1 genotypes were skewed in vitro to a Th1 phenotype for 5 days. (A) The expansion of NR1 cells was compared for the three genotypes. (B) The activation of live NR1 cells was assessed by gating for CD25+CD44+ cells. (C) TNF-α and IFN-γ production was examined using intracellular cytokine staining. (D) After 5 days of in vitro stimulation, 105 NR1 cells were transferred intravenously into naïve hosts. The following day, mice were transcervically infected with 5x106 IFU of C. trachomatis. Five days after infection, the genital tract was harvested and genomic DNA was purified. The levels of Chlamydia 16S DNA relative to the levels of host GADPH were quantified using qPCR. Shown are representative results from one of three independent experiments. * p < 0.05 and ** p < 0.01.
Upon showing that the various genotypes of NR1 cells had normal effector phenotypes, we then tested whether deficiency in either the β1 or β7 chain would adversely affect the protective capacity of Chlamydia-specific CD4+ T cells in vivo. Based on our previous results from antibody blocking experiments, we hypothesized that β1-deficient NR1 cells would be unable to protect the genital mucosa from C. trachomatis infection. We transferred 105 wildtype, β1−/− or β7−/− Th1-skewed NR1 cells into naïve mice 1 day prior to transcervical infection with C. trachomatis (Fig 5D). Five days after infection, we harvested the genital tract and quantified the levels of C. trachomatis using qPCR. As expected, mice that received wildtype NR1 cells showed a significant reduction in C. trachomatis burden compared to mice that received no transfer. Mice that received β1−/− NR1 cells were not protected against C. trachomatis infection as indicated by bacterial burdens similar to mice receiving no transferred NR1 cells. In line with our previous findings using antibody blockade, we also found that mice receiving β7−/− NR1 cells were significantly protected against C. trachomatis infection relative to mice that received no transfer, and trended toward being even more protective than wildtype NR1 cells. These findings definitively show that integrin β7 on NR1 cells is dispensable for protecting the uterus from C. trachomatis infection. In summary, integrin β1, but not β7, is necessary for Chlamydia-specific CD4+ T cells to protect against infection in the genital mucosa.
Integrin β1 deficiency impairs C. trachomatis-specific CD4+ T cell homing to the uterus
We next sought to understand the mechanisms responsible for the loss of protective capacity in β1−/− Chlamydia-specific CD4+ T cells. Our previous data using antibody blocking showed that T cell trafficking to the genital mucosa was inhibited and therefore provided limited protection. Here we used a competitive homing assay to test the trafficking potential of integrin-deficient Chlamydia-specific CD4+ T cells. We directly compared the migration of integrin-sufficient and deficient NR1 cells under identical conditions within the same infected host. We transferred an equal number of CD45.2/CD90.2 β1−/−, β7−/− or wildtype NR1 cells and CD45.2/CD90.1 wildtype NR1 cells into congenically mismatched CD45.1 recipients. The next day, we infected mice transcervically with C. trachomatis. Seven days after infection, we isolated and processed tissues to quantify the numbers of both NR1 populations using flow cytometry. We found that β1−/− NR1 cells were far less efficient than wildtype NR1 cells in trafficking to the genital mucosa, while β7−/− NR1 cells outcompeted their wildtype counterparts (Fig 6A). None of the experimental groups showed defects in their migration to the draining lymph node. Interestingly, we noted a higher percentage of β1−/− NR1 cells in the lymph nodes compared to the wildtype NR1 cells, inverse of what we observed in the genital mucosa. These results are likely due to a decreased ability of β1−/− NR1 cells to leave the circulation to enter the infected genital mucosa. To normalize for mouse-to-mouse variation in absolute T cell numbers, we calculated a migration index for each mouse by comparing the ratio of the two transferred NR1 cell populations in the uterus (integrin deficient CD90.2% : integrin sufficient CD90.1%) to the ratio of the transferred populations in the draining lymph nodes or spleen within the same animal (Fig 6B). A smaller migration index indicated less efficient trafficking of integrin-deficient NR1 cells specifically to the uterus relative to the circulation. We found a dramatically lower migration index for β1−/− NR1 cells demonstrating that trafficking to the uterus during infection was significantly impaired relative to wildtype NR1 cells. Intriguingly, the migration index for β7−/− NR1 cells was significantly higher than wildtype, demonstrating enhanced homing of Chlamydia-specific CD4+ T cells to the uterus in the absence of integrin β7. These results suggest that integrin β7 is not only dispensable, but that deficiency of β7 enhances antigen-specific CD4+ T cell migration to the genital tract during C. trachomatis infection. Our findings collectively reveal that integrin β1 plays a crucial in trafficking of CD4+ T cells to the genital mucosa and that absence of β1 negatively affects the protective capacity of C. trachomatis-specific CD4+ T cells.
FIGURE 6.
β1 deficient Chlamydia-specific CD4+ T cells are unable to traffic efficiently to the genital tract following infection. An equivalent number of CD45.2/CD90.1 integrin wildtype NR1 cells and CD45.2/CD90.2 wildtype, β1−/−, or β7−/− NR1 cells were transferred intravenously into CD45.1/CD90.2 host mice. The next day, mice were infected transcervically with 106 IFU of C. trachomatis. The genital tract, draining lymph nodes and spleen were harvested 7 days after infection and prepared for flow cytometry. We examined the recruitment of NR1 cells by pre-gating on live Vα2+CD4+CD45.2+ cells. We then differentiated the competing NR1 populations by examining the number of CD90.1+ or CD90.2+ cells. (A) Shown are representative plots of integrin sufficient and deficient NR1 cells in the draining lymph node (top) and genital mucosa (bottom). (B) The migration index within each group was calculated by comparing the percentage of CD90.2+ to CD90.1+ NR1 cells in the uterus to the CD90.2+ to CD90.1+ NR1 cells in the draining lymph nodes (left) or spleen (right). A lower migration index indicates less efficient trafficking of integrin-deficient NR1 cells specifically to the uterus relative to the circulation. Shown are representative results from one of three independent experiments. ** p < 0.01, *** p < 0.001, and **** p < 0.0001.
Discussion
The orchestration of events required for a successful T cell response determines whether an intracellular pathogen will be eliminated from the host. In the case of C. trachomatis infection, a robust CD4+ Th1 cell population that homes to the genital tract provides the vigorous IFN-γ response necessary for bacterial clearance (10). The integrin receptors important for C. trachomatis-specific CD4+ T cell-mediated protection have not been previously described. Using C. trachomatis-specific CD4+ T cells, we demonstrated that perturbing integrin α4β1 but not α4β7, through antibody blockade or genetic deletion, results in impaired T cell trafficking to the genital tract and a loss of protective capacity following C. trachomatis infection. Together these observations demonstrate that integrin α4β1 is necessary for CD4+ T cell-mediated protection against C. trachomatis infection in the murine genital tract.
We previously demonstrated that the chemokine receptors CXCR3 and CCR5 are required for Chlamydia-specific CD4+ T cells to home to and protect the genital mucosa following infection with C. trachomatis (22). Here we extended these studies to identify integrin receptors that are important for CD4+ T cell-mediated protection. When we examined the surface expression of β1 and β7 on both bulk and antigen-specific CD4+ T cells, we found that relative β1 expression was dramatically increased on the majority of CD4+ T cells found in the uterus following C. trachomatis infection (Fig 1A and 2A). These findings are in line with Perry et al who demonstrated that surface β1 is upregulated on bulk lymphocytes localized in the genital tract but not in the intestinal mucosa following infection with the mouse-adapted species Chlamydia muridarum (25). Although most CD4+ T cells were expressing high levels of surface integrin β1, this report also noted a minor percentage of β7+ CD4+ T cells in the genital tract (25). Separate reports suggested that α4β7 was the dominant integrin receptor expressed on CD4+ T cells in the genital tract. However, these studies had significant experimental constraints that limit their interpretation. For example, Kelly et al did not interrogate the integrin profile during the primary response or differentiate between antigen-specific and bystander CD4+ T cells in the genital tract (30). Another report examined the surface integrin receptors on a memory CD4+ T cell line in culture following stimulation but did not assess in vivo integrin dynamics (23). Memory CD4+ T cells can exert effector activities with greater ease than primary T cells (31), therefore examining memory cells is not indicative of the trafficking properties of T cells in a primary response. Because these studies did not examine naïve, antigen-specific CD4+ T cells they were unable to recapitulate the initial activation events during primary C. trachomatis infection. In addition, these reports did not examine how perturbation of distinct integrin complexes alters the protective capacity of CD4+ T cells responding to genital C. trachomatis infection.
The results presented here show that α4β1 drives Chlamydia-specific CD4+ T cells to the infected genital mucosa (Fig 4 and 6) and is required to mediate protective immunity following C. trachomatis infection (Fig 3 and 5). While α4β1 was the dominant integrin in the genital mucosa we did identify a second β7+ population that when perturbed did not alter immunity to Chlamydia infection. The existence of two NR1 cell populations with distinct integrin profiles and identical TCR specificity shows that integrin levels are not hardwired but rather imprinted during T cell activation as has been suggested previously (32). Our results also suggest that only CD4+ T cells with the correct integrin profile can extravasate into the infected genital mucosa. We speculate that while α4β1-expressing T cells can enter the infected tissue efficiently, the α4β7+ CD4+ T cells remain in the circulation and fail to provide protection in the genital mucosa. Because the entire uterus, including its associated vasculature, was harvested in all experiments it was not possible to distinguish between those T cells present in blood vessels and those that had completed transendothelial migration. It is possible that the α4β7+ CD4+ T cell population still contributes to immunity elsewhere in the mouse, but this remains to be tested. Interestingly, one recent report uncovered a mechanism that drives distinct integrin expressing T cell populations in the lungs of mice. Ruane et al found that a subset of residing dendritic cells (DCs) can imprint lung T cells to express α4β7. The T cells expressing α4β7 do not mediate protection in the lungs but rather provide gut mucosal immunity (33). Given that a subset of lung DCs can imprint a population of T cells to express α4β7, we speculate that a similar process also occurs in the genital tract. While the majority of CD4+ T cells are primed to express α4β1, which mediates immunity in the genital mucosa during C. trachomatis infection, perhaps a subset of uterine DCs also imprints α4β7 on a fraction of the CD4+ T cell population that may provide systemic mucosal immunity. This hypothesis will need to be examined in future experiments.
It is well established that integrin α4β1 adheres to the addressin VCAM-1 and the extracellular matrix protein fibronectin (34, 35). Surface VCAM-1 increases on endothelial cells lining microvessels following inflammation (27). For example, patients with CNS autoimmune disorders are often treated with natalizumab, an anti-α4 antibody that blocks α4β1 and α4β7 interactions with their ligands. It is thought that natalizumab treatment decreases undesirable inflammation in the CNS by interfering with α4β1-mediated immune cell recruitment to this sensitized area in the body (36). Previous studies have also shown that surface VCAM-1 becomes abundant on murine and human genital mucosa following Chlamydia infection. In contrast, the expression of the mucosal addressin cell adhesion molecule-1 (MAdCAM-1), the binding partner for α4β7, has been reported to be expressed robustly in the gut but only modestly in the genital tract (25, 37). The noticeable increase of surface VCAM-1 in the genital mucosa following Chlamydia infection corresponds to the upregulation of surface α4β1 on Chlamydia-specific CD4+ T cells in the uterus observed in this study.
Several other signals stimulate the rapid increase of VCAM-1 on vaginal epithelial cells including IFN-γ treatment and herpes simplex virus infection (38). Given that VCAM-1 can be selectively upregulated on both endothelial and non-endothelial cells, responding CD4+ T cells may require α4β1 signaling for multiple steps in order to provide protection. Previous studies have characterized the importance of α4β1 to slow/arrest lymphocytes in the blood vessel, but α4β1 may also mediate the interactions between effector CD4+ T cells and infected epithelial cells as has been suggested to occur during Chlamydia infection (39). It remains unknown whether C. trachomatis-specific CD4+ T cells directly interact with infected epithelial cells in the genital mucosa or if their antimicrobial effects occur by altering the cytokine milieu at the site of infection. In this study we observed that integrin β1 and β7 were dispensable for proliferation and differentiation to a Th1 phenotype (Fig 5A, B and C). Consequently, we conclude that the function of β1 in mediating protection (Fig 5D) is to allow successful Chlamydia-specific CD4+ T cell trafficking to the uterus (Fig 6) rather than playing a role in activation or production of IFN-γ. Future studies must further explore the interaction between IFN-γ-producing CD4+ T cells and the infected epithelium to determine whether β1 is required for cellular interactions in vivo within the genital mucosa. Additional studies will need to determine if T cell recruitment and effector activity, mediated by α4β1, contributes to genital tract pathology following Chlamydia infection. Although integrin β1 is found primarily in complex with α4 on T cells, our findings do not entirely eliminate the possibility that β1 can form additional heterodimers on CD4+ T cell and that these heterodimers may play a role in the retention of T cells in the genital tract following infection.
The results obtained through this study further elucidate the essential homing receptors required for an effective CD4+ T cell defense in the genital mucosa. It remains to be determined whether CD4+ T cells responding to other pathogens in the genital tract also require the homing receptors we have identified (CXCR3, CCR5 and α4β1) as required for successful trafficking during C. trachomatis infection. Generating a robust and long-lived protective T cell response is crucial to clear infection and avoid recurrent cycles of inflammation and associated pathology. Vaccine efforts against intracellular pathogens should examine whether the appropriate T cell population, with the necessary homing molecules, is being generated to ensure protection and minimize pathology. In addition to the current treatments for autoimmune diseases in the gut and the CNS, integrin and chemokine receptor targeted therapies could be used to selectively shape the recruitment of desired T cells to other mucosal tissues.
Supplementary Material
Acknowledgements
We thank members of the Starnbach lab for helpful discussions.
Footnotes
This work was supported by grants AI062827 and AI39558 from the National Institutes of Health.
Disclosures
The authors declare no financial conflicts of interest.
References
- 1.Brunham R, Rey-Ladino J. Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nature reviews. Immunology. 2005;5:149–161. doi: 10.1038/nri1551. [DOI] [PubMed] [Google Scholar]
- 2.Cohen C, Brunham R. Pathogenesis of Chlamydia induced pelvic inflammatory disease. Sexually transmitted infections. 1999;75:21–24. doi: 10.1136/sti.75.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roan N, Starnbach M. Immune-mediated control of Chlamydia infection. Cellular microbiology. 2008;10:9–19. doi: 10.1111/j.1462-5822.2007.01069.x. [DOI] [PubMed] [Google Scholar]
- 4.Perry L, Su H, Feilzer K, Messer R, Hughes S, Whitmire W, Caldwell H. Differential sensitivity of distinct Chlamydia trachomatis isolates to IFN-gamma-mediated inhibition. Journal of immunology (Baltimore, Md. : 1950) 1999;162:3541–3548. [PubMed] [Google Scholar]
- 5.Thomas S, Garrity L, Brandt C, Schobert C, Feng G, Taylor M, Carlin J, Byrne G. IFN-gamma-mediated antimicrobial response. Indoleamine 2,3-dioxygenase-deficient mutant host cells no longer inhibit intracellular Chlamydia spp. or Toxoplasma growth. Journal of immunology (Baltimore, Md. : 1950) 1993;150:5529–5534. [PubMed] [Google Scholar]
- 6.Mayer J, Woods M, Vavrin Z, Hibbs J. Gamma interferon-induced nitric oxide production reduces Chlamydia trachomatis infectivity in McCoy cells. Infection and Immunity. 1993;61:491–497. doi: 10.1128/iai.61.2.491-497.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson D, Virok D, Wood H, Roshick C, Johnson R, Whitmire W, Crane D, Steele-Mortimer O, Kari L, McClarty G, Caldwell H. Chlamydial IFN-gamma immune evasion is linked to host infection tropism. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10658–10663. doi: 10.1073/pnas.0504198102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coers J, Bernstein-Hanley I, Grotsky D, Parvanova I, Howard J, Taylor G, Dietrich W, Starnbach M. Chlamydia muridarum evades growth restriction by the IFN-gamma-inducible host resistance factor Irgb10. Journal of immunology (Baltimore, Md. : 1950) 2008;180:6237–6245. doi: 10.4049/jimmunol.180.9.6237. [DOI] [PubMed] [Google Scholar]
- 9.Gondek D, Roan N, Starnbach M. T cell responses in the absence of IFN-gamma exacerbate uterine infection with Chlamydia trachomatis. Journal of immunology (Baltimore, Md. : 1950) 2009;183:1313–1319. doi: 10.4049/jimmunol.0900295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gondek D, Olive A, Stary G, Starnbach M. CD4+ T cells are necessary and sufficient to confer protection against Chlamydia trachomatis infection in the murine upper genital tract. Journal of immunology (Baltimore, Md. : 1950) 2012;189:2441–2449. doi: 10.4049/jimmunol.1103032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johansson M, Lycke N. Immunological memory in B-cell-deficient mice conveys long-lasting protection against genital tract infection with Chlamydia trachomatis by rapid recruitment of T cells. Immunology. 2001;102:199–208. doi: 10.1046/j.1365-2567.2001.01167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Springer T. Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell. 1994;76:301–314. doi: 10.1016/0092-8674(94)90337-9. [DOI] [PubMed] [Google Scholar]
- 13.Richard OH. IntegrinsBidirectional, Allosteric Signaling Machines. Cell. 2002;110 [Google Scholar]
- 14.Makgoba M, Sanders M, Ginther Luce G, Dustin M, Springer T, Clark E, Mannoni P, Shaw S. ICAM-1 a ligand for LFA-1-dependent adhesion of B, T and myeloid cells. Nature. 1988;331:86–88. doi: 10.1038/331086a0. [DOI] [PubMed] [Google Scholar]
- 15.Springer T, Dustin M, Kishimoto T, Marlin S. The lymphocyte function-associated LFA-1, CD2, and LFA-3 molecules: cell adhesion receptors of the immune system. Annual review of immunology. 1987;5:223–252. doi: 10.1146/annurev.iy.05.040187.001255. [DOI] [PubMed] [Google Scholar]
- 16.Mora J, Bono M, Manjunath N, Weninger W, Cavanagh L, Rosemblatt M, Von Andrian U. Selective imprinting of gut-homing T cells by Peyer's patch dendritic cells. Nature. 2003;424:88–93. doi: 10.1038/nature01726. [DOI] [PubMed] [Google Scholar]
- 17.Yednock T, Cannon C, Fritz L, Sanchez-Madrid F, Steinman L, Karin N. Prevention of experimental autoimmune encephalomyelitis by antibodies against alpha 4 beta 1 integrin. Nature. 1992;356:63–66. doi: 10.1038/356063a0. [DOI] [PubMed] [Google Scholar]
- 18.Bauer M, Brakebusch C, Coisne C, Sixt M, Wekerle H, Engelhardt B, Fässler R. Beta1 integrins differentially control extravasation of inflammatory cell subsets into the CNS during autoimmunity. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1920–1925. doi: 10.1073/pnas.0808909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feagan B, Greenberg G, Wild G, Fedorak R, Paré P, McDonald J, Dubé R, Cohen A, Steinhart A, Landau S, Aguzzi R, Fox I, Vandervoort M. Treatment of ulcerative colitis with a humanized antibody to the alpha4beta7 integrin. The New England journal of medicine. 2005;352:2499–2507. doi: 10.1056/NEJMoa042982. [DOI] [PubMed] [Google Scholar]
- 20.Steinman L. Blocking adhesion molecules as therapy for multiple sclerosis: natalizumab. Nature reviews. Drug discovery. 2005;4:510–518. doi: 10.1038/nrd1752. [DOI] [PubMed] [Google Scholar]
- 21.Roan N, Gierahn T, Higgins D, Starnbach M. Monitoring the T cell response to genital tract infection. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12069–12143. doi: 10.1073/pnas.0603866103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olive A, Gondek D, Starnbach M. CXCR3 and CCR5 are both required for T cell-mediated protection against C. trachomatis infection in the murine genital mucosa. Mucosal immunology. 2011;4:208–216. doi: 10.1038/mi.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hawkins R, Rank R, Kelly K. Expression of mucosal homing receptor alpha4beta7 is associated with enhanced migration to the Chlamydia-infected murine genital mucosa in vivo. Infection and Immunity. 2000;68:5587–5594. doi: 10.1128/iai.68.10.5587-5594.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly K, Chan A, Butch A, Darville T. Two different homing pathways involving integrin β7 and E-selectin significantly influence trafficking of CD4 cells to the genital tract following Chlamydia muridarum infection. American journal of reproductive immunology (New York, N.Y. : 1989) 2009;61:438–445. doi: 10.1111/j.1600-0897.2009.00704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perry L, Feilzer K, Portis J, Caldwell H. Distinct homing pathways direct T lymphocytes to the genital and intestinal mucosae in Chlamydia-infected mice. Journal of immunology (Baltimore, Md. : 1950) 1998;160:2905–2914. [PubMed] [Google Scholar]
- 26.Di Genova G, Savelyeva N, Suchacki A, Thirdborough S, Stevenson F. Bystander stimulation of activated CD4+ T cells of unrelated specificity following a booster vaccination with tetanus toxoid. European journal of immunology. 2010;40:976–985. doi: 10.1002/eji.200940017. [DOI] [PubMed] [Google Scholar]
- 27.von Andrian U, Mackay C. T-cell function and migration. Two sides of the same coin. The New England journal of medicine. 2000;343:1020–1034. doi: 10.1056/NEJM200010053431407. [DOI] [PubMed] [Google Scholar]
- 28.Fässler R, Meyer M. Consequences of lack of beta 1 integrin gene expression in mice. Genes & development. 1995;9:1896–1908. doi: 10.1101/gad.9.15.1896. [DOI] [PubMed] [Google Scholar]
- 29.DeNucci C, Pagán A, Mitchell J, Shimizu Y. Control of alpha4beta7 integrin expression and CD4 T cell homing by the beta1 integrin subunit. Journal of immunology (Baltimore, Md. : 1950) 2010;184:2458–2467. doi: 10.4049/jimmunol.0902407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelly K, Rank R. Identification of homing receptors that mediate the recruitment of CD4 T cells to the genital tract following intravaginal infection with Chlamydia trachomatis. Infection and Immunity. 1997;65:5198–5208. doi: 10.1128/iai.65.12.5198-5208.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Croft M, Bradley L, Swain S. Naive versus memory CD4 T cell response to antigen. Memory cells are less dependent on accessory cell costimulation and can respond to many antigen-presenting cell types including resting B cells. Journal of immunology (Baltimore, Md. : 1950) 1994;152:2675–2685. [PubMed] [Google Scholar]
- 32.Iwata M, Hirakiyama A, Eshima Y, Kagechika H, Kato C, Song S-Y. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 2004;21:527–538. doi: 10.1016/j.immuni.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 33.Ruane D, Brane L, Reis B, Cheong C, Poles J, Do Y, Zhu H, Velinzon K, Choi J-H, Studt N, Mayer L, Lavelle E, Steinman R, Mucida D, Mehandru S. Lung dendritic cells induce migration of protective T cells to the gastrointestinal tract. The Journal of experimental medicine. 2013;210:1871–1888. doi: 10.1084/jem.20122762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mariano JE, Laurelee O, Yoshikazu T, Carol C, Stefan L, Martin EH, Roy RL. VCAM-1 on activated endothelium interacts with the leukocyte integrin VLA-4 at a site distinct from the VLA-4/Fibronectin binding site. Cell. 1990;60 doi: 10.1016/0092-8674(90)90661-w. [DOI] [PubMed] [Google Scholar]
- 35.Alon R. The integrin VLA-4 supports tethering and rolling in flow on VCAM-1. The Journal of Cell Biology. 1995;128 doi: 10.1083/jcb.128.6.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Andrian U, Engelhardt B. Alpha4 integrins as therapeutic targets in autoimmune disease. The New England journal of medicine. 2003;348:68–72. doi: 10.1056/NEJMe020157. [DOI] [PubMed] [Google Scholar]
- 37.Johansson E, Rudin A, Wassén L, Holmgren J. Distribution of lymphocytes and adhesion molecules in human cervix and vagina. Immunology. 1999;96:272–277. doi: 10.1046/j.1365-2567.1999.00675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parr M, Parr E. Interferon-gamma up-regulates intercellular adhesion molecule-1 and vascular cell adhesion molecule-1 and recruits lymphocytes into the vagina of immune mice challenged with herpes simplex virus-2. Immunology. 2000;99:540–545. doi: 10.1046/j.1365-2567.2000.00980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jayarapu K, Kerr M, Ofner S, Johnson RM. Chlamydia-specific CD4 T cell clones control Chlamydia muridarum replication in epithelial cells by nitric oxide-dependent and -independent mechanisms. J Immunol. 2010;185:6911–6920. doi: 10.4049/jimmunol.1002596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.