Abstract
A key feature of Alzheimer’s disease (AD) is deposition of extracellular amyloid plaque comprised chiefly of the amyloid β (Aβ) peptide. Studies of Aβ have shown that it may be catabolized by proteolysis or cleared from brain via members of the low-density lipoprotein receptor family. Alternatively, Aβ can undergo a conformational transition from α-helix to β-sheet, a conformer that displays a propensity to self-associate, oligomerize and form fibrils. Furthermore, β-sheet conformers catalyze conversion of other α-helical Aβ peptides to β-sheet, feeding the oligomer and fibril assembly process. A factor that influences the fate of Aβ in the extracellular space is apolipoprotein (apo) E. Polymorphism at position 112 or 158 in apoE give rise to three major isoforms. One isoform in particular, apoE4 (Arg at 112 and 158), has generated considerable interest since the discovery that it is the major genetic risk factor for development of late onset AD. Despite this striking correlation, the molecular mechanism underlying apoE4’s association with AD remains unclear. A tertiary structural feature distinguishing apoE4 from apoE2 and apoE3, termed domain interaction, is postulated to affect the conformation and orientation of its’ two independently folded domains. This feature has the potential to influence apoE4’s interaction with Aβ, its sensitivity to proteolysis or its lipid accrual and receptor binding activities. Thus, domain interaction may constitute the principal molecular feature of apoE4 that predisposes carriers to late onset AD. By understanding the contribution of apoE4 to AD at the molecular level new therapeutic or prevention strategies will emerge.
Keywords: Alzheimer’s disease, apolipoprotein E, isoform, domain interaction, amyloid beta peptide
I. ALZHEIMER’s DISEASE: INTRODUCTION AND OVERVIEW
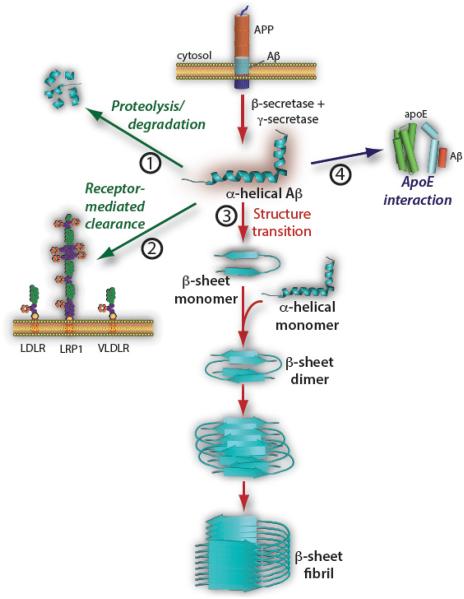
Alzheimer’s disease (AD) is the most frequent cause of senile dementia, affecting 40% of Americans over age 85. Moreover, as the proportion of aged individuals increases, the burden of this disease is certain to worsen [1]. AD is characterized by progressive neurodegeneration associated with extracellular deposition of amyloid beta (Aβ) peptide as plaque and accumulation of hyperphosphorylated Tau protein as intracellular neurofibrillary tangles. Aβ is a collection of peptides ranging in length from 12 to 42 amino acids derived from proteolytic processing of the transmembrane amyloid precursor protein by the consecutive action of β-and γ-secretases (Fig. 1). The major species, Aβ (1-40) and Aβ (1-42), are hydrophobic peptides that accumulate in amyloid plaque deposits that represent a clinical hallmark of AD. A central tenet of the “amyloid hypothesis” is that disease results from a persistent imbalance between Aβ production and clearance. Failure to efficiently degrade Aβ, or clear it from the extracellular space [2], appears to contribute to initiation and progression of disease. Under physiological conditions, Aβ-degrading proteases, including neprolysin and insulin degrading enzyme, function to digest Aβ directly; Fig. 1, path 1). Alternatively, Aβ may be subject to receptor-mediated clearance through interaction with members of the low density lipoprotein receptor (LDLR) family, including the LDLR, the low-density lipoprotein receptor related protein 1 (LRP1) or the very low density lipoprotein receptor (VLDLR) [3, 4]; (Fig. 1, path 2). Both proteolytic degradation and receptor mediated endocytosis represent physiological Aβ catabolic paths that counterbalance Aβ production. A third path (Fig. 1, path 3) involves the structural transition of Aβ peptides mentioned above. This process, considered to be a fundamental contributor to fibrillization and plaque formation, is initiated when Aβ transitions from α-helix to β-sheet secondary structure. This event precedes the pathological sequence of self-association, oligomerization and fibril formation, an end point that constitutes the structural basis of Aβ plaque deposits [5-7]. In addition, once produced, β-sheet conformers are capable of catalyzing the α-helix → β-sheet transition of other Aβ monomers [8, 9]. In this way, the pathological process is sustained by continuous production of β-sheet conformers that supply building blocks for fibril growth.
Fig. (1). Pathways of Aβ metabolism.
Soluble amyloid beta (Aβ) peptide is generated from proteolytic processing of amyloid precursor protein (APP) by the successive action of β- and γ-secretases in brain cells (top center). The level of Aβ production is counterbalanced by its degradation via protease digestion (Path 1) and receptor mediated endocytosis (Path 2). Alternatively, soluble α-helical Aβ may undergo a pathological transition to β-sheet conformer that promotes self-association and oligomerization (Path 3). How interaction between apolipoprotein (apoE) and Aβ (Path 4) influences Aβ metabolic fate is the subject of this review.
Also impacting the metabolic fate of Aβ are myriad extraneous factors that influence its propensity to form plaque deposits. One factor, in particular, that has a profound effect on Aβ metabolism is apolipoprotein (apo) E (Fig. 1, path 4) and the present review focuses on the unique and perplexing relationship between this protein, Aβ metabolism, lipid flux and AD.
II. THE apoE CONNECTION
Although it was originally discovered and characterized for its role in plasma lipoprotein metabolism, apoE is now recognized to have a major impact on neuronal function [10]. Human apoE is a 299 amino acid secreted protein that exists as one of three isoforms that differ by a single amino acid. The parent isoform, apoE3, possesses Cys at position 112 and Arg at position 158. ApoE2 has Cys at both these sites while apoE4 has Arg. Structural studies of apoE have shown it is comprised of two independently folded domains, a 22 kDa N-terminal (NT) four helix bundle domain and a 10 kDa C-terminal (CT) domain that are connected by an unstructured hinge segment (Fig. 2) [11, 12]. Cell types and tissues that express apoE include liver, macrophages and brain, predominantly astrocytes and microglia. In brain, apoE is secreted as a lipid-poor protein that accrues lipid to form brain specific lipoprotein particles [13]. ApoE has been shown to bind Aβ and studies comparing apoE3 and apoE4 have documented differences in their respective Aβ binding properties [14-18]. Such differences are entirely consistent with the revelation that apoE4 is the major genetic risk factor for late-onset AD [19]. Indeed, individuals with a single copy of APOE4 manifest a 5 fold increased chance of developing AD while those with two copies have an estimated 20 fold increased risk [20]. The positive predictive value for symptomatic AD in patients who carry at least one APOE4 allele is >95%. Thus, early in the clinical course of dementia, when diagnosis may be ambiguous, the presence of APOE4 raises the diagnostic accuracy of AD [21]. A fundamental question emerging from this striking genetic association relates to the molecular basis of this effect. A plausible explanation is that structural differences among apoE isoforms affect their respective interactions with Aβ. This may include isoform specific differences in Aβ binding or a differential ability to affect the conformational status of Aβ. Unfortunately, there is not enough information or experimental results available to answer these issues in a definitive manner. Indeed, it is not known whether Aβ binding to apoE is conformation specific or if it displays a differential binding affinity for Aβ monomers versus oligomers.
Fig. (2). Two-domain structural model of apoE.
Full-length apoE (299 amino acids) is composed of two independently folded structural domains. The 4-helix bundle structure of the N-terminal (NT) domain (X-ray crystal structure PDB ID:1LPE, Wilson et al., 1991) is connected to the modeled C-terminal (CT) domain by a flexible hinge segment that is sensitive to proteolysis. The NT domain contains the LDL receptor family binding recognition sequence (residues 136-150) while the CT domain is responsible for lipid binding and Aβ interaction.
Regardless of the precise nature of its binding interaction with Aβ, it is generally recognized that structural differences among apoE isoforms underlie the pathology associated with apoE4 [22]. Moreover, if apoE3 is considered neutral, apoE2 is regarded as protective against AD [23, 24]. On the basis of in vitro binding assays, Strittmatter et al. [14] documented isoform specific differences between apoE3 and apoE4 in terms of Aβ binding. Noting that these isoforms differ from apoE4 by single amino acid substitutions within the NT helix bundle domain, it may be anticipated that the NT domain alone constitutes the critical part of the apoE molecule that is associated with AD pathology. This, however, appears not to be the case. The finding that Aβ interacts with the CT domain of apoE [17, 25-27] indicates isoform specific differences in Aβ-dependent AD pathology likely involves some form of communication between the NT and CT domains. Toward this end, a phenomenon known as “domain interaction” has emerged as a unique structural feature that distinguishes apoE4 from other isoforms.
III. DOMAIN INTERACTION
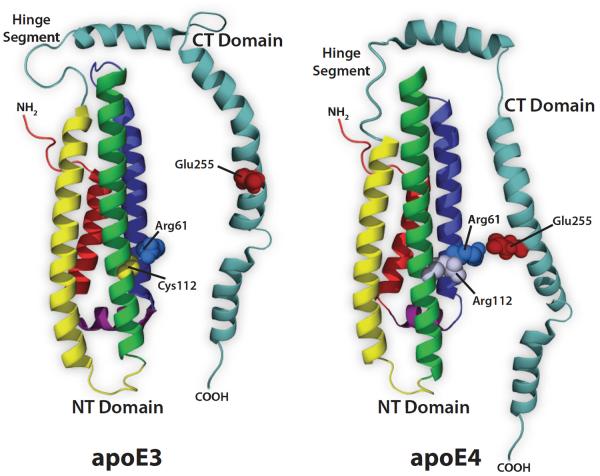
The concept of domain interaction in apoE4 emerged from studies of plasma lipoprotein metabolism. Briefly, Gregg et al. [28] found that apoE4 distributes abnormally among lipoproteins in plasma. Whereas apoE3 localizes to high-density lipoproteins (HDL), apoE4 displays a preference for larger, VLDL particles. In pursuing this, Weisgraber found that apoE4’s preference for VLDL was directly related to the Arg for Cys substitution at position 112 in the NT domain of this isoform [29]. On the basis of an in vitro lipoprotein binding preference assay, X-ray crystallography and site directed mutagenesis, evidence was obtained that Arg112 affects the spatial orientation of Arg61, such that its positively charged side chain forms a salt bridge with the negatively charged side chain of Glu255 in the CT domain (Fig. 3) [30, 31]. Since Arg61 and Glu255 reside in different domains of apoE, the term “domain interaction” was coined to describe this phenomenon. A question emerging from these results is “how could domain interaction in apoE4 manifest pathophysiological consequences associated with AD”? One possibility is that it imposes a structural constraint [32] that affects how apoE4, or apoE4-Aβ complexes, are processed. For example, if domain interaction alters the lipid binding and accrual properties [27] or protease sensitivity of apoE4 [33], an impact on Aβ metabolic fate would be anticipated.
Fig. (3). Putative isoform specific differences in NT – CT domain interaction.
The NT domain 4-helix bundle is from the X-ray crystal structure of the isolated apoE3 and apoE4 domains (PDB ID:1NFN and PDB ID:1B68, respectively). The CT domain and hinge segment have been modeled for illustration (adapted from Zhong and Weisgraber (2009a)). Key residues, known to be involved in the isoform specific structural differences between apoE3 and apoE4 are indicated. In apoE4, the presence of Arg112 (compared with Cys112 in apoE3) on helix 3 changes the conformation of Arg61 on helix 2 to allow for greater NT and CT domain interaction via an Arg61-Glu255 salt bridge.
Evidence suggests domain interaction alters the orientation or alignment of CT domain α-helices such that the protein is attracted to more planar lipid surfaces [30]. Insofar as brain possesses only HDL particles with a high degree of surface curvature [34-36], it is conceivable that domain interaction alters the relative affinity of apoE4 for brain lipoproteins. If so, it may be that a higher proportion of apoE4 exists in a lipid-poor state. Considering that other apolipoproteins (e.g. apoA-I) are rapidly degraded if they are unable to accrue lipid [37], it follows that domain interaction-induced structural constraints that lead to defective lipid accrual would result in a lower concentration of apoE4 compared to other isoforms. In keeping with this postulate, Bales et al. [38] found that lower levels of apoE4 in brain are associated with increased Aβ accumulation, suggesting a domino effect on Aβ clearance capacity [39, 40]. If less apoE is available for interaction with Aβ, it follows that the probability A will enter the pathological path toward fibril formation will increase. The corollary to this, that increasing apoE expression facilitates Aβ clearance [39, 41, 42], is currently under intense investigation.
IV. THE EFFECT OF apoE LIPIDATION ON Aβ METABOLIC FATE
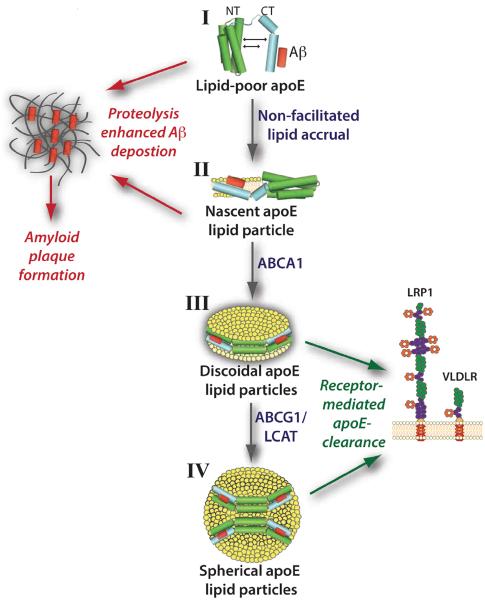
ApoE secreted from astrocytes and microglia gradually accrues lipid, ultimately forming a mature spherical lipoprotein particle. As shown in (Fig. 4), the process of apoE lipidation can be quantized into discrete stages. The first stage, lipid-poor apoE, is likely a transient species that rapidly associates with phospholipid and cholesterol through active transport (i.e. via ATP-binding cassette transporter A1 (ABCA1) [43, 44] or indirectly through passive accumulation. The latter mode is operative in abca1 (-/-) mice where central nervous system lipoproteins exist as small, lipid-poor particles ∼8 nm in diameter [45, 46]. These apoE-lipid complexes (Fig. 4, stage II) exist as discrete entities that manifest unique properties. A defining feature of these particles is the manner in which apoE physically contacts the lipid substrate. As mentioned above, apoE is comprised of two structural domains. Whereas the 10 kDa CT domain has a high affinity for lipid surfaces and initiates contact with lipid [47, 48], the 22 kDa NT domain four-helix bundle is exceptionally stable [49, 50] and displays a weak affinity for lipid surfaces [29]. Thus, when ABCA1 is absent or efflux accessible lipid is limiting, the CT domain sequesters available lipid, generating apoE lipid particles in which the NT domain maintains a four-helix bundle conformation. Insofar as the CT domain drives apoE interaction with ABCA1 [51], it may be envisioned that, when lipid availability is low, ABCA1-mediated cholesterol and phospholipid efflux generates stage II apoE lipid particles. Importantly, neither stage I nor stage II apoE particles are capable of functioning as ligands for members of the LDLR family [11, 52]. This is because critical positively charged amino acid residues in helix 4 of the NT domain (residues 136 – 150) do not adopt a receptor competent conformation at this stage of lipid particle maturation [53]. Interaction of lipid-poor or nascent apoE lipid particles (i.e. Fig. 4, stage I or II) with ABCA1 under conditions where lipid availability is not limiting will generate discoidal particles (Fig. 4, stage III) that serve as precursors of mature spheroidal lipoproteins present in brain. In stage III particles, the NT domain “opens” about a hinge region connecting helical segments essentially substituting helix-helix interactions that stabilize the four-helix bundle for helix-lipid interactions [54, 55]. On these particles, apoE adopts an extended conformation that circumscribes the perimeter of a disk-shaped lipid bilayer, contacting and stabilizing the edge of the disk [56]. This interaction is possible because of the amphipathic nature of apoE helix segments [57]. Such helices bind lipid surfaces via their hydrophobic face while their polar face is directed toward the aqueous milieu. Conversion of discoidal particles to spherical HDL requires the combined action of ABCA1 and ABCG1 [13], which funnel cholesterol and phospholipid into the particle as lipid modifying enzymes and transfer proteins remodel their composition. Lecithin:cholesterol acyltransferase (LCAT) catalyzes the transfer of an acyl moiety from the sn-2 position of phosphatidylcholine to the β-OH group of cholesterol, generating cholesteryl ester [58]. The cholesteryl ester product partitions between leaflets of the bilayer, creating a particle core that expands as a function of continued efflux and LCAT activity. Remodeling proteins, such as phospholipid transfer protein, redistribute lipid between lipoproteins and/or membranes as part of the maturation process [59]. Both stage III and stage IV apoE lipid particles manifest full LDLR family binding activity [47]. Thus, if Aβ associates with the CT domain of apoE, in order for these complexes to be cleared via LDLR family members, lipid particle maturation must be achieved.
Fig. (4). Putative effects of apoE lipidation on Aβ metabolism.
Lipid-poor apolipoprotein (apo) E is secreted from astrocytes and glial cells in brain and is lipidated in discrete stages (I-IV) by the collective action of ATP-binding cassette (ABC) transporter proteins (ABCA1 and ABCG1), lipid modifying enzymes and transfer proteins (e.g. lecithin:cholesterol acyltransferase (LCAT)). Poorly lipidated apoE (stage I) and nascent apoE particles (stage II) are more susceptible to proteolysis which may lead to greater Aβ deposition and enhanced plaque formation due to impaired clearance, whereas lipidated discoidal (stage III) and spherical (stage IV) apoE-containing lipoproteins are ligands for LDL receptor family members, potentially leading to enhanced binding and clearance of apoE-Aβ complexes from brain.
In the absence of timely lipid particle maturation it is conceivable that apoE susceptibility to proteolysis increases. Toward this end, Huang et al. [60] showed that domain interaction enhances protease sensitivity in the CT domain of apoE4, generating truncated species that contribute to AD pathology. Harris et al. [61] went on to identify a chymotrypsin-like serine protease with a preference for apoE4 that generates “toxic” CT truncated fragments. More recently, Jones et al. [62] hypothesized that domain interaction in apoE4 alters protease sensitivity in the hinge segment connecting the NT and CT domains (see Fig. 2). If proteolysis of apoE occurs in this hinge segment, Aβ associated with the CT domain cannot be cleared via apoE-dependent LDLR family interactions. In this case, Aβ may have increased opportunity to interact with other Aβ molecules in the extracellular space and, as such, be subject to pathological folding events analogous to prion disease (see Fig. 1) [9, 63]. The extent to which Aβ interaction with different apoE isoforms affects its sensitivity to protease digestion has yet to be investigated in detail.
Insofar as apoE-Aβ metabolism is affected by multiple factors, scenarios may exist where apoE is either beneficial or detrimental. Because lipidation state is a major factor affecting apoE-Aβ metabolic fate, it is reasonable to consider that, when lipid is limiting, a greater proportion of apoE-Aβ will exist in a lipid-poor or nascent lipid particle state and, as a result, Aβ susceptibility to pathological folding will increase. On the other hand, if efflux accessible lipid is abundant, ABC transporter dependent lipidation of apoE-Aβ complexes will generate mature lipid particles that expedite Aβ clearance via LDLR family member interactions. This concept has gained support from studies in an Alzheimer’s susceptible mouse model over-expressing ABCA1, wherein Aβ deposition as plaque was dramatically decreased, presumably a result of increased apoE lipidation and enhanced Aβ metabolism [64]. More recently, Youmans et al. [65] employed a mouse model of AD to show that less apoE4 is lipoprotein associated (and possibly present in a less lipidated state) compared to apoE2 and apoE3. Importantly, these isoform specific differences correlate with differences in the relative abundance of soluble and oligomeric Aβ. In another study [66], in vivo and in vitro evidence suggests apoE4 impacts the formation of Aβ oligomers through interaction with its C-terminal domain in a manner that is dependent upon its lipidation state.
V. ApoE: IS MORE BETTER?
An obvious way to examine the effect of apoE on neurodegeneration is to assess the impact of its overexpression or gene disruption. Whereas the apoE null mouse appears normal [67], cognitive deficits and other phenotypic changes do occur with age [68]. It is noteworthy, however, that brain tissue possesses compensatory mechanisms, such as up-regulation of apoJ (also known as Clusterin), that are capable of fulfilling functions normally carried out by apoE including lipid association [69, 70], Aβ binding and receptor (e.g. LRP2/Megalin) mediated clearance [71-73]. Thus, the extent to which compensation by other proteins occurs must be kept in mind when interpreting experiments designed to manipulate apoE levels.
Studies by Holtzman et al. [74] found that expression of human apoE3 or apoE4 in a mouse model of AD reduced Aβ deposition. At the same time, Buttini et al. [75] reported differential effects on neuronal integrity in apoE null mice expressing human apoE3 versus apoE4. Another model has emerged from studies targeting nuclear hormone receptors that regulate apoE expression. Agonists of the liver X receptor [76, 77], the retinoid X receptor (RXR) or peroxisome proliferator-activated receptor gamma (PPAR-γ) lead to increased apoE production [78, 79]. Transactivation using the RXR agonist, bexarotene, LXR agonists (e.g. TO901317 or GW3965) or the PPAR-γ agonist, pioglitazone, results in coordinated up-regulation of apoE, ABCA1 and ABCG1 in brain [80-84]. In mouse models of AD, administration of such agonists leads to enhanced Aβ clearance and reversal of cognitive deficits associated with disease. The intriguing result that bexarotene had no effect when administered to apoE null mice [75] is consistent with the concept that increased expression of apoE promotes enhanced binding and clearance of soluble Aβ, effectively diverting it from path 3 to path 4 in (Fig. 1). While these results support a “chaperone” role for apoE in Aβ metabolism, they do not explain isoform-specific effects of human apoE on this process [85]. Moreover, it is reasonable to consider that, given apoE4’s strong connection to AD pathology, overexpression of this isoform may ultimately be detrimental, despite the fact that short-term up-regulation via nuclear hormone receptor activation improves Aβ clearance.
Contrary to data indicating up-regulation of apoE confers therapeutic benefit, others report that decreasing the level of apoE in brain improves Aβ clearance [86, 87]. If apoE levels are limiting while cells are replete with efflux accessible lipid, a greater proportion of the apoE protein pool will achieve a mature lipidation state, such that greater flux of apoE-Aβ through LDLR family member mediated endocytosis will occur. A deleterious scenario may exist, however, if there is an abundance of apoE but insufficient lipid availability. In this case a greater proportion of apoE will be unable to attain a mature lipidation state in a timely manner, resulting in longer residence time in the extracellular space, increased susceptibility to proteolysis and aberrant Aβ metabolism (i.e. pathological folding). An example of disrupted apoE lipidation is the ABCA1 null mouse, where the absence of this transporter led to an 80% decrease in apoE levels and a corresponding increase in amyloid load when the mice were crossed into an Alzheimer’s susceptible background [88-90]. The presence of apoE4 may exacerbate issues created by insufficient lipid availability due to its unique Aβ interaction properties, susceptibility to proteolysis and/or altered lipid accrual kinetics [85]. Thus, whereas pharmacological “tuning” of ABCA1 and apoE levels may provide therapeutic promise, knowing the exact contribution and role of the different apoE isoforms in lipidation and Aβ metabolizing pathways is required before proceeding with a unilateral approach. It is also worth noting that apoE expression levels are also affected by single nucleotide polymorphisms (SNPs) in the promoter region of APOE [91]. Furthermore, the finding that an “Aβ interacting domain” exists within the APOE promoter region [92] suggests Aβ itself may serve as a transcription factor capable of influencing apoE gene expression.
VI. FUTURE DIRECTIONS
AD is a complex, progressive disease with multiple contributing factors. The strong positive correlation between the ε4 allele of APOE and AD has driven a concerted research effort. This has led to the postulate that domain interaction in apoE4 is a cardinal feature that distinguishes this isoform from apoE3 and apoE2. Mechanism-based research, designed to explain how domain interaction in apoE4 increases the risk of late onset AD, presumably by influencing the metabolic fate of Aβ, has progressed steadily.
Another promising area of research, with the potential to give rise to therapeutic intervention strategies, is activation of nuclear hormone receptors that regulate production of apoE and ABC transporters [79]. By up-regulating these proteins, increased lipid flux drives apoE toward a mature lipidation state. In so doing, associated Aβ will be cleared via LDLR family members and degraded [64]. A factor that must be considered, however, is lipid availability/supply for efflux dependent apoE particle maturation. Under physiological conditions, activation of nuclear hormone receptors serves as a mechanism to limit free cholesterol in tissues experiencing high cholesterol flux. As such, simply making more apoE and ABC transporters may have a negative impact if insufficient lipid is available to fuel the pathway. Indeed, it is likely that, in the absence of an efflux accessible lipid pool, increased production of apoE will result in a larger proportion of this protein present in a lipid-poor or nascent lipid particle state (stages I and II in Fig. 4). In this case, apoE association with Aβ may be pathological, driving Aβ toward fibril formation. Further confounding the balance between lipid availability, Aβ binding and apoE concentration, are isoform specific differences in apoE. While many interpretations are possible, the most prevalent is that apoE4, perhaps owing to domain interaction, is defective in one or more of lipid accrual, Aβ binding or protease sensitivity. Thus, the idea that more apoE4 is better is not necessarily true, especially long term.
Given the evidence that domain interaction may be directly related to pathological consequences associated with apoE4, pharmacological disruption of this structural feature has been pursued as an approach to abrogate the negative impact of this isoform. Using mitochondrial dysfunction as a model of AD pathology, Chen et al. [93] showed that treatment of apoE4 expressing neuro2a cells with a molecule capable of disrupting domain interaction effectively restores mitochondrial respiratory complex 4 levels. Subsequently, Chen et al. [94] used a high throughput assay to identify a phthalazinone “structure corrector” that reversed impairments in mitochondrial motility and neurite outgrowth. Taken together, these data suggest that pharmacological disruption of domain interaction in apoE4 has the potential to ameliorate its pathological effects in vivo.
As the search for “magic bullet” small molecules capable of specifically disrupting domain interaction in apoE4 progresses, it is important to focus on the structural basis of this phenomenon as well as the precise nature of isoform-specific differences. At present, the sole defining feature of domain interaction is a salt bridge between Arg61 and Glu255 in apoE4 but not in apoE3 (see Fig. 3) [12]. However, despite the apparent lack of an Arg61 – Glu255 salt bridge in apoE3 [95], Narayanaswami et al. [96], Hatters et al. [97] and Chen et al. [64] have reported that the NT and CT domains in apoE3 are proximal to one another. Thus, it appears that subtle differences in the relative strength of domain interaction may distinguish these isoforms.
In summary, ongoing work on apoE-Aβ interactions has led to testable hypotheses that should yield definitive answers. We anticipate that a combination of structure-based studies, cell and model organism investigations as well as pharmacological intervention, will lead to new strategies for the diagnosis, prevention and treatment of this growing epidemic.
ACKNOWLEDGEMENTS
We would like to acknowledge Drs. Andrzej Witkowski and Jens Simonsen for careful reading of the manuscript. Supported by a grant from NIH (HL 64159).
Footnotes
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflicts of interest.
References
- 1.Selkoe DJ. Preventing Alzheimer’s disease. Science. 2012;337(6101):1488–92. doi: 10.1126/science.1228541. [DOI] [PubMed] [Google Scholar]
- 2.Mawuenyega KG, Sigurdson W, Ovod V, Munsell L, Kasten T, Morris JC, et al. Decreased clearance of CNS beta-amyloid in Alzheimer’s disease. Science. 2010;330(6012):1774. doi: 10.1126/science.1197623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deane R, Bell RD, Sagare A, Zlokovic BV. Clearance of amyloid-beta peptide across the blood-brain barrier: implication for therapies in Alzheimer’s disease. CNS Neurol Disord Drug Targets. 2009;8(1):16–30. doi: 10.2174/187152709787601867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holtzman DM, Herz J, Bu G. Apolipoprotein e and apolipoprotein e receptors: normal biology and roles in Alzheimer disease. Cold Spring Harb Perspect Med. 2012;2(3):a006312. doi: 10.1101/cshperspect.a006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu J, Zhang JZ, Xiang Y. Molecular Dynamics Simulation and Computational Two-dimensional Infrared Spectroscopic Study of Model Amyloid ss-peptide Oligomers. J Phys Chem A. 2013 doi: 10.1021/jp403748z. [DOI] [PubMed] [Google Scholar]
- 6.Cerf E, Sarroukh R, Tamamizu-Kato S, Breydo L, Derclaye S, Dufrene YF, et al. Antiparallel beta-sheet: a signature structure of the oligomeric amyloid beta-peptide. Biochem J. 2009;421(3):415–23. doi: 10.1042/BJ20090379. [DOI] [PubMed] [Google Scholar]
- 7.Sarroukh R, Cerf E, Derclaye S, Dufrene YF, Goormaghtigh E, Ruysschaert JM, et al. Transformation of amyloid beta(1-40) oligomers into fibrils is characterized by a major change in secondary structure. Cell Mol Life Sci. 2011;68(8):1429–38. doi: 10.1007/s00018-010-0529-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prusiner SB. Cell biology. A unifying role for prions in neurodegenerative diseases. Science. 2012;336(6088):1511–3. doi: 10.1126/science.1222951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stohr J, Watts JC, Mensinger ZL, Oehler A, Grillo SK, DeArmond SJ, et al. Purified and synthetic Alzheimer’s amyloid beta (Abeta) prions. Proc Natl Acad Sci U S A. 2012;109(27):11025–30. doi: 10.1073/pnas.1206555109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hauser PS, Narayanaswami V, Ryan RO. Apolipoprotein E: from lipid transport to neurobiology. Prog Lipid Res. 2011;50(1):62–74. doi: 10.1016/j.plipres.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weisgraber KH. Apolipoprotein E: structure-function relationships. Adv Protein Chem. 1994;45:249–302. doi: 10.1016/s0065-3233(08)60642-7. [DOI] [PubMed] [Google Scholar]
- 12.Zhong N, Weisgraber KH. Understanding the association of apolipoprotein E4 with Alzheimer disease: clues from its structure. J Biol Chem. 2009;284(10):6027–31. doi: 10.1074/jbc.R800009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan J, Donkin J, Wellington C. Greasing the wheels of Abeta clearance in Alzheimer’s disease: the role of lipids and apolipoprotein E. Biofactors. 2009;35(3):239–48. doi: 10.1002/biof.37. [DOI] [PubMed] [Google Scholar]
- 14.Holtzman DM, Bales KR, Tenkova T, Fagan AM, Parsadanian M, Sartorius LJ, et al. Apolipoprotein E isoform-dependent amyloid deposition and neuritic degeneration in a mouse model of Alzheimer’s disease. Proc Natl Acad Sci U S A. 2000;97(6):2892–7. doi: 10.1073/pnas.050004797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LaDu MJ, Falduto MT, Manelli AM, Reardon CA, Getz GS, Frail DE. Isoform-specific binding of apolipoprotein E to beta-amyloid. J Biol Chem. 1994;269(38):23403–6. [PubMed] [Google Scholar]
- 16.Sanan DA, Weisgraber KH, Russell SJ, Mahley RW, Huang D, Saunders A, et al. Apolipoprotein E associates with beta amyloid peptide of Alzheimer’s disease to form novel monofibrils. Isoform apoE4 associates more efficiently than apoE3. J Clin Invest. 1994;94(2):860–9. doi: 10.1172/JCI117407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strittmatter WJ, Weisgraber KH, Huang DY, Dong LM, Salvesen GS, Pericak-Vance M, et al. Binding of human apolipoprotein E to synthetic amyloid beta peptide: isoform-specific effects and implications for late-onset Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90(17):8098–102. doi: 10.1073/pnas.90.17.8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tokuda T, Calero M, Matsubara E, Vidal R, Kumar A, Permanne B, et al. Lipidation of apolipoprotein E influences its isoform-specific interaction with Alzheimer’s amyloid beta peptides. Biochem J. 2000;348(Pt 2):359–65. [PMC free article] [PubMed] [Google Scholar]
- 19.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261(5123):921–3. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 20.Strittmatter WJ. Medicine. Old drug, new hope for Alzheimer’s disease. Science. 2012;335(6075):1447–8. doi: 10.1126/science.1220725. [DOI] [PubMed] [Google Scholar]
- 21.Roses AD. Apolipoprotein E and Alzheimer’s disease. The tip of the susceptibility iceberg. Ann N Y Acad Sci. 1998;855:738–43. doi: 10.1111/j.1749-6632.1998.tb10653.x. [DOI] [PubMed] [Google Scholar]
- 22.Mahley RW, Weisgraber KH, Huang Y. Apolipoprotein E4: a causative factor and therapeutic target in neuropathology, including Alzheimer’s disease. Proc Natl Acad Sci U S A. 2006;103(15):5644–51. doi: 10.1073/pnas.0600549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benjamin R, Leake A, McArthur FK, Ince PG, Candy JM, Edwardson JA, et al. Protective effect of apoE epsilon 2 in Alzheimer’s disease. Lancet. 1994;344(8920):473. doi: 10.1016/s0140-6736(94)91804-x. [DOI] [PubMed] [Google Scholar]
- 24.Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC, Jr., et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet. 1994;7(2):180–4. doi: 10.1038/ng0694-180. [DOI] [PubMed] [Google Scholar]
- 25.Cho HS, Hyman BT, Greenberg SM, Rebeck GW. Quantitation of apoE domains in Alzheimer disease brain suggests a role for apoE in Abeta aggregation. J Neuropathol Exp Neurol. 2001;60(4):342–9. doi: 10.1093/jnen/60.4.342. [DOI] [PubMed] [Google Scholar]
- 26.Phu MJ, Hawbecker SK, Narayanaswami V. Fluorescence resonance energy transfer analysis of apolipoprotein E C-terminal domain and amyloid beta peptide (1-42) interaction. J Neurosci Res. 2005;80(6):877–86. doi: 10.1002/jnr.20503. [DOI] [PubMed] [Google Scholar]
- 27.Tamamizu-Kato S, Cohen JK, Drake CB, Kosaraju MG, Drury J, Narayanaswami V. Interaction with amyloid beta peptide compromises the lipid binding function of apolipoprotein E. Biochemistry. 2008;47(18):5225–34. doi: 10.1021/bi702097s. [DOI] [PubMed] [Google Scholar]
- 28.Gregg RE, Zech LA, Schaefer EJ, Stark D, Wilson D, Brewer HB., Jr. Abnormal in vivo metabolism of apolipoprotein E4 in humans. J Clin Invest. 1986;78(3):815–21. doi: 10.1172/JCI112645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weisgraber KH. Apolipoprotein E distribution among human plasma lipoproteins: role of the cysteine-arginine interchange at residue 112. J Lipid Res. 1990;31(8):1503–11. [PubMed] [Google Scholar]
- 30.Dong LM, Weisgraber KH. Human apolipoprotein E4 domain interaction. Arginine 61 and glutamic acid 255 interact to direct the preference for very low density lipoproteins. J Biol Chem. 1996;271(32):19053–7. doi: 10.1074/jbc.271.32.19053. [DOI] [PubMed] [Google Scholar]
- 31.Dong LM, Wilson C, Wardell MR, Simmons T, Mahley RW, Weisgraber KH, et al. Human apolipoprotein E. Role of arginine 61 in mediating the lipoprotein preferences of the E3 and E4 isoforms. J Biol Chem. 1994;269(35):22358–65. [PubMed] [Google Scholar]
- 32.Zhong N, Scearce-Levie K, Ramaswamy G, Weisgraber KH. Apolipoprotein E4 domain interaction: synaptic and cognitive deficits in mice. Alzheimers Dement. 2008;4(3):179–92. doi: 10.1016/j.jalz.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barbier A, Clement-Collin V, Dergunov AD, Visvikis A, Siest G, Aggerbeck LP. The structure of human apolipoprotein E2, E3 and E4 in solution 1. Tertiary and quaternary structure. Biophys Chem. 2006;119(2):158–69. doi: 10.1016/j.bpc.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 34.DeMattos RB, Brendza RP, Heuser JE, Kierson M, Cirrito JR, Fryer J, et al. Purification and characterization of astrocyte-secreted apolipoprotein E and J-containing lipoproteins from wild-type and human apoE transgenic mice. Neurochem Int. 2001;39(5-6):415–25. doi: 10.1016/s0197-0186(01)00049-3. [DOI] [PubMed] [Google Scholar]
- 35.Ladu MJ, Reardon C, Van Eldik L, Fagan AM, Bu G, Holtzman D, et al. Lipoproteins in the central nervous system. Ann N Y Acad Sci. 2000;903:167–75. doi: 10.1111/j.1749-6632.2000.tb06365.x. [DOI] [PubMed] [Google Scholar]
- 36.Pitas RE, Boyles JK, Lee SH, Foss D, Mahley RW. Astrocytes synthesize apolipoprotein E and metabolize apolipoprotein E-containing lipoproteins. Biochim Biophys Acta. 1987;917(1):148–61. doi: 10.1016/0005-2760(87)90295-5. [DOI] [PubMed] [Google Scholar]
- 37.Oram JF, Vaughan AM. ABCA1-mediated transport of cellular cholesterol and phospholipids to HDL apolipoproteins. Curr Opin Lipidol. 2000;11(3):253–60. doi: 10.1097/00041433-200006000-00005. [DOI] [PubMed] [Google Scholar]
- 38.Bales KR, Liu F, Wu S, Lin S, Koger D, DeLong C, et al. Human APOE isoform-dependent effects on brain beta-amyloid levels in PDAPP transgenic mice. J Neurosci. 2009;29(21):6771–9. doi: 10.1523/JNEUROSCI.0887-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riddell DR, Zhou H, Atchison K, Warwick HK, Atkinson PJ, Jefferson J, et al. Impact of apolipoprotein E (ApoE) polymorphism on brain ApoE levels. J Neurosci. 2008;28(45):11445–53. doi: 10.1523/JNEUROSCI.1972-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong N, Weisgraber KH. Understanding the Basis for the Association of Apoe4 with Alzheimer’s Disease: Opening the Door for Therapeutic Approaches. Curr Alzheimer Res. 2009 doi: 10.2174/156720509789207921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bales KR. Brain lipid metabolism, apolipoprotein E and the pathophysiology of Alzheimer’s disease. Neuropharmacology. 2010;59(4-5):295–302. doi: 10.1016/j.neuropharm.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 42.Sullivan PM, Han B, Liu F, Mace BE, Ervin JF, Wu S, et al. Reduced levels of human apoE4 protein in an animal model of cognitive impairment. Neurobiol Aging. 2011;32(5):791–801. doi: 10.1016/j.neurobiolaging.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 43.Yokoyama S. Assembly of high-density lipoprotein. Arterioscler Thromb Vasc Biol. 2006;26(1):20–7. doi: 10.1161/01.ATV.0000195789.39418.e8. [DOI] [PubMed] [Google Scholar]
- 44.Koldamova R, Fitz NF, Lefterov I. The role of ATP-binding cassette transporter A1 in Alzheimer’s disease and neurodegeneration. Biochim Biophys Acta. 2010;1801(8):824–30. doi: 10.1016/j.bbalip.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wahrle SE, Jiang H, Parsadanian M, Legleiter J, Han X, Fryer JD, et al. ABCA1 is required for normal central nervous system ApoE levels and for lipidation of astrocyte-secreted apoE. J Biol Chem. 2004;279(39):40987–93. doi: 10.1074/jbc.M407963200. [DOI] [PubMed] [Google Scholar]
- 46.Fan J, Stukas S, Wong C, Chan J, May S, DeValle N, et al. An ABCA1-independent pathway for recycling a poorly lipidated 8.1 nm apolipoprotein E particle from glia. J Lipid Res. 2011;52(9):1605–16. doi: 10.1194/jlr.M014365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Narayanaswami V, Ryan RO. Molecular basis of exchangeable apolipoprotein function. Biochim Biophys Acta. 2000;1483(1):15–36. doi: 10.1016/s1388-1981(99)00176-6. [DOI] [PubMed] [Google Scholar]
- 48.Saito H, Dhanasekaran P, Nguyen D, Holvoet P, Lund-Katz S, Phillips MC. Domain structure and lipid interaction in human apolipoproteins A-I and E, a general model. J Biol Chem. 2003;278(26):23227–32. doi: 10.1074/jbc.M303365200. [DOI] [PubMed] [Google Scholar]
- 49.Wetterau JR, Aggerbeck LP, Rall SC, Jr., Weisgraber KH. Human apolipoprotein E3 in aqueous solution. I. Evidence for two structural domains. J Biol Chem. 1988;263(13):6240–8. [PubMed] [Google Scholar]
- 50.Wilson C, Wardell MR, Weisgraber KH, Mahley RW, Agard DA. Three-dimensional structure of the LDL receptor-binding domain of human apolipoprotein E. Science. 1991;252(5014):1817–22. doi: 10.1126/science.2063194. [DOI] [PubMed] [Google Scholar]
- 51.Vedhachalam C, Narayanaswami V, Neto N, Forte TM, Phillips MC, Lund-Katz S, et al. The C-terminal lipid-binding domain of apolipoprotein E is a highly efficient mediator of ABCA1-dependent cholesterol efflux that promotes the assembly of high-density lipoproteins. Biochemistry. 2007;46(10):2583–93. doi: 10.1021/bi602407r. [DOI] [PubMed] [Google Scholar]
- 52.Raussens V, Fisher CA, Goormaghtigh E, Ryan RO, Ruysschaert JM. The low density lipoprotein receptor active conformation of apolipoprotein E. Helix organization in n-terminal domain-phospholipid disc particles. J Biol Chem. 1998;273(40):25825–30. doi: 10.1074/jbc.273.40.25825. [DOI] [PubMed] [Google Scholar]
- 53.Gupta V, Narayanaswami V, Budamagunta MS, Yamamato T, Voss JC, Ryan RO. Lipid-induced extension of apolipoprotein E helix 4 correlates with low density lipoprotein receptor binding ability. J Biol Chem. 2006;281(51):39294–9. doi: 10.1074/jbc.M608085200. [DOI] [PubMed] [Google Scholar]
- 54.Chen J, Li Q, Wang J. Topology of human apolipoprotein E3 uniquely regulates its diverse biological functions. Proc Natl Acad Sci U S A. 2011;108(36):14813–8. doi: 10.1073/pnas.1106420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fisher CA, Ryan RO. Lipid binding-induced conformational changes in the N-terminal domain of human apolipoprotein E. J Lipid Res. 1999;40(1):93–9. [PubMed] [Google Scholar]
- 56.Narayanaswami V, Maiorano JN, Dhanasekaran P, Ryan RO, Phillips MC, Lund-Katz S, et al. Helix orientation of the functional domains in apolipoprotein e in discoidal high density lipoprotein particles. J Biol Chem. 2004;279(14):14273–9. doi: 10.1074/jbc.M313318200. [DOI] [PubMed] [Google Scholar]
- 57.Segrest JP, Garber DW, Brouillette CG, Harvey SC. Anantharamaiah GM. The amphipathic alpha helix: a multifunctional structural motif in plasma apolipoproteins. Adv Protein Chem. 1994;45:303–69. doi: 10.1016/s0065-3233(08)60643-9. [DOI] [PubMed] [Google Scholar]
- 58.Hirsch-Reinshagen V, Donkin J, Stukas S, Chan J, Wilkinson A, Fan J, et al. LCAT synthesized by primary astrocytes esterifies cholesterol on glia-derived lipoproteins. J Lipid Res. 2009;50(5):885–93. doi: 10.1194/jlr.M800584-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Demeester N, Castro G, Desrumaux C, De Geitere C, Fruchart JC, Santens P, et al. Characterization and functional studies of lipoproteins, lipid transfer proteins, and lecithin:cholesterol acyltransferase in CSF of normal individuals and patients with Alzheimer’s disease. J Lipid Res. 2000;41(6):963–74. [PubMed] [Google Scholar]
- 60.Huang Y, Liu XQ, Wyss-Coray T, Brecht WJ, Sanan DA, Mahley RW. Apolipoprotein E fragments present in Alzheimer’s disease brains induce neurofibrillary tangle-like intracellular inclusions in neurons. Proc Natl Acad Sci U S A. 2001;98(15):8838–43. doi: 10.1073/pnas.151254698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harris FM, Brecht WJ, Xu Q, Tesseur I, Kekonius L, Wyss-Coray T, et al. Carboxyl-terminal-truncated apolipoprotein E4 causes Alzheimer’s disease-like neurodegeneration and behavioral deficits in transgenic mice. Proc Natl Acad Sci U S A. 2003;100(19):10966–71. doi: 10.1073/pnas.1434398100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jones PB, Adams KW, Rozkalne A, Spires-Jones TL, Hshieh TT, Hashimoto T, et al. Apolipoprotein E: isoform specific differences in tertiary structure and interaction with amyloid-beta in human Alzheimer brain. PLoS One. 2011;6(1):e14586. doi: 10.1371/journal.pone.0014586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297(5580):353–6. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 64.Wahrle SE, Jiang H, Parsadanian M, Kim J, Li A, Knoten A, et al. Overexpression of ABCA1 reduces amyloid deposition in the PDAPP mouse model of Alzheimer disease. J Clin Invest. 2008;118(2):671–82. doi: 10.1172/JCI33622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Youmans KL, Tai LM, Nwabuisi-Heath E, Jungbauer L, Kanekiyo T, Gan M, et al. APOE4-specific changes in Abeta accumulation in a new transgenic mouse model of Alzheimer disease. J Biol Chem. 2012;287(50):41774–86. doi: 10.1074/jbc.M112.407957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hashimoto T, Serrano-Pozo A, Hori Y, Adams KW, Takeda S, Banerji AO, et al. Apolipoprotein E, especially apolipoprotein E4, increases the oligomerization of amyloid beta peptide. J Neurosci. 2012;32(43):15181–92. doi: 10.1523/JNEUROSCI.1542-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anderson R, Barnes JC, Bliss TV, Cain DP, Cambon K, Davies HA, et al. Behavioural, physiological and morphological analysis of a line of apolipoprotein E knockout mouse. Neuroscience. 1998;85(1):93–110. doi: 10.1016/s0306-4522(97)00598-8. [DOI] [PubMed] [Google Scholar]
- 68.Masliah E, Samuel W, Veinbergs I, Mallory M, Mante M, Saitoh T. Neurodegeneration and cognitive impairment in apoE-deficient mice is ameliorated by infusion of recombinant apoE. Brain Res. 1997;751(2):307–14. doi: 10.1016/s0006-8993(96)01420-5. [DOI] [PubMed] [Google Scholar]
- 69.Koldamova RP, Lefterov IM, Lefterova MI, Lazo JS. Apolipoprotein A-I directly interacts with amyloid precursor protein and inhibits A beta aggregation and toxicity. Biochemistry. 2001;40(12):3553–60. doi: 10.1021/bi002186k. [DOI] [PubMed] [Google Scholar]
- 70.Paula-Lima AC, Tricerri MA, Brito-Moreira J, Bomfim TR, Oliveira FF, Magdesian MH, et al. Human apolipoprotein A-I binds amyloid-beta and prevents Abeta-induced neurotoxicity. Int J Biochem Cell Biol. 2009;41(6):1361–70. doi: 10.1016/j.biocel.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 71.Harris-White ME, Frautschy SA. Low density lipoprotein receptor-related proteins (LRPs), Alzheimer’s and cognition. Curr Drug Targets CNS Neurol Disord. 2005;4(5):469–80. doi: 10.2174/156800705774322102. [DOI] [PubMed] [Google Scholar]
- 72.Nuutinen T, Suuronen T, Kauppinen A, Salminen A. Clusterin: a forgotten player in Alzheimer’s disease. Brain Res Rev. 2009;61(2):89–104. doi: 10.1016/j.brainresrev.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 73.Wu ZC, Yu JT, Li Y, Tan L. Clusterin in Alzheimer’s disease. Adv Clin Chem. 2012;56:155–73. doi: 10.1016/b978-0-12-394317-0.00011-x. [DOI] [PubMed] [Google Scholar]
- 74.Holtzman DM, Bales KR, Wu S, Bhat P, Parsadanian M, Fagan AM, et al. Expression of human apolipoprotein E reduces amyloid-beta deposition in a mouse model of Alzheimer’s disease. J Clin Invest. 1999;103(6):R15–R21. doi: 10.1172/JCI6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Buttini M, Orth M, Bellosta S, Akeefe H, Pitas RE, Wyss-Coray T, et al. Expression of human apolipoprotein E3 or E4 in the brains of Apoe-/- mice: isoform-specific effects on neurodegeneration. J Neurosci. 1999;19(12):4867–80. doi: 10.1523/JNEUROSCI.19-12-04867.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Donkin JJ, Stukas S, Hirsch-Reinshagen V, Namjoshi D, Wilkinson A, May S, et al. ATP-binding cassette transporter A1 mediates the beneficial effects of the liver X receptor agonist GW3965 on object recognition memory and amyloid burden in amyloid precursor protein/presenilin 1 mice. J Biol Chem. 2010;285(44):34144–54. doi: 10.1074/jbc.M110.108100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fitz NF, Cronican A, Pham T, Fogg A, Fauq AH, Chapman R, et al. Liver X receptor agonist treatment ameliorates amyloid pathology and memory deficits caused by high-fat diet in APP23 mice. J Neurosci. 2010;30(20):6862–72. doi: 10.1523/JNEUROSCI.1051-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Landreth G, Jiang Q, Mandrekar S, Heneka M. PPARgamma agonists as therapeutics for the treatment of Alzheimer’s disease. Neurotherapeutics. 2008;5(3):481–9. doi: 10.1016/j.nurt.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mandrekar-Colucci S, Landreth GE. Nuclear receptors as therapeutic targets for Alzheimer’s disease. Expert Opin Ther Targets. 2011;15(9):1085–97. doi: 10.1517/14728222.2011.594043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cramer PE, Cirrito JR, Wesson DW, Lee CY, Karlo JC, Zinn AE, et al. ApoE-directed therapeutics rapidly clear beta-amyloid and reverse deficits in AD mouse models. Science. 2012;335(6075):1503–6. doi: 10.1126/science.1217697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Koldamova RP, Lefterov IM, Staufenbiel M, Wolfe D, Huang S, Glorioso JC, et al. The liver X receptor ligand T0901317 decreases amyloid beta production in vitro and in a mouse model of Alzheimer’s disease. J Biol Chem. 2005;280(6):4079–88. doi: 10.1074/jbc.M411420200. [DOI] [PubMed] [Google Scholar]
- 82.Mandrekar-Colucci S, Karlo JC, Landreth GE. Mechanisms underlying the rapid peroxisome proliferator-activated receptor-gamma-mediated amyloid clearance and reversal of cognitive deficits in a murine model of Alzheimer’s disease. J Neurosci. 2012;32(30):10117–28. doi: 10.1523/JNEUROSCI.5268-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fujiyoshi M, Ohtsuki S, Hori S, Tachikawa M, Terasaki T. 24Shydroxycholesterol induces cholesterol release from choroid plexus epithelial cells in an apical- and apoE isoform-dependent manner concomitantly with the induction of ABCA1 and ABCG1 expression. J Neurochem. 2007;100(4):968–78. doi: 10.1111/j.1471-4159.2006.04240.x. [DOI] [PubMed] [Google Scholar]
- 84.Liang Y, Lin S, Beyer TP, Zhang Y, Wu X, Bales KR, et al. A liver X receptor and retinoid X receptor heterodimer mediates apolipoprotein E expression, secretion and cholesterol homeostasis in astrocytes. J Neurochem. 2004;88(3):623–34. doi: 10.1111/j.1471-4159.2004.02183.x. [DOI] [PubMed] [Google Scholar]
- 85.Jiang Q, Lee CY, Mandrekar S, Wilkinson B, Cramer P, Zelcer N, et al. ApoE promotes the proteolytic degradation of Abeta. Neuron. 2008;58(5):681–93. doi: 10.1016/j.neuron.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bien-Ly N, Gillespie AK, Walker D, Yoon SY, Huang Y. Reducing human apolipoprotein E levels attenuates age-dependent Abeta accumulation in mutant human amyloid precursor protein transgenic mice. J Neurosci. 2012;32(14):4803–11. doi: 10.1523/JNEUROSCI.0033-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim J, Jiang H, Park S, Eltorai AE, Stewart FR, Yoon H, et al. Haploinsufficiency of human APOE reduces amyloid deposition in a mouse model of amyloid-beta amyloidosis. J Neurosci. 2011;31(49):18007–12. doi: 10.1523/JNEUROSCI.3773-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hirsch-Reinshagen V, Maia LF, Burgess BL, Blain JF, Naus KE, McIsaac SA, et al. The absence of ABCA1 decreases soluble ApoE levels but does not diminish amyloid deposition in two murine models of Alzheimer disease. J Biol Chem. 2005;280(52):43243–56. doi: 10.1074/jbc.M508781200. [DOI] [PubMed] [Google Scholar]
- 89.Koldamova R, Staufenbiel M, Lefterov I. Lack of ABCA1 considerably decreases brain ApoE level and increases amyloid deposition in APP23 mice. J Biol Chem. 2005;280(52):43224–35. doi: 10.1074/jbc.M504513200. [DOI] [PubMed] [Google Scholar]
- 90.Wahrle SE, Jiang H, Parsadanian M, Hartman RE, Bales KR, Paul SM, et al. Deletion of Abca1 increases Abeta deposition in the PDAPP transgenic mouse model of Alzheimer disease. J Biol Chem. 2005;280(52):43236–42. doi: 10.1074/jbc.M508780200. [DOI] [PubMed] [Google Scholar]
- 91.Maloney B, Ge YW, Petersen RC, Hardy J, Rogers JT, Perez-Tur J, et al. Functional characterization of three single-nucleotide polymorphisms present in the human APOE promoter sequence: Differential effects in neuronal cells and on DNA-protein interactions. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(1):185–201. doi: 10.1002/ajmg.b.30973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Maloney B, Lahiri DK. The Alzheimer’s amyloid beta-peptide (Abeta) binds a specific DNA Abeta-interacting domain (AbetaID) in the APP, BACE1, and APOE promoters in a sequence-specific manner: characterizing a new regulatory motif. Gene. 2011;488(1-2):1–12. doi: 10.1016/j.gene.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen HK, Ji ZS, Dodson SE, Miranda RD, Rosenblum CI, Reynolds IJ, et al. Apolipoprotein E4 domain interaction mediates detrimental effects on mitochondria and is a potential therapeutic target for Alzheimer disease. J Biol Chem. 2011;286(7):5215–21. doi: 10.1074/jbc.M110.151084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen HK, Liu Z, Meyer-Franke A, Brodbeck J, Miranda RD, McGuire JG, et al. Small molecule structure correctors abolish detrimental effects of apolipoprotein E4 in cultured neurons. J Biol Chem. 2012;287(8):5253–66. doi: 10.1074/jbc.M111.276162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xu Q, Brecht WJ, Weisgraber KH, Mahley RW, Huang Y. Apolipoprotein E4 domain interaction occurs in living neuronal cells as determined by fluorescence resonance energy transfer. J Biol Chem. 2004;279(24):25511–6. doi: 10.1074/jbc.M311256200. [DOI] [PubMed] [Google Scholar]
- 96.Narayanaswami V, Szeto SS, Ryan RO. Lipid association-induced N- and C-terminal domain reorganization in human apolipoprotein E3. J Biol Chem. 2001;276(41):37853–60. doi: 10.1074/jbc.M102953200. [DOI] [PubMed] [Google Scholar]
- 97.Hatters DM, Budamagunta MS, Voss JC, Weisgraber KH. Modulation of apolipoprotein E structure by domain interaction: differences in lipid-bound and lipid-free forms. J Biol Chem. 2005;280(40):34288–95. doi: 10.1074/jbc.M506044200. [DOI] [PubMed] [Google Scholar]