Abstract
The human intestine harbors a diverse community of microbes that promote metabolism and digestion in their symbiotic relationship with the host. Disturbance of its homeostasis can result in disease. We review factors that disrupt intestinal homeostasis and contribute to non-alcoholic fatty liver disease (NAFLD), steatohepatitis (NASH), alcoholic liver disease, and cirrhosis. Liver disease has long been associated with qualitative and quantitative (overgrowth) dysbiotic changes in the intestinal microbiota. Extrinsic factors, such as the Western diet and alcohol, contribute to these changes. Dysbiosis results in intestinal inflammation, a breakdown of the intestinal barrier, and translocation of microbial products in animal models. However, the contribution of the intestinal microbiome to liver disease goes beyond simple translocation of bacterial products that promote hepatic injury and inflammation. Microbial metabolites produced in a dysbiotic intestinal environment and host factors are equally important in the pathogenesis of liver disease. We review how the combination of liver insult and disruptions in intestinal homeostasis contribute to liver disease.
Keywords: Microbiota, endotoxin, alcoholic steatohepatitis
Introduction
The intestine and its microbiota (bacteria and other microbes) have a symbiotic relationship. The microbiota contributes to digestion, synthesis of vitamins, and resistance to intestinal colonization by pathogens, but also contains potentially pathogenic bacteria. Disruption of intestinal homeostasis and alterations in the intestinal microbiome contribute to the pathogenesis of many disorders, including liver disease. How do disturbances in the intestinal microbiome (detected by analyses of its metagenome and metabolome) contribute to liver disease? We discuss how host and dietary factors, including alcohol, affect the composition of the intestinal microbiome and development of liver diseases, and in turn, how liver disease can alter the enteric microbiome. We review alterations in the composition of the intestinal microbiome associated with several liver diseases. These studies were performed primarily in patients with disease (see Tables 1 and 2). The functional consequences of the intestinal microbiome for each of these diseases, based primarily on animal models, are then reviewed in the following section.
Table 1.
Changes in the intestinal microbiota associated with NAFLD and NASH in humans
| Disease | Comparison1 | Implicated Microbiota 2 | Methodology | Ref. 3 | ||||
|---|---|---|---|---|---|---|---|---|
| Phylum | Class | Order | Family | Genus | ||||
| Healthy (n=30) NAFLD (n=30) |
Healthy vs NAFLD | Firmicutes | Bacilli | Lactobacillales | Lactobacillaceae ↑ | Lactobacillus ↑ | 16S rRNA gene Pyrosequencing Stool sample |
5 |
| Clostridia | Clostridiales | Lachnospiraceae ↑ | Robinsoniella ↑, Roseburia ↑, Dorea ↑ | |||||
| Clostridia | Clostridiales | Ruminococcaceae ↓ | ||||||
| Oscillospiraceae | Oscillibacter ↓ | |||||||
| Children: Healthy (n=16) Obese (n=25) NASH (n=22) |
Healthy vs Obese | Bacteroidetes ↑ | Prevotellaceae ↑ | Prevotella ↑ | 16S rRNA gene Pyrosequencing Stool sample |
7 | ||
| Rikenellaceae ↓ | Alistipes ↓ | |||||||
| Firmicutes ↓ | Lachnospiraceae ↓ | Blautia ↓, Coprococcus ↓, Roseburia ↓ | ||||||
| Eubacteriaceae | Eubacterium ↓ | |||||||
| Ruminococcaceae ↓ | ||||||||
| Healthy vs NASH | Bacteroidetes ↑ | Prevotellaceae ↑ | Prevotella ↑ | |||||
| Rikenellaceae ↓ | Alistipes ↓ | |||||||
| Firmicutes ↓ | Lachnospiraceae ↓ | Blautia ↓, Coprococcus ↓ | ||||||
| Eubacteriaceae | Eubacterium ↓ | |||||||
| Ruminococcaceae ↓ | Oscillospira ↓ | |||||||
| Actinobacteria ↓ | Bifidobacteriaceae ↓ | Bifidobacterium ↓ | ||||||
| Proteobacteria ↑ | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae ↑ | Escherichia ↑ | ||||
| Obese vs NASH | Proteobacteria ↑ | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae ↑ | Escherichia ↑ | |||
| Healthy (n=17) Steatosis (n=11) NASH (n=22) |
Healthy vs NASH | Bacteroidetes ↓ | Quantitative realtime PCR Stool sample |
6 | ||||
| Proteobacteria | Enterobacteriaceae | Escherichia coli ns | ||||||
| Steatosis vs NASH | Bacteroidetes ↓ | |||||||
| Firmicutes | Lachnospiraceae | Clostridium coccoides ↑ | ||||||
| Proteobacteria | Enterobacteriaceae | Escherichia coli ns | ||||||
Comparison condition A vs condition B: ↑ Increase in condition B relative to condition A, ↓ Decrease in condition B relative to condition A, ns not significant
Taxonomy was updated using the NCBI Taxonomy Browser
References focus on microbiota changes associated with liver disease rather than obesity or the metabolic syndrome
Table 2.
Changes in the intestinal microbiota associated with liver cirrhosis in humans
| Disease | Comparison1 | Implicated Microbiota2 | Methodology | Ref. | ||||
|---|---|---|---|---|---|---|---|---|
| Phylum | Class | Order | Family | Genus | ||||
| Healthy (n=32) HBV cirrhotics (n=31) | Healthy vs HBV cirrhotics | Bacteroidetes | Prevotella ↓ | Quantitative realtime PCR Stool sample |
27 | |||
| Firmicutes | Enterococcus faecalis ↑ | |||||||
| Faecalibacterium prausnitzii ↓, Clostridium clusters XI ↓, Clostridium clusters XIV ↓ | ||||||||
| Lactic acid bacteria ↓ (including Lactobacillus, Pediococcus, Leuconostoc, and Weissella) | ||||||||
| Actinobacteria | Bifidobacterium ↓ | |||||||
| Proteobacteria | Enterobacteriaceae ↑ | |||||||
| Healthy (n=15) HBV cirrhotics (n=16) |
Healthy vs HBV cirrhotics | Actinobacteria | Bifidobacterium catenulatum group ↓ | Quantitative realtime PCR Stool sample |
29 | |||
| Healthy (n=38) HBV cirrhotics (n=61) |
Healthy vs HBV cirrhotics | Firmicutes | Lactobacillus acidophilus ↓, Lactobacillus rhamnosus ↓, Lactobacillus reuteri ↓, Lactobacillus gasseri ↑ | Quantitative realtime PCR Stool sample |
30 | |||
| Healthy (n=24) HBV cirrhotics (n=24) Alcoholic cirrhotics (n=12) |
Healthy vs Cirrhotics | Bacteroidetes ↓ | Bacteroidia ↓ | Bacteroidaceae ↓ | 16S rRNA gene Pyrosequencing, Quantitative realtime PCR Stool sample |
28 | ||
| Firmicutes | Enterococcus faecalis ↑ | |||||||
| Bacilli ↑ | Streptococcaceae ↑ | |||||||
| Clostridia | Lachnospiraceae ↓ | |||||||
| Negativicutes | Veillonellaceae ↑ | |||||||
| Clostridium clusters XI ↑ | ||||||||
| Proteobacteria ↑ | Gammaproteobacteria ↑ | Enterobacteriaceae ↑, Pasteurellaceae ↑ | ||||||
| Fusobacteria ↑ | Fusobacteriia ↑ | Fusobacteriaceae ↑ | ||||||
| Healthy vs Alcoholic cirrhotics | Bacteroidetes | Prevotellaceae ↑ | ||||||
| HBV cirrhosis vs Alcoholic cirrhotics | Bacteroidetes | Prevotellaceae ↑ | ||||||
| Healthy (n=10) Cirrhotics2 (n=25) |
Healthy vs Cirrhotics | Firmicutes | Lachnospiraceae ↓, Ruminococcaceae ↓, Clostridium Incertae sedis XIV ↓, Leuconostocaceae ↑, Lactobacillaceae ↑ | 16S rRNA gene Pyrosequencing Stool sample |
32 | |||
| Proteobacteria | Enterobacteriaceae ↑, Alcaligenaceae ↑ | |||||||
| Fusobacteria | Fusobacteriaceae ↑ | |||||||
| Healthy (n=17) Cirrhotics2 (n=36) |
Mucosal samples - Healthy vs Cirrhotics | Firmicutes | Clostridiaceae | Clostridium ↑ | 16S rRNA gene Pyrosequencing Stool sample, Rectosigmoid mucosal biopsy |
31 | ||
| Lachnospiraceae | Dorea ↓ | |||||||
| Ruminococcaceae | Subdoligranulum ↓ | |||||||
| Acidaminococcaceae | Acidaminococcus ↑ | |||||||
| Enterococcaceae | Enterococcus ↑ | |||||||
| Proteobacteria | Burkholderiaceae | Burkholderia ↑, Ralstonia↑ | ||||||
| Enterobacteriaceae | Proteus ↑ | |||||||
| Mucosal samples Cirrhotics vs Stool samples Cirrhotics | Firmicutes | Veillonellaceae | Veillonella ↑ | |||||
| Lachnospiraceae | Roseburia ↑, Blautia ↓ | |||||||
| Leuconostocaceae | Leuconostoc ↑ | |||||||
| Actinobacteria | Propionibacteriaceae | Propionibacterium ↓ | ||||||
| Streptomycetaceae | Streptomyces ↓ | |||||||
| Proteobacteria | Vibrionaceae | Vibrio ↓ | ||||||
Comparison condition A vs condition B: ↑ Increase in condition B relative to condition A, ↓ Decrease in condition B relative to condition A, ns not significant
Taxonomy was updated using the NCBI Taxonomy Browser
Mixed etiology
HBV, Hepatitis B virus
Intestinal Microbiome Composition and Liver Disease
Non-alcoholic Fatty Liver Disease (NAFLD) and Steatohepatitis (NASH)
NAFLD is the hepatic manifestation of the metabolic syndrome. NAFLD is generally a benign disease; approximately one third of the US population has hepatic steatosis1. The prevalence of NASH among a general medical population diagnosed with NAFLD is 30%2. NASH is characterized by the development of liver inflammation and fibrosis. Patients with NASH have a high likelihood of developing advanced fibrosis and cirrhosis; it has been estimated that about one third of cases of early-stage NASH will progress to stage 3 or 4 fibrosis (cirrhosis) over 5–10 years3.
Dietary factors and changes in diet are determinants of the composition of the microbiome4. Although patients with NAFLD are often obese and insulin resistant, we focus on published studies of patients with documented liver disease, rather than obesity. The fecal microbiota in NAFLD and NASH patients has been assessed using culture-independent techniques such as quantitative PCR and deep sequencing of a conserved region in the bacterial 16S rRNA gene5–7. Details about dysbiosis associated with NAFLD and NASH are summarized in Table 1. Microbiota samples from patients with NAFLD or NASH have a lower proportion of members of the family Ruminococcaceae than healthy subjects. Escherichia is the only abundant genus of bacteria in the intestinal microbiota that is significantly disproportionate between obese children and pediatric patients with NASH7. In contrast, adult patients with NASH had a significantly higher percentage of Clostridium coccoides than patients with biopsy-proven NAFLD6. However, studies comparing the bacterial taxonomic composition of patients with NAFLD vs those with NASH produced variable and even contradictory findings. Possible reasons for discrepant results include small number of subjects included in the studies, differences in cohorts (age, sex, ethnicity, geographical location, medication use), insufficient documentation of liver disease, and differences in methodology. To determine whether patients with NAFLD and NASH have distinct compositions of the intestinal microbiome, studies (ideally longitudinal) are needed of larger, better characterized cohorts. Identifying specific microbial compositions of these patients could improve our understanding of intestine–liver interactions and lead to fecal biomarkers for NAFLD and/or NASH.
Small bowel bacterial overgrowth is a disorder in which abnormally large numbers of bacteria grow in the small intestine. Patients with obesity or NAFLD have a higher prevalence small intestinal bacterial overgrowth8, 9. Intestinal permeability and bacterial overgrowth correlate with severity of steatosis, but not fibrosis or hepatic inflammation, based on liver biopsy analysis8. Small intestinal bacterial overgrowth was also present in 50% of patients with NASH, which is significantly higher than in healthy controls, matched for sex and age10. In these studies, patients with small intestinal bacterial overgrowth were identified by breath tests. However, researchers have debated whether breath tests accurately detect this disorder. Total bacterial counts in the feces, based on real-time PCR, did not differ between healthy subjects and persons with NAFLD or NASH6. Further studies are needed to determine whether fecal bacterial counts actually correlate with the amount of microbes present in the small intestine. Culture-and breath test-independent methods are needed to reassess the prevalence of intestinal bacterial overgrowth in patients with NAFLD or NASH.
Alcoholic Liver Disease
Alcohol abuse is one of the leading causes of chronic liver disease. Chronic alcoholic liver disease may progress from simple steatosis to steatohepatitis, liver fibrosis, and in 15%–40% of patients, cirrhosis. Patients with only alcoholic fatty liver disease usually do not present with any clinical symptoms and their liver continues to function well11, 12.
Research into the role of the microbiome in alcoholic liver disease is unfortunately not as advanced as that for obesity or fatty liver disease. The mucosa-associated bacterial taxonomy was evaluated in patients with alcoholic cirrhosis and alcoholics without liver disease using 16S rRNA gene sequencing. The proportion of Bacteroidaceae was lower in samples from alcoholic patients than from non-alcoholic individuals13.
Although microbiome studies in humans are important to associate distinct compositions of the intestinal microbiome with different disease states, studies in animal models, under carefully controlled conditions, offer some advantages. Preclinical studies allow researchers to control for age, sex, environment, diet, and genetic background. Littermates can be compared in mouse studies. Pups are typically colonized with the microbes they first encounter, typically from their mothers,14 so littermates usually have the same microbiota composition. Changes in the microbiota can be monitored in response to different environmental factors, and compared among mice that had the same initial microbial composition.
For example, in the Tsukamoto-French model of alcoholic liver disease, mice are placed on specific liquid diets and given intragastric infusions of ethanol, whereas control littermates are placed on the same diet but instead given an isocaloric amount of dextrose. Using this system, researchers have been able to detect quantitative and qualitative changes in the microbiome associated with ethanol intake. Bacterial overgrowth was observed along almost the entire gastrointestinal tract; the dysbiosis was characterized by significant reductions in proportions of probiotic bacteria such as Lactobacillus, Pediococcus, Leuconostoc and Lactococcus15. An alcohol-associated decrease in the number of intestinal Lactobacillus, confirmed by quantitative real-time PCR,16 was also observed in the Lieber DeCarli diet model of alcohol feeding for 8 weeks (unpublished data). Alternatively, several studies have reported that administration of probiotic Lactobacillus reduces features of alcoholic liver disease in animal models17–19. A small clinical trial has also demonstrated improve alcohol-induced liver injury in patients taking probiotics20. Similar to observations made in animal models, aerobic and anaerobic bacterial cultures of jejunal aspirates from patients who chronically abuse alcohol were found to have bacterial overgrowth21. Excessive alcohol intake is therefore accompanied by dysbiosis and an increased intestinal bacterial load, based on clinical and preclinical studies.
Multiple factors are likely to contribute to changes in the intestinal microbiome during development of alcoholic liver disease. These might include small intestinal dysmotility22, changes in gastric acid secretion,23 and alterations to the intestinal innate immune response. Antimicrobial molecules, which are part of the innate immune response, are secreted from enterocytes or intestinal Paneth cells. In particular, the antimicrobial molecules regenerating islet-derived (Reg)3b and Reg3g are reduced in the small intestines of mice following 3 weeks of intragastric ethanol feeding15. Further studies are needed to determine if and to what extent an impaired innate immune response contributes to disease progression. The commensal microbiota not only produces ethanol, but also metabolizes it24. It is not clear whether ethanol, as a dietary component or as an energy source for certain bacterial strains, directly alters the microbiota.
Cirrhosis
Liver fibrosis may result in end-stage liver disease or cirrhosis, which eventually disrupts the metabolic functions of the liver. Although patients with hepatic fibrosis are often asymptomatic, development of cirrhosis in these patients is the major determinant of morbidity and mortality25. Major clinical complications are infections, ascites, renal failure, variceal hemorrhage, and hepatic encephalopathy. Patients with these complications have a poor prognosis and liver transplantation is often indicated26.
Several studies assessed the taxonomic composition of the intestinal microbiota in patients with cirrhosis27–32 (see Table 2). A common feature of cirrhosis is an increase of potentially pathogenic bacteria, accompanied by reduced proportions of beneficial bacteria. Fecal microbial communities are similar among patients with cirrhosis of different etiologies. Therefore, features of end-stage liver disease, such as reduced bile flow, might determine the shape of the intestinal microbiome. However, fecal microbial communities from patients with alcoholic cirrhosis have significant increases in the family of Prevotellaceae compared to patients with hepatitis B-related cirrhosis or healthy individuals, based on sequencing of the common 16S rRNA gene region of bacteria28. Etiology (particularly an alcohol association) therefore appears to contribute to the composition of the intestinal microbiome in patients with end-stage liver disease.
Most patients with cirrhosis have intestinal bacterial overgrowth, demonstrated by quantitative analyses of bacterial cultures from jejunal aspirates33, 34. So, they not only have taxonomic differences in microbial communities, compared to people without cirrhosis, but also an increased intestinal burden of bacteria. Several factors contribute to intestinal bacterial overgrowth in patients with cirrhosis. These include impaired motility of the small intestine35, reduced bile flow27, and altered secretion of immunoglobulin A27 and antimicrobial molecules36. In rats with cirrhosis, ascites, and translocation of viable bacteria to mesenteric lymph nodes, Paneth cells produce lower levels of defensins and Reg3 molecules, compared to those without bacterial translocation. This reduction is accompanied by reduced antimicrobial activity against Enterobacteriaceae36. Little is known about how Paneth cell function is impaired during development of cirrhosis. Compromised intestinal host defense might therefore contribute to qualitative and quantitative changes in the enteric microbiome associated with end-stage liver disease. New sequencing techniques to analyze the microbiome should help determine the contribution of these factors to compositional changes in the microbiota.
Functional Consequences of Changes in the Intestinal Microbiome
NAFLD and NASH
Most patients with NAFLD are obese and diabetic. Obesity and insulin resistance are risk factors for fatty liver disease and are associated with changes in the intestinal microbiome37, 38. The intestinal microbiome is an important factor in the development of obesity; germ-free mice are protected from high fat diet-induced weight gain and obesity39, 40.
Changes in bacterial taxonomy might not be as important as changes in bacterial genes (metagenomics and metatranscriptomics) in the development of NAFLD and NASH41. Obesity is accompanied by an intestinal metagenome that has an increased capacity to collect energy from the host diet. Bacterial enzymes aid in digestion of otherwise indigestible dietary polysaccharides and extraction of calories from them37. In addition, enteric bacteria suppress the synthesis of fasting-induced adipocyte factor (Fiaf; also known as angiopoietin-like 4) and secretion from the small intestine, resulting in increased activity of lipoprotein lipase (LPL) and increased accumulation of triglycerides in the liver39, 40. This process provides a direct link between the intestinal microbiome and fat deposition in the liver.
Complex interactions between the enteric microbiome and the host are often mediated by metabolites. Several changes in bacterial metabolites have been associated with obesity, and in particular, with NAFLD. One of these metabolites is ethanol, a product of the intestinal microbiome. Obese animals have higher blood concentrations of ethanol, determined by breath tests, than lean animals42. Alcohol is absorbed and reaches the liver via the portal blood. Ethanol causes triglyceride accumulation in hepatocytes43, and might also provide a second hit to livers that have already accumulated fat, via production of reactive oxygen species and initiation of liver inflammation. Obese children with NAFLD do not have increased blood levels of ethanol, whereas pediatric patients with NASH do7. Meta-transcriptomic and metabolomic studies of intestinal contents are needed for further analysis and confirmation.
Choline is another important metabolite that has been implicated in the pathogenesis of NAFLD and NASH. Choline deficiency in the diet has been linked to liver disease for a long time44. Choline-deficient diets are used to create rodent models of NASH. However, until recently, it was not known that choline deficiency could occur under pathophysiologic conditions. High-fat diets lead to formation of intestinal microbiota that convert dietary choline into methylamines, reducing circulating plasma levels of phosphatidylcholine to produce similar effects of choline-deficient diets and causing NASH45. Phosphatidylcholine is necessary for the assembly and secretion of very-low-density lipoprotein (VLDL)46. Microbiota-induced choline deficiency therefore results in triglyceride accumulation in hepatocytes, secondary to lower hepatic secretion of VLDL, whereas the increase of plasma level of trimethylamine (TMA) and its hepatic metabolism to trimethylamine-N-oxide (TMAO) have been linked to atherosclerosis and cardiovascular disease47, 48. People with choline-deficient diets develop fatty liver, based on MRI imaging studies. However, a single-nucleotide polymorphism in the promoter region of PEMT (rs12325817), which affects de novo synthesis of phosphatidylcholine, is required for development of a fatty liver49. Taken together, diet-induced changes in the intestinal microbiome can produce dramatic changes in metabolites in the host.
Microbial products contribute to the pathogenesis of NAFLD and NASH. Children with NAFLD had markedly higher serum concentrations of endotoxin than control subjects50. Endotoxemia was also observed in patients with NASH51. The most convincing evidence for the importance of translocated microbial products comes from preclinical studies of NAFLD. Signaling via toll-like receptor (TLR)4, a receptor for lipopolysaccharide (LPS), in hematopoietic-derived cells is required for the development of liver steatosis, but not for the development of obesity in mice52. Mice deficient in sensing pathogen-associated molecular patterns (PAMPs) or downstream signaling are resistant to NASH53, 54. Microbial products reach the liver via the portal circulation and cause inflammation, among other effects. Genetically obese mice are more sensitive to endotoxin-induced hepatotoxicity and develop steatohepatitis after exposure to low doses of LPS55. Increased intestinal permeability and disruption of the mucosal barrier are required for microbial products to translocate from the intestinal lumen to extra-intestinal space. Patients with NAFLD have significantly increased intestinal permeability and alterations in the intestinal tight junctions, compared to healthy individuals8. Bacterial overgrowth is particularly important in patients with a leaky gut because it increases the luminal amount of PAMPs.
But what causes the onset of intestinal barrier dysfunction? Intestinal inflammation might cause intestinal leakage and translocation of microbial products. Obesity is accompanied by inflammation in the colorectal mucosa; in obese individuals, diet-induced weight loss reduces colorectal inflammation and alters expression of inflammatory and cancer-related genes56. In mice, high-fat diets increase activity of the transcription factor NFκB and expression of tumor necrosis factor (TNF)α in the small intestine57. Intestinal inflammation depends on the enteric microbiota; germ-free mice are protected from inflammation of the small intestine57. More direct evidence for dysbiosis-induced intestinal inflammation and bacterial translocation has come from studies of mice deficient for Nlrp3 and Nlrp6. These mice cannot form cytoplasmic multi-protein complexes composed of nucleotide-binding domain and leucine-rich repeat containing proteins (NLRPs), called inflammasomes. Inflammasomes are sensors of exogenous PAMPs or endogenous damage-associated molecular patterns (DAMPs) that regulate cleavage of precursors to inflammatory cytokines such as pro-interleukin (IL)1β and pro-IL18. In mice, loss of Nlrp3 and Nlrp6 inflammasomes is associated with intestinal dysbiosis and eventual inflammation of the colon, via the chemokine CCL5. Dysbiosis is characterized by an increase in Prevotella58. Subsequent microbe translocation leads to increased accumulation of bacterial products such as LPS and bacterial DNA in the portal vein. These bacterial products induce an inflammatory response in the liver that promotes progression of NAFLD to NASH. Importantly, this phenotype can be transmitted, by co-housing wild type and NASH-prone mice58. Dysbiosis therefore induces colonic inflammation and bacterial translocation, causing simple hepatic steatosis to turn into NASH. Dysbiosis is therefore a significant contributor to liver disease. This study also demonstrated how features of the host can determine the composition of the microbiome.
In summary, diet- and host-induced changes in the intestinal microbiota contribute to the onset of NAFLD and NASH (Figure 1). Changes in the intestinal microbiota can affect the liver via translocated microbial products or absorbed bacterial metabolites. Alternatively, direct host–microbiota interactions in the intestine alter intestinal homeostasis, affecting the liver as distant organ.
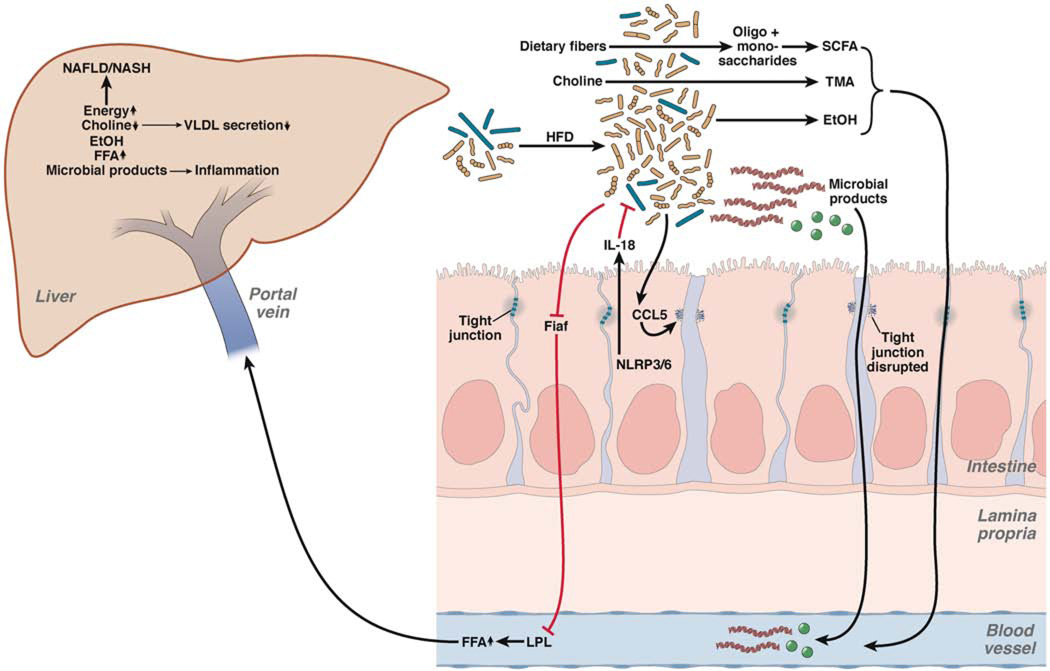
Figure 1. Effects of the Intestinal Microbiota on NAFLD and Progression to Steatohepatitis.
High-fat diets (HFD) result in dysbiosis and intestinal bacterial overgrowth. Alterations in the intestinal microbiota increase energy extraction and fermentation of dietary fibers into oligosaccharides, monosaccharides, and short chain fatty acids (SCFA), respectively. Dietary choline is metabolized by the intestinal microbiota to TMA, resulting in choline deficiency. Hepatic choline deficiency results in decreased VLDL efflux, producing hepatic steatosis. Changes in the microbiota also produce ethanol (EtOH), which is absorbed and metabolized in the liver. The intestinal microbiota suppresses gene expression of Fiaf in intestinal epithelial cells, resulting in enhanced activity of LPL and increased levels of free fatty acids (FFA). NLRPs regulate microbial composition via changes of the effector protein IL18. Dysbiosis, in turn, causes CCL5-mediated disruption of tight junctions in enterocytes. Increased intestinal permeability leads to translocation of microbial products to the liver and causes inflammation by activating TLRs.
Alcoholic Liver Disease
A prominent feature of alcohol abuse is disruption of the intestinal barrier. Animal models of alcoholic liver disease have leaky gut16, and patients have impairments to the intestinal barrier. There is debate over which marker is reliable for identification of patients with leaky intestine. Arguably the best method to assess increased intestinal permeability is direct measurement of bacterial products that originate only from the intestinal lumen, and must therefore have translocated into the extra-intestinal space, blood, and organs. Alcoholics with no evidence of liver disease and patients with alcoholic hepatitis or alcohol-associated cirrhosis have higher plasma levels of endotoxin than healthy controls59.
Does the intestinal microbiome initiate and mediate an increase in intestinal permeability? Intestinal sterilization protects against alcohol-induced intestinal barrier leakage and prevents bacterial translocation60, 61. Although mice that express nonfunctional Tlr4 are protected from experimental alcoholic liver disease, levels of endotoxin in the portal vein increase to levels similar to those of mice that express wild-type Tlr4, indicating that Tlr4 does not control intestinal permeability62.
So how might the intestinal microbiota increase intestinal permeability? Intestinal bacteria metabolize ethanol and produce acetaldehyde63. Ethanol and its metabolic derivative, acetaldehyde, disrupt tight junction integrity64. The intestinal microbiome also synthesizes ethanol, which might have deleterious effects on the intestinal barrier. Alternatively, a decrease in commensal probiotics could contribute to loss of the protective tight junction barrier65. A mast cell membrane stabilizer prevents ethanol-induced epithelial barrier alteration in vivo61. Intestinal inflammatory cells such as mast cells might therefore contribute to the onset of a leaky intestine, and could be activated by qualitative and/or quantitative changes in the microbiome. Further metagenomic and metabolomic studies are needed to determine the functional contribution of the intestinal microbiome to barrier dysfunction and thereby alcoholic liver disease. Alcohol-induced liver disease itself could decrease the intestinal barrier, by increasing systemic levels of IL1β or TNFα, which disrupt tight junctions66. It is not clear to what extent changes in liver function contribute to mucosal barrier defects.
Intestinal bacterial overgrowth and dysbiosis are important factors in the pathogenesis of alcoholic liver disease in patients with leaky intestine. Increased intestinal permeability facilitates translocation of microbial products from the intestinal lumen to extraintestinal organs. Mice that are protected from bacterial overgrowth have decreased alcohol-induced liver disease despite leakier guts16. Similarly, rats fed non-absorbable antibiotics that reduce the load of Gram-negative bacteria in the intestine have lower systemic levels of endotoxin and develop less-severe liver disease following ethanol administration60. Ethanol-induced liver inflammation and injury were also significantly lower in mice that express nonfunctional Tlr4 compared with mice that express the wild-type protein62, providing further evidence for the role of bacterial products in alcoholic liver disease.
Microbial products translocate from the intestine to the liver in humans and animal models after ethanol intake. In patients or animals with leaky intestine, the total intraluminal load of enteric bacteria determines the amount of translocated bacterial products. PAMPs reach the liver via the portal system. Alcohol, as an initiating liver insult, and microbial products might synergize to promote progression of liver disease. Changes in the intestinal microbiome (particularly bacterial overgrowth) and increased bacterial translocation contribute to alcoholic liver disease (see Figure 2).
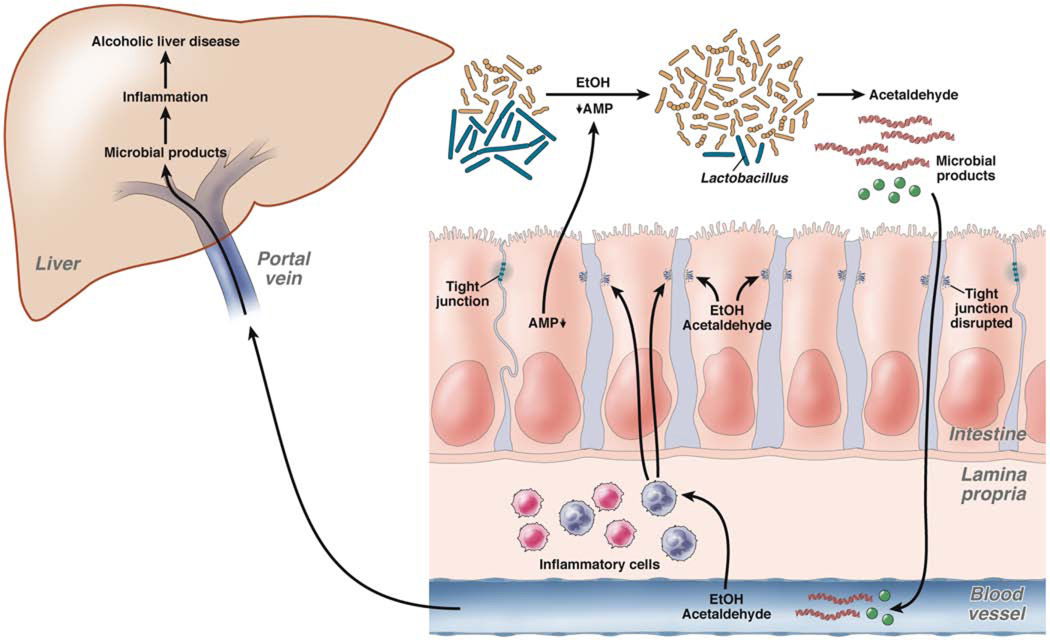
Figure 2. Effects of the Intestinal Microbiota on Alcoholic Liver Disease.
Suppressed secretion of antimicrobial peptides and proteins (AMP), and possibly EtOH itself, contribute to bacterial overgrowth and dysbiosis. Qualitative changes in the microbiota are characterized by decreased Lactobacilli in experimental models of alcohol-induced liver disease. An altered intestinal microbiota is able to produce ethanol and metabolize it into acetaldehyde. Luminal or systemic ethanol and acetaldehyde disrupt tight junctions and increase intestinal permeability. An influx of microbial products into the liver via the portal vein results in hepatic inflammation, which synergizes with ethanol to induce alcoholic liver disease. EtOH and/or acetaldehyde-induced inflammation in the intestinal lamina propria might contribute to dysfunctional tight junctions and reduced production of antimicrobial peptides and proteins by enterocytes.
End-stage Liver Disease
The intestinal microbiome has been implicated in complications of cirrhosis such as hepatic encephalopathy and infections. Increased levels of endotoxin, systemic inflammation, and production of ammonia (a bacterial byproduct) contribute to pathogenesis of hepatic encephalopathy67. Intestinal decontamination with non-absorbable antibiotics such as rifaximin is effective treatment for subclinical and overt hepatic encephalopathy68, 69.
Bacterial translocation occurs in healthy individuals and is important for immune system development, but can also be harmful. Translocation of microbial products contributes to progression of NAFLD, NASH, and alcoholic liver disease. In patients with cirrhosis, bacterial translocation induces inflammation and hemodynamic derangement70, and can cause serious infections, with reported 38% mortality71. Infections such as spontaneous bacterial peritonitis and bacteremia develop in patients with end-stage liver disease, caused by migration of intestinal bacteria into the peritoneal cavity or circulation.
Several mechanisms contribute to intestinal translocation of bacteria in patients with cirrhosis, such as a leaky intestinal barriers and immune deficits72. Some of these mechanisms are closely related to the content of the intestinal microbiome. Most infections (approximately 80%) are caused by Gram-negative bacilli–especially Escherichia coli70. Interestingly, bacteria from the family Enterobacteriaceae (including E coli, Klebsiella, Proteus, and Enterobacter) increase in the microbiota of patients with cirrhosis27, 28, 31, 32. Small intestinal bacterial overgrowth in patients with cirrhosis was associated with systemic endotoxemia and found to predispose animal models of cirrhosis to bacterial translocation30, 31, 70. Overgrowth of a dysbiotic microbiota might make a major contribution to translocation of viable bacteria and thereby infections. A small observational study highlighted the clinical importance of intestinal bacterial overgrowth in patients with decompensated cirrhosis. Intestinal decontamination with the non-absorbable antibiotics rifaximin reduced endotoxemia and reduced the severity of liver disease73. Randomized placebo-controlled trials are needed to confirm these preliminary findings.
Other factors that contribute to bacterial translocation include intestinal inflammation and changes in intestinal immune surveillance. Patients with liver cirrhosis were found to have inflammation of the duodenum, which could promote leakiness74. Rats with cirrhosis have increased numbers of activated CD103+ dendritic cells in the lamina propria and mesenteric lymph nodes, with detectable bacterial DNA but no viable bacteria in the mesenteric lymph nodes. In contrast, in rats with viable bacteria in mesenteric lymph nodes, CD103+ dendritic cells did not appear to be activated, indicating tolerance and exhaustion. Intestinal sterilization prevented bacterial translocation, and reduced the activation and function of CD103+ dendritic cells, indicating that the intestinal microbiome, rather than the host, seems to mediate the effects of bacterial translocation75.
Although the intestinal microbiota contributes to progression of liver disease in pre-cirrhotic states, the pathologic functions of the intestinal microbiota change during advanced stages of liver disease. Translocated viable bacteria and microbial products make important contributions to clinical complications associated with end-stage liver disease. Bacterial translocation, rather than failed liver function, could be the major determinant of mortality in this patient cohort.
Interactions Between Liver and Intestine via Bile Acids
Bile acids mediate communication between the liver and intestine. They are produced as glycine or taurine conjugates in the liver, from cholesterol, for secretion into the small intestine. Conjugated bile acids are absorbed in the terminal ileum to return to the liver. Intestinal bacteria in the large intestine generate secondary bile acids by deconjugation and dehydroxylation. Bile acids are important not only for the absorption of dietary fats and vitamins, they are also ligands for the nuclear receptor farnesoid X receptor (FXR) and the G-protein coupled receptor TGR5. The intestine therefore communicates with the liver via the entero-hepatic circulation.
Not surprisingly, germ-free rats have an altered bile acid profile, characterized predominately by taurine-conjugated bile acids with relatively lower amounts of unconjugated and glycine-conjugated bile acids76. Fecal samples from patients with cirrhosis have reduced total bile acids, likely due to decreased bile flow. Interestingly, the ratio between secondary and primary fecal bile acids is also decreased, possibly from reduced microbial deconjugation77—bacterial gene expression studies are needed to confirm this. Levels of conjugated and unconjugated bile acids are higher in the serum samples from patients with cirrhosis—especially those with advanced-stage disease77. Changes in serum bile acids have also been reported in experimental models of NASH and alcoholic liver disease78,79. It is not clear whether the microbiota contributes to these changes.
Bile acids have direct bacteriostatic effects; intestinal bacterial overgrowth might result from the decrease in total fecal bile acids in patients with cirrhosis77. Administration of conjugated bile acids to rats with cirrhosis normalized bile secretion and reduced intestinal bacterial overgrowth and translocation80. Conjugated bile acids bind to FXR in intestinal epithelial cells, which increases production of the antimicrobial proteins angiogenin 1 and RNase family member 4. These prevent bacterial overgrowth and promote epithelial cell integrity81. Bile acids therefore inhibit bacterial proliferation directly and indirectly, by modulating host cells expression of antimicrobial genes.
Microbial modification of bile acids is an important mechanism by which the microbiota can interact with the host and affect not only liver disease, but other organs and metabolic pathways. FXR and TGR5 have been implicated in the metabolic syndrome. Fxr-deficient mice are protected from genetic and diet-induced obesity, but not hepatic steatosis82. The FXR agonist obeticholic acid reduced markers of liver inflammation and fibrosis in patients with type 2 diabetes mellitus and NAFLD in a phase 2 clinical trial83. Interestingly, cholestatic liver fibrosis following bile duct ligation is lower in Fxr-deficient mice, but the absence of Fxr had no effect on toxin-induced liver fibrosis84. Activation of Tgr5 by bile acids in brown adipose tissue and muscle increased energy expenditure and attenuated diet-induced obesity in mice85. The Tgr5 agonist INT-777 caused release of intestinal glucagon-like peptide-1, and reduced adiposity and hepatic steatosis in mice placed on high-fat diets86.
Therefore, the intestinal microbiota might contribute to liver disease by modifying intestinal bile acids and regulating FXR and TGR5 signaling. Future studies should investigate how changes in expression of bacterial genes and the bile acid profile affect the host via modulation of FXR and TGR5 and contribute to liver disease.
Future Directions
The intestinal microbiome contributes to the onset and progression of alcoholic liver disease and NAFLD, and mediates complications in end-stage liver disease. There appears to be an association between intestinal dysbiosis and liver disease in patients. Changes in the intestinal microbiome were found to cause liver disease mostly in animal models, and few have been associated with metabolic and immunologic features of patients with NAFLD and NASH. Future studies should assess microbial gene expression, proteins, and metabolites, and focus on patients in particular. Increasing our understanding of the delicate homeostasis between the intestine and its microbes could lead to new insights into the pathogenesis of liver disease and therapeutic strategies. There is sufficient evidence to justify a rationale attempt to modulate the intestinal microbiome to treat liver disease. The ultimate goal is to restore eubiosis, which might restore intestinal homeostasis.
Acknowledgements
The study was supported in part by NIH grants K08 DK081830, R01 AA020703 to BS, U01 AA021856 to BS and DAB, and 2 P42 ES010337 to DAB.
Abbreviations
- DAMP
damage-associated molecular pattern
- Fiaf
fasting induced adipocyte factor
- FXR
farnesoid X receptor
- IL
interleukin
- LPL
lipoprotein lipase
- LPS
lipopolysaccharide
- NAFLD
non-alcoholic fatty liver disease
- NASH
non-alcoholic steatohepatitis
- NLRP
NLR family, pyrin domain containing
- PAMP
pathogen-associated molecular pattern
- Reg3
regenerating islet-derived 3
- TLR
Toll-like receptor
- TNF
tumor necrosis factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: The authors declare the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Browning JD, Szczepaniak LS, Dobbins R, Nuremberg P, Horton JD, Cohen JC, Grundy SM, Hobbs HH. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40:1387–1395. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- 2.Williams CD, Stengel J, Asike MI, Torres DM, Shaw J, Contreras M, Landt CL, Harrison SA. Prevalence of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis among a largely middle-aged population utilizing ultrasound and liver biopsy: a prospective study. Gastroenterology. 2011;140:124–131. doi: 10.1053/j.gastro.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 3.Caldwell S, Argo C. The natural history of non-alcoholic fatty liver disease. Dig Dis. 2010;28:162–168. doi: 10.1159/000282081. [DOI] [PubMed] [Google Scholar]
- 4.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raman M, Ahmed I, Gillevet PM, Probert CS, Ratcliffe NM, Smith S, Greenwood R, Sikaroodi M, Lam V, Crotty P, Bailey J, Myers RP, Rioux KP. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic Fatty liver disease. Clin Gastroenterol Hepatol. 2013;11:868–875. doi: 10.1016/j.cgh.2013.02.015. e3. [DOI] [PubMed] [Google Scholar]
- 6.Mouzaki M, Comelli EM, Arendt BM, Bonengel J, Fung SK, Fischer SE, McGilvray ID, Allard JP. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology. 2013;58:120–127. doi: 10.1002/hep.26319. [DOI] [PubMed] [Google Scholar]
- 7.Zhu L, Baker SS, Gill C, Liu W, Alkhouri R, Baker RD, Gill SR. Characterization of the gut microbiome in non-alcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology. 2013;57(2):601–609. doi: 10.1002/hep.26093. [DOI] [PubMed] [Google Scholar]
- 8.Miele L, Valenza V, La Torre G, Montalto M, Cammarota G, Ricci R, Masciana R, Forgione A, Gabrieli ML, Perotti G, Vecchio FM, Rapaccini G, Gasbarrini G, Day CP, Grieco A. Increased intestinal permeability and tight junction alterations in nonalcoholic fatty liver disease. Hepatology. 2009;49:1877–1887. doi: 10.1002/hep.22848. [DOI] [PubMed] [Google Scholar]
- 9.Sabate JM, Jouet P, Harnois F, Mechler C, Msika S, Grossin M, Coffin B. High prevalence of small intestinal bacterial overgrowth in patients with morbid obesity: a contributor to severe hepatic steatosis. Obes Surg. 2008;18:371–377. doi: 10.1007/s11695-007-9398-2. [DOI] [PubMed] [Google Scholar]
- 10.Wigg AJ, Roberts-Thomson IC, Dymock RB, McCarthy PJ, Grose RH, Cummins AG. The role of small intestinal bacterial overgrowth, intestinal permeability, endotoxaemia, and tumour necrosis factor alpha in the pathogenesis of non-alcoholic steatohepatitis. Gut. 2001;48:206–211. doi: 10.1136/gut.48.2.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bode C, Bode JC. Activation of the innate immune system and alcoholic liver disease: effects of ethanol per se or enhanced intestinal translocation of bacterial toxins induced by ethanol? Alcohol Clin Exp Res. 2005;29:166S–1671S. doi: 10.1097/01.alc.0000189280.19073.28. [DOI] [PubMed] [Google Scholar]
- 12.Yan AW, Schnabl B. Bacterial translocation and changes in the intestinal microbiome associated with alcoholic liver disease. World J Hepatol. 2012;4:110–118. doi: 10.4254/wjh.v4.i4.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mutlu EA, Gillevet PM, Rangwala H, Sikaroodi M, Naqvi A, Engen PA, Kwasny M, Lau CK, Keshavarzian A. Colonic microbiome is altered in alcoholism. Am J Physiol Gastrointest Liver Physiol. 2012;302:G966–G978. doi: 10.1152/ajpgi.00380.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schnabl B. Linking intestinal homeostasis and liver disease. Curr Opin Gastroenterol. 2013;29:264–270. doi: 10.1097/MOG.0b013e32835ff948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan AW, Fouts DE, Brandl J, Stärkel P, Torralba M, Schott E, Tsukamoto H, Nelson KE, Brenner DA, Schnabl B. Enteric Dysbiosis Associated with a Mouse Model of Alcoholic Liver Disease. Hepatology. 2011;53:96–105. doi: 10.1002/hep.24018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartmann P, Chen P, Wang HJ, Wang L, McCole DF, Brandl K, Starkel P, Belzer C, Hellerbrand C, Tsukamoto H, Ho SB, Schnabl B. Deficiency of intestinal mucin-2 ameliorates experimental alcoholic liver disease in mice. Hepatology. 2013;58:108–119. doi: 10.1002/hep.26321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nanji AA, Khettry U, Sadrzadeh SM. Lactobacillus feeding reduces endotoxemia and severity of experimental alcoholic liver (disease) Proc Soc Exp Biol Med. 1994;205:243–247. doi: 10.3181/00379727-205-43703. [DOI] [PubMed] [Google Scholar]
- 18.Forsyth CB, Farhadi A, Jakate SM, Tang Y, Shaikh M, Keshavarzian A. Lactobacillus GG treatment ameliorates alcohol-induced intestinal oxidative stress, gut leakiness, and liver injury in a rat model of alcoholic steatohepatitis. Alcohol. 2009;43:163–172. doi: 10.1016/j.alcohol.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Liu Y, Sidhu A, Ma Z, McClain C, Feng W. Lactobacillus rhamnosus GG culture supernatant ameliorates acute alcohol-induced intestinal permeability and liver injury. Am J Physiol Gastrointest Liver Physiol. 2012;303:G32–G41. doi: 10.1152/ajpgi.00024.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirpich IA, Solovieva NV, Leikhter SN, Shidakova NA, Lebedeva OV, Sidorov PI, Bazhukova TA, Soloviev AG, Barve SS, McClain CJ, Cave M. Probiotics restore bowel flora and improve liver enzymes in human alcohol-induced liver injury: a pilot study. Alcohol. 2008;42:675–682. doi: 10.1016/j.alcohol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bode JC, Bode C, Heidelbach R, Durr HK, Martini GA. Jejunal microflora in patients with chronic alcohol abuse. Hepatogastroenterology. 1984;31:30–34. [PubMed] [Google Scholar]
- 22.Wegener M, Schaffstein J, Dilger U, Coenen C, Wedmann B, Schmidt G. Gastrointestinal transit of solid-liquid meal in chronic alcoholics. Dig Dis Sci. 1991;36:917–923. doi: 10.1007/BF01297141. [DOI] [PubMed] [Google Scholar]
- 23.Bode C, Bode JC. Alcohol's role in gastrointestinal tract disorders. Alcohol Health Res World. 1997;21:76–83. [PMC free article] [PubMed] [Google Scholar]
- 24.Salaspuro V, Nyfors S, Heine R, Siitonen A, Salaspuro M, Jousimies-Somer H. Ethanol oxidation and acetaldehyde production in vitro by human intestinal strains of Escherichia coli under aerobic, microaerobic, and anaerobic conditions. Scand J Gastroenterol. 1999;34:967–973. doi: 10.1080/003655299750025057. [DOI] [PubMed] [Google Scholar]
- 25.Poynard T, Ratziu V, Benhamou Y, Opolon P, Cacoub P, Bedossa P. Natural history of HCV infection. Baillieres Best Pract Res Clin Gastroenterol. 2000;14:211–228. doi: 10.1053/bega.1999.0071. [DOI] [PubMed] [Google Scholar]
- 26.Gines P, Cardenas A, Arroyo V, Rodes J. Management of cirrhosis and ascites. N Engl J Med. 2004;350:1646–1654. doi: 10.1056/NEJMra035021. [DOI] [PubMed] [Google Scholar]
- 27.Lu H, Wu Z, Xu W, Yang J, Chen Y, Li L. Intestinal microbiota was assessed in cirrhotic patients with hepatitis B virus infection. Intestinal microbiota of HBV cirrhotic patients. Microb Ecol. 2011;61:693–703. doi: 10.1007/s00248-010-9801-8. [DOI] [PubMed] [Google Scholar]
- 28.Chen Y, Yang F, Lu H, Wang B, Lei D, Wang Y, Zhu B, Li L. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 2011;54:562–572. doi: 10.1002/hep.24423. [DOI] [PubMed] [Google Scholar]
- 29.Xu M, Wang B, Fu Y, Chen Y, Yang F, Lu H, Chen Y, Xu J, Li L. Changes of fecal Bifidobacterium species in adult patients with hepatitis B virus-induced chronic liver disease. Microb Ecol. 2012;63:304–313. doi: 10.1007/s00248-011-9925-5. [DOI] [PubMed] [Google Scholar]
- 30.Wu ZW, Lu HF, Wu J, Zuo J, Chen P, Sheng JF, Zheng SS, Li LJ. Assessment of the Fecal Lactobacilli Population in Patients with Hepatitis B Virus-Related Decompensated Cirrhosis and Hepatitis B Cirrhosis Treated with Liver Transplant. Microb Ecol. 2012;63(4):929–937. doi: 10.1007/s00248-011-9945-1. [DOI] [PubMed] [Google Scholar]
- 31.Bajaj JS, Hylemon PB, Ridlon JM, Heuman DM, Daita K, White MB, Monteith P, Noble NA, Sikaroodi M, Gillevet PM. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am J Physiol Gastrointest Liver Physiol. 2012;303:G675–G685. doi: 10.1152/ajpgi.00152.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bajaj JS, Ridlon JM, Hylemon PB, Thacker LR, Heuman DM, Smith S, Sikaroodi M, Gillevet PM. Linkage of gut microbiome with cognition in hepatic encephalopathy. Am J Physiol Gastrointest Liver Physiol. 2012;302:G168–G175. doi: 10.1152/ajpgi.00190.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bauer TM, Schwacha H, Steinbruckner B, Brinkmann FE, Ditzen AK, Aponte JJ, Pelz K, Berger D, Kist M, Blum HE. Small intestinal bacterial overgrowth in human cirrhosis is associated with systemic endotoxemia. Am J Gastroenterol. 2002;97:2364–2370. doi: 10.1111/j.1572-0241.2002.05791.x. [DOI] [PubMed] [Google Scholar]
- 34.Bauer TM, Steinbruckner B, Brinkmann FE, Ditzen AK, Schwacha H, Aponte JJ, Pelz K, Kist M, Blum HE. Small intestinal bacterial overgrowth in patients with cirrhosis: prevalence and relation with spontaneous bacterial peritonitis. Am J Gastroenterol. 2001;96:2962–2967. doi: 10.1111/j.1572-0241.2001.04668.x. [DOI] [PubMed] [Google Scholar]
- 35.Chang CS, Chen GH, Lien HC, Yeh HZ. Small intestine dysmotility and bacterial overgrowth in cirrhotic patients with spontaneous bacterial peritonitis. Hepatology. 1998;28:1187–1190. doi: 10.1002/hep.510280504. [DOI] [PubMed] [Google Scholar]
- 36.Teltschik Z, Wiest R, Beisner J, Nuding S, Hofmann C, Schoelmerich J, Bevins CL, Stange EF, Wehkamp J. Intestinal bacterial translocation in rats with cirrhosis is related to compromised Paneth cell antimicrobial host defense. Hepatology. 2012;55:1154–1163. doi: 10.1002/hep.24789. [DOI] [PubMed] [Google Scholar]
- 37.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 38.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Backhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci U S A. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cope K, Risby T, Diehl AM. Increased gastrointestinal ethanol production in obese mice: implications for fatty liver disease pathogenesis. Gastroenterology. 2000;119:1340–1347. doi: 10.1053/gast.2000.19267. [DOI] [PubMed] [Google Scholar]
- 43.Hartmann P, Chen WC, Schnabl B. The intestinal microbiome and the leaky gut as therapeutic targets in alcoholic liver disease. Front Physiol. 2012;3:402. doi: 10.3389/fphys.2012.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blumberg H, McCollum EV. The prevention by choline of liver cirrhosis in rats on high fat, low protein diets. Science. 1941;93:598–599. doi: 10.1126/science.93.2425.598. [DOI] [PubMed] [Google Scholar]
- 45.Dumas ME, Barton RH, Toye A, Cloarec O, Blancher C, Rothwell A, Fearnside J, Tatoud R, Blanc V, Lindon JC, Mitchell SC, Holmes E, McCarthy MI, Scott J, Gauguier D, Nicholson JK. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci U S A. 2006;103:12511–12516. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang XC, Li Z, Liu R, Yang XP, Pan M, Lagrost L, Fisher EA, Williams KJ. Phospholipid transfer protein deficiency impairs apolipoprotein-B secretion from hepatocytes by stimulating a proteolytic pathway through a relative deficiency of vitamin E and an increase in intracellular oxidants. J Biol Chem. 2005;280:18336–18340. doi: 10.1074/jbc.M500007200. [DOI] [PubMed] [Google Scholar]
- 47.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ, Hazen SL. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368:1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spencer MD, Hamp TJ, Reid RW, Fischer LM, Zeisel SH, Fodor AA. Association Between Composition of the Human Gastrointestinal Microbiome and Development of Fatty Liver With Choline Deficiency. Gastroenterology. 2011;140:976–986. doi: 10.1053/j.gastro.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alisi A, Manco M, Devito R, Piemonte F, Nobili V. Endotoxin and plasminogen activator inhibitor-1 serum levels associated with nonalcoholic steatohepatitis in children. J Pediatr Gastroenterol Nutr. 2010;50:645–649. doi: 10.1097/MPG.0b013e3181c7bdf1. [DOI] [PubMed] [Google Scholar]
- 51.Farhadi A, Gundlapalli S, Shaikh M, Frantzides C, Harrell L, Kwasny MM, Keshavarzian A. Susceptibility to gut leakiness: a possible mechanism for endotoxaemia in non-alcoholic steatohepatitis. Liver Int. 2008;28:1026–1033. doi: 10.1111/j.1478-3231.2008.01723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saberi M, Woods NB, de Luca C, Schenk S, Lu JC, Bandyopadhyay G, Verma IM, Olefsky JM. Hematopoietic cell-specific deletion of toll-like receptor 4 ameliorates hepatic and adipose tissue insulin resistance in high-fat-fed mice. Cell Metab. 2009;10:419–429. doi: 10.1016/j.cmet.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miura K, Kodama Y, Inokuchi S, Schnabl B, Aoyama T, Ohnishi H, Olefsky JM, Brenner DA, Seki E. Toll-like receptor 9 promotes steatohepatitis by induction of interleukin-1beta in mice. Gastroenterology. 2010;139:323–334. doi: 10.1053/j.gastro.2010.03.052. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rivera CA, Adegboyega P, van Rooijen N, Tagalicud A, Allman M, Wallace M. Toll-like receptor-4 signaling and Kupffer cells play pivotal roles in the pathogenesis of non-alcoholic steatohepatitis. J Hepatol. 2007;47:571–579. doi: 10.1016/j.jhep.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang SQ, Lin HZ, Lane MD, Clemens M, Diehl AM. Obesity increases sensitivity to endotoxin liver injury: implications for the pathogenesis of steatohepatitis. Proc Natl Acad Sci U S A. 1997;94:2557–2562. doi: 10.1073/pnas.94.6.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pendyala S, Neff LM, Suarez-Farinas M, Holt PR. Diet-induced weight loss reduces colorectal inflammation: implications for colorectal carcinogenesis. Am J Clin Nutr. 2011;93:234–242. doi: 10.3945/ajcn.110.002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ding S, Chi MM, Scull BP, Rigby R, Schwerbrock NM, Magness S, Jobin C, Lund PK. High-fat diet: bacteria interactions promote intestinal inflammation which precedes and correlates with obesity and insulin resistance in mouse. PLoS One. 2010;5:e12191. doi: 10.1371/journal.pone.0012191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, Thaiss CA, Kau AL, Eisenbarth SC, Jurczak MJ, Camporez JP, Shulman GI, Gordon JI, Hoffman HM, Flavell RA. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parlesak A, Schafer C, Schutz T, Bode JC, Bode C. Increased intestinal permeability to macromolecules and endotoxemia in patients with chronic alcohol abuse in different stages of alcohol-induced liver disease. J Hepatol. 2000;32:742–747. doi: 10.1016/s0168-8278(00)80242-1. [DOI] [PubMed] [Google Scholar]
- 60.Adachi Y, Moore LE, Bradford BU, Gao W, Thurman RG. Antibiotics prevent liver injury in rats following long-term exposure to ethanol. Gastroenterology. 1995;108:218–224. doi: 10.1016/0016-5085(95)90027-6. [DOI] [PubMed] [Google Scholar]
- 61.Ferrier L, Berard F, Debrauwer L, Chabo C, Langella P, Bueno L, Fioramonti J. Impairment of the intestinal barrier by ethanol involves enteric microflora and mast cell activation in rodents. Am J Pathol. 2006;168:1148–1154. doi: 10.2353/ajpath.2006.050617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uesugi T, Froh M, Arteel GE, Bradford BU, Thurman RG. Toll-like receptor 4 is involved in the mechanism of early alcohol-induced liver injury in mice. Hepatology. 2001;34:101–108. doi: 10.1053/jhep.2001.25350. [DOI] [PubMed] [Google Scholar]
- 63.Elamin EE, Masclee AA, Dekker J, Jonkers DM. Ethanol metabolism and its effects on the intestinal epithelial barrier. Nutr Rev. 2013;71:483–499. doi: 10.1111/nure.12027. [DOI] [PubMed] [Google Scholar]
- 64.Rao RK, Seth A, Sheth P. Recent Advances in Alcoholic Liver Disease I. Role of intestinal permeability and endotoxemia in alcoholic liver disease. Am J Physiol Gastrointest Liver Physiol. 2004;286:G881–G884. doi: 10.1152/ajpgi.00006.2004. [DOI] [PubMed] [Google Scholar]
- 65.Wang Y, Kirpich I, Liu Y, Ma Z, Barve S, McClain CJ, Feng W. Lactobacillus rhamnosus GG treatment potentiates intestinal hypoxia-inducible factor, promotes intestinal integrity and ameliorates alcohol-induced liver injury. Am J Pathol. 2011;179:2866–2875. doi: 10.1016/j.ajpath.2011.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 67.Tranah TH, Vijay GK, Ryan JM, Shawcross DL. Systemic inflammation and ammonia in hepatic encephalopathy. Metab Brain Dis. 2013;28:1–5. doi: 10.1007/s11011-012-9370-2. [DOI] [PubMed] [Google Scholar]
- 68.Sidhu SS, Goyal O, Mishra BP, Sood A, Chhina RS, Soni RK. Rifaximin improves psychometric performance and health-related quality of life in patients with minimal hepatic encephalopathy (the RIME Trial) Am J Gastroenterol. 2011;106:307–316. doi: 10.1038/ajg.2010.455. [DOI] [PubMed] [Google Scholar]
- 69.Bass NM, Mullen KD, Sanyal A, Poordad F, Neff G, Leevy CB, Sigal S, Sheikh MY, Beavers K, Frederick T, Teperman L, Hillebrand D, Huang S, Merchant K, Shaw A, Bortey E, Forbes WP. Rifaximin treatment in hepatic encephalopathy. N Engl J Med. 2010;362:1071–1081. doi: 10.1056/NEJMoa0907893. [DOI] [PubMed] [Google Scholar]
- 70.Bellot P, Frances R, Such J. Pathological bacterial translocation in cirrhosis: pathophysiology, diagnosis and clinical implications. Liver Int. 2013;33:31–39. doi: 10.1111/liv.12021. [DOI] [PubMed] [Google Scholar]
- 71.Arvaniti V, D'Amico G, Fede G, Manousou P, Tsochatzis E, Pleguezuelo M, Burroughs AK. Infections in patients with cirrhosis increase mortality four-fold and should be used in determining prognosis. Gastroenterology. 2010;139:1246–1256. doi: 10.1053/j.gastro.2010.06.019. e1–5. [DOI] [PubMed] [Google Scholar]
- 72.Wiest R, Lawson M, Geuking M. Pathological bacterial translocation in liver cirrhosis. J Hepatol. 2013 doi: 10.1016/j.jhep.2013.07.044. [DOI] [PubMed] [Google Scholar]
- 73.Kalambokis GN, Tsianos EV. Rifaximin reduces endotoxemia and improves liver function and disease severity in patients with decompensated cirrhosis. Hepatology. 2012;55:655–656. doi: 10.1002/hep.24751. [DOI] [PubMed] [Google Scholar]
- 74.Du Plessis J, Vanheel H, Janssen CE, Roos L, Slavik T, Stivaktas PI, Nieuwoudt M, van Wyk SG, Vieira W, Pretorius E, Beukes M, Farre R, Tack J, Laleman W, Fevery J, Nevens F, Roskams T, Van der Merwe SW. Activated intestinal macrophages in patients with cirrhosis release NO and IL-6 that may disrupt intestinal barrier function. J Hepatol. 2013;58:1125–1132. doi: 10.1016/j.jhep.2013.01.038. [DOI] [PubMed] [Google Scholar]
- 75.Munoz L, Jose Borrero M, Ubeda M, Lario M, Diaz D, Frances R, Monserrat J, Pastor O, Aguado-Fraile E, Such J, Alvarez-Mon M, Albillos A. Interaction between intestinal dendritic cells and bacteria translocated from the gut in rats with cirrhosis. Hepatology. 2012;56:1861–1869. doi: 10.1002/hep.25854. [DOI] [PubMed] [Google Scholar]
- 76.Swann JR, Want EJ, Geier FM, Spagou K, Wilson ID, Sidaway JE, Nicholson JK, Holmes E. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4523–4530. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, Takei H, Muto A, Nittono H, Ridlon JM, White MB, Noble NA, Monteith P, Fuchs M, Thacker LR, Sikaroodi M, Bajaj JS. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 2013;58:949–955. doi: 10.1016/j.jhep.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xie G, Zhong W, Li H, Li Q, Qiu Y, Zheng X, Chen H, Zhao X, Zhang S, Zhou Z, Zeisel SH, Jia W. Alteration of bile acid metabolism in the rat induced by chronic ethanol consumption. FASEB J. 2013;27:3583–3593. doi: 10.1096/fj.13-231860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tanaka N, Matsubara T, Krausz KW, Patterson AD, Gonzalez FJ. Disruption of phospholipid and bile acid homeostasis in mice with nonalcoholic steatohepatitis. Hepatology. 2012;56:118–129. doi: 10.1002/hep.25630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lorenzo-Zuniga V, Bartoli R, Planas R, Hofmann AF, Vinado B, Hagey LR, Hernandez JM, Mane J, Alvarez MA, Ausina V, Gassull MA. Oral bile acids reduce bacterial overgrowth, bacterial translocation, and endotoxemia in cirrhotic rats. Hepatology. 2003;37:551–557. doi: 10.1053/jhep.2003.50116. [DOI] [PubMed] [Google Scholar]
- 81.Inagaki T, Moschetta A, Lee YK, Peng L, Zhao G, Downes M, Yu RT, Shelton JM, Richardson JA, Repa JJ, Mangelsdorf DJ, Kliewer SA. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A. 2006;103:3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Prawitt J, Abdelkarim M, Stroeve JH, Popescu I, Duez H, Velagapudi VR, Dumont J, Bouchaert E, van Dijk TH, Lucas A, Dorchies E, Daoudi M, Lestavel S, Gonzalez FJ, Oresic M, Cariou B, Kuipers F, Caron S, Staels B. Farnesoid X receptor deficiency improves glucose homeostasis in mouse models of obesity. Diabetes. 2011;60:1861–1871. doi: 10.2337/db11-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mudaliar S, Henry RR, Sanyal AJ, Morrow L, Marschall HU, Kipnes M, Adorini L, Sciacca CI, Clopton P, Castelloe E, Dillon P, Pruzanski M, Shapiro D. Efficacy and safety of the farnesoid x receptor agonist obeticholic Acid in patients with type 2 diabetes and nonalcoholic Fatty liver disease. Gastroenterology. 2013;145:574–582. doi: 10.1053/j.gastro.2013.05.042. e1. [DOI] [PubMed] [Google Scholar]
- 84.Fickert P, Fuchsbichler A, Moustafa T, Wagner M, Zollner G, Halilbasic E, Stoger U, Arrese M, Pizarro M, Solis N, Carrasco G, Caligiuri A, Sombetzki M, Reisinger E, Tsybrovskyy O, Zatloukal K, Denk H, Jaeschke H, Pinzani M, Trauner M. Farnesoid X receptor critically determines the fibrotic response in mice but is expressed to a low extent in human hepatic stellate cells and periductal myofibroblasts. Am J Pathol. 2009;175:2392–2405. doi: 10.2353/ajpath.2009.090114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, Messaddeq N, Harney JW, Ezaki O, Kodama T, Schoonjans K, Bianco AC, Auwerx J. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 86.Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, Macchiarulo A, Yamamoto H, Mataki C, Pruzanski M, Pellicciari R, Auwerx J, Schoonjans K. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10:167–177. doi: 10.1016/j.cmet.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]