Abstract
Invasive, life-threatening fungal infections are an important cause of morbidity and mortality, particularly for patients with compromised immune function. The number of therapeutic options for the treatment of invasive fungal infections is quite limited when compared with those available to treat bacterial infections. Indeed, only three classes of molecules are currently used in clinical practice and only one new class of antifungal drugs has been developed in the last 30 years. Here we summarize the unmet clinical needs of current antifungal therapy, discuss challenges inherent to antifungal drug discovery and development, and review recent developments aimed at addressing some of these challenges.
Only one new class of antifungal drugs has been developed in the last 30 years. Meanwhile, the number of patients at risk for fungal infections (e.g., HIV/AIDS patients) has been increasing.
Over the past 30 years, the importance of antifungal drugs to the practice of modern medicine has increased dramatically. Because the vast majority of life-threatening fungal infections affect people with altered immune function, the increased incidence of invasive fungal infections can be correlated with an expansion in the number of people living with conditions or treatments that affect immune function, examples of which include HIV/AIDS, primary immune deficiency, cancer chemotherapy, hematologic and solid organ transplantation, prematurity, and immune-modulatory medications (Richardson 2005). It is, therefore, sobering to consider that two of the three classes of antifungal drugs (azoles and polyenes) in current use had already been introduced into the clinics by 1980 and the third class (echinocandins) had been discovered (Butts and Krysan 2012). Furthermore, even with these newest therapies, the clinical outcomes for most invasive fungal infections are far from ideal. Indeed, infections caused by species of molds for which there is no reliable medical therapy are emerging as are strains of the more common organisms such as Candida albicans and Candida glabrata that are resistant to currently used drugs. It therefore seems fairly clear that the tempo of antifungal drug development has not kept pace with the clinical needs. In this review, we further discuss the unmet clinical needs of medical mycology, the challenges inherent to new antifungal drug development, and new strategies to meet some of these challenges (Brown et al. 2012a,b).
THE CLINICAL PROBLEM
Medically important fungal infections can be broadly classified into two types (Richardson 2005). The first group is mycoses of superficial surfaces such as skin, skin structures, and mucosa. Specific examples include thrush, oropharyngeal candidiasis, and dermatophyte infections of various regions of the body. Although immunocompromised people may have increased rates or severity of disease, superficial mycoses are common among those with intact immune function. The second major group is invasive fungal infections, which, by definition, involve sterile body sites such as the bloodstream, central nervous system, or organs including lung, liver, and kidneys. Many of the fungi that cause invasive disease either infect, or colonize, most human beings but the vast majority of the clinically significant disease occurs in people with compromised immune function. For example, C. albicans is a normal part of the flora of the human gastrointestinal tract and, although it causes superficial disease in immunocompetent individuals, invasive disease occurs almost exclusively in the setting of immune dysfunction or the use of invasive interventions to support life (Pfaller et al. 2012). Furthermore, serological evidence of infection with Cryptococcus neoformans is common over the age of 2 yr (Goldman et al. 2001), but this organism almost never causes clinically significant disease unless the patient develops deficits in cell-mediated immunity.
In resource-rich regions of the world, most clinically significant invasive fungal disease occurs as a complication of prematurity, surgery, chemotherapy, hematopoietic or solid organ transplantation, or immunomodulatory therapies. Patients with primary immunodeficiencies are also at risk for fungal infections but these conditions are quite rare (Lionakis 2012). In general, resource-rich regions also have good access to combined antiretroviral therapy and, consequently, the rates of opportunistic infections associated with HIV/AIDS have decreased significantly. In the U.S. healthcare setting, C. albicans is the fourth most common cause of healthcare-associated bloodstream infections (Lewis 2009). The incidence of mold infections is much lower than candidiasis; however, mold infections are a significant cause of morbidity and mortality among immunocompromised patients. The most common invasive mold infections are attributable to Aspergillus fumigatus (Steinbach et al. 2012). Infections caused by difficult-to-treat molds such as Mucor spp. (Lanternier et al. 2012) and Fusarium (Muhammed et al. 2011), for example, are increasing in incidence. In resource-limited regions, cryptococcosis is a major health problem and causes more deaths in people living with HIV/AIDS than tuberculosis (Park et al. 2009). Because cryptococcal meningitis is frequently the first indication that a person has HIV/AIDS, many people need to survive cryptococcosis if they are to take advantage of highly effective antiretroviral therapy.
CURRENT ANTIFUNGAL THERAPIES
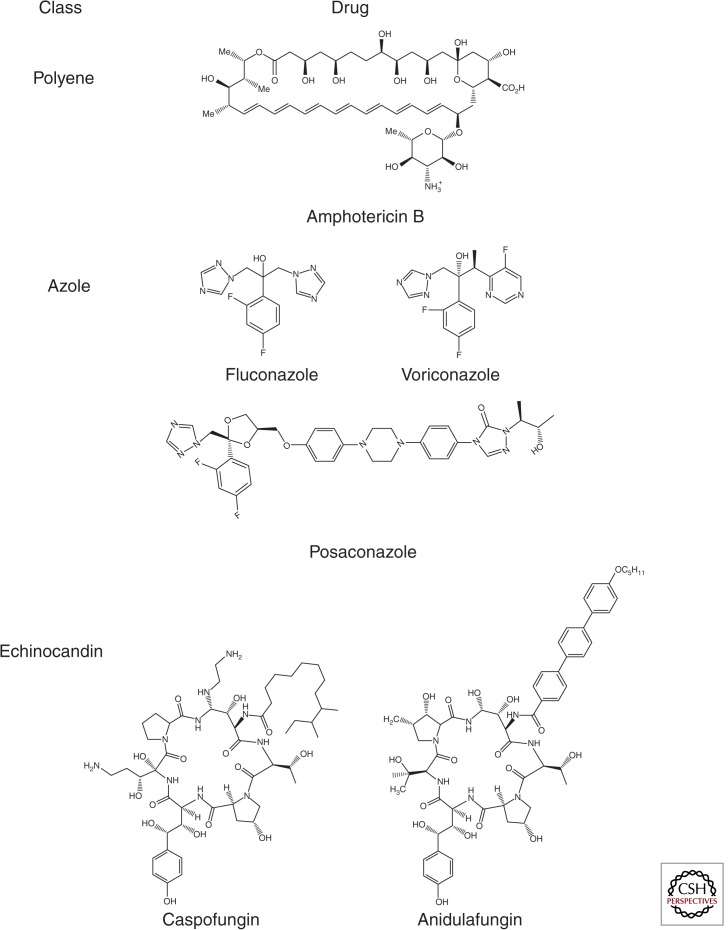
The therapeutic options for invasive fungal infections are quite limited and include only three structural classes of drugs: polyenes, azoles, and echinocandins (Fig. 1). Indeed, there are now more classes of antiretroviral drugs than antifungals. The oldest class of antifungal drugs is the polyenes, of which amphotericin B is the only example used to treat systemic infections. Amphotericin B binds to ergosterol, a membrane sterol that is unique to fungi, as part of its mechanism of action (Gray et al. 2012). Amphotericin B is fungicidal and is the most broad spectrum antifungal available. One of the primary drawbacks of polyenes is their significant toxicity, although the development of lipid formulations of amphotericin B has reduced this problem significantly (Hamill 2013); such formulations are quite expensive and not available in some regions. Amphotericin B, in combination with the adjunctive drug 5-flucytosine, is the treatment of choice for cryptococcal meningitis (Day et al. 2013) as well as for a wide range of less common invasive mycoses. For many of the most common invasive fungal infections, the better tolerated azoles and echinocandins have emerged as first-line agents.
Figure 1.
Classes and representative examples of antifungal drugs in current use.
The azoles are the most widely used class of antifungal drugs (Lass-Flörl 2011). Although there are many azoles currently available (Fig. 2), fluconazole, voriconazole, and posaconazole are most commonly used to treat invasive fungal infections. Azoles inhibit ergosterol biosynthesis and, in general, are fungistatic; an important exception is that voriconazole is fungicidal toward A. fumigatus (Meletiadis et al. 2007). Azoles are extremely well tolerated, although they interfere with the metabolism of a number of other drugs owing to their ability to inhibit cytochrome P450. In general, fluconazole has broad activity against clinically relevant yeast including Candida spp. and Cryptococcus. Many isolates of C. glabrata and Candida krusei, however, are intrinsically less susceptible. Because amphotericin B and 5-flucytosine are not available in many resource-limited regions, fluconazole is widely used to treat cryptococcal meningitis despite the fact that it is less effective. Importantly, fluconazole has essentially no clinically relevant activity against molds. In contrast, itraconazole, voriconazole, and posaconazole all have activity against yeast and molds (Lass-Flörl 2011). Voriconazole is currently the treatment of choice for Aspergillus based on its superiority to amphotericin B in a head-to-head clinical trial (Herbrecht et al. 2002). Posaconazole is distinguished from the other azoles by its in vitro activity against Mucor spp., organisms with reduced susceptibility to other drugs (Luo et al. 2013).
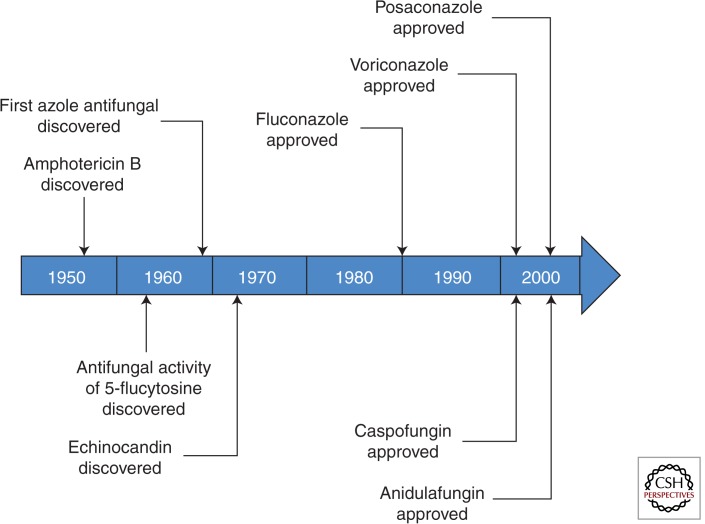
Figure 2.
Time line depicting key milestones of antifungal drug development.
The echinocandins are the most recent addition to the antifungal pharmacopeia (Fig. 1) with the first example, caspofungin, introduced into clinical use a decade ago (Mukherjee et al. 2011). The echinocandins inhibit 1,3-β-glucan synthesis (GS), a key component of the fungal cell wall. The echinocandins have broad fungicidal activity against Candida spp. and have emerged as an important therapeutic option for candidiasis. By growth assays and in vivo studies, the echinocandians are fungistatic toward Aspergillus; however, vital dye studies indicate that caspofungin, for example, kills growing Aspergillus fumigatus (Bowman et al. 2002). At this time, the echinocandins are considered a second-line salvage therapy for those infections. The echinocandins do not have clinically useful activity against Cryptococcus or Zygomycetes.
UNMET CLINICAL NEED
The small number of available antifungal drugs, in and of itself, would not be a problem if the outcomes for invasive fungal infections were satisfactory. By and large, however, this is not the case. For example, 90-day survival following the diagnosis of candidemia varies between 55% and 70%, depending on the underlying condition of the patient and the specific species causing infection (Pfaller et al. 2012); it is important to note that one of the challenges in studying the outcomes of fungal infections is separating mortality attributed to the infection from mortality owing to comorbidities. The outcomes are even worse for aspergillosis despite the use of voriconazole (Herbrecht et al. 2002). In resource-rich regions with access to amphotericin B and 5-flucytosine, the 1-year mortality owing to cryptococcosis is approximately 25% (Bratton et al. 2013), whereas in resource-limited regions where fluconazole is the only available therapy the mortality is much higher (Sloan et al. 2009). As pointed out in an excellent essay on the state of the art for the diagnosis and treatment of fungal disease, the poor outcomes for invasive fungal infections are most likely owing to relatively poor diagnostic methods as well as the need for more effective therapy (Brown et al. 2012). However, it is important to point out that the newest class of antifungal drugs, the echinocandins, was discovered in the 1970s and took 30 years to enter clinical practice (Fig. 2). Similarly, the best therapy for cryptococcosis is based on two drugs that are more than 50 years old. Because medical innovations and the use of immunomodulatory therapies continue to increase, the numbers of patients at risk for fungal infections are almost certain to increase. Thus, the current pace of antifungal drug development is unlikely to keep up with the clinical needs, particularly as resistance to current agents is being reported more and more frequently.
CHALLENGES OF ANTIFUNGAL DRUG DEVELOPMENT
In comparison to the development of new antimicrobials targeting bacteria, antifungal drug development faces a key fundamental challenge in that fungal pathogens are more closely related to the host. For example, the success of Saccharomyces cerevisiae as a model eukaryotic organism is owing to the fact that many fundamental biochemical and cell biological processes are conserved from fungi to humans. Consequently, many small molecules that are toxic to yeast are also toxic to humans. As such, it is therefore not surprising that the three major classes of antifungal drugs target structures that are unique to fungi. In addition to scientific challenges affecting the identification of new lead compounds, the evaluation of new antifungal agents also presents a number of challenges with respect to clinical trial design that further complicate development (Rex et al. 2001). Unfortunately, these fundamental challenges are in addition to the well-documented scientific, economic, and regulatory challenges that face the development of anti-infectives, in general (Boucher et al. 2009). Taken together, it is perhaps not surprising that the pace of new antifungal drug development lags considerably when compared with other therapeutic areas. To address this significant gap in the anti-infective pipeline, creative approaches to the problems discussed above will be required. The remaining sections will present work toward that goal.
ANTIFUNGAL DRUG DISCOVERY: PROCESS AND NEW APPROACHES
Here, we summarize recent developments in approaches and technologies that have improved and are likely to further facilitate the discovery of new antifungal small molecules. Historically, the most common approach to identifying antifungal small molecules has been to screen large libraries of synthetic small molecules or natural products for their ability to inhibit the growth of a selected fungus. In recent years, the importance of the chemical characteristics and origin of the molecules within the library has become better understood. As high-throughput screening has emerged as a tool for both drug discovery and biological investigation, there has also been an explosion in the number of libraries of synthetic small molecules available commercially. The vast majority of the molecules within these libraries has been designed or collected using criteria that maximize their “drug-like” properties with respect to mammalian targets and physiology. Unfortunately, successful anti-infective molecules have physicochemical properties very different from molecules designed for other clinical indications; this is owing in part to the requirement that the molecule traverse microbial cell walls (Lewis 2013). Thus, new screening efforts for antibacterials or antifungals may benefit from the use of libraries focused on an alternate set of “drug-like” properties.
Two out of the three major classes of currently used antifungals are derived from natural products (polyene and echinocandins) (Ostrosky-Zeichner et al. 2010). Indeed, natural product-based screening has led to the discovery of the majority of clinically used antibiotics as well. As interest in the discovery of new anti-infectives waned in the pharmaceutical industry, so did natural product-based screening. In addition, many anti-infective screening campaigns simply rediscovered the same basic scaffolds from screens of seemingly diverse libraries. More recently, interest in natural product-based screening, however, has enjoyed a renaissance. This has been driven not only by the recognition of the valuable features of natural product hits, but by improvements in structure-identification, separation, and target identification (see below). An important survey of antifungal screening efforts at Merck serves to highlight many of these issues and provide insights that may prove useful for future campaigns (Roemer et al. 2011a). For example, the mechanism of action for molecules derived from fungal isolates was much more likely to be target specific (e.g., cell wall or ergosterol biosynthesis), whereas those from actinomycete-rich isolates were more likely to have molecules with nonspecific mechanisms of action (i.e., ionophores, mitochondrial respiration inhibitors, DNA intercalators, and alkylating agents) (Roemer et al. 2011b). Interestingly, the most abundant target-specific antifungals identified from natural product sources in the Merck screening campaign corresponded to new and known structural classes of GS inhibitors; similarly, azoles were the most widely abundant class of target-selective antifungals identified when screening synthetic chemical libraries. As such, it is likely not a coincidence these drug classes were discovered early and that much of the antifungal “low hanging fruit” may have already been picked. If true, these data emphasize the need for truly innovative antifungal discovery approaches that holistically address the chemical libraries, targets, and pathways screened as well as superior methodologies and technologies to rapidly and rigorously determine the precise mechanism of action of those rare but future antifungal leads.
The assay used most commonly to identify antifungal and antibacterial small molecule is the traditional broth or microbroth growth-inhibition assay in which microbial growth is measured by optical density of the culture. Like all assays, these growth-inhibition assays have limitations and a number of these dramatically reduce its utility in antifungal drug discovery. First, the fact that many pathogenic fungi grow as filaments (e.g., Aspergillus) leads to a poor correlation between organism growth and optical density (Bowman et al. 2002). Second, traditional growth assays are not useful for identifying molecules active against fungal biofilms, a medically important growth phase of these pathogens (Pierce et al. 2008; LaFleur et al. 2011; Srinivasan et al. 2013). Third, traditional growth assays are unable to distinguish between molecules that inhibit growth and those that directly kill the organism, a feature that is particularly important for the treatment of some fungi (e.g., Cryptococcus) (Bicanic et al. 2009).
To address the limitations of traditional growth assay screening in antifungal drug discovery, a number of screening assays using alternative readouts have been developed in recent years. A very productive strategy has been to adapt eukaryotic cell viability assays to fungi. The most widely used approach has been to use reporters of metabolic activity such as Alamar Blue and XTT. These dyes are converted to fluorescent molecules when metabolized by viable organisms. These types of assays were initially applied to antifungal molecules in the context of alternative methods for susceptibility testing (Pfaller and Barry 1994), but have been extended to the screening arena by a number of groups. XTT and Alamar Blue–based assays have been particularly useful for screening against C. albicans biofilms, a growth phase that is not amenable to growth-based assay in the context of high-throughput screening. Indeed, straightforward and reproducible protocols for such screens have been developed by the Lopez-Ribot (Pierce et al. 2008) and Lewis groups (LaFleur et al. 2011) and have led to the identification of novel molecules with activity against C. albicans biofilms. Screening for molecules active against molds presents many of the same technical challenges posed by biofilms. Monteiro et al. (2012) have, accordingly, developed an assay for screening A. fumigatus using the metabolism of the dye resazurin and applied this to high-throughput screening. Important characteristics of the metabolic activity-based assays are that they perform well in the high-throughput screening setting with Z′ scores well above 0.5, which is generally accepted as the minimum required for a useful assay; are applicable to clinical isolates without genetic manipulation; and use readily available low-cost reagents and equipment.
As mentioned above, traditional growth assays do not differentiate between fungistatic and fungicidal molecules. In principle, fungicidal molecules would appear to be preferred over fungistatic agents because most patients with invasive fungal infections have compromised immunity and, thus, are more dependent on the antifungal agent to clear infections. In the setting of cryptococcal meningitis (Bicanic et al. 2009), early fungicidal activity has been shown to correlate with clinical outcome, providing a mechanism for the superiority of fungicidal amphotericin B–based therapy when compared with treatment with fungistatic fluconazole. A second type of viability assay has been applied to the specific identification of fungicidal agents: the detection of extracellular adenylate kinase as a reporter of cell lysis (DiDone et al. 2010). Adenylate kinase is an intracellular enzyme that converts two molecules of ADP to ATP and AMP. When the cell loses membrane integrity, adenylate kinase is released into the growth medium; the adenylate kinase activity in the extracellular medium is detected by coupling ATP formation to luciferase activity using a commercially available assay (DiDone et al. 2010). Fungicidal molecules such as the echinocandins generate robust signal, whereas no extracellular adenylate kinase activity is detectable in cultures of fluconazole-treated cells. The assay has been adapted to a 384-well format and applied to S. cerevisiae, C. albicans, and C. neoformans. Finally, it is more sensitive than growth assays in that it can detect the lytic activity of echinocandins at concentrations 10-fold lower than the minimum inhibitory concentration. In addition to the AK-based method, Rabjohns et al. (2013) have also devised a useful, Alamar Blue–based protocol to identify molecules with fungicidal activity against C. neoformans that is applicable to high-throughput screening.
Recently, broad interest in identifying molecules that synergize with existing classes of antibacterial and antifungal drugs as an approach to improve efficacy has emerged (Mukherjee et al. 2005; Ejim et al. 2011). In part, this enthusiasm is based on the shown therapeutic superiority of coadministering amphotericin B and 5-flucytosine (5-FC) to treat cryptococcal meningitis (Day et al. 2013). It is further based on the extensive network of synthetic lethal genetic interactions identified between loss-of-function (or hypomorphic) mutations otherwise singly tolerated by S. cerevisiae (Costanzo et al. 2010). Of particular interest are such mutants that exacerbate Erg11 or GS activity as they provide a genetic prediction that small molecule inhibitors of such targets would display chemical synergy in combination with azoles or echinocandins and thus could be developed as adjuvants to improve the potency and spectrum of existing antifungal agents (Lesage et al. 2004; Costanzo et al. 2010). Most of the work in this regard has focused on either improving activity of, or reversing resistance to, fluconazole. Until recently, the majority of published reports describing molecules that interact with fluconazole involved characterization of a single molecule or class. However, the direct screening for molecules that potentiate fluconazole activity has been reported and led to the identification of known chemical probes (e.g., brefeldin A), previously approved drugs that synergize with fluconazole as well as novel molecules that overcome fluconazole resistance (Spitzer et al. 2011; Kaneko et al. 2013). For example, a screen performed as part of the NIH-Molecular Libraries and Probes Screening Network project identified a class of indole derivatives that restore fluconazole susceptibility to resistant C. albicans isolates (Youngsaye et al. 2012).
The dedicated search for molecules that improve the activity has also included screens of previously approved molecules as potential agents for repurposing to antifungal indications. For example, Spitzer et al. identified a set of previously approved drugs that synergize with fluconazole in vitro and used chemical-genetic analysis to explore their mode of action. In addition, they showed that the antidepressant sertraline combined with fluconazole provides improved activity relative to either drug alone in an invertebrate model of cryptococcosis (Spitzer et al. 2011). Concurrent work in the Lin laboratory showed similar activity in a mouse model of disseminated cryptococcosis (Zhai et al. 2012). Most intriguingly, Cowen and colleagues have shown that inhibitors of the molecular chaperone Hsp90, including the natural products geldanamycin and radicicol, possess potent in vitro synergy in combination with azoles and echinocandins, that this synergy extends across both C. albicans and A. fumigatus, and that it is observed in an invertebrate model of candidiasis (Cowen et al. 2009; Singh et al. 2009; Shapiro et al. 2011). Therefore, ongoing chemical modification of geldanamycin as a novel oncology agent offers opportunities to “repurpose” new geldanamycin analogs as adjuvants for existing antifungals.
EMERGING TARGETS AND MOLECULAR SCAFFOLDS
It is impossible to predict which (if any) of the lead molecules currently being explored will emerge as the next clinically useful antifungal. The science and business of antimicrobial drug development is an unpredictable and fickle world, as highlighted by those responsible for the discovery and development of Cancidas, the first echinocandin antifungal drug to reach the clinic (Mukherjee et al. 2011). In this section, we present a selection of novel antifungal molecules that have emerged from a variety of research programs.
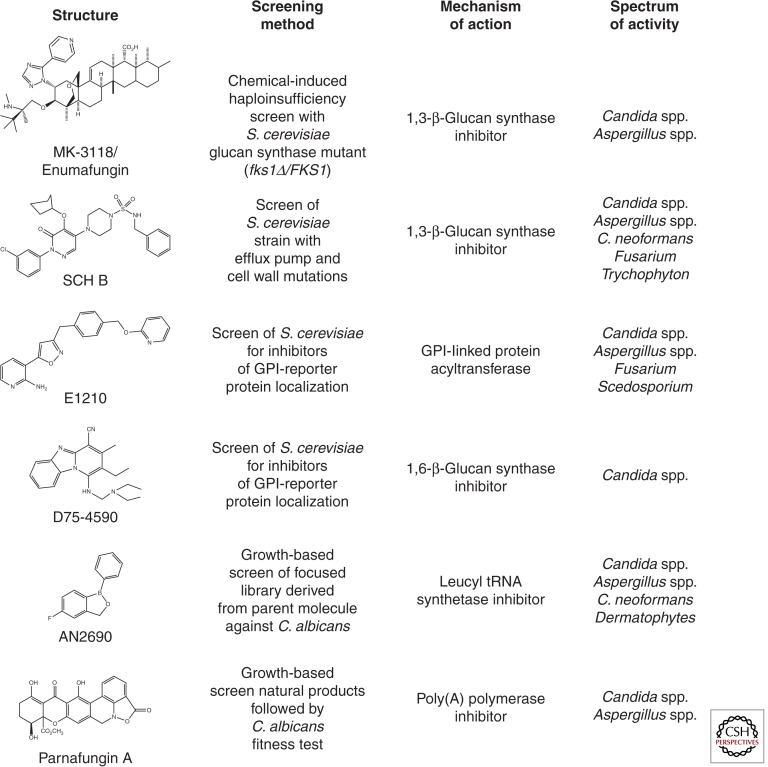
From first principles, one of the most attractive antifungal drug targets is the cell wall; this structure is absent from host cells and, in a sense, represents a histological common denominator between the eukaryotic fungi and prokaryotic bacteria. Because bacterial cell wall-targeted molecules (penicillins, cephlasporins, carbapenam, and glycopepties such as vancomycin) are staples of our antibacterial pharmacopeia, it follows that cell wall-targeted antifungal drugs should be similarly useful. Additionally, the success of the echinocandins further emphasizes the potential of molecules that target cell wall-related processes and significant effort has been directed to identifying and developing new antifungal drug leads targeting GS. Enfumifungin represents a structurally distinct natural product class of GS inhibitors. Originally discovered in Merck by screening natural product extracts to which a S. cerevisiae fks1 heterozygote deletion mutant displayed hypersensitivity, enfumafungin and several related acidic terpenoids (ascosterocide, arundifungin, and ergokonin A) were identified (Onishi et al. 2000). The current development candidate MK-3118 (Fig. 3) is an orally active, semisynthetic derivative of enfumafungin with potent in vitro GS activity (Heasley et al. 2012) with potent in vivo activity against Candida and Aspergillus spp. (Pfaller et al. 2013a,b). Importantly, although echinocandins and enfumafungin both target the GS enzyme (encoded by the Fks1-encoding catalytic subunit and GTPase regulatory subunit Rho1), drug-resistant mutations to each GS inhibitor class map to fks1 but do not display cross-resistance, emphasizing that the two molecules have distinct mechanisms of GS inhibition. Schering-Plough, taking a similar S. cerevisiae whole cell screening approach but reliant on a synthetic chemical library and a sensitized strain deleted of major efflux pumps and certain cell wall biosynthetic genes identified a series of piperazinyl-pyridazinones (SCH A–D) also shown to inhibit GS activity (Walker et al. 2011). Although not a clinical development candidate, SCH C possesses potent in vitro activity against Candida and Aspergillus spp. as well as anti-Cryptococcus and antidermatophyte activity. Significant oral efficacy was achieved in a murine infection model of C. glabrata when treated with SCH B (Fig. 3). Again, drug resistance mutants to these inhibitors correspond to distinct regions of Fks1 and no cross-resistance was observed between these fks1 mutants and echinocandin or enfumafungin class GS inhibitors.
Figure 3.
Examples of antifungal small molecules in development. The structure, method of identification, mechanism of action, and spectrum of antifungal activity are listed for each molecule.
Efforts to identify new antifungal drug leads targeting other essential processes critical to fungal cell wall biogenesis have also yielded early success. Recently, two groups have reported classes of molecules that inhibit glycosylphosphatidylinositol (GPI) biosynthesis. GPI-modified proteins are essential for the construction of the yeast cell wall as well as for proper membrane homeostasis. The first GPI-anchor biosynthesis inhibitors were discovered by Tsukuba Laboratories and emerged from a screen for molecules that interfered with the cell wall localization of a GPI-anchored reporter protein: a nongrowth assay (Tsukahara et al. 2003). The initial hit from this screen was 1-[4-butylbenzyl]isoquinoline (BIQ) and mechanism of action studies identified the acyl transferase Gwt1 as the target. Medicinal chemical-based optimization of this initial hit led to the pyridine-2-amine-based molecule E1210 (Fig. 3), an oral, broad-spectrum antifungal molecule with broad activity against both yeast and mold infections (Hata et al. 2011). E1210 has activity at ng/mL concentrations against Candida spp., Aspergillus spp., and the difficult-to-treat molds Fusarium and Scedosporium. Importantly, the molecule is well tolerated and proved efficacious in murine models of oropharyngeal candidiasis, disseminated candidiasis, aspergillosis, and fusariosis. It is also active against echinocandin-resistant C. albicans. In 2012, a second chemical scaffold with activity against Gwt1 was identified in the course of a high-throughput screening campaign (McLellan et al. 2012). The molecule is a phenyoxyacetanilide and the target was identified by screening an ordered library of S. cerevisiae strains overexpressing each open reading frame (ORF) and comparing the data to a chemically induced haploinsufficiency screen of a set of S. cerevisiae heterozygous deletion mutants; GWT1 was the only ORF that hit in both screens. Subsequently, the target was confirmed by biochemical assays. These two efforts nicely illustrate the use of nontraditional screening approaches followed by chemical genetics-based target identification leading to novel targets and scaffolds.
A second class of cell wall–targeted molecules, β-1,6-glucan synthesis inhibitors, has also been identified by specifically screening for molecules that interfere with cell wall construction (Kitamura et al. 2009a). GPI-linked cell wall proteins frequently are cross-linked within the cell wall through β-1,6-glucan; however, development of specific inhibitors of β-1,6-glucan synthesis has been hampered by the fact that no specific protein or catalytic activity has been directly linked to β-1,6-glucan synthesis. Similar to the screening assay described above, the group at Daiichi Sankyo identified the pyridobenzimidazole (Fig. 3) scaffold by initially screening for agents that disrupted cell wall localization of a reporter construct. The target for this class was identified by traditional screening for UV-generated resistant mutants followed by cloning. A mutation in the KRE6 gene, a gene known to be involved in β-1,6-glucan synthesis, was isolated. In addition, biochemical analysis of the cell wall material isolated from drug-treated cells showed that β-1,6-glucan levels were reduced. The spectrum of activity of this class is not as broad as E1210 and is largely limited to Candida spp.; it has no activity against A. fumigatus and neither E1210 nor the pyridobenzimidazoles have activity against C. neoformans. One example of this scaffold, D21-6076, displayed weak in vitro activity against C. albicans but good in vitro activity against C. glabrata. However, the molecule was equally effective in murine models of disseminated C. albicans and C. glabrata infection. Its activity against the former species seems to be owing to its ability to inhibit C. albicans hyphal morphogenesis and tissue invasion (Kitamura et al. 2009b).
Although GS (and presumably additional aspects of cell wall biogenesis) is clinically validated as a therapeutic target suitable for antifungal development, other essential processes of fungal growth and cell viability should not be ignored. The clinical reliance of azoles with potent and highly specific inhibition of ergosterol biosynthesis, targeting lanosterol 14-α demethylase (Erg11) over its human ortholog, emphasizes this point. Natural product-derived parnafungins (Fig. 3), which inhibit poly(A) polymerase, serve as a salient example (Bills et al. 2009). Parnafungins display potent broad spectrum activity against all clinically relevant Candida spp. (including azole and echinocandin-resistant isolates), anti-Aspergillus activity (albeit best detected under conditions in which poly(A) polymerase enzyme activity is partially depleted by genetic means), and, most importantly, significant therapeutic efficacy in a murine infection model of candidiasis without any obvious indication of cytotoxicity in mice or human cell lines tested (Jiang et al. 2008). Similarly, the leucyl tRNA synthase inhibitor AN2690 (Fig. 3) shows high selectivity against Trichophyton spp. (Rock et al. 2007; Seiradake et al. 2009) and is in clinical development to treat onychomycoses, commonly referred to as toenail fungal infections. Finally, a broadening set of additional antifungal inhibitors targeting the 26S proteasome (fellutamides), translational elongation (yefafungin), cAMP homeostasis (campafungin), microtubule dynamics (12-deoxy-hamigerone), and other basic eukaryotic processes including fatty acid, ergosterol, and ribosome biosynthesis highlight the largely unexploited opportunities to identify fungal-specific agents (Roemer et al. 2011b; Xu et al. 2011). Such agents may fortuitously possess fungal specificity by inhibiting fungal-specific protein domains and/or target unique ligand-binding sites; differences in substrate specificities between fungal and human enzymes may also result in fungal specificity of such agents. Finally, issues of differential cell permeability or prodrug activation (as in the case with 5-FC) may maximize antifungal activity while mitigating host toxicity.
FUTURE DIRECTIONS
The global burden of fungal disease is significant and, as a number of investigators have emphasized, relatively underappreciated and underfunded relative to other diseases (Brown et al. 2012a,b). Currently, the gold standard therapy for cryptococcosis, one of the most prevalent invasive life-threatening fungal infections on the planet, is based on drugs that were developed in the 1950s, when penicillin was a state-of-the-art anti-infective. Since the introduction of amphotericin, only two additional classes of antifungals have been developed. This rate of antifungal drug discovery is unlikely to be sufficient for future demands. This is particularly true because the number of patients at risk for fungal infections is increasing as immunomodulatory therapies continue to expand and our ability to support highly immunocompromised patients improves. Consequently, we are faced with the challenge of an expanding set of at-risk patients, increasing the prevalence of difficult-to-treat organisms, and a slow pace of new drug development.
To meet this challenge, a renewed and resolute commitment by the pharmaceutical industry partnering with academic laboratories, combining innovative screening strategies and novel chemical libraries is required to achieve success. As has been shown and universally accepted within the antibacterial discovery community, in vitro–based high-throughput screening of individual antibacterial targets has been unsuccessful and is unlikely to provide a different outcome for antifungal lead discovery (Payne et al. 2007). Rather, the traditionally successful “compound-centric” approach of empiric screening for small molecules with desirable whole cell bioactivity, cidal activity, and requisite spectrum against clinically relevant pathogens remains warranted (Roemer and Boone 2013; Walsh and Wencewicz 2013). However, combining this classic approach with genomics-era technologies that accelerate discovery time lines is essential. For example, forward genetics platforms such as the S. cerevisiae haploinsufficiency profiling (HOP) (Shoemaker et al. 1996; Giaever et al. 1999, 2002, 2004; Roemer et al. 2011a) or C. albicans fitness test (Xu et al. 2007; Jiang et al. 2008; Roemer et al. 2011b) offer whole cell target-specific assays for essentially all possible drug targets in yeast and has proven enormously successful in the discovery and mechanism of action (MOA) determination of novel antifungal agents (Roemer et al. 2011a, 2012). As routinely performed in antibacterial discovery (Mann et al. 2013; Roemer and Boone 2013; Wang et al. 2013), next-generation sequencing also offers greater speed and resolution in determining the MOA of potential antifungal leads. Whole genomes of drug-resistant mutants derived from haploid S. cerevisiae, C. glabrata, C. neoformans, or newly derived haploid C. albicans strains (Hickman et al. 2013), and even A. fumigatus are now (or soon to be) routinely sequenced to map causal mutations, thereby definitively identifying the drug target by genetic means. Genetic strategies based on systems-level synthetic lethality networks (Costanzo et al. 2010; Roemer and Boone 2013) also offer important opportunities to identify antifungal adjuvants targeting nonessential proteins that may be paired with existing antifungals to enhance their spectrum or restore their therapeutic effects against drug-resistant strains. Short of completely “new and improved” chemical libraries to screen—an important but challenging request—future antifungal discovery success will require these and other innovative approaches to screening existing chemical matter.
ACKNOWLEDGMENTS
We thank Melanie Wellington (University of Rochester) for assistance with the figures. This work is supported in part by the following National Institutes of Health grants to D.J.K.: 1R01AI091422-03 and 1R01AI097142-02.
Footnotes
Editors: Arturo Casadevall, Aaron P. Mitchell, Judith Berman, Kyung J. Kwon-Chung, John R. Perfect, and Joseph Heitman
Additional Perspectives on Human Fungal Pathogens available at www.perspectivesinmedicine.org
REFERENCES
- Bicanic T, Muzoora C, Brouwer AE, Meintjes G, Longley N, Taseera K, Rebe K, Loyse A, Jarvis J, Bekker LG, et al. 2009. Independent association of clearance of infection and clinical outcome of HIV-associated cryptococcal meningitis: Analysis of combined cohort of 262 patients. Clin Infect Dis 49: 702–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bills GF, Platas G, Overy DP, Collado J, Fillola A, Jiménez MR, Martín J, del Val AG, Vicente F, Tormo JR, et al. 2009. Discovery of the parnafungins, antifungal metabolites that inhibit mRNA polyadenylation, from the Fusarium larvarum complex and other Hypocrealean fungi. Mycologia 101: 449–472 [DOI] [PubMed] [Google Scholar]
- Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J 2009. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48: 1–12 [DOI] [PubMed] [Google Scholar]
- Bowman JC, Hicks PS, Kurtz MB, Rosen J, Schmatz DM, Liberator PA, Douglas CM 2002. The antifungal echinocandin caspofungin acetate kills growing cells of Aspergillus fumigatus in vitro. Antimicrob Agents Chemother 46: 3001–3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratton EW, El Husseini N, Chastain CA, Lee MS, Poole C, Stürmer T, Weber DJ, Juliano JJ, Perfect JR 2013. Approaches to antifungal therapies and their effectiveness in patients with cryptococcosis. Antimicrob Agents Chemother 57: 2485–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TG 2012a. Hidden killers: Human fungal infections. Sci Transl Med 4: 165rv13. [DOI] [PubMed] [Google Scholar]
- Brown GD, Denning DW, Levitz SM 2012b. Tackling human fungal infections. Science 11: 647. [DOI] [PubMed] [Google Scholar]
- Butts A, Krysan DJ 2012. Antifungal drug discovery: Something old and something new. PLoS Pathog 8: e1002870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. 2010. The genetic landscape of a cell. Science 327: 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowen LE, Singh SD, Köhler JR, Collins C, Zaas AK, Schell WA, Aziz H, Mylonakis E, Perfect JR, Whitesell L, et al. 2009. Harnessing Hsp90 function as a powerful, broadly effective therapeutic strategy for fungal infectious disease. Proc Natl Acad Sci 106: 2818–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JN, Chau TT, Wolbers M, Mai PP, Dung NT, Mai NH, Phu NH, Nghia HD, Phong ND, Thai CQ, et al. 2013. Combination therapy for cryptococcal meningitis. N Engl J Med 368: 1291–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDone L, Scrimale T, Baxter BK, Krysan DJ 2010. A high-throughput assay of yeast cell lysis for drug discovery and genetic analysis. Nat Protoc 5: 1107–1114 [DOI] [PubMed] [Google Scholar]
- Ejim L, Faha MA, Falconer SB, Wildenhain J, Coombes BK, Tyers M, Brown ED, Wright G 2011. Combinations of antibiotic and nonantibiotic drugs enhance antimicrobial efficacy. Nat Chem Biol 7: 348–350 [DOI] [PubMed] [Google Scholar]
- Giaever G, Shoemaker DD, Jones TW, Liang H, Winzeler EA, Astromoff A, Davis RW 1999. Genomic profiling of drug sensitivities via induced haploinsufficiency. Nat Genet 21: 278–283 [DOI] [PubMed] [Google Scholar]
- Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B, et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391 [DOI] [PubMed] [Google Scholar]
- Giaever G, Flaherty P, Kumm J, Proctor M, Nislow C, Jaramillo DF, Chu AM, Jordan MI, Arkin AP, Davis RW 2004. Chemogenomic profiling: Identifying the functional interactions of small molecules in yeast. Proc Natl Acad Sci 101: 793–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman DL, Khine H, Abadi J, Lindenberg DJ, Pirofski L-S, Niang R, Casadevall A 2001. Serologic evidence for Cryptococcus neoformans infection in early childhood. Pediatrics 107: e66. [DOI] [PubMed] [Google Scholar]
- Gray KC, Palacios DS, Dailey I, Endo ME, Uno BE, Wilcock BC, Burke MC 2012. Amphotericin primarily kills yeast by simply binding ergoesterol. Proc Natl Acad Sci 109: 2234–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill RJ 2013. Amphotericin B formulations: A comparative review of efficacy and toxicity. Drugs 73: 919–934 [DOI] [PubMed] [Google Scholar]
- Hata K, Horii T, Miyazaki M, Watanabe NA, Okubo M, Sonoda J, Nakamoto K, Tanaka K, Shirotori S, Murai N, et al. 2011. Efficacy of oral E1210, a new broad-spectrum antifungal with a novel mechanism of action, in murine models of candidiasis, aspergillosis, and fusariosis. Antimicrob Agents Chemother 55: 4543–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasley BH, Pacofsky GJ, Mamai A, Liu H, Nelson K, Coti G, Peel MR, Balkovec JM, Greenlee ML, Liberator P, et al. 2012. Synthesis and biological evaluation of antifungal derivatives of enfumafungin as orally bioavailable inhibitors of β-1,3-glucan synthase. Bioorg Med Chem Lett 22: 6811–6816 [DOI] [PubMed] [Google Scholar]
- Herbrecht R, Denning DW, Patterson TF, Bennett JE, Greene RE, Oestmann JW, Kern WV, Marr KA, Ribaud P, Lortholary O, et al. 2002. Voriconazole versus amphotericin B for primary therapy of invasive aspergillosis. N Engl J Med 347: 408–415 [DOI] [PubMed] [Google Scholar]
- Hickman MA, Zeng G, Forche A, Hirakawa MP, Abbey D, Harrison BD, Wang YM, Su CH, Bennett RJ, Wang Y, et al. 2013. The “obligate diploid” Candida albicans forms mating-competent haploids. Nature 494: 55–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B, Xu D, Allocco J, Parish C, Davison J, Veillette K, Sillaots S, Hu W, Rodriguez-Suarez R, Trosok S, et al. 2008. PAP inhibitor with in vivo efficacy identified by Candida albicans genetic profiling of natural products. Chem Biol 15: 363–374 [DOI] [PubMed] [Google Scholar]
- Kaneko Y, Fukazawa H, Ohno H, Miyazaki Y 2013. Combinatory effect of fluconazole and FDA-approved drugs against Candida albicans. J Infect Chemother 19: 1141–1145 [DOI] [PubMed] [Google Scholar]
- Kitamura A, Someya K, Hata M, Nakajima R, Takemura M 2009a. Discovery of a small-molecule inhibitor of β-1,6-glucan synthesis. Antimicrob Agents Chemother 53: 670–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura A, Higuchi S, Hata M, Kawakami K, Yoshida K, Namba K, Nakajima R 2009b. Effect of β-1,6-glucan synthesis inhibitors on the invasion process of Candida albicans: Potential mechanism of their in vivo efficacy. Antimicrob Agents Chemother 53: 3963–3971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFleur MD, Lucumi E, Napper AD, Diamond SL, Lewis K 2011. Novel high-throughput screen against Candida albicans identifies antifungal potentiators and agents effective against biofilms. J Antimicrob Ther 66: 820–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanternier F, Sun H-Y, Ribaud P, Sing N, Kontoyiannis, Lortholary O 2012. Mucormycosis in organ and stem cell transplant patients. Clin Infect Dis 54: 1629–1636 [DOI] [PubMed] [Google Scholar]
- Lass-Flörl C 2011. Triazole antifungal agents in invasive fungal infections: A comparative review. Drugs 71: 2405–2419 [DOI] [PubMed] [Google Scholar]
- Lesage G, Sdicu AM, Ménard P, Shapiro J, Hussein S, Bussey H 2004. Analysis of β-1,3-glucan assembly in Saccharomyces cerevisiae using a synthetic interaction network and altered sensitivity to caspofungin. Genetics 167: 35–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RE 2009. Overview of the changing epidemiology of candidemia. Curr Med Res Opin 25: 1732–1740 [DOI] [PubMed] [Google Scholar]
- Lewis K 2013. Platforms for antibiotic discovery. Nat Rev Microbiol 12: 371–387 [DOI] [PubMed] [Google Scholar]
- Lionakis MS 2012. Genetic susceptibility to fungal infections in humans. Curr Fungal Infect Rep 6: 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G, Gebremariam T, Lee H, French SW, Wiederhold NP, Patterson TF, Filler SG, Ibrahim AS 2013. Efficacy of liposomal amphotericin B and posaconazole in intratracheal models of murine mucormycosis. Antimicrob Agents Chemother 57: 3340–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann PA, Müller A, Xiao L, Pereira PM, Yang C, Ho Lee S, Wang H, Trzeciak J, Schneeweis J, Dos Santos MM, et al. 2013. Murgocil is a highly bioactive Staphylococcal-specific inhibitor of the peptidoglycan glycosyltransferase enzyme MurG. ACS Chem Biol 8: 2442–2451 [DOI] [PubMed] [Google Scholar]
- McLellan CA, Whitesell L, King OD, Lancaster AK, Mazitschek R, Lindquist S 2012. Inhibiting GPI anchor biosynthesis in fungi stresses the endoplasmic reticulum and enhances immunogenicity. ACS Chem Biol 7: 1520–1528 [DOI] [PubMed] [Google Scholar]
- Meletiadis J, Antachopoulos C, Stergiopoulou T, Pournaras S, Roilides E, Walsh TJ 2007. Differential fungicidal activities of amphotericin B and voriconazole against Aspergillus species determined by microbroth methodology. Antimicrob Agents Chemother 51: 3329–3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro MC, de la Cruz M, Cantizani J, Moreno C, Tormo JR, Mellado E, De Lucas JR, Asensio F, Valiante V, Brakhage AA, et al. 2012. A new approach to drug discovery: High-throughput screening of natural extracts against Aspergillus fumigatus using resazurin. J Biomol Screen 17: 542–549 [DOI] [PubMed] [Google Scholar]
- Muhammed M, Coleman JJ, Carneiro HA, Mylonakis E 2011. The challenge of managing fusariosis. Virulence 2: 91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee PK, Sheehan DJ, Hitchcock CA, Ghannoum MA 2005. Combination treatment of invasive fungal infections. Clin Microbiol Rev 8: 163–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee PK, Sheehan D, Puzniak L, Schlamm H, Ghannoum MA 2011. Echinocandins: Are they all the same? J Chemother 23: 319–325 [DOI] [PubMed] [Google Scholar]
- Onishi J, Meinz M, Thompson J, Curotto J, Dreikorn S, Rosenbach M, Douglas C, Abruzzo G, Flattery A, Kong L, et al. 2000. Discovery of novel antifungal (1,3)-β-d-glucan synthase inhibitors. Antimicrob Agents Chemother 44: 368–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrosky-Zeichner L, Casadevall A, Galgiani JN, Odds FC, Rex JH 2010. An insight into the antifungal pipeline: Selected new molecules and beyond. Nat Rev Drug Discov 9: 719–727 [DOI] [PubMed] [Google Scholar]
- Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM 2009. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS 23: 525–530 [DOI] [PubMed] [Google Scholar]
- Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL 2007. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat Rev Drug Discov 6: 29–40 [DOI] [PubMed] [Google Scholar]
- Pfaller MA, Barry ML 1994. Evaluation of a novel colorimetric broth microdilution method for antifungal susceptibility testing of yeast isolates. J Clin Microbiol 32: 1992–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller M, Neofytos D, Diekema D, Azie N, Meier-Krisesche H-U, Quan S-P, Horn D 2012. Epidemiology and outcomes of candidemia in 3648 patients: Data for the Prospective Antifungal Therapy (PATH Alliance®) registry, 2004–2008. Diagn Microbiol Infect Dis 74: 323–331 [DOI] [PubMed] [Google Scholar]
- Pfaller MA, Messer SA, Motyl MR, Jones RN, Castanheira M 2013a. Activity of MK-3118, a new oral glucan synthase inhibitor, tested against Candida spp. by two international methods (CLSI and EUCAST). J Antimicrob Chemother 68: 858–863 [DOI] [PubMed] [Google Scholar]
- Pfaller MA, Messer SA, Motyl MR, Jones RN, Castanheira M 2013b. In vitro activity of a new oral glucan synthase inhibitor (MK-3118) tested against Aspergillus spp. by CLSI and EUCAST broth microdilution methods. Antimicrob Agents Chemother 57: 1065–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce CG, Uppuluri P, Tristan AR, Wormley FL Jr, Mowat E, Ramage G, Lopez-Ribot JL 2008. A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat Protoc 3: 1494–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabjohns JLA, Park Y-D, Dehdashti Sun W, Henderson C, Zelazny A, Metallo SJ, Zheng W, Williamson PR 2013. A high-throughput screening assay for fungicidal compounds against Cryptococcus neoformans. J Biomol Screen 19: 270–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex JH, Walsh TJ, Nettleman M, Anaissie EJ, Bennett JE, Bow EJ, Carillo-Munoz AJ, Chavanet P, Cloud GA, Denning DW, et al. 2001. Need for alternative trial designs and evaluation strategies for therapeutic studies of invasive mycoses. Clin Infect Dis 33: 95–106 [DOI] [PubMed] [Google Scholar]
- Richardson MR 2005. Changing patterns and trends in systemic fungal infections. J Antimicrob Chemother 56: S5–S11 [DOI] [PubMed] [Google Scholar]
- Rock FL, Mao W, Yaremchuk A, Tukalo M, Crépin T, Zhou H, Zhang YK, Hernandez V, Akama T, Baker SJ, et al. 2007. An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science 316: 1759–1761 [DOI] [PubMed] [Google Scholar]
- Roemer T, Boone C 2013. Systems-level antimicrobial drug and drug synergy discovery. Nat Chem Biol 9: 222–231 [DOI] [PubMed] [Google Scholar]
- Roemer T, Xu D, Singh SB, Paish CA, Harris G, Wang G, Wang H, Davies JE, Bills GF 2011a. Confronting the challenges of natural product-based antifungal discovery. Chem Biol 18: 148–164 [DOI] [PubMed] [Google Scholar]
- Roemer T, Davies J, Giaever G, Nislow C 2011b. Bugs, drugs and chemical genomics. Nat Chem Biol 8: 46–56 [DOI] [PubMed] [Google Scholar]
- Seiradake E, Mao W, Hernandez V, Baker SJ, Plattner JJ, Alley MR, Cusack S 2009. Crystal structures of the human and fungal cytosolic Leucyl-tRNA synthetase editing domains: A structural basis for the rational design of antifungal benzoxaboroles. J Mol Biol 10: 196–207 [DOI] [PubMed] [Google Scholar]
- Shapiro RS, Robbins N, Cowen LE 2011. Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol Mol Biol Rev 75: 213–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker DD, Lashkari DA, Morris D, Mittmann M, Davis RW 1996. Quantitative phenotypic analysis of yeast deletion mutants using a highly parallel molecular bar-coding strategy. Nat Genet 14: 450–456 [DOI] [PubMed] [Google Scholar]
- Singh SD, Robbins N, Zaas AK, Schell WA, Perfect JR, Cowen LE 2009. Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via calcineurin. PLoS Path 5: e1000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DJ, Dedicoat MJ, Lalloo DG 2009. Treatment of cryptococcal meningitis in resource-limited settings. Curr Opin Infect Dis 22: 455–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer M, Griffiths E, Blakely KM, Wildenhain J, Ejim L, Rossi L, De Pascale G, Curak J, Brown E, Tyers M, et al. 2011. Cross-species discovery of syncretic drug combinations that potentiate the antifungal fluconazole. Mol Syst Biol 7: 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan A, Leung KP, Lopez-Ribot JL, Ramasubramanian AK 2013. High-throughput nano-biofilm microarray for antifungal drug discovery. mBio 4: e00331-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbach WJ, Marr KA, Anaissie EJ, Azie N, Quan S-P, Meier-Kriesche H-U, Apewokin S, Horn DL 2012. Clinical epidemiology of 969 patients with invasive aspergillosis from the PATH alliance registry. J Infect 65: 453–464 [DOI] [PubMed] [Google Scholar]
- Tsukahara K, Hata K, Nakamoto K, Sagane K, Watanabe NA, Kuromitsu J, Kai J, Tsuchiya M, Ohba F, Jigami Y, et al. 2003. Medicinal genetics approach towards identifying the molecular target of a novel inhibitor of fungal cell wall assembly. Mol Microbiol 48: 1029–1042 [DOI] [PubMed] [Google Scholar]
- Walker SS, Xu Y, Triantafyllou I, Waldman MF, Mendrick C, Brown N, Mann P, Chau A, Patel R, Bauman N, et al. 2011. Discovery of a novel class of orally active antifungal β-1,3-d-glucan synthase inhibitors. Antimicrob Agents Chemother 55: 5099–5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CT, Wencewicz TA 2013. Prospects for new antibiotics: A molecule-centered perspective. J Antibiot (Tokyo) 67: 7–22 [DOI] [PubMed] [Google Scholar]
- Wang H, Gill CJ, Lee SH, Mann P, Zuck P, Meredith TC, Murgolo N, She X, Kales S, Liang L, et al. 2013. Discovery of wall teichoic acid inhibitors as potential anti-MRSA β-lactam combination agents. Chem Biol 20: 272–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Jiang B, Ketela T, Lemieux S, Veillette K, Martel N, Davison J, Sillaots S, Trosok S, Bachewich C, et al. 2007. Genome-wide fitness test and mechanism-of-action studies of inhibitory compounds in Candida albicans. PLoS Path 3: e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Ondeyka J, Harris GH, Zink D, Kahn JN, Wang H, Bills G, Platas G, Wang W, Szewczak AA, et al. 2011. Isolation, structure, and biological activities of Fellutamides C and D from an undescribed Metulocladosporiella (Chaetothyriales) using the genome-wide Candida albicans fitness test. J Nat Prod 26: 1721–1730 [DOI] [PubMed] [Google Scholar]
- Youngsaye W, Dockendorff C, Vincent B, Hartland CL, Bittker JA, Dandapani S, Palmer M, Whitesell L, Lindquist S, Schrieber SL, et al. 2012. Overcoming fluconazole resistance in Candida albicans clinical isolates with tetracyclic indoles. Bioorg Med Chem Lett 22: 3362–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai B, Wu C, Wang L, Sachs MS, Lin X 2012. The antidepressant sertraline provides a promising therapeutic option for neurotropic cryptococcal infections. Antimicrob Agents Chemother 56: 3758–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]