Abstract
MYC’s tumorigenic potential involves increased ribosome biogenesis and translational capacity, which supply the cell with protein required for enhanced cell growth and subsequent cell division. In addition to activation of protein-encoding genes transcribed by RNA polymerase II, MYC must stimulate transcription by RNA polymerase I and RNA polymerase III to meet this synthetic demand. In the past decade our knowledge of the mechanisms and importance of MYC regulation of RNA polymerases I and III has flourished. Here we discuss MYC’s influence on transcription by these “odd” RNA polymerases and the physiological impact of this regulation is evaluated with relevance to cancer development and treatment.
MYC stimulates transcription by RNA polymerases I and III, enhancing ribosome biogenesis and cell growth. Cancer therapies that target these transcriptional activities may impede tumor development.
ENHANCED RIBOSOME BIOGENESIS AND PROTEIN SYNTHESIS REQUIRES RNA POLYMERASE I AND III
In eukaryotes there are three major nuclear RNA polymerases. They are large multisubunit complexes that each use specific cofactors to drive transcription in vivo. RNA polymerase II (Pol II) transcribes thousands of protein-encoding genes into mRNA. RNA polymerase I (Pol I) produces only one transcript, the 47S immature ribosomal RNA (pre-rRNA), from the rDNA gene repeats. RNA polymerase III (Pol III) synthesizes 5S rRNA and other short noncoding RNAs, such as transfer RNAs (tRNAs). Pol I and Pol III activities account for ∼ 80% of nuclear transcription and their activity is altered dynamically in response to developmental and environmental signals. Consistent with their role in cellular growth, Pol I and Pol III products and cofactors are overexpressed in many cancer cells (White 2008; Hannan et al. 2013). In recent years, much has been elucidated of the control of Pol I and Pol III transcription by oncogenes and tumor suppressors (White 2008; Hannan et al. 2013). In particular, the manufacture of Pol I and Pol III products is highly regulated by MYC. In multiple cell types, MYC stimulates ribosome biogenesis, RNA processing and cell growth (accumulation of mass) (Ji et al. 2011), processes that rely heavily on Pol I and Pol III function. Protein synthesis determines the rate at which a cell accumulates mass and under most conditions proliferation is coupled to cell growth. The rate of ribosome biogenesis is precisely controlled to allow the requisite protein synthesis. This requires the coordinated activity of all three major nuclear RNA polymerases to synthesize ribosomal components. Pol I synthesizes 47S pre-rRNA, which undergoes processing into mature 18S, 5.8S, and 28S rRNA, and together with 5S rRNA (transcribed by Pol III) constitutes the nucleic acid backbone and catalytic activity of the ribosome. Pol III also contributes the tRNA that is essential for translation. Pol II produces mRNAs encoding a large number of ribosomal proteins and accessory factors, such as those required for translation elongation. In cancer, the metabolic appetite of a cell is increased as more macromolecules are required to sustain enhanced growth and subsequent cell division. As Pol I and Pol III products are core ribosomal components, it is perhaps not surprising that these enzymes, their cofactors and products are found to be up-regulated in cancer.
MYC EXPRESSION PROMOTES RIBOSOME BIOGENESIS AND INCREASED rRNA LEVELS
A principal function of MYC is to influence ribosome biogenesis. Genes with ribosome, nucleolar, or RNA processing functions comprise a core signature of MYC-responsive genes in mammals (Ji et al. 2011). In agreement with studies in mammalian systems, many of the genes up-regulated by Drosophila Myc (dMyc) encode products involved in ribosome biogenesis (Grewal et al. 2005). There is much evidence that augmented ribosome activity and translational capacity are tumor-promoting and that enhanced capacity for protein synthesis may be critical for MYC’s oncogenic effect (Ruggero 2009; van Riggelen et al. 2010).
In resting MYC-null rat fibroblasts, rRNA levels are reduced by ∼20% relative to wild-type. On cell cycle stimulation, rRNA accumulation is delayed twofold, with a similar reduction in proliferative rate (Mateyak et al. 1997). Restoration of MYC leads to rapid rRNA gene transcription (Shiue et al. 2009). Therefore, MYC expression correlates with levels of rRNA synthesis. It seems plausible that any growth stimulus would up-regulate ribosome biogenesis, yet there is specificity in the MYC effect, as overexpression of either insulin receptor (InR) or the PI3K subunit dp110 (which increase cell growth in Drosophila) did not elevate rRNA synthesis, unlike dMyc expression (Grewal et al. 2005).
EFFECTS OF MYC ON TRANSCRIPTION BY POL I
Pol I is a 14 subunit enzyme that enlists a small number of cofactors to efficiently transcribe the 47S pre-rRNA, illustrated in Figure 1. In brief, UBF (upstream binding factor) binds to multiple sites throughout rDNA and allows preinitiation complex assembly and remodeling of rDNA chromatin. Selectivity factor 1 (SL1/TIF-1B) is composed of the TATA-binding protein (TBP) and TBP-associated factors (TAFs). SL1 interacts with UBF and binds rDNA promoter sequences. Rrn3 (also known as TIF-1A) interacts with the UBF/SL1 complex and recruits Pol I, allowing transcription to commence (Russell and Zomerdijk 2005).
Figure 1.

Assembly of Pol I apparatus on rDNA. The SL1 complex, UBF, and Rrn3 proteins are basal Pol I transcription factors. Arrow begins at transcription start site.
MYC can regulate Pol I activity through influencing the abundance of UBF, a factor that is rate-limiting for Pol I transcription under some circumstances (Poortinga et al. 2004, 2011). When activated in fibroblasts using hydroxytamoxifen, a MYC-ER fusion protein (in which MYC is regulated by the ligand-binding domain of the estrogen receptor [ER]) bound to E-box sequences in the UBF promoter and raised UBF expression (Poortinga et al. 2004). UBF binding to rDNA was enriched in proliferating cells and MYC induction of rRNA required UBF (Poortinga et al. 2004). MYC regulation of Pol I transcription is not secondary to cell cycle changes, as these effects also occurred in noncycling cells (Poortinga et al. 2004). In Drosophila, dMyc increases the expression of Pol I subunits and cofactors (Grewal et al. 2005). Induction of dMyc in early third-instar larvae, mitotic cells of imaginal wing disc or salivary gland cells all resulted in increased rDNA transcription and ribosome biogenesis (Grewal et al. 2005). Conversely, decreased dMyc expression in first-instar larvae downregulated rDNA transcription and decreased nucleolar size, evidencing decreased ribosome production (Grewal et al. 2005). These effects appear to reflect dMyc/dMax binding, as E-box sequences are found in the promoters of the dMyc-responsive Pol I subunits and cofactors (Grewal et al. 2005) and dMyc requires dMax to increase rRNA expression (Steiger et al. 2008).
In addition to regulating UBF expression, MYC can also influence the expression of other Pol-I-specific subunits and transcription factors. In line with this, ENCODE ChIP-seq data annotates the promoters of all subunits of SL1, as well as Rrn3 and UBF, as sites of MYC binding (Rosenbloom et al. 2013). Induction of MYC in mouse promyelocytes increased the expression of 79% of genes encoding Pol-I-specific subunits or cofactors, including UBF and Rrn3 (Poortinga et al. 2011). Direct binding of MYC to the UBF and Rrn3 promoters was confirmed by ChIP. MYC also raised expression of 43% of the genes encoding Pol-III-specific subunits and transcription factors. In contrast, only 14% of genes encoding Pol-II-specific subunits and factors were responsive to MYC under these conditions (Poortinga et al. 2011). The importance of up-regulation of Pol I expression for MYC’s oncogenic program should not be underestimated. A Pol I subunit is among 51 protein-encoding genes that comprise a MYC core signature across numerous cancer cell lines and thousands of microarray samples (Ji et al. 2011).
Although MYC can clearly regulate the Pol-II-dependent expression of Pol I apparatus (summarized in Table 1), this is not the only mechanism it uses for regulation of rRNA synthesis. MYC-induced increase in rRNA levels can occur even in the presence of concentrations of α-amanitin that specifically inhibit synthesis of mRNAs, including those encoding the subunits of Pol I and its cofactors (Arabi et al. 2005; Grandori et al. 2005). This observation, along with the rapidity of induction of rDNA expression, suggests a direct effect of MYC on rDNA transcription (Arabi et al. 2005; Grandori et al. 2005). Indeed, in mammalian cells the 5.8S, 18S, and 28S rRNAs are encoded by genes bound and activated by MYC (Arabi et al. 2005; Grandori et al. 2005; Lin et al. 2012). These rRNA genes occur in repeats that cluster in the nucleolus where they are transcribed. On either serum stimulation of resting cells or in cells treated with proteasome inhibitors, MYC colocalizes in the nucleolus with rDNA genes (Arabi et al. 2005; Grandori et al. 2005). Human rDNA genes are enriched for E-box MYC-binding motifs and ChIP assay showed that MYC binds to rDNA genes in regions containing noncanonical E boxes (Arabi et al. 2005; Grandori et al. 2005). Furthermore, MYC associates with components of the SL1 complex and increases SL1 and Pol I loading (Grandori et al. 2005). The transactivation domain of MYC is required for increased rDNA transcription (Grandori et al. 2005), whereas carboxy-terminal regions including the basic region at residues 355–368 are important for nucleolar localization (Arabi et al. 2005).
Table 1.
MYC-regulated Pol II transcribed genes that encode Pol I/Pol III subunits or cofactors
| Subunits or cofactors | References |
|---|---|
| Pol I | |
| UBF | Poortinga et al. 2004 |
| TIF-1A, ZPR1, RPI135, RPI1, CG18600 | Grewal et al. 2005 |
| POLR1A, POLR1B, POLR1C,a POLR1D, RPA43, PAF49, PAF53, POLR2E,b POLR2F,b POLR2H,b POLR2Lb RRN3, UBF, SL1 (TAF1C, TAF1D, TBPb) | Poortinga et al. 2011 |
| POLR1B | Ji et al. 2011 |
| Pol III | |
| BN51/RPC4 | Greasley et al. 2000 |
| TFIIIC (GTF3C4, GTF3C5) | Li et al. 2003 |
| CG12267, RPIII128, CG3756 | Grewal et al. 2005 |
| TFIIIB (BRF1) | Sansom et al. 2007 |
| POLR3E, POLR3F, POLR3G, POLR3H, POLR3K, POLR1C,a POLR1D,a POLR2E,b POLR2F,b POLR2H,b POLR2Lb TFIIIC (GTF3C3, GTF3C4), TFIIIB (TBPb) | Poortinga et al. 2011 |
aSubunits common to Pol I and Pol III.
bSubunits common to Pol I, II, and III.
MYC can influence rRNA synthesis by recruiting histone acetyltransferases to alter the chromatin structure of rDNA. MYC induction increased levels of acetylated histones H3 and H4 at rDNA (Grandori et al. 2005). Proliferative stimulation of lymphocytes rapidly induced MYC levels and binding to rDNA, along with increased presence of Pol I, acetylated histone H4 and the transformation/transcription domain associated protein (TRRAP), a coactivator that associates with histone acetyltransferases (Arabi et al. 2005). Although induction of ribosome biogenesis (RiBi) genes is thought to be an ancient function of MYC (Brown et al. 2008), direct activation of Pol I transcription by MYC appears to have been acquired relatively late during evolution. Human and rat rDNA both contain multiple E-box sequences, but their number and spacing are not conserved (Arabi et al. 2005; Grandori et al. 2005; Shiue et al. 2009). Furthermore, Drosophila rDNA does not contain canonical E boxes nor could dMyc binding to rDNA be detected (Grewal et al. 2005). MYC-dependent alterations in higher order chromatin structure may also contribute to activation of rDNA gene expression. On serum stimulation of quiescent rat fibroblasts, MYC is necessary for full Pol I activity and MYC induction not only facilitated binding of Pol I and TBP to rDNA promoter and upstream enhancer sequences, but also resulted in their recruitment to non-transcribed downstream sequences (Shiue et al. 2009). In the presence of MYC, serum stimulation rapidly led to the formation of gene loop structures of rDNA that may enhance transcriptional reinitiation by linking promoter and terminator sequences (Shiue et al. 2009).
About half of the ∼200 copies of rDNA repeat are transcribed by Pol I in mammalian cells, the remainder being associated with chromatin states that are not conducive to transcription (McStay and Grummt 2008). A model has been proposed in which MYC induction of Pol II transcription occurs by amplifying the output of the existing gene program, rather than switching on additional genes (Lin et al. 2012; Nie et al. 2012). In keeping with this scenario, activation of a MYC-ER construct with hydroxytamoxifen in terminally differentiated murine granulocytes lead to a twofold increase in Pol I transcription without any appreciable change in the fraction of active rDNA repeats (Poortinga et al. 2011). However, when MYC-ER was activated before differentiation was complete, the ratio of active to inactive repeats was shifted by ∼10% (Poortinga et al. 2011). The investigators suggested that this may reflect induction of UBF, which antagonizes the epigenetic silencing of rDNA.
MYC also influences Pol I product output at the level of rRNA processing that occurs in the nucleolus. A screen using a human B-cell line and rat MYC null fibroblasts, both with conditional activation of MYC, identified new target genes, the largest class of which had nucleolar function (Schlosser et al. 2003). MYC expression induced both the synthesis of 47S rRNA precursor and its subsequent processing to mature 5.8S, 18S, and 28S rRNA ribosomal components (Schlosser et al. 2003). This likely reflects the fact that many direct MYC targets encode proteins involved in rRNA processing and maturation in the nucleolus. Examples include fibrillarin, dyskeratin, nucleolin, and nucleophosmin (Schlosser et al. 2003). In addition to endoribonuclease activity that contributes to pre-rRNA processing, nucleophosmin has also been shown to promote SL1 recruitment to rRNA gene promoters, thereby stimulating Pol I transcription (Bergstralh et al. 2007). This effect may be mediated, at least in part, by the ability of nucleophosmin to draw MYC into the nucleolus (Li and Hann 2013). Accordingly, overexpression of nucleophosmin significantly enhances the increase in pre-rRNA synthesis invoked by MYC-ER activation, whereas the response is compromised if nucleophosmin is depleted by RNAi (Li and Hann 2013). Thus, the ability of MYC to directly stimulate transcription of the nucleophosmin gene may serve to amplify its direct effects on rRNA expression.
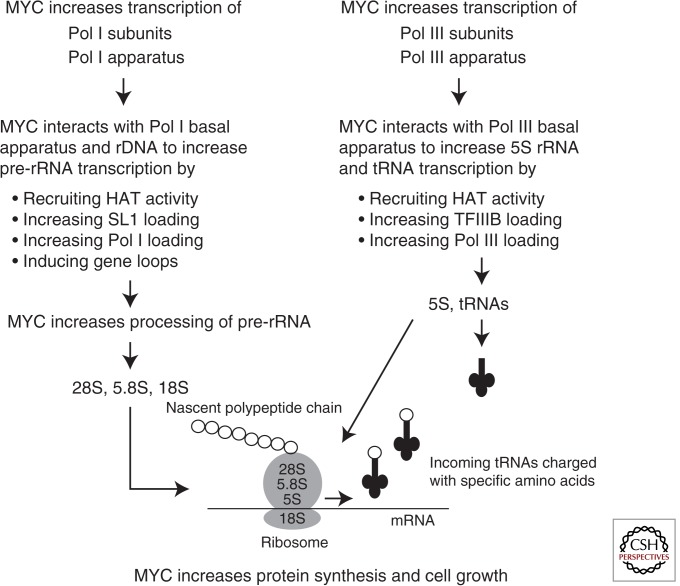
As summarized in Figure 3, MYC enhances rRNA production through multiple mechanisms and at multiple levels. MYC up-regulates the expression of Pol I and Pol I cofactors, directly binds to rDNA in mammalian cells, and improves chromatin accessibility through histone acetylation. MYC interacts with Pol-I-specific transcription factors to further enhance Pol I recruitment to its targets and also alters the higher order chromatin structure of rDNA, creating gene loops to maximize Pol I output. Finally, MYC augments the processing of the Pol I product into its mature form.
Figure 3.
MYC regulates Pol I and Pol III at multiple levels to stimulate protein synthesis and cell growth.
MYC INTERACTIONS WITH POL III
Pol III is the largest RNA polymerase, comprising 17 subunits, and also uses specific transcription factors to drive expression of its target genes, illustrated in Figure 2. This requires the ordered recruitment of basal Pol-III-specific transcription factors TFIIIC and TFIIIB before polymerase loading. TFIIIC is composed of six subunits and binds promoter DNA located within the transcribed region of Pol III target genes. TFIIIC does not have high affinity for the 5S rRNA promoter and at this gene TFIIIA must be bound first to recruit TFIIIC. TFIIIB is composed of 3 subunits; TBP, Bdp1, and Brf1 or Brf2. Once TFIIIC is bound, TFIIIB is able to recognize its binding site upstream of the transcriptional start site. TFIIIB recruits Pol III and transcription can start (Schramm and Hernandez 2002).
Figure 2.
Assembly of Pol III apparatus on tRNA and 5S rRNA genes. TFIIIB and TFIIIC complexes are basal Pol III transcription factors. TFIIIA is required for TFIIIC recruitment to the 5S rRNA gene. Arrow begins at transcription start site. Shaded areas in tRNA and 5S rRNA genes indicate internal promoter regions.
The level of Pol III products such as B2 RNA and tRNAs are decreased in fibroblasts deficient for MYC, but rapidly up-regulated on MYC-ER induction (Gomez-Roman et al. 2003; Kenneth et al. 2007). Similarly, increased MYC results in enhanced synthesis of tRNA and/or 5S rRNAs in B cells, cardiomyocytes and hepatocytes (Goodfellow et al. 2006; Lin et al. 2012; Shukla and Kumar 2012). Induction of tRNA synthesis was also seen in vivo when MYC-ER was activated in transgenic mice (Goodfellow et al. 2006). Conversely, depletion of MYC by RNAi led to decreased expression of Pol III products (Felton-Edkins et al. 2003; Owen et al. 2010). In insect cells, dMyc overexpression stimulated Pol III transcription, whereas dMyc knockdown had the opposite effect (Steiger et al. 2008). Like Pol I and Pol II transcribed genes, there is much evidence to show that Pol III templates are direct MYC targets. MYC has been detected at tRNA, 5S rRNA, and other Pol III-transcribed genes in numerous cell types (Felton-Edkins et al. 2003; Gomez-Roman et al. 2003; Raha et al. 2010; Lin et al. 2012). Endogenous MYC has been detected at 74% of genes occupied by Pol III in the human K562 erythroleukemia cell line (Raha et al. 2010). Therefore, in multiple cell types the level of MYC parallels Pol III activity and this corresponds with MYC presence at Pol III-transcribed genes.
The transactivation domain of MYC is required to regulate Pol III transcription, as induction of MYC-S (lacking the amino-terminus of MYC) did not increase B2 RNA levels and deletion of MYC residues 106-143 prevented tRNA induction (Gomez-Roman et al. 2003). Contained within this region is the motif MBII, and TRRAP coactivator binding to MBII provides a mechanistic explanation for elevated Pol III transcription (Kenneth et al. 2007). This is somewhat similar to Pol II transcription, in that MYC facilitates recruitment of the coactivator TRRAP, which associates with the histone acetyltransferase GCN5 to increase histone acetylation (Kenneth et al. 2007). However, unlike many Pol II targets, there was increased histone H3 but not H4 acetylation at Pol III-transcribed genes on MYC recruitment (Kenneth et al. 2007). This selectivity could be caused by high basal acetylation of histone H4 at these genes or perhaps because the Tip60 histone acetyltransferase was not detected at Pol III target genes (Kenneth et al. 2007). Under conditions of ribosomal stress 5S and tRNA expression is reduced and released ribosomal protein L11 can be detected at these genes (Dai et al. 2010). L11 binding to MYC prevents TRRAP recruitment to Pol III transcribed genes in a similar feedback mechanism as at Pol I and Pol II target genes (Dai et al. 2010). Therefore, the amino-terminal region of MYC allows alteration of histone acetylation status at Pol III target genes to facilitate Pol III loading. Although MYC does not typically increase Pol II occupancy at its target genes, elevated Pol III binding was clearly seen in fibroblasts in response to MYC (Ernens et al. 2006; Kenneth et al. 2007). With one exception, each mammalian tRNA is encoded by a family of redundant genes, many of which are silent (White 2011). It is not yet clear whether MYC can activate silent tRNA genes, or serves exclusively to amplify expression of the active tRNA genes, as suggested for Pol II transcription (Lin et al. 2012; Nie et al. 2012).
As most Pol III target genes are not close to canonical DNA sequences that are recognized by MYC/MAX heterodimers, how is MYC recruited to these promoters? MYC binds TBP, a component of the basal transcription apparatus for all three multisubunit RNA polymerases (Hateboer et al. 1993; McEwan et al. 1996). Both TBP and Brf1 TFIIIB subunits interact with a GST-fusion protein containing the amino-terminal 262 residues of MYC, but not the carboxy-terminal 92 amino acids (Gomez-Roman et al. 2003). MYC coimmunoprecipitated with Brf1 in nonproliferative rat neonatal cardiomyocytes (Ernens et al. 2006). Brf and dMyc also coimmunoprecipitated in extracts from insect cells and this interaction was maintained even when the carboxy-terminal region of dMyc was absent (Steiger et al. 2008). dMyc not only binds Brf but a genetic interaction has also been shown. Heterozygous loss of Brf in a dMyc mutant background resulted in reduced eye size and more frequent eye defects (Steiger et al. 2008). Therefore, it seems likely that MYC is recruited to Pol III target genes via interaction with components of TFIIIB.
TFIIIC may also attract MYC to Pol III templates. A proteomic screen identified four of six TFIIIC subunits associated with MYC in transformed human embryonic kidney cells and a colorectal cancer cell line (Koch et al. 2007). It is conceivable that MYC is differentially recruited to specific Pol III templates in different cell types via TFIIIB or TFIIIC. Alternatively, the primary interaction of MYC could be with TFIIIB, as this has been mapped using in vitro pull-down assay (Gomez-Roman et al. 2003). Perhaps the detection of TFIIIC interaction with MYC is indicative of a MYC/TFIIIB/TFIIIC complex as direct binding of MYC to TFIIIC has not been reported (Koch et al. 2007). Another possibility is that TFIIIC recruits MYC, and associated histone acetlytransferase activity, which then promotes TFIIIB binding. This is consistent with the observation that MYC induction increases TFIIIB presence at target genes, whereas TFIIIC recruitment is unchanged (Kenneth et al. 2007). Regardless of which of these scenarios proves true, it is clear that interaction with Pol III cofactors at the promoters of Pol III target genes offers one mechanism by which MYC regulates Pol III activity.
Does MYC activate expression of Pol III subunits or cofactors? MYC activation of Pol III transcription certainly does not require up-regulation of Pol III subunits or transcription factors, as inhibition of Pol II transcription with α-amanitin did not prevent rapid induction of pol III-transcribed genes by MYC-ER (Gomez-Roman et al. 2003). Induction of MYC-ER in fibroblasts did not alter expression of a subset of TFIIIB, TFIIIC, and Pol III subunits that were investigated (Gomez-Roman et al. 2003; Kenneth et al. 2007). Although the early time points used in these experiments were relevant to the induction of Pol III target gene expression, they do not exclude a secondary indirect effect through later up-regulation of these proteins. It seems pertinent that five of six TFIIIC subunits, as well as the Pol-III-specific components of TFIIIB, have all been shown to have MYC bound in the vicinity of their promoters in ChIP-seq experiments (Rosenbloom et al. 2013). Furthermore, dMyc overexpression up-regulated three genes implicated in Pol III transcription (Grewal et al. 2005) and Pol III-subunits and cofactors have been identified as MYC-regulated genes in mammalian cells (see Table 1). MYC is capable of robust induction of Pol III target genes (e.g., 12-fold induction of pre-tRNALeu by MYC-ER in primary human fibroblasts) (Gomez-Roman et al. 2003), whereas MYC induction of Pol II target genes is generally more conservative. This is perhaps reflective of MYC stimulating Pol III activity at multiple levels, summarized in Figure 3. MYC can bind to TFIIIB and recruit histone acetyltransferase activity to Pol III templates to enhance Pol III loading. MYC can up-regulate the expression of Pol III subunits and cofactors, which allows potential for further stimulation of activity, although the requirement for this in vivo has still to be shown. In these respects, MYC regulation of Pol I and Pol III is quite similar. This seems logical, as their products must act in concert to facilitate MYC-driven cell growth (Fig. 3). It is not known whether MYC can influence the higher-order chromatin structure at Pol III target genes into gene loops as documented at rDNA. tRNA genes, like rDNA, are dispersed throughout the genome, often occurring in clusters, and it would appear an efficient mechanism to coordinate their expression by grouping in higher-order structures.
INVOLVEMENT OF THE MAX NETWORK IN REGULATING POL I AND POL III
In mammalian cells, MAX colocalizes with MYC in the nucleolus (the site of rRNA synthesis) following proteasome inhibition (Arabi et al. 2005) or serum stimulation of resting cells (Grandori et al. 2005). MAX can be cross-linked to the rDNA repeats (Arabi et al. 2005). Inhibitors that disrupt the MAX/MYC interaction can reduce the rRNA content of activated B cells (Arabi et al. 2005; Nie et al. 2012) and tRNA expression in serum-stimulated cardiomyocytes (Goodfellow et al. 2006). This suggests that MAX is involved in regulating Pol I and Pol III, in which case other components of the MAX network may contribute. Indeed, mice deficient for MAD1 have enlarged granulocytes and nucleoli, as well as increased levels of RNA (Poortinga et al. 2004). MAD1 can decrease Pol III-dependent expression of UBF and the Pol III subunit BN51/RPC4 (Greasley et al. 2000; Poortinga et al. 2004). Increased expression of dMnt in Drosophila decreases rDNA transcription (Grewal et al. 2005). In Drosophila, dMax is required for activation of Pol I transcription, probably through increases in Pol I subunits and transcription factors, as E-box sequences are present in their promoters but not in Drosophila rDNA (Grewal et al. 2005; Steiger et al. 2008). However, a mutant dMyc that is incapable of interacting with dMax or binding to E-box sequences can still activate expression of Pol III-transcribed genes (Steiger et al. 2008). In fact, stimulation of Pol III target genes by dMyc is actually enhanced in the absence of dMax (Steiger et al. 2008). Thus, Pol III regulation by MYC may require MAX in mammals but not in flies. However, the situation in mammalian cells has yet to be fully characterized and it remains unclear to what extent transcription by Pol I and Pol III is controlled by other proteins of the MAX network besides MYC.
UP-REGULATION OF POL I AND POL III TRANSCRIPTION BY MYC DRIVES CELL GROWTH
Both Pol I and Pol III activities are important for MYC-driven cell growth, which can be uncoupled from cell division in Drosophila (Johnston et al. 1999). Expression of a mutant form of Pol I decreased cell size of developing wing disc clones with elevated dMyc (Grewal et al. 2005). Comparison of dMyc mutants, whose only identified differential gene expression effects were on Pol III target genes, suggested that dMyc’s ability to up-regulate Pol III activity resulted in increased survival and growth (Steiger et al. 2008). In mammalian cells, MYC regulation of Pol I and Pol III is also linked to cell growth. As granulocytes differentiate into neutrophils they become smaller in the absence of cell division. When this was modeled in culture by stimulating mouse promyelocytes to differentiate, it was found that MYC levels decreased as did the expression of many Pol I and Pol III subunits/cofactors (Poortinga et al. 2011). A decrease in MYC binding to rDNA and in rRNA expression was also observed (Poortinga et al. 2011). Interestingly, induction of MYC in these differentiated cells was sufficient to increase cell size and resulted in cells that appeared less mature and in which Pol I and Pol III machinery were up-regulated (Poortinga et al. 2011). Therefore, in terminally differentiated granulocytes, MYC controls cell size and Pol I /Pol III activity.
Increased protein synthesis capacity and activity of Pol I and Pol III are found in cardiomyocyte hypertrophy, which has been associated with cardiac pathologies (Goodfellow et al. 2006). Cardiomyocytes require increased Pol I transcription to grow (Brandenburger et al. 2001) and hypertrophic signaling in these terminally differentiated cells induces MYC expression, which was necessary and sufficient for resultant growth without division (Goodfellow et al. 2006). Increased Pol III transcription was observed in cardiomyocytes when MYC was elevated, and incubation with a chemical inhibitor of MYC/MAX binding prevented both increased Pol III target gene expression and increased cell volume on hypertrophic stimulation, suggesting that MYC is required for both (Goodfellow et al. 2006). This is supported by preliminary data showing that hypertrophic enlargement of cardiomyocytes can be blocked with tagetin, a selective inhibitor of Pol III (Goodfellow and White 2007). The level of the TFIIIB subunit Brf1 is rate-limiting for Pol III transcription in these cells and hypertrophy also induces Brf1 expression (Ernens et al. 2006; Goodfellow et al. 2006). As Brf1 and MYC physically interact in cardiomyocytes, it seems likely that MYC exerts its influence on Pol III transcription in this context via direct interaction with the Pol III transcriptional apparatus (Ernens et al. 2006; Goodfellow et al. 2006), but secondary mechanisms such as up-regulation of Pol III subunits or cofactors may also occur. This is in accord with mechanisms already identified for MYC’s regulation of Pol III transcription in proliferating cells. Therefore, increases in Pol I and Pol III activity are not merely a consequence of increased proliferation driven by MYC, but also occur in the absence of cell division to facilitate MYC-driven cell growth.
DOES MYC UP-REGULATION OF POL I AND POL III DRIVE TUMORIGENESIS?
Increased levels of Pol I and Pol III subunits, transcription factors and products are a feature of numerous cancers, but does MYC need elevated Pol I and Pol III activity to drive tumorigenesis? In Eμ-MYC mouse models, the MYC translocation found in Burkitt lymphoma leads to constitutive MYC transgene expression in the B-cell lineage and lymphoma development (Adams et al. 1985; Harris et al. 1988). B cells from Eμ-MYC mice show enhanced proliferation, increased protein synthesis and cell size (Iritani and Eisenman 1999). In B cells from young healthy Eμ-MYC mice, Pol I subunits and cofactors are up-regulated, showing that increased Pol I activity precedes MYC-induced transformation (Bywater et al. 2012). Enhanced protein synthesis seems a potent component of MYC’s transforming ability in this transgenic mouse model, as restoration of protein synthesis to normal levels in Eμ-MYC mice via ribosomal protein L24 haploinsufficiency markedly delayed tumorigenesis (Barna et al. 2008). Reduced Pol I or Pol III activity might therefore impede tumor development in this system. Inhibition of Pol I transcription with the small molecule inhibitor CX-5461, specifically induced cell death in Eμ-MYC lymphoma cells without toxicity to normal B lymphocytes or discernible adverse effect on the health of mice (Bywater et al. 2012). A one-third reduction in Pol I activity resulted in nucleolar disruption, p53 activation and rapid apoptosis (Bywater et al. 2012). CX-5461 was effective against an array of hematopoietic tumor cell lines in culture, a mouse acute myeloid leukemia model of AML1/ETO gene fusion and in a xenotransplant with human myelomonocytic leukemia cells (Bywater et al. 2012). Deregulated MYC may be a key factor in each of these examples and it is possible that aberrant MYC activity primes these cells for death by Pol I inhibition. CX-5461 is ∼300-fold more potent at killing Eμ-MYC lymphoma cells than Nutlin-3A, a small molecule p53 activator that is currently in clinical trials (Bywater et al. 2012). In mice bearing Eμ-MYC lymphomas, prolonged dosing with CX-5461 resulted in a significant period of disease remission, although resistance is eventually acquired (Bywater et al. 2012). This groundbreaking work encourages optimism concerning the potential use of Pol I inhibitors in cancer therapy and a clinical trial using CX-5461 in leukemia and lymphoma patients has commenced (Hannan et al. 2013).
Although analysis is less advanced for Pol III, there are grounds to hope that it too might prove a useful target. BRF1 and GTF3C4, encoding subunits of TFIIIB and TFIIIC respectively, were identified as genes whose knockdown had a MYC-specific synthetic lethal effect in human mammary epithelial cells (Kessler et al. 2012). Both of these Pol-III-specific transcription factors have been found to physically interact with MYC (Gomez-Roman et al. 2003; Koch et al. 2007) and have also been identified as MYC target genes (Li et al. 2003; Sansom et al. 2007). In rat fibroblasts, partial knockdown of Brf1, to abrogate the up-regulation of Pol III activity induced by MYC transformation, did not impair proliferation but restricted MYC-driven anchorage-independent growth and tumor formation in mice (Johnson et al. 2008). This suggests that enhanced Pol III transcription contributes to the oncogenic functions of MYC in this context. Similarly, anchorage-independent growth of immortalized human fibroblasts induced by MYC can be suppressed using tagetin to inhibit Pol III (Shukla and Kumar 2012). Therefore, Pol III transcription may make an attractive therapeutic target in cancers with deregulated MYC.
CONCLUDING REMARKS
Aberrant MYC activity and increased Pol I and Pol III transcription are frequently seen in tumor cells. These observations are not merely coincidental or a consequence of cell cycle effects, as MYC exerts control over Pol I and Pol III transcription in multiple ways, across species and even in nondividing cells. MYC’s requirement for increased Pol I and Pol III products may leave cancer cells vulnerable to cancer therapies that target their transcription.
ACKNOWLEDGMENTS
K.J.C. is supported by a Dorothy Hodgkin Fellowship from the Royal Society. We thank Joanna Birch and Sarah Dowding for useful comments on this manuscript.
Footnotes
Editors: Chi V. Dang and Robert N. Eisenman
Additional Perspectives on MYC and the Pathway to Cancer available at www.perspectivesinmedicine.org
REFERENCES
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL 1985. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 318: 533–538 [DOI] [PubMed] [Google Scholar]
- Arabi A, Wu S, Ridderstrale K, Bierhoff H, Shiue C, Fatyol K, Fahlen S, Hydbring P, Soderberg O, Grummt I, et al. 2005. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol 7: 303–310 [DOI] [PubMed] [Google Scholar]
- Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, Rao PH, Ruggero D 2008. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature 456: 971–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstralh DT, Conti BJ, Moore CB, Brickey WJ, Taxman DJ, Ting JP 2007. Global functional analysis of nucleophosmin in Taxol response, cancer, chromatin regulation, and ribosomal DNA transcription. Exp Cell Res 313: 65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenburger Y, Jenkins A, Autelitano DJ, Hannan RD 2001. Increased expression of UBF is a critical determinant for rRNA synthesis and hypertrophic growth of cardiac myocytes. FASEB J 15: 2051–2053 [DOI] [PubMed] [Google Scholar]
- Brown SJ, Cole MD, Erives AJ 2008. Evolution of the holozoan ribosome biogenesis regulon. BMC Genomics 9: 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bywater MJ, Poortinga G, Sanij E, Hein N, Peck A, Cullinane C, Wall M, Cluse L, Drygin D, Anderes K, et al. 2012. Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell 22: 51–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai MS, Sun XX, Lu H 2010. Ribosomal protein L11 associates with c-Myc at 5 S rRNA and tRNA genes and regulates their expression. J Biol Chem 285: 12587–12594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernens I, Goodfellow SJ, Innes F, Kenneth NS, Derblay LE, White RJ, Scott PH 2006. Hypoxic stress suppresses RNA polymerase III recruitment and tRNA gene transcription in cardiomyocytes. Nucleic Acids Res 34: 286–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felton-Edkins ZA, Kenneth NS, Brown TR, Daly NL, Gomez-Roman N, Grandori C, Eisenman RN, White RJ 2003. Direct regulation of RNA polymerase III transcription by RB, p53 and c-Myc. Cell Cycle 2: 181–184 [PubMed] [Google Scholar]
- Gomez-Roman N, Grandori C, Eisenman RN, White RJ 2003. Direct activation of RNA polymerase III transcription by c-Myc. Nature 421: 290–294 [DOI] [PubMed] [Google Scholar]
- Goodfellow SJ, White RJ 2007. Regulation of RNA polymerase III transcription during mammalian cell growth. Cell Cycle 6: 2323–2326 [DOI] [PubMed] [Google Scholar]
- Goodfellow SJ, Innes F, Derblay LE, MacLellan WR, Scott PH, White RJ 2006. Regulation of RNA polymerase III transcription during hypertrophic growth. EMBO J 25: 1522–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ 2005. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol 7: 311–318 [DOI] [PubMed] [Google Scholar]
- Greasley PJ, Bonnard C, Amati B 2000. Myc induces the nucleolin and BN51 genes: Possible implications in ribosome biogenesis. Nucleic Acids Res 28: 446–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SS, Li L, Orian A, Eisenman RN, Edgar BA 2005. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat Cell Biol 7: 295–302 [DOI] [PubMed] [Google Scholar]
- Hannan KM, Sanij E, Rothblum LI, Hannan RD, Pearson RB 2013. Dysregulation of RNA polymerase I transcription during disease. Biochim Biophys Acta 1829: 342–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris AW, Pinkert CA, Crawford M, Langdon WY, Brinster RL, Adams JM 1988. The Eμ-myc transgenic mouse. A model for high-incidence spontaneous lymphoma and leukemia of early B cells. J Exp Med 167: 353–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hateboer G, Timmers HT, Rustgi AK, Billaud M, van’t Veer LJ, Bernards R 1993. TATA-binding protein and the retinoblastoma gene product bind to overlapping epitopes on c-Myc and adenovirus E1A protein. Proc Natl Acad Sci 90: 8489–8493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iritani BM, Eisenman RN 1999. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc Natl Acad Sci 96: 13180–13185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Wu G, Zhan X, Nolan A, Koh C, De Marzo A, Doan HM, Fan J, Cheadle C, Fallahi M, et al. 2011. Cell-type independent MYC target genes reveal a primordial signature involved in biomass accumulation. PLoS ONE 6: e26057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SA, Dubeau L, Johnson DL 2008. Enhanced RNA polymerase III-dependent transcription is required for oncogenic transformation. J Biol Chem 283: 19184–19191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P 1999. Drosophila myc regulates cellular growth during development. Cell 98: 779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenneth NS, Ramsbottom BA, Gomez-Roman N, Marshall L, Cole PA, White RJ 2007. TRRAP and GCN5 are used by c-Myc to activate RNA polymerase III transcription. Proc Natl Acad Sci 104: 14917–14922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler JD, Kahle KT, Sun T, Meerbrey KL, Schlabach MR, Schmitt EM, Skinner SO, Xu Q, Li MZ, Hartman ZC, et al. 2012. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science 335: 348–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch HB, Zhang R, Verdoodt B, Bailey A, Zhang CD, Yates JR 3rd, Menssen A, Hermeking H 2007. Large-scale identification of c-MYC-associated proteins using a combined TAP/MudPIT approach. Cell Cycle 6: 205–217 [DOI] [PubMed] [Google Scholar]
- Li Z, Hann SR 2013. Nucleophosmin is essential for c-Myc nucleolar localization and c-Myc-mediated rDNA transcription. Oncogene 32: 1988–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B 2003. A global transcriptional regulatory role for c-Myc in Burkitt’s lymphoma cells. Proc Natl Acad Sci 100: 8164–8169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA 2012. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151: 56–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateyak MK, Obaya AJ, Adachi S, Sedivy JM 1997. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ 8: 1039–1048 [PubMed] [Google Scholar]
- McEwan IJ, Dahlman-Wright K, Ford J, Wright AP 1996. Functional interaction of the c-Myc transactivation domain with the TATA binding protein: Evidence for an induced fit model of transactivation domain folding. Biochemistry 35: 9584–9593 [DOI] [PubMed] [Google Scholar]
- McStay B, Grummt I 2008. The epigenetics of rRNA genes: From molecular to chromosomal biology. Annu Rev Cell Dev Biol 24: 131–157 [DOI] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. 2012. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151: 68–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen TJ, O’Neill JD, Dawson CW, Hu C, Chen X, Yao Y, Wood VH, Mitchell LE, White RJ, Young LS, et al. 2010, Epstein-Barr virus-encoded EBNA1 enhances RNA polymerase III-dependent EBER expression through induction of EBER-associated cellular transcription factors. Mol Cancer 9: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poortinga G, Hannan KM, Snelling H, Walkley CR, Jenkins A, Sharkey K, Wall M, Brandenburger Y, Palatsides M, Pearson RB, et al. 2004. MAD1 and c-MYC regulate UBF and rDNA transcription during granulocyte differentiation. EMBO J 23: 3325–3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poortinga G, Wall M, Sanij E, Siwicki K, Ellul J, Brown D, Holloway TP, Hannan RD, McArthur GA 2011. c-MYC coordinately regulates ribosomal gene chromatin remodeling and Pol I availability during granulocyte differentiation. Nucleic Acids Res 39: 3267–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raha D, Wang Z, Moqtaderi Z, Wu L, Zhong G, Gerstein M, Struhl K, Snyder M 2010. Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc Natl Acad Sci 107: 3639–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, Wong MC, Maddren M, Fang R, Heitner SG, et al. 2013. ENCODE data in the UCSC Genome Browser: Year 5 update. Nucleic Acids Res 41: D56–D63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D 2009. The role of Myc-induced protein synthesis in cancer. Cancer Res 69: 8839–8843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J, Zomerdijk JC 2005. RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem Sci 30: 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom OJ, Meniel VS, Muncan V, Phesse TJ, Wilkins JA, Reed KR, Vass JK, Athineos D, Clevers H, Clarke AR 2007. Myc deletion rescues Apc deficiency in the small intestine. Nature 446: 676–679 [DOI] [PubMed] [Google Scholar]
- Schlosser I, Holzel M, Murnseer M, Burtscher H, Weidle UH, Eick D 2003. A role for c-Myc in the regulation of ribosomal RNA processing. Nucleic Acids Res 31: 6148–6156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm L , Hernandez N 2002. Recruitment of RNA polymerase III to its target promoters. Genes Dev 16: 2593–2620 [DOI] [PubMed] [Google Scholar]
- Shiue CN, Berkson RG, Wright AP 2009. c-Myc induces changes in higher order rDNA structure on stimulation of quiescent cells. Oncogene 28: 1833–1842 [DOI] [PubMed] [Google Scholar]
- Shukla SK, Kumar V 2012. Hepatitis B virus X protein and c-Myc cooperate in the upregulation of ribosome biogenesis and in cellular transformation. FEBS J 279: 3859–3871 [DOI] [PubMed] [Google Scholar]
- Steiger D, Furrer M, Schwinkendorf D, Gallant P 2008. Max-independent functions of Myc in Drosophila melanogaster. Nature Genet 40: 1084–1091 [DOI] [PubMed] [Google Scholar]
- van Riggelen J, Yetil A, Felsher DW 2010. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 10: 301–309 [DOI] [PubMed] [Google Scholar]
- White RJ 2008. RNA polymerases I and III, non-coding RNAs and cancer. Trends in Genetics 24: 622–629 [DOI] [PubMed] [Google Scholar]
- White RJ 2011. Transcription by RNA polymerase III: More complex than we thought. Nature Rev Genet 12: 459–463 [DOI] [PubMed] [Google Scholar]