Abstract
Objectives. To teach first-year (P1) pharmacy students to apply the principles of pharmacogenomics underlying clinical pharmacotherapeutics to cancer patients.
Design. Using polymerase chain reaction (PCR) and high-resolution melting analysis of deoxyribonucleic acid (DNA) from colorectal cancer cell lines to determine the presence of somatic mutations for an oncogenic marker, students formulated the proper course of treatment for a patient with similar tumor genomics.
Assessment. In a postintervention survey, students highly rated the effectiveness of the laboratory session for learning pharmacogenomics, and subsequent examination scores reflected retention of principles and understanding of clinical application.
Conclusion. The pharmacogenomic laboratory exercise prepared students to understand how genetic markers give clinical insight into the appropriate application of drugs in oncology pharmacotherapy. Further, the session inspired their interest in learning more about pharmacogenomics and their professional roles in personalized medicine.
Keywords: pharmacogenomics, genomics, oncology, cancer, pharmacotherapy, active learning
INTRODUCTION
Personalized medicine is a rapidly growing field in which healthcare providers can determine appropriate pharmacotherapy for cancer patients by using the genetic markers within their tumor DNA. Discoveries regarding the importance of pathways in cancer have resulted in a change in cancer treatments from conventional chemotherapy and radiation therapy to a molecular targeted approach to treatment. In particular, the field of oncology is using identification of specific mutations within tumor cells to determine selective pharmacotherapeutic targeting of the tumor cells, which results in less damage to the patient’s normal cells.
It is important for pharmacy educators to teach future pharmacists to optimize personalized treatment based on patients’ genomics with the hope of increasing patient survival. Our pharmacogenomics laboratory exercise incorporates the Accreditation Council for Pharmacy Education competencies specific for pharmacists to be able to use knowledge of the biomedical sciences and emerging technologies to provide pharmaceutical care as a member of the health care team.1 It also addresses CAPE competency 1.1.4, Application of knowledge in foundational sciences to solve therapeutic problems and advance patient-centered care.2
In the last 30 years, researchers have discovered the importance of somatic mutations in tumor cells, which are driving the development of the tumor.3 In general, somatic mutations can result in alterations of proteins involved in regulatory pathways such as the ERK/MAPK (extracellular signal-regulated kinases/mitogen-activated protein kinases) pathway, giving cells a growth advantage, thus driving carcinogenesis. Epidermal growth factor receptor (EGFR), v-Raf murine sarcoma viral oncogene homolog B1 (BRAF), and V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS) genes are examples of known oncogenes, which encode for proteins that are involved in the ERK/MAPK pathway. A somatic mutation altering any 1 of the 3 oncogenic proteins in a manner that stimulates this signaling pathway results in proliferation of tumor cells.4
To block signaling of the cancer promoting mutant proteins, 2 types of pharmacologic inhibitors have been developed to suppress the activation of the ERK/MAPK pathway. Cetuximab (Erbitux) and panitumumab (Vectibix) are monoclonal antibodies that block activation of the oncogenic EGFR protein.5-8 These antagonists impede receptor signaling by either attaching to the ligand binding site, or by hindering receptor homodimerization. Another type of inhibitor, small molecule kinase inhibitors, directly binds to the mutated proteins, inhibiting their signaling function. Sorafenib (Nexavar) is a nonselective inhibitor that attaches to B-raf and C-raf proteins, maintaining these kinase proteins in an inactive form.9,10 Vemurafenib (Zelboraf) inhibits by selectively binding to the mutated form of the B-raf protein.11 Although somatic alteration of the oncogenes are informative for tailored treatment, the Food and Drug Administration (FDA) requires EGFR protein expression analysis for the monoclonal antibody therapies, but only recommends genetic testing of the BRAF mutation for vemurafenib.12-14
Healthcare professionals may not use targeted therapies because of the lack of knowledge about appropriate molecular tests to identify these specific markers.15 Pharmacists who can identify the appropriate markers and apply the clinically available genetic tests can improve patient care by helping define optimal targeted therapies. Understanding the principles of pharmacogenomics is a key element in bridging the gap between bench and bedside.
The purpose of this laboratory exercise was to introduce P1 pharmacy students to the fundamental concepts and application of pharmacogenomics early in their training. Concurrently, P1 students were enrolled in Human Biochemistry and Medical Immunology courses, which provided important foundational concepts that were integrated into the Practice Integrated Laboratory Sequence (PILS) exercises. This P1 pharmacogenomics exercise incorporated 6 of the 12 core competencies of the Presbyterian College School of Pharmacy (PCSP) curriculum: communication, problem prevention and solving, evidence-based decisions, interprofessional interaction and teamwork, advancement of pharmacy and healthcare, and promoting of health and public welfare.
DESIGN
The P1 students obtained a foundational knowledge of cancer genetics, types of mutations, and monoclonal antibody therapies for cancer treatment in their basic science courses, including in Human Biochemistry and Medical Immunology. This pharmacogenomics exercise in the PILS demonstrated and applied these concepts of genetics and medication therapy using an active-learning approach in a student-centered learning environment. The expected outcomes of the pharmacogenomics laboratory exercise were for students to be able to: comprehend the scientific basis of genomic mutations; identify specific mutations in the tumor DNA; analyze whether a specific mutation is a drug target; and evaluate the appropriate pharmacotherapy based on the tumor genome. The goal of this pharmacogenomics laboratory exercise was to illustrate the importance of determining the proper treatment for a specific patient through the identification of genetic markers, in this case using the known somatic marker in the BRAF gene of colorectal cancer patients.16
Somatic mutation of the BRAF gene is observed in approximately 5%-10% of all colorectal cancer cases.16,17 BRAF gene encodes for a serine/threonine-protein kinase, which is an activating protein in the ERK/MAPK pathway. In 90% of somatic BRAF mutations, the nucleotide change is a thymidine to adenosine in position 1799. This mutational change translates into a substitution of a valine (V) to a glutamic acid (E) in the protein at position 600. The missense mutation (V600E) results in an activating event, increasing kinase signaling which stimulates proliferation through the ERK/MAPK pathway.18 The somatic BRAF V600E mutation has been linked to alterations in the response to several cancer treatments, such as cetuximab, panitumumab, sorafinib, and vemurafenib. Because the immunotherapies target the Egf receptor protein in the ERK/MAPK pathway in tumor DNA, a somatic V600E mutation in the downstream effector B-raf protein of tumor cells impairs the responsiveness to cetuximab or panitumumab for colorectal patients.19-21 Studies showed that the immunotherapies were more effective in patients who expressed the unaltered (wild type) B-raf protein. For EGFR-positive colorectal patients with the somatic BRAF V600E mutation, a targeted approach could be considered using the small Raf kinase inhibitors sorafinib and vemurafenib, in conjunction with the immunotherapies.9, 22 This multi-targeted approach to oncogenic proteins more effectively suppresses the activation of the ERK/MAPK pathway in patients who possess a somatic BRAF V600E mutation and overexpress the EGFR protein in their tumor cells.
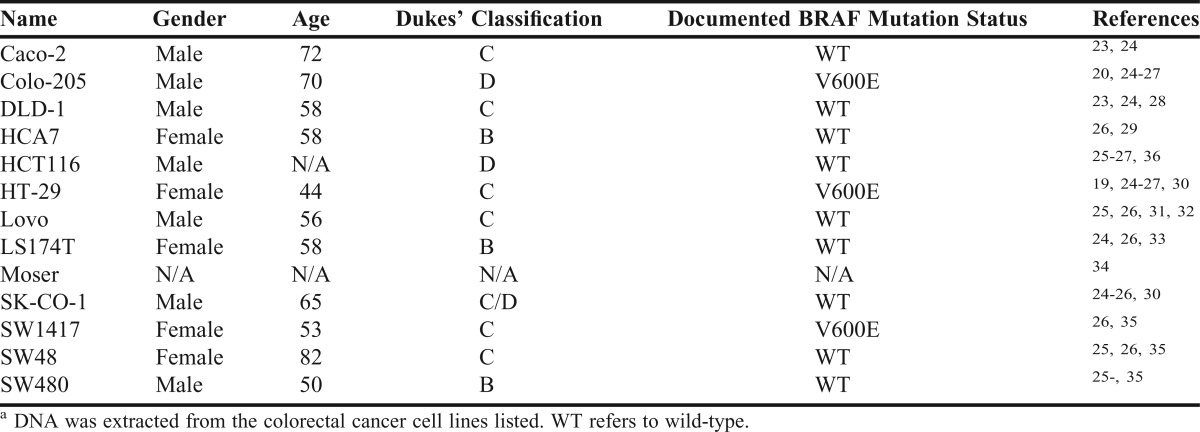
Because the somatic BRAF V600E mutation is found in colorectal tumor DNA, we used 13 commercially available colorectal cancer cell lines in the laboratory exercise. We provided clinical and genetic information to students about each of the cell lines from the ATCC website, research articles, and online databases (Table 1).
Table 1.
Description of Colorectal Adenomacarcinoma Cell Linesa
Isolation of DNA
In preparation for the laboratory exercise, the cell lines were grown using standard methods as described in the ATCC cell culture procedures. The cell culture of the colorectal cancer cells was a major challenge, as growing the cells was labor intensive because of the number of cell lines. The culturing also needed to be coordinated because of the individual culturing requirements for each cell line. Confluent cell lines were harvested with lysis buffer/Proteinase K from the Agencourt GENFIND v2 DNA isolation kit (Beckman Coulter Genomics, Danvers MA). An aliquot of each cell lysate was used to confirm mutation status, while the remainder was retained for the laboratory exercise. The status of the V600E mutation in the cell lines was identified with Sanger Sequencing and Scorpion probes. For the pharmacogenomics exercise, the high resolution melting (HRM) analysis was used as the appropriate screening technique, because the test is high-throughput and affordable.37
The class of 78 students was divided into 2 laboratory sections of 39 each, which were further organized into individual work groups. Two pharmaceutical sciences faculty members who had a strong background in molecular biology and pharmacogenomics were required to prepare and teach the exercise. The 2200-square-foot biotechnology laboratory has an overhead camera for instruction of students in laboratory techniques.
The protocol for the DNA isolation and the notes for the pharmacogenomic exercise were available online a week prior to the laboratory exercise. The students were provided with a 1-hour lecture on the background of mutations and polymerase chain reaction (PCR) methods, in which the procedures for DNA isolation and HRM analysis were discussed. They were also given the clinical information about the patient from whom the cancer cell lines were derived, including the gender and age of the patient, and tumor staging information. The staging information was based on the Dukes classification, which describes the extent of the tumor growth. The majority of the tumors were classified as stage C or D, which describes the cancer as spreading beyond the primary tumor. The students were told that patients with late-stage cancers would need additional therapy after surgery. By identifying the genetic markers, the appropriate pharmacotherapy treatment could be applied to the cancer patient.
In addition to the lecture material, the students reviewed 2 relevant papers on BRAF and discussed the implications of knowing the BRAF status of the tumor cell lines and how this knowledge could benefit individuals with late-stage colorectal cancer.19,38
Extraction and Quantitation of Tumor DNA
Each group of 5 students was provided a lysate from 1 of the 13 cancer cell lines, and directed to extract DNA using a protocol provided with the laboratory instruction materials. The DNA extraction protocol was adapted from the manufacturer’s protocol provided with the Agencourt GENFIND v2 DNA isolation kit (Beckman Coulter Genomics, Danvers MA). Tumor DNA samples were eluted with purified nuclease-free water. Quantitation of tumor DNA was done using a Thermo Scientific NanoDrop Lite Spectrophotometer and all samples were diluted to a final concentration of 2.5 ng/μL.
Amplification of the BRAF Marker
To identify the somatic mutation at nucleotide position 1799, a real-time polymerase chain reaction (RT-PCR) was performed using primers specific for a 100 bp segment of the BRAF gene, designed using the Primer3 primer design tool (available at http://bioinfo.ut.ee/primer3-0.4.0/) (forward, 5’-TTCATGAAGACCTCACAGTAAAA-3’; reverse, 5’-TCAGGGCCAAAAATTTAATCA-3’). PCR amplifications were performed using a BioRad C1000 Touch Thermal Cycler and CFX96 Touch Real-Time PCR Detection System (BioRad, Hercules, CA). The students were instructed to make a real-time PCR master mix containing 10μl BioRad Ssofast EvaGreen Mastermix, 6μl of nuclease-free water, 2μl of 5μM BRAF primers to which they added 5ng of tumor DNA. The PCR mixes were loaded into a 96-well plate, which included a control for the somatic V600E mutation and a control for the wild type BRAF marker (commercially available DNA, Promega). The PCR protocol was designed using the BioRad CFX Manager (BioRad, Hercules, CA), and consisted of a 98°C for 2 min; 98°C for 5 sec, and 60°C for 10 sec; (40 cycles); 95°C for 1min and 70°C for 1min; Melt Curve 70°C to 95°C at 0.2 °C/10 sec.
Mutational Analysis
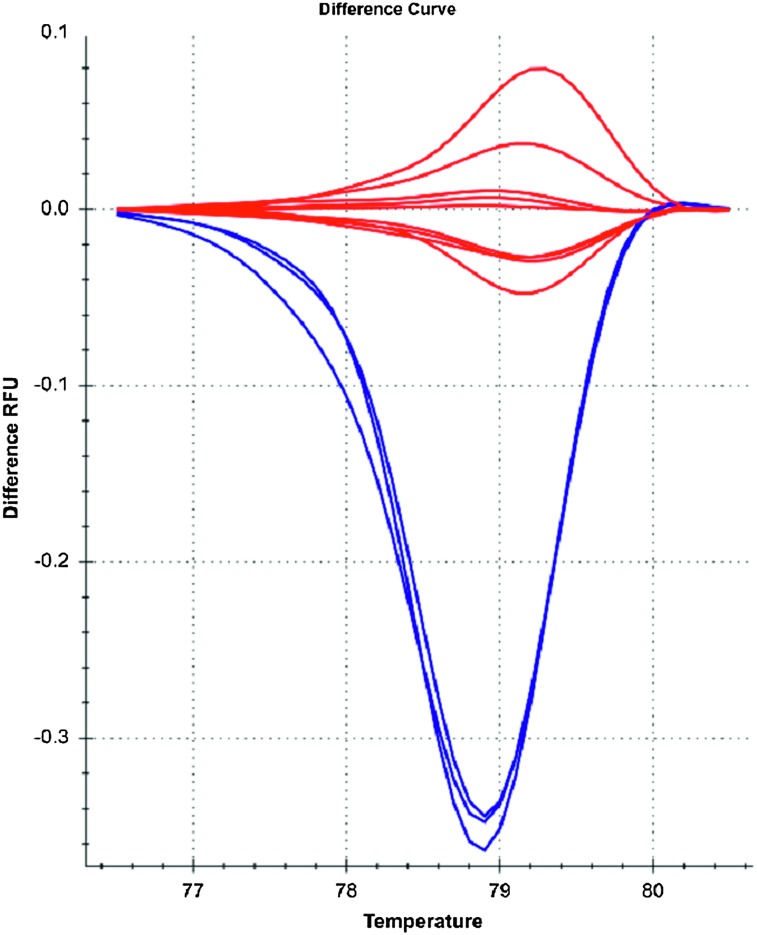
The identification of the somatic mutations was done using HRM software analysis (BioRad Precision Melt Analysis). The HRM analysis clustered the curves into 2 distinct variants with different melt peaks. The students were shown the somatic V600E mutation and wild type controls with the HRM results. Each laboratory group was given the opportunity to identify the status of the colorectal cancer cell lines. Ten of the cell lines demonstrated melt peaks consistent with the wild type BRAF, and the somatic V600E mutations were observed in 3 of the cell lines (Figure 1). After students were given the BRAF status and clinical information for each colorectal cell line, the laboratory groups were asked to explain the treatment options for each cell line as if they were treating a colorectal cancer patient. The Presbyterian College Institutional Review Board determined this research was exempt.
Figure 1.
Results of Laboratory Exercise. The BRAF Status of V600 for the pharmacogenomic exercise was performed using the high resolution melting (HRM) analysis. The V600E mutation curves for the 3 samples are identified in blue and the wild-type curves for the 10 samples are in red.
EVALUATION AND ASSESSMENT
We defined success of the laboratory exercise as an increase in understanding of pharmacogenomic principles by a majority of the students as reflected in their performance on a summative examination and their positive perception of the learning experience. A week prior to the pharmacogenomics laboratory exercise, the students were asked to complete an in-class, preexercise survey instrument that was anonymous and voluntary. During the week following the laboratory exercise, the students were asked to complete an in-class, postexercise survey instrument to evaluate the effectiveness of the laboratory exercise.
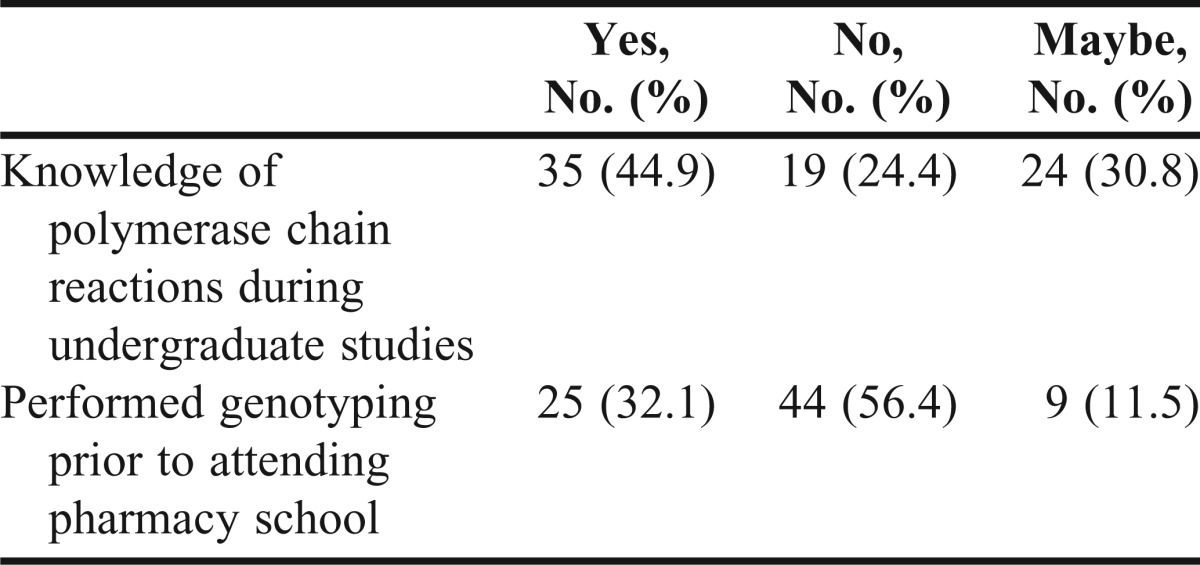
The survey completion rate was 100% for the preexercise survey instrument and 98.7% for the postexercise survey instrument (data not shown). Prior to the laboratory exercise, fewer than half of the students reported having previously learned about PCR. Additionally, less than a third of the students reported genotyping experience prior to the laboratory exercise (Table 2).
Table 2.
First-Year Pharmacy Students’ Self-Assessment of Knowledge/Experience Prior to Laboratory Exercise
Survey questions were included to determine the students’ knowledge of the application of the laboratory techniques in the area of pharmacogenomics. Prior to the laboratory exercise, the average rating on understanding how somatic mutations influence tumor growth was 3.7 out of 5. After the laboratory exercise, the average score for this question was 4.6 out of 5. Similarly, the average rating on the application of somatic mutations in altering drug response or as a drug target increased from 2.9 before to 4.5 out of 5 following the laboratory exercise (Table 3).
Table 3.
Rating of Knowledge of Pharmacogenomics Before and After Exerciseb

Students were also asked to rate the effectiveness of the laboratory exercise as an educational tool using 5 questions. The average rating for all 5 assessment questions was 4 or greater out of 5. The overall rating of the laboratory exercise was favorable (Table 4) and the feedback that was given on the open-ended assessment questions (data not shown) was positive as well.
Table 4.
Assessment of Effectiveness of Laboratory Exercisec

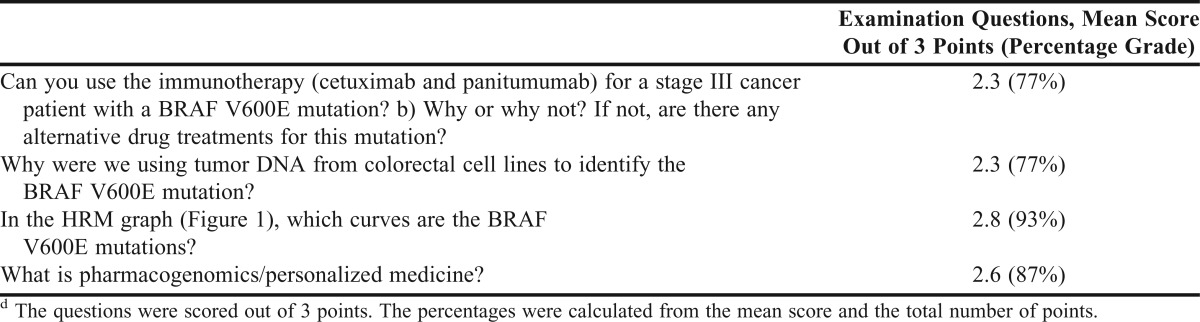
Students’ knowledge of pharmacogenomics was evaluated using short-answer application and knowledge questions included on the PILS Biotechnology Laboratory final examination. Most of the students demonstrated an understanding of how the identification of BRAF marker applies to oncology treatment (Table 5). Students scored especially well on their knowledge of how to distinguish a wild type sequence from the V600E mutation using the HRM graph. In their overall performance on the final examination questions, more than 77% of the students showed they had developed an understanding of pharmacogenomic principles and their application to the pharmacotherapy of cancer.
Table 5.
Scoring of the Examination Questions From the Pharmacogenomic Laboratory Exercised
DISCUSSION
Pharmacy educators are exploring best practices to teach crucial topics of pharmacogenomics and targeted oncogenic therapy.39-41 In the PCSP curriculum, we have developed a program that emphasizes the importance of genomics from the beginning of the student’s pharmacy education. Responses on the initial survey instrument administered showed that many students entering the pharmacy program had a limited background in genomics; thus, it is critical to begin bridging that gap as soon as possible. By incorporating principles of genomics into Medical Immunology and Human Biochemistry courses in their first academic semester, we have been able to reinforce these principles with active-learning laboratory experience in the PILS.
The students gain practical experience from these laboratory exercises, and this enables them to move from basic scientific discovery of mutations to clinical application of the data for a patient. This pharmacogenomic exercise emphasizes the scientific foundation for understanding the role somatic mutations play in human disease and therapeutics, using a well-understood BRAF somatic mutation. Responses to examination questions showed students’ level of comprehension of pharmacogenomic principles and how to apply them. Students’ responses on the presurvey and postsurvey instruments demonstrate the value of this exercise. Not only did students’ perceived level of knowledge of pharmacogenomics increase, but also their interest in the subject was stimulated. Students’ examination scores reflected improved comprehension of both pharmacogenomic principles and their clinical application.
The pharmacogenomics laboratory exercise was designed to address several core competencies for genetics education established in 2007 for all health professionals by the National Coalition for Health Professional Education in Genetics (NCHPEG):
• 1.3 - Understanding how identification of disease-associated genetic variations facilitates development of prevention, diagnosis, and treatment options,
• 1.5 - Understanding the interaction of genetic, environmental, and behavioral factors in predisposition to disease, onset of disease, response to treatment, and maintenance of health.
• 1.6 - Understanding the difference between clinical diagnosis of disease and identification of genetic predisposition to disease.42
Our pharmacogenomics laboratory exercise incorporated each of these principles. As students move through the PCSP curriculum, they establish a solid scientific foundation to apply to patient pharmacotherapeutics. Courses in the curriculum that incorporate pharmacogenetic principles and their application include Pathophysiology in the P1 year; Pharmacology and Medicinal Chemistry, Pharmacogenomics, and Measuring Therapeutic Parameters in the P2 year; and Oncology, and Infectious Disease Medication Therapy Management courses in the P3 year. By incorporating a comprehensive program of pharmacogenomics throughout the PCSP curriculum, these future pharmacists will be at the forefront of meeting the challenges of personalized medicine in the 21st century.
In this era of the patient-centered medical home, it is important for healthcare practitioners to understand the scientific basis for clinical decisions. Pharmacists who participate in clinical therapeutic modalities have a critical role in optimizing patient outcomes. In the next decade, a patient’s genome likely will be the crucial piece of the puzzle in personalized medicine. Pharmacists in a wide range of practice sites will need to understand and apply pharmacogenomics in order to maintain best practice for the benefit of their patients.
SUMMARY
As personalized medicine and medication therapy management become more central parts of standardized patient care, pharmacists will need to understand how the identification of somatic mutations will be used in tailoring the appropriate cancer therapy. The pharmacogenomic exercise in the PILS course was designed to teach students about the basics of personalized oncology therapy using a well-documented somatic mutation associated with specific targeted therapies. Students actively participated in the isolation of tumor DNA, the identification of a somatic mutation, and the determination of a course of clinical treatment appropriate to a patient with a specific mutation. Student responses to the survey instruments and their performance on summative examination questions confirmed that the laboratory exercise effectively demonstrated the importance of personalized medicine and pharmacogenomics for oncology treatment, while increasing their interests in learning more about the clinical application of pharmacogenomics.
ACKNOWLEDGEMENTS
We appreciate the input of Dr. Meg Franklin regarding the data analysis. We would also like to thank Dr. Dan Dixon for providing colorectal cell lines for the laboratory exercise.
REFERENCES
- 1.Accreditation Council for Pharmacy Education. Accreditation standards and guidelines for the professional program in pharmacy leading to the doctor of pharmacy degree. February 2006. https://www.acpe-accredit.org/pdf/ACPE_Revised_PharmD_Standards_Adopted_Jan152006.pdf. Accessed October 1, 2013.
- 2.Turner CJ, Altier R, Fish D, et al. An assessment system for mapping CAPE outcomes in an advanced pharmacy practice experience program. Am J Pharm Educ. 2006;70(3):Article 60. doi: 10.5688/aj700360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314(5797):268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 4.Moroni M, Veronese S, Benvenuti S, et al. Gene copy number for epidermal growth factor receptor (EGFR) and clinical response to antiEGFR treatment in colorectal cancer: a cohort study. Lancet Oncol. 2005;6(5):279–286. doi: 10.1016/S1470-2045(05)70102-9. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham D, Humblet Y, Siena S, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351(4):337–345. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 6.Herbst RS, Shin DM. Monoclonal antibodies to target epidermal growth factor receptor-positive tumors: a new paradigm for cancer therapy. Cancer. 2002;94(5):1593–1611. doi: 10.1002/cncr.10372. [DOI] [PubMed] [Google Scholar]
- 7.Lynch DH, Yang XD. Therapeutic potential of ABX-EGF: a fully human anti-epidermal growth factor receptor monoclonal antibody for cancer treatment. Semin Oncol. 2002;29(1 Suppl 4):47–50. doi: 10.1053/sonc.2002.31522. [DOI] [PubMed] [Google Scholar]
- 8.Van Cutsem E, Peeters M, Siena S, et al. Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J Clin Oncol. 2007;25(13):1658–1664. doi: 10.1200/JCO.2006.08.1620. [DOI] [PubMed] [Google Scholar]
- 9.Al-Marrawi MY, Saroya BS, Brennan MC, Yang Z, Dykes TM, El-Deiry WS. Off-label use of cetuximab plus sorafenib and panitumumab plus regorafenib to personalize therapy for a patient with V600E BRAF-mutant metastatic colon cancer. Cancer Biol Ther. 2013;14(8):703–710. doi: 10.4161/cbt.25191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilhelm SM, Carter C, Tang L, et al. BAY 43-9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res. 2004;64(19):7099–7109. doi: 10.1158/0008-5472.CAN-04-1443. [DOI] [PubMed] [Google Scholar]
- 11.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Highlights of prescribing information for Erbitux (cetuximab) http://www.accessdata.fda.gov/drugsatfda_docs/label/2009/125084s168lbl.pdf . Accessed October 1, 2013.
- 13.Highlights of prescribing information for Vectibix (panitumumab) http://pi.amgen.com/united_states/vectibix/vectibix_pi.pdf. Accessed October 1, 2013.
- 14.Highlights of prescribing information for Zelboraf (vemurafenib) http://www.accessdata.fda.gov/drugsatfda_docs/label/2011/202429s000lbl.pdf . Accessed October 1, 2013.
- 15.McCullough KB, Formea CM, Berg KD, et al. Assessment of the pharmacogenomics educational needs of pharmacists. Am J Pharm Educ. 2011;75(3):Article 51. doi: 10.5688/ajpe75351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 17.McCubrey JA, Steelman LS, Chappell WH, et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim Biophys Acta. 2007;1773(8):1263–1284. doi: 10.1016/j.bbamcr.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wan PT, Garnett MJ, Roe SM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116(6):855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 19.Di Nicolantonio F, Martini M, Molinari F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26(35):5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 20.De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11(8):753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 21.Laurent-Puig P, Cayre A, Manceau G, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol. 2009;27(35):5924–5930. doi: 10.1200/JCO.2008.21.6796. [DOI] [PubMed] [Google Scholar]
- 22.Yang H, Higgins B, Kolinsky K, et al. Antitumor activity of BRAF inhibitor vemurafenib in preclinical models of BRAF-mutant colorectal cancer. Cancer Res. 2012;72(3):779–789. doi: 10.1158/0008-5472.CAN-11-2941. [DOI] [PubMed] [Google Scholar]
- 23.Oikonomou E, Koc M, Sourkova V, Andera L, Pintzas A. Selective BRAFV600E inhibitor PLX4720, requires TRAIL assistance to overcome oncogenic PIK3CA resistance. PloS One. 2011;6(6):e21632. doi: 10.1371/journal.pone.0021632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trainer DL, Kline T, McCabe FL, et al. Biological characterization and oncogene expression in human colorectal carcinoma cell lines. Int J Cancer. 15. 1988;41(2):287–296. doi: 10.1002/ijc.2910410221. [DOI] [PubMed] [Google Scholar]
- 25.Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forbes SA, Tang G, Bindal N, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38(Database issue):D652–657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikediobi ON, Davies H, Bignell G, et al. Mutation analysis of 24 known cancer genes in the NCI-60 cell line set. Mol Cancer Ther. 2006;5(11):2606–2612. doi: 10.1158/1535-7163.MCT-06-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen TR, Dorotinsky CS, McGuire LJ, Macy ML, Hay RJ. DLD-1 and HCT-15 cell lines derived separately from colorectal carcinomas have totally different chromosome changes but the same genetic origin. Cancer Genet Cytogenet. 1995;81(2):103–108. doi: 10.1016/0165-4608(94)00225-z. [DOI] [PubMed] [Google Scholar]
- 29.Kirkland SC. Dome formation by a human colonic adenocarcinoma cell line (HCA-7) Cancer Res. 1985;45(8):3790–3795. [PubMed] [Google Scholar]
- 30.Fogh J, Fogh JM, Orfeo T. One hundred and twenty-seven cultured human tumor cell lines producing tumors in nude mice. J Natl Cancer Inst. 1977;59(1):221–226. doi: 10.1093/jnci/59.1.221. [DOI] [PubMed] [Google Scholar]
- 31.Jarry A, Masson D, Cassagnau E, Parois S, Laboisse C, Denis MG. Real-time allele-specific amplification for sensitive detection of the BRAF mutation V600E. Mol Cell Probes. 2004;18(5):349–352. doi: 10.1016/j.mcp.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 32.Drewinko B, Romsdahl MM, Yang LY, Ahearn MJ, Trujillo JM. Establishment of a human carcinoembryonic antigen-producing colon adenocarcinoma cell line. Cancer Res. 1976;36(2 Pt 1):467–475. [PubMed] [Google Scholar]
- 33.Tom BH, Rutzky LP, Jakstys MM, Oyasu R, Kaye CI, Kahan BD. Human colonic adenocarcinoma cells. I. Establishment and description of a new line. In Vitro. 1976;12(3):180–191. doi: 10.1007/BF02796440. [DOI] [PubMed] [Google Scholar]
- 34.Chakrabarty S, Jan Y, Brattain MG, Tobon A, Varani J. Diverse cellular responses elicited from human colon carcinoma cells by transforming growth factor-beta. Cancer Res. 15. 1989;49(8):2112–2117. [PubMed] [Google Scholar]
- 35.Leibovitz A, Stinson JC, McCombs WB, 3rd, McCoy CE, Mazur KC, Mabry ND. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 1976;36(12):4562–4569. [PubMed] [Google Scholar]
- 36.Brattain MG, Fine WD, Khaled FM, Thompson J, Brattain DE. Heterogeneity of malignant cells from a human colonic carcinoma. Cancer Res. 1981;41(5):1751–1756. [PubMed] [Google Scholar]
- 37.Pichler M, Balic M, Stadelmeyer E, et al. Evaluation of high-resolution melting analysis as a diagnostic tool to detect the BRAF V600E mutation in colorectal tumors. J Mol Diagn. 2009;11(2):140–147. doi: 10.2353/jmoldx.2009.080100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483(7387):100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 39.Brazeau DA, Brazeau GA. A required course in human genomics, pharmacogenomics, and bioinformatics. Am J Pharm Educ. 2006;70(6):Article 125. doi: 10.5688/aj7006125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Latif DA, McKay AB. Pharmacogenetics and pharmacogenomics instruction in colleges and schools of pharmacy in the United States. Am J Pharm Educ. 2005;69(2):Article 23. [Google Scholar]
- 41.Zdanowicz MM, Huston SA, Weston G. Pharmacogenomics in the professional pharmacy curriculum: Content, presentation and importance. Int J Pharm Educ. 2006;2(Fall):1–12. [Google Scholar]
- 42.National Coalition for Health Professional Education in Genetics. Core competencies for all health professionals. http://www.nchpeg.org/index.php?option=com_content&view=article&id=237&Itemid=84. Accessed June 1, 2013. [DOI] [PubMed]