Abstract
Autophagy is a process traditionally known to contribute to cellular cleaning through the removal of intracellular components in lysosomes. In recent years, the intensive scrutiny that autophagy has been subjected to at the molecular level, has also contributed to expand our understanding of the physiological role of this pathway. Added to the well-characterized role in quality control, autophagy has proven important in the maintenance of cellular homeostasis and of the energetic balance, in cellular and tissue remodeling and in the cellular defense against extracellular insults and pathogens. It is not a surprise that in light of this growing number of physiological functions, connections between autophagic malfunctioning and human pathologies have also been strengthened. In this review, we focus on several pathological conditions associated to primary or secondary defects in autophagy, and comment on a recurring theme for many of them, that is the fact that autophagy can often exert both beneficial and aggravating effects on the progression of disease. Elucidating the factors that determine the switch between these dual functions of autophagy in disease has become a priority when considering the potential therapeutic implications of the pharmacological modulation of autophagy in many of these pathological conditions.
Keywords: cancer, cardiomyopathy, immunity, lysosomes, neurodegeneration, protein aggregation, proteolysis, proteotoxicity, T-cell function
Introduction to autophagy
The Greek term autophagy (‘self-eating’) was coined almost half a century ago by Christian DeDuve [1]. Today, this intensely investigated pathway has come to be recognized as an evolutionarily conserved intracellular process through which cytosolic components - ranging from proteins, lipids, sugars and nucleotides to whole organelles and invading pathogens - are targeted for lysosomal degradation [2,3]. Autophagy contributes both, to the removal of damaged long-lived proteins and organelles and to the normal turnover of these intracellular components as part of its role in cellular quality control. In addition, autophagy also serves as a cellular adaptive response to compromised conditions, such as metabolic stress, when degradation of intracellular materials through this pathway becomes an alternative source of energy [4].
In recent years, a growing number of functions have been added to these two basic cellular functions of the autophagic process – quality control and maintenance of the cellular energetic balance – helping to shape the physiological relevance of this pathway. Furthermore, a better understanding of the molecular mechanisms that mediate the autophagic process has helped establish connections between autophagy and the pathogenesis of different disorders and of aging [3,5,6]. In this review, we briefly describe the main characteristics and molecular components of the different autophagic variants to then provide a general view of the multiple physiological functions of this cellular process. In the second part, we comment on specific autophagy-related pathologies, which either result from a primary defect in the autophagic process, or for which changes in autophagy have been described to exert a modulatory effect on the course of the disease.
The molecular dissection of the different autophagic variants
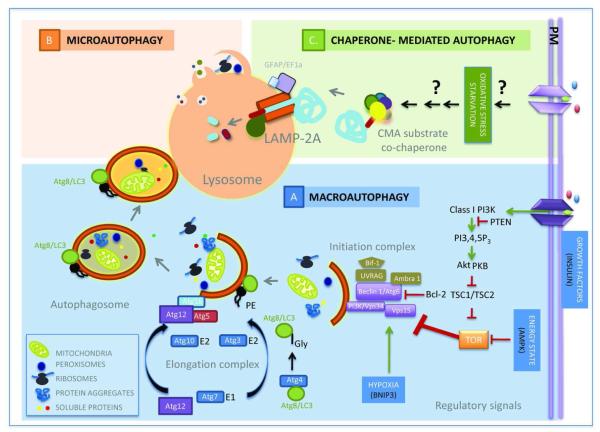
To date, three basic forms of autophagy have been described, namely, macroautophagy, microautophagy and chaperone-mediated autophagy (CMA), which primarily differ in the way cytosolic components (cargo) are delivered to lysosomes [2,7,8] (Fig. 1). Macroautophagy and microautophagy were both first described as mechanisms for ‘in bulk’ sequestration of cytoplasmic components (including entire organelles) into vesicular compartments. In macroautophagy, these vesicles form de novo from the growth in the cytosol, of a limiting membrane of non-lysosomal origin that seals upon itself to form free-standing double membrane carriers known as autophagosomes [9,10]. Fusion of lysosomes with the limiting membrane of the autophagosome grants lysosomal hydrolases access to the sequestered cargo assuring its complete degradation. In the case of microautophagy, the engulfment of cargo occurs through deformation of the lysosomal membrane itself, to form vesicles that invaginate towards the lysosomal lumen [11,12]. Pinch-off of these vesicles from the lysosomal membrane into the lumen and the subsequent degradation of their limiting membrane by the lysosomal hydrolases precede cargo degradation.
Figure 1. Autophagic pathways.
Three different general mechanisms of lysosomal delivery of cargo set the basis for the different types of autophagy described in mammalian cells (clockwise). A. Macroautophagy: Different extracellular and intracellular signals activate the recruitment of the macroautophagy initiation complex to the sites of autophagosome formation. Shuttling of proteins and lipids to these regions and posttranslational modifications of the lipids initiate the formation of a limiting membrane that grows through the assembly of proteins conjugated to proteins or lipids while it sequesters components of the cytosol. Once the membrane seals to form the autophagosome, this double membrane vesicle is delivered to lysosomes where upon membrane fusion, lysosomal hydrolases gain access to cargo. B. Microautophagy: Through stimuli yet poorly identified, cytosolic soluble proteins and organelles are directly sequestered by invaginations in the surface of lysosomes and late endosomes. Cargo internalized in the small luminal vesicles is degraded after the vesicles pinch-off from the limiting membrane. Although most microautophagy probably occurs in bulk, selective targeting by hsc70 of cytosolic proteins to forming microvesicles has been described. C. Chaperone-mediated autophagy: It is induced by stimuli such as prolonged starvation, oxidative stress and other conditions resulting in protein damage, but the signaling mechanism activated by these stimuli remain unknown. When CMA is activated, selective cytosolic proteins bearing a targeting motif are recruited by hsc70 and co-chaperones to the surface of lysosomes. Upon binding to the receptor protein LAMP-2A, substrates cross the membrane through the LAMP-2A-dependent translocation complex and are then rapidly degraded in the lumen.
Both macroautophagy and microautophagy were initially described in mammalian cells, but their molecular characterization has been mostly carried out in yeast, taking advantage of the genetic analysis that can be performed in this model organism. These genetic screenings have identified that more than 30 different genes (known as ATG or autophagy-related genes) participate in the execution and regulation of macroautophagy [2,13] (Fig. 1). The protein products of these genes organize into functional complexes that mediate the nucleation of the limiting membrane, its elongation, sealing and fusion with lysosomes. Nucleation is attained through post-translational modifications of pre-existing lipids in the membranes of different intracellular organelles (ER, mitochondria, Golgi) as well as in the plasma membrane [14-16]. Coordinated recruitment of lipid modifying enzymes and proteins that constitute the building blocks at the place of autophagosome formation gives rise to the limiting membrane [17]. Growth of this membrane occurs through two enzyme regulated processes that mediate conjugation of a protein – light chain protein 3 or LC3 - to one of the constituent membrane lipids and of two cytosolic proteins (Atg5 and Atg12) between them [9,18]. SNARE-like proteins, molecular motors, and additional lipid-modifying enzymes (phosphatases and kinases) participate in autophagosome-lysosome fusion. Most of the regulatory components exert their action on the nucleation complexes. For example, the Target of Rapamycin, a well-characterized negative regulator of this pathway, prevents the recruitment and interaction of specific Atgs of the nucleation complex to the site of autophagosome formation [19-21]. Although less is known about the regulation of later steps of the autophagic process, recent studies have identified transcription factor EB as a global regulator of Atgs that can upregulate initiation, elongation and fusion, as well as lysosomal biogenesis in response to nutritional stress [22]. Homologues for most Atgs have been identified in many other species (Drosophila [23], C.elegans [24], Dictyostelium [25], Arabidopsis [26], Trypanosoma [27]) and in mammals, where these genes often have several variants with diverse function [28].
The number of yeast genes shown to participate in microautophagy does not exceed a dozen, but this process also partially depends on Atgs shared with macroautophagy [29]. Microautophagy-specific proteins contribute to formation of and sealing of the vacuolar membrane (equivalent of the lysosome in yeast) [30]. Failure to identify mammalian homologues of the yeast microautophagy genes initially led to propose that microautophagy was not evolutionarily conserved. However, recent studies show otherwise, as a microautophagy-like process has been described to occur in mammalian late endosomes. This process, known as endosomal-microautophagy, has adopted the machinery involved in biogenesis of multivesicular bodies to form the invaginating vesicles that sequester cytosolic material for degradation [31].
In recent years, in addition to this “in bulk” non selective degradation of cytosolic components, specific sequestration of cargo has also been described for both macro- and microautophagy. In the case of selective macroautophagy, the distinctive characteristic is the presence of a single type of cytosolic material in autophagosomes, in clear contrast to the “in bulk” process where heterogeneous cargo occupies the lumen of the autophagosome. Selective forms of macroautophagy are named depending on the sequestered material, giving rise to names such as mitophagy [32,33], pexophagy [34,35], reticulophagy [36,37], lipophagy [38], ribophagy [39] and aggrephagy [40]. Similar criteria apply to selective forms of microautophagy, such as micropexophagy [34,35] micronucleophagy [41,42] and microglycophagy [43]. All these selective forms of autophagy utilize the same essential components of the autophagic machinery described for their “in bulk” variants but they add an extra step related to cargo recognition. A specific subset of proteins known as cargo-recognition proteins, have been involved in this step and include, among others, p62, neighbour of BRCA1 (NBR1), Nix and the PINK/Parkin pair [44]. These adaptor proteins connect the degradation machinery with particular components – often ubiquitin moieties - on the surface of the cytosolic component to be sequestered [44-46]. Cytosolic chaperones, in particular the heat shock cognate protein of 70 kDa or hsc70 and BAG-3, have also been recently described to contribute to selective recognition of aggregates for macroautophagy in chaperone-assisted selective autophagy (CASA) [47].
Selectivity is the distinctive feature of CMA, the third type of mammalian autophagy, wherein particular soluble proteins bearing in their sequence a pentapeptide targeting motif biochemically related to KFERQ, are selectivity recognized by hsc70 that mediates their delivery to the lysosomes for direct translocation across the lysosomal membrane [8,48] (Fig. 1). Although yeast genetic approaches could not be applied to the molecular dissection of CMA, since this pathway has only been described in mammals so far, about 8 different proteins have been identified for this pathway by biochemical procedures. Most of the identified CMA components have promiscuous functions and are shared with other intracellular processes. Such is the case for the cytosolic chaperones hsc70 and hsp90 [49,50] and the novel pair of regulators the glial fibrillary acidic protein and the elongation factor 1α [51]. Interestingly, lysosome-specific variants have been identified for each of these proteins, suggesting that post-translational modifications of the cytosolic proteins determines their association with this degradative compartment and their commitment to CMA. So far, the only CMA-exclusive component is the lysosome-associated membrane protein type 2A (LAMP-2A) which acts as a receptor for substrates of this pathway [52]. Once the cytosolic proteins destined for CMA degradation bind to this single transmembrane protein, they promote its assembly into a higher - order molecular complex required for translocation of the CMA substrates into the lysosomal lumen [50].
Interested readers are referred to more focused reviews [2,3,8,29,53], for a comprehensive description of the molecular apparatus executing these different autophagic pathways. For the purposes of this review, the term ‘autophagy’ will represent macroautophagy, unless otherwise mentioned.
Physiological relevance of autophagy
As described in the introduction, a growing number of cellular functions have been attributed to autophagy. We highlight here some of the best characterized functions that have contributed to expanding the physiological relevance of this catabolic process. The role of autophagy in cellular defense by participating in innate and adaptive immunity will be discussed in detail in the section dedicated to autophagy and the immune function.
Autophagy in the cellular energetic balance
Both macroautophagy and CMA are induced as an acute adaptive response to a variety of metabolic stressors including, among others, nutrient starvation, growth factors withdrawal, high lipid content challenges or hypoxia [4,54-56]. Under these conditions, free amino acids (especially branched-chain amino acids) released by autophagic proteolysis of intracellular proteins and organelles, are recycled to maintain protein synthesis even when nutrients are scarce. Autophagy in the liver converts this organ into a main source of amino acids that are then delivered to other organs through the blood stream during starvation [5,55,57,58]. In fact, the low plasma concentrations of essential amino acids and decreased ATP levels in most tissues observed in autophagy defective Atg5−/− and Atg7−/− neonates confirm the importance of this pathway during nutritional deprivation, in this case as a result of the cease in transplacental nutrient supply at birth [59,60]. Amino acids resulting from autophagic breakdown could also be utilized for production of cellular ATP through direct oxidation or by fueling the TCA cycle and gluconeogenesis with intermediates such as oxaloacetate. Although this contribution of amino acids to energy homeostasis has been confirmed in specific conditions such as in the case of interleukin-3-deprived hematopoietic cells, where addition of TCA cycle substrates such as methylpyruvate is enough to preserve cellular viability [61], overall amino acids are not an efficient source of energy. However, the array of energy stores mobilized by autophagy has expanded towards more energetically efficient molecules such as lipids, glycogen and nucleic acids. The hydrolytic products of these molecules, free fatty acids, glucose and nucleotides can be funneled into, the TCA cycle, gluconeogenesis or glycolysis to produce ATP [4,43].
This capability of autophagy to maintain ATP production and support macromolecular synthesis makes it a pro-survival pathway of particular importance in organs with high energetic requirements such as, the heart or skeletal muscles. Alterations of this specific autophagic function also constitute the basis of some common metabolic disorders.
Autophagy and cellular quality control
The proteome and cellular organelles are susceptible to different toxic insults that can lead to the generation of misfolded or modified soluble proteins, protein cross-linking and oligomerization into high order irreversible structures or aggregates and accumulation of defective, no longer functional organelles, that could become harmful for the cells [62-64]. Basal autophagy – often referred to as quality control autophagy - is integral to the cellular surveillance machinery responsible for recognition and removal of these aberrant structures [65,66]. Basal autophagy is particularly important in non-dividing post-mitotic cells that cannot dilute the cellular damage through division. For example, conditional knock-out of the essential autophagy genes atg7 or atg5 in hepatocytes, neurons and cardiomyocytes leads to marked accumulation of ubiquitin-positive protein aggregates and damaged organelles, even in the absence of an added toxic challenge [59,67,68].
Exposure to stressors such as oxidative stress, ER stress or other conditions resulting in massive amounts of unfolded proteins and organelle damage elicits activation of inducible forms of autophagy [69-72]. In this context, activation of CMA contributes to the selective removal of soluble (aggregate-prone) proteins whereas macroautophagy facilitates clearance of protein aggregates [40,62,73,74] and whole organelles [33,36,37]. However, it is possible that macroautophagy ameliorates proteotoxicity not only through aggregate removal but also by the in-bulk sequestration of ‘still-soluble’ forms of the pathogenic proteins before they aggregate [67,75,76]. Autophagy is also important to restore organelle homeostasis [36,71,77] and eliminate damaged organelles after stress [37,78] and assist cells to adapt their organelle content to the changing environmental conditions (for example, it controls peroxisome number during nutrient auxotrophy or total mitochondria mass for energetic balance) [79].
Autophagy-mediated control of protein and organelle quality as well as organelle number is essential for the maintenance of cellular homeostasis and to guarantee cellular survival during stress.
Cellular remodeling and autophagy
Cellular differentiation often requires elimination of large subsets of proteins, nucleic acids or organelles in order to switch into the next developmental stage or final differentiated state. Furthermore, cellular remodeling is an energy demanding process often associated to nutrient-deprived developmental phases. It is no surprise therefore, that autophagy has emerged through evolution, as one of the favored cellular tools to accomplish these developmental remodeling tasks. The capability to eliminate whole regions of the cytosol, unique to some of the autophagy variants, and its recycling properties underlie the important role that this process plays during development. Autophagy has been shown to be necessary in yeast during sporulation [80], in C.elegans during dauer formation [24], in Drosophila during the transition from larval to pupal stage [81] and in Dictyostelium for switching from amoeba to fruiting body [25]. Examples of autophagy-mediated differentiation events in mammalian cells include among others, the removal of maternal macromolecules during early embryogenesis [82] and the clearance of mitochondria during erythrocyte [83-85], lymphocyte [86,87] and adipocyte [88,89] differentiation.
Autophagy in cellular death and cell survival
As indicated in the previous sections, the ability of autophagy to assist cells to adapt to the changing environment, to protect them against the damage caused by toxic insults and to defend them from pathogens has granted the classification of this pathway as a cell-survival mechanism. However, interplay between autophagy and mechanisms related to cell death have also been described. The best accepted example of autophagy as cell death effector is the degradation of the salivary glands during pupal transition in Drosophila [81,90]. In contrast, direct evidence of a similar role for autophagy in mammals is sparse [91]. In fact, many of the original claims of “cell death by autophagy” have been currently revised and reformulated as “cell death with autophagy” since the main support for autophagic involvement in many of these cases was the observation of autophagic vacuoles in the dying cell which often result from blockage in the autophagic flux [92]. However, as described in the following sections, once in a disease context, interactions between autophagy and apoptosis (or type I cell death) are widely contemplated (i.e. tumorigenesis, cardiomyopathies, metabolic failure of the pancreatic beta-cell etc.) [93-95]. In fact, autophagy and apoptosis share some molecular components and appear to be in a delicate balance with each other in most systems examined [96,97].
Pathology of Autophagy
The broad array of physiological functions attributed to autophagy justifies why alterations in this catabolic process lead to cellular malfunctioning and often cell death, and has set the basis for its contribution to the pathogenesis of severe human disorders. Rather than providing an exhaustive list of disease conditions in which autophagy has been shown to be altered, in the following sections, we have selected four common pathological conditions – cancer, neurodegeneration, heart dysfunction and immune and infectious diseases – to comment on their interplay with the autophagic system. The reason behind this selection is the fact that in all of these pathologies growing evidence supports a dual role of autophagy in their pathogenesis and disease progression, which has led often to wonder: “should we inhibit or activate autophagy in these pathologies?”.
Autophagy and cancer
Cancer was the first human pathology connected to autophagy through the discovery that Beclin1 (Atg6/VPS30 homolog), a core component of the autophagosome nucleation complex was monoallelically deleted in 40-75% of sporadic human breast, ovarian and prostate cancers [98]. Independent studies verified that heterozygous Beclin1+/− mice develop spontaneous tumors including lymphomas, lung carcinomas, hepatocellular carcinomas and mammary precancerous lesions [99,100]. Analogous generation of spontaneous hyper-proliferating tumors was also reported later, upon deletion of other autophagy related genes such as the ultraviolet resistance associated gene (UVRAG), the Bax interacting factor-1 (Bif-1) and the LC3 (Atg8 homolog) processing protease Atg4c [101-103]. Furthermore, common oncogenes like class I PI3K, PKB, TOR, Bcl-2 have been shown to act as autophagy repressors whereas tumor suppressor genes such as p53, PTEN, DAPk, TSC1/TSC2 exert a stimulatory effect on autophagy [3,97,104]. Although in light of this opposite effect of oncogenes and tumor suppressors on autophagy, this process was initially classified as an anti-oncogenic mechanism, this conclusion has been challenged by experimental evidences supporting that under certain conditions autophagy can also be pro-oncogenic (Fig. 2).
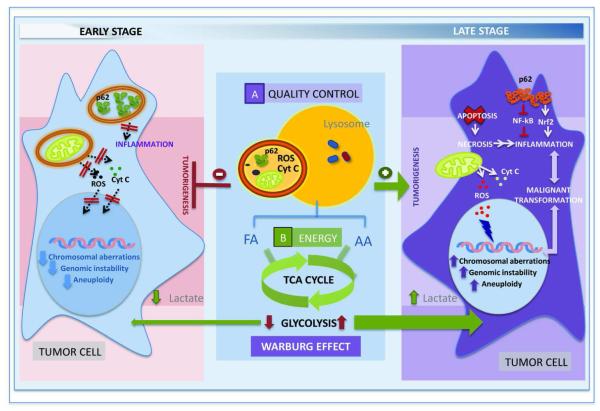
Figure 2. Autophagy and Cancer.
Autophagy may play opposite roles in the oncogenic process. Anti-tumoral effect (left): Active maintenance of cellular quality control for cytosolic pro-oncogenic proteins such as p62 prevents malignant transformation of non-tumoral cells. In addition, the supply of energy provided through macroautophagy activation, reduces the dependence on glycolysis while assuring the energy required for maintenance of a stable genome, further preventing oncogenesis. Pro-oncogenic effect (right): The reduction in macroautophagic activity in early stages of the oncogenic process favors malignant transformation as the accumulation of molecules such as p62 activates signaling mechanisms that promote necrosis and inflammation. Poor quality control as a result of diminished macroautophagy can also result in accumulation of defective mitochondria with the subsequent release of harmful molecules (cytochrome C and reactive oxygen species) that contribute to further alter genome maintenance. However, as the tumor progresses, activation of macroautophagy is observed in many oncogenic process in part to compensate for the poor nutritional supply associated to rapidly growing tumors and defend cancer cells against damage induced by anti-oncogenic therapies. In addition., enhanced mitochondrial degradation in this stage may contribute to the upregulation of glycolysis to maintain the energetic balance (Warburg effect) characteristic of malignant cells.
Thus, it has been proposed that in the early stages of cancer development, quality control by autophagy, particularly over genome maintenance, inhibits tumorigenesis conferring this pathway with anti-oncogenic functions. In addition, autophagy plays a role in the maintenance or entry of cells into the G0 phase, and consequently, proper autophagy prevents spontaneous hyper-proliferation of cells [105,106]. In contrast, in the late stages of oncogenesis, autophagy is necessary for cancer survival, as it contributes both energy for the rapidly dividing cancer cells and quality control functions to eliminate intracellular damage caused by the aggressive tumor microenvironment and by anti-oncogenic interventions (Fig. 2). In fact, consistent with the pro-survival function of autophagy, compromise of this pathway (i.e. Beclin 1 interference) in certain tumor cells impairs their survival in metabolic stress conditions in vitro and in vivo [107]. In general, blockage of autophagy sensitizes cells to the metabolic stress leading often to necrotic cell death accompanied by inflammation [108-110]. Metabolic stress is intrinsic to rapidly growing tumors in which poor vascularization results in lack of nutrients and oxygen for long periods of time [111,112]. Activation of macroautophagy under these conditions provides cancer cells with amino acids, free fatty acids and glucose required to generate energy through the TCA cycle and via β-oxidation (Fig. 2). Macroautophagy has also been recently shown to facilitate the unique utilization of glucose by cancer cells – known as the “Warburg effect” [113,114] by which cells favor anaerobic glycolysis to accelerate ATP production and generate glycolysis intermediates required for the transformed cells [115,116]. This effect is, in part, obtained through activation of the removal of mitochondria via autophagy (mitophagy) to force energy dependence on glycolysis [115,116] (Fig. 2). Blockage of autophagy would interrupt anaerobic glycolysis eliminating the energetic advantage of the cancer cells. Furthermore, reduction in cellular energy upon autophagic compromise reduces fidelity of cellular processes like DNA replication and mitosis resulting in genetic aberrations [117]. The occurrence of these aberrations in tumor cells with defective quality control of organelles and toxic proteins due to the autophagic compromise can precipitate death of the cancer cells. In this respect, accumulation of the protein p62 (normally degraded by macroautophagy) when this pathway is compromised, leads to formation of p62 aggregates capable of stimulating inflammation by inhibition of the nuclear factor-κB (NF-κB) and activation of NF-E2 related factor 2 (Nrf-2) [118-120] (Fig. 2).
Little is known till date about the contribution of other forms of autophagy (CMA and microautophagy) to cancer biology. Recent studies have revealed that CMA contributes to the turnover of inactive forms of the M2 isoform of pyruvate kinase, a key enzyme in the maintenance of anaerobic glycolysis in cancer cells [121]. A number of additional glycolytic enzymes have been recognized as CMA substrates including, glyderaldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase, aspartate aminotransferase and aldolase B, strengthening the possible link between CMA and the energetic balance of cancer cells [48,122].
Collectively, the relationship between autophagy and cancer has proved to be a very complex one. This dual capability of autophagy to facilitate tumor progression and to diminish malignant transformation has reinforced the need to critically evaluate autophagy in a disease context- and stage-specific manner, especially during the design of targeted therapy, as autophagy function is tightly related to these factors [77,93].
Autophagy and Neurodegeneration
Maintenance of cellular homeostasis is essential in neurons, a typical example of non-dividing differentiated cells. The importance of autophagy in neuronal homeostasis has now been well supported, despite original disagreement on whether autophagy was active in neurons. The reason for this contention was the fact that autophagosomes, the morphological feature of macroautophagy, were rarely observed in neurons. However, recent studies support that this pathway is constitutively active and that it can be further upregulated in neurons in response to very different stressors [123-125]. Perhaps, the most conclusive support to the existence of basal autophagy in neurons and of its contribution to maintenance of neuronal survival came from the studies in transgenic mouse models conditionally knocked out for essential autophagy genes in central nervous system [67,68]. These animals, even in the absence of any added stressor, displayed marked neurodegeneration associated with accumulation of intracellular protein inclusions and neuronal loss. The efficient capability of the lysosomal system to remove newly formed autophagosomes could be behind the low occurrence of detectable autophagosomes in neurons in a given moment.
As with many other cells, neurons upregulate autophagy in response to common stressors, but also in defense against neuron-specific injuries such as axotomy, ischemia or excitory toxicity [126-128] (Fig. 3). Often, failure to upregulate autophagy under these conditions or primary compromise of autophagic activity during the stress, precipitates neuronal death post-injury [129]. However, activation of autophagy in neurons may have a relatively narrow window of opportunity because recent reports have revealed that genetic or chemical inhibition of autophagy becomes beneficial at some specific states post-injury [104]. These findings suggest that either excessive upregulation of autophagy or upregulation of this pathway under conditions in which autophagosome formation or clearance cannot be guaranteed may be harmful to neurons [130]. In further support of the possible detrimental effect of massive upregulation of autophagy, recent studies have shown that autophagosomes under specific conditions can become a source of reactive oxygen species and thus aggravate neurotoxicity [131].
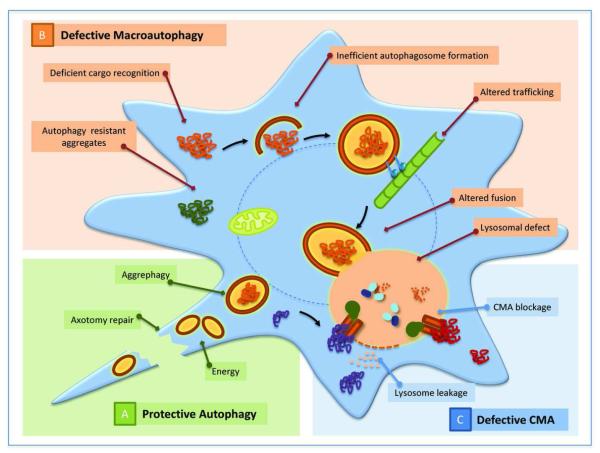
Figure 3. Autophagy and Neurodegeneration.
A. Protective autophagy: Macroautohagy Macroautophagy and CMA both contribute to maintenance of neuronal homeostasis and are necessary in the defense of neurons against injury and stressors. B. Defective macroautophagy: Defects in macroautophagy have been described to occur at very different levels in neurodegenerative diseases. Some of the possible steps affected in this process are highlighted in the model and the conditions in which they have been observed are described in the text. C. Defective CMA: Primary defects in CMA have been described both in Parkinson’s disease and in certain tauopathies. While in the former condition, pathogenic proteins such as α-synuclein can block access of other cytosolic proteins to lysosomes via CMA by abnormally binding to the translocation machinery, in tauopathies the accumulation at the surface of lysosomes of oligomeric forms of pathogenic tau targeted via CMA destabilizes the lysosomal membrane and results in leakage of lysosomal enzymes in the cytosol which often triggers cellular death.
Changes in autophagic activity have been described in many protein conformational disorders and among them neurodegenerative diseases have received particular attention. The feature shared by all these conditions is the presence of pathogenic proteins that accumulate inside neurons organized in the form of oligomeric or multimeric structures. The disease originates from both, the loss of function of the pathogenic protein as well as the toxicity associated with the presence of the abnormal protein structures that often leads to neuronal death. The first connection between neurodegeneration and autophagy originated from the observation that protein aggregates can be eliminated by autophagy [40,73] (Fig. 3). In fact, pharmacological activation of this catabolic process in experimental animal models of Huntington’s disease, reduced cellular toxicity and slowed down progression of the disease [132]. After these initial reports, the degradation of many other pathogenic proteins by autophagy and the beneficial effect of upregulation of this pathway have proven true in experimental models of many other neurodegenerative conditions [133]. In fact, not only the acute pharmacological activation of autophagy but even chronic conditions that lead to a progressive maintained upregulation of this process, such as in the maintained ER stress, have been shown to have a positive effect [134]. These findings have now opened the possibility of utilizing modulators of autophagy as the basis for possible therapeutic approaches for these types of pathologies, and in fact, ongoing chemical screenings aim at identifying chemical modulators of this pathway with higher selectivity and potency [135].
However, there are certain limitations to the universal use of upregulation of macroautophagy with anti-neurodegenerative purposes. On one side, it has been reported that not all protein-aggregates are recognized by the macroautophagic machinery. Despite the current dissection of molecules that participate in autophagic recognition of aggregates (p62, NBR1 and several cytosolic chaperones including hsc70 and BAG-3), the presence of these proteins in an aggregate, although necessary, does not seem to be sufficient. for recognition. Thus, some protein inclusions positive for these cargo-recognition proteins remain unnoticed by the autophagic machinery, making superfluous any attempt to enhance autophagosome formation as a way to eliminate these toxic protein products [136]. Further studies are needed to address whether expression of other cellular components or specific post-translational modifications in the aggregate proteins could enhance their recognition by the autophagic systems.
A second scenario in which chemical upregulation of macroautophagy may not be effective against neurodegeneration is in those conditions in which macroautophagy itself is compromised. Understanding the macroautophagy step or steps defective in specific neurodegenerative disorders is a priority if autophagy enhancers are to be moved forward in the treatment of these conditions. Examples of defects in almost each of the steps of macroautophagy have been described in different neurodegenerative disorders [65] (Fig. 3). For example, initiation of autophagy may be compromised because of altered signaling through the insulin or mTOR pathways, which is tightly bound to activation of autophagy. Also, conditions resulting in lower content of available Atgs will reduce neuronal autophagic capability, such as the recently described depletion of LC3-II through abnormal interaction and aggregation with p62 in the presence of dopaminergic neurotoxin [137]. Along the same lines, abnormal interaction between mutant α-synuclein (the protein that accumulates in protein inclusions in Parkinson’s disease), with Rab1 has been shown to mislocalize Atg9, a protein required for the formation of autophagosomes out of the endoplasmic reticulum membrane [138]. In other conditions, problems arise at the level of cargo recognition either because of alterations in the organelle-specific markers for degradation or in the autophagic machinery. A primary defect at the level of the limiting membrane that seems to prevent selective recognition of cargo has been recently described in cellular and animal models of Huntington’s disease [139]. In these cases, mutant huntingtin binds tightly to the inner surface of the forming autophagosome resulting in an abnormally high interaction with p62 in this compartment that affects its ability to bind cargo. In other conditions autophagosomes form and sequester the pertinent cargo but they fail to be cleared from the cytosol. Problems with clearance could result from alterations at very different levels. For example, problems with vesicular trafficking could indirectly interfere with the mobilization of autophagosomes toward the lysosomal compartment [140]. Pathogenic proteins can also interfere with the fusion step, which although still not completely elucidated at the molecular level, is known to depend on different SNARE proteins, the actin cytoskeleton and the histone deacetylase C 6 (HDAC6) [141]. Consequently, changes in any of these components could lead to intracellular accumulation of autophagosomes. Lastly, primary defects in the lysosomal compartment also have a negative impact in clearance of autophagosomes in different neurological disorders. The reasons for lysosomal failure could be multiple. In principle, most lysosomal storage disorders could compromise autophagosome clearance because often the accumulation of the undegraded product inside lysosomes limits their degradative capacity [142]. In addition, other conditions that alter lysosomal membrane stability, decrease lysosomal biogenesis or change lysosomal pH could also alter autophagosome clearance. In fact, defective acidification of lysosomes because of compromised targeting of a component of the proton-pump from the ER to the lysosomal membrane has been already identified as causative of the reduced rates of autophagy in some models of Alzheimer’s disease [143]. As functional analysis of autophagy becomes more broadly utilized in the study of neurodegenerative diseases, it is likely that new connections between these pathologies and autophagy will become evident.
Alterations of CMA have also been described in some neurodegenerative disorders (Fig. 3). Although in many of them, this pathway becomes upregulated as a consequence of the failure in macroautophagy, different pathogenic proteins have been shown to directly interfere with CMA activity [144]. Mutant forms of α-synuclein can be targeted to lysosomes via CMA but, in contrast to the wild-type protein that binds to the lysosomal receptor and rapidly reaches the lumen for degradation, the mutant variants fail to translocate. Persistance of mutant synuclein tightly bound to the CMA receptor at the lysosomal membrane, inhibits lysosomal degradation of other cytosolic proteins via this pathway [145]. A similar effect was later described for wild-type synuclein upon modification by dopamine, which makes CMA blockage relevant for sporadic forms of Parkinson’s disease, more common than the familial forms [146]. Later studies have also proposed interference of CMA by other Parkinson’s disease-related proteins such as mutant forms of UCH-L1 [147]. Higher degree of lysosomal compromise as a result of lysosomal targeting of pathogenic proteins via CMA has been described in certain tauopathies, where the mutant forms of the protein not only interact abnormally with CMA components blocking this pathway, but they also disrupt the lysosomal membrane as they organize into oligomeric toxic species on their surface [148].
To date, systematic studies analyzing the contribution of alterations in microautophagy to neurodegeneration have not been performed. However, the fact that protein unfolding is not a prerequisite for microautophagy makes the possibility of the delivery of micro-aggregates to this pathway for degradation an attractive one.
Autophagy and the failing heart
The heart is comprised of long-lived, post-mitotic cells with little regenerative capacity, which are continually subject to stress such as ischemia, pressure overload and ischemia-reperfusion injury. Adaptation to these conditions is attained through remodeling (myocytes elongate and undergo hypertrophy) and failure to do so frequently constitutes the basis of coronary artery disease, hypertension and congestive heart failure [149,150]. Evidence for the role of basal autophagy in quality control and housekeeping of cardiomyocytes was first obtained through genetic models of LAMP-2 knockout mice (defective for autophagosome-lysosome fusion) that demonstrated cardiomyopathy and abnormal accumulation of autophagic vacuoles, similar to that observed in Danon disease patients [151,152]. In fact, mutations in the LAMP-2 gene were subsequently described in these patients [153]. Studies in cardiomyocyte-specific Atg5 and Atg7 knockout models have reiterated the need for functional autophagy to preserve normal cardiac function both under basal conditions and in response to stress load [154]. In fact, pronounced loss of autophagy in cardiomyocytes with age seems behind different forms of age-related cardiomyopathies [155].
Added to the key role of basal macroautophagy in cardiomyocyte homeostasis, growing evidence also supports an important contribution of this pathway in heart in the response to stressors. As described in the brain, macroautophagy has also proven essential for prevention of cardiomyocyte proteinopathies such as those arising from desmin and α-β-crystallin accumulation [156,157]. Autophagy induced in response to cardiac stress (especially during ischemia/reperfusion injury) also plays a cytoprotective role in the heart. In fact, in support of this prosurvival function, different studies have demonstrated that pharmacological inhibition of autophagy during mild ischemic stress enhances cardiomyocyte death [158,159] whereas macroautophagy activation is cardioprotective [160,161]. However, induction of macroautophagy in the stressed heart can also be detrimental and contribute to heart failure judging by the fact that pharmacological or genetic blockage of macroautophagy enhances cell survival post ischemia/reperfusion injury in specific settings [162]. Timing and the condition that induced cardiac stress may be essential for the switch in the effect of macroautophagy. Thus, some studies suggest that macroautophagy is cardioprotective during ischemic recovery, but maladaptive during reperfusion recovery [163]. It is possible that during reperfusion the need for removal of dysfunctional mitochondria and oxidatively damaged cellular structures that accumulated during the ischemic period, could drive the autophagic response to be more aggressive than desired, resulting in cell death. In fact, genetic upregulation of autophagy in response to the hemodynamic stress-induced hypertrophic growth response has also proven to be maladaptive and to result in pathological cardiac hypertrophy [164].
In summary, as in the case of tumor biology and neurodegeneration, the role of autophagy in heart pathology is also context-dependent and excessive or insufficient macroautophagy is often associated to disease. However, when maintained under strict control, upregulation of macroautophagy by compounds such as resveratrol has proven to be a useful cardioprotective strategy during ischemia/reperfusion injury [165-167].
Although the heart is one of the organs where CMA is upregulated most rapidly in response to starvation [168], the contribution of CMA to cardiomyocyte homeostasis and to heart pathology has not been explored in depth. As mentioned above, mutations in LAMP-2 occur in patients with Danon cardiomyopathy, but the phenotype seems for the most part, to be related to the presence of large autophagic vacuoles in muscle reflective of compromised macroautophagy. This could be explained by the fact that the lamp2 gene undergoes splicing giving rise to three protein variants A, B and C [169] and that mutations in only the B variant, tightly related to macroautophagy, are enough to reproduce the full vacuolar phenotype [153], whereas changes in LAMP-2A, the variant required for CMA, do not affect macroautophagy [170].
Autophagy, infectious disorders and autoimmunity
The autophagy machinery can act as a cell-autonomous defense against invading pathogens through a process known as xenophagy [171]. Activation of this process has been extensively reported for example in response to Group A streptococcus infection in epithelial cells (Fig. 4). The fact that Atg5 deficiency allows bacteria to survive and multiply robustly, supports the normal contribution of autophagy in the elimination of this pathogen [172]. Autophagic surveillance is not limited to bacteria as in fact, a protective role for autophagy has been also demonstrated for the vesicular stomatitis virus, herpes simplex virus 1 or HIV-1 among others [173-175].
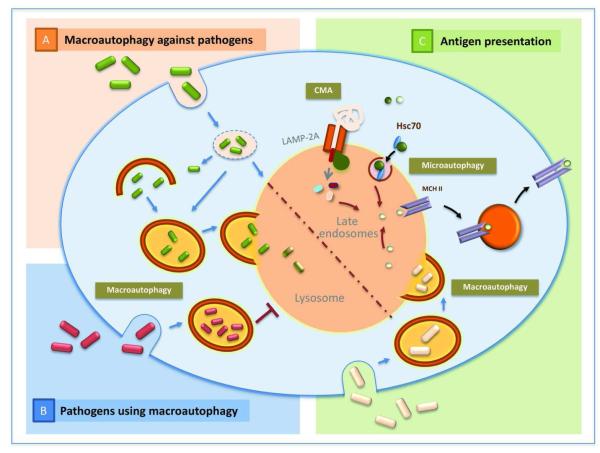
Figure 4. Autophagy and the immune system.
A. Macroautophagy against pathogens: Macroautophagy contributes to the elimination of different types of pathogens – bacteria and viruses – when they escape to the cytosol after internalization in the phagosome. In certain conditions fusion of autophagosomes with phagosomes is required before degradation can occur. B. Pathogens using macroautophagy: Growing evidence supports that certain pathogens have evolved to utilize autophagosomes as a site of replication and can actively prevent the fusion of this compartment with lysosomes to guarantee their survival. C. Antigen presentation: All three forms of autophagy, macroautophagy, CMA and microautophagy have been shown to contribute to antigen loading of MCH class II molecules for presentation of antigens to activate T cells.
Cytosolic autophagy often becomes a second surveillance point for pathogens such as L. monocytogenes that can escape phagosomes by puncturing their membrane to replicate in the cytoplasm. However, once in the cytoplasm, the autophagic surveillance mechanism is activated to entrap the escaped bacteria and deliver them to endocytic and lysosomal compartments for degradation [176]. Pathogens that survive inside vesicular compartments can also be controlled by macroautophagy. For example, M. tuberculosis resides in phagosomes by avoiding their fusion to lysosomes. Interestingly, this blockage can be overcome by induction of autophagy, leading to the degradation of this pathogen [177].
Despite this active involvement of autophagy in pathogen elimination, some pathogens have evolved to take advantage of the different compartments of the autophagy pathway to establish replicative niches [178,179] (Fig. 4). Evidence that the autophagosome compartment is required for the replication of certain pathogens such as P. gingivalis or C. burnetti was provided by demonstrating that treatment with 3-methyladenine, a well-characterized inhibitor of autophagosome formation, was efficient in decreasing the survival of these pathogens [180,181]. In fact, some of these microorganisms that utilize compartments of the autophagic system have developed mechanisms to expand the size and number of these compartments. For example, S. aureus is capable of secreting a factor which activates autophagy [182]. Seclusion inside the autophagosomes may be beneficial for pathogens as a way to escape the cytosolic surveillance mechanisms, but also these compartments may contain cytosolic materials that can be utilized as an energy source by the microorganisms.
This bivalent role of autophagy in pathogen defense suggests that although it should be possible to pharmacologically manipulate the autophagic system to eliminate different pathogens, it is essential to first understand the characteristics of the pathogen-autophagy interaction in each of the individual instances [183,184].
Apart from its role in cell-autonomous immunity, autophagy plays a role in activation of adaptive immunity through its recently described involvement in antigen processing and presentation to lymphocytes [185-187] (Fig. 4). In professional APCs, phagosome-processed extracellular antigens are presented through MHC class II molecules to CD4 T cells. However, several cytosolic and nuclear proteins have been found associated to MHC II molecules [188,189]. In fact, pharmacological inhibition of macroautophagy has been shown to be efficient in reducing MHC class II intracellular antigen presentation [190-192] whereas activation of autophagy promotes this type of presentation [193]. Later studies have confirmed the presence of intracellular antigens in autophagosomes, and marked reduction of MHC II presentation in lymphoblastoid cell lines upon knock-down of essential Atgs as well as in vivo in Atg5-deficient dendritic cells [194,195]. Interestingly, presentation of self-antigens by class II molecules is not limited to macroautophagy but it also may involve other types of autophagy. In fact, overexpression of LAMP-2A, the receptor for chaperone-mediated autophagy, has been shown to enhance MCH II presentation of intracellular antigens [196], and a new microautophagy-dependent process of antigen delivery into late endosomes in dendritic cells has also been recently characterized [31]. Furthermore, participation of autophagy in antigen presentation may not be limited to presentation through MCH II. Although it is traditionally accepted that MHC class I presents intracellular antigens processed through the proteasomal pathway, growing evidence supports that macroautophagy could also assist in MHC I presentation by collaborating with the proteasome in the processing of intracellular proteins [197,198].
Also of interest is the recently proposed contribution of autophagy in the establishment of tolerance to self antigens. Self-reactive T-cells are usually actively eliminated through MCH class II presentation in epithelial cells of the thymus to allow T-cell tolerance to self antigens [199]. Proof that autophagy regulates central T cell tolerance has been provided by a recent study showing that transplantation into athymic nude hosts of Atg5-defienct thymus results in defects in positive and negative selection and the development of autoimmunity [200]. Interestingly, the antigen-loading compartments for MHC class II in immature dendritic cells, which are involved in peripheral tolerance, continuously receive input from autophagosomes [201]. Furthermore, genetic polymorphisms in two different autophagy genes (ATG16L1 and IRGM) have been linked to Crohn’s disease [202,203]. Compromised autophagic function may result in insufficient induction of tolerance against commensals or self-antigens in the gut, [202,203], although it may also affect the secretion of antimicrobial proteins by Paneth cells [7].
This novel role of autophagy in the establishment and maintenance of tolerance also suggests a possible role for autophagy in autoimmunity [204]. In this respect, autophagy is known to contribute to apoptotic-cell clearance [205], and defective clearance of apoptotic cells has been proposed to lead to autoimmune diseases, such as systemic lupus erythematosus [206].
As any other cell type, cells of the immune system also depend on autophagy for the maintenance of their homeostasis [207], but changes in autophagy in T cells in paticular have also been described to contribute to modulation of specific cell functions. Thus, the activation of macroautophagy observed upon T cell activation is required to promote T cell differentiation and survival and allows cells to accommodate the bioenergetic requirements of these conditions [208-210].
Overall, recent studies support that autophagy defends the cell in a bimodal fashion; first, it directly eliminates invading pathogens [211] and second, it simultaneously assists the immune system of the host organism to mount a specialized immune response against the invader by processing pathogenic antigens for presentation to T cells [6,171,212]. Therefore autophagy can be envisaged as an immediate as well as a persistent mechanism of cellular defense.
Conclusions and future perspectives
The better molecular characterizations of the different autophagic pathways as well as the possibility to genetically manipulate these cellular processes have helped establish tight connections between autophagy malfunctioning and disease. The initial excitement about the possibility of modulating autophagy with therapeutic purposes in different disorders has been followed by some degree of confusion as to whether autophagy should be upregulated or down regulated in these conditions. As described in this review, both upregulation and downregulation of macroautophagy have been shown to be protective against cancer, neurodegeneration, infectious diseases and ischemic insult in heart. How to decide what to do? Further analysis in most of these conditions is required to fully understand the contribution of autophagy malfunctioning to the disease. Questions such as when in the course of the disease autophagic function becomes compromised, what changes in each autophagic pathway and whether these changes are primary or compensatory to failure in other systems need to be answered before autophagy modulators can be systematically used in the treatment of most of these conditions.
A considerable advance in this respect has been the introduction of functional read outs of autophagy to complement the initial morphological analysis. For example, the presence of a higher number of autophagosomes in many disease conditions, initially interpreted as an increase in macroautophagy, is now more cautiously analyzed, because blockage in the late steps of this pathway can also have a similar morphological signature. Thus, conditions initially labeled as having “too much autophagy” are being currently revised as having “a blockage in autophagic clearance”. Blocking autophagosome formation may only be beneficial then, when excessive autophagosome content and autophagic clogging contribute to the pathology, but not when autophagy is upregulated in the disease to compensate for failure in other systems. Along the same lines, the ultimate goal may not be to repress autophagosome formation in some of these cases, but to instead facilitate their clearance by directly repairing possible defects in the late steps of the autophagic process.
In addition to determining the right timing for modulating autophagy in many of these diseases, and whether this process should be upregulated or downregulated, further expansion of the chemical options to manipulate autophagy is needed for it to become a routine therapeutic target. To date, most of the macroautophagy inhibitors used in clinical trials act at the late steps of this process, on the lysosomal compartment, which is also shared by other autophagic pathways. In contrast, for activators, most of the available drugs have an effect on initiation and autophagosome formation, but few compounds have been shown to be effective at increasing the overall autophagic flux. Ongoing chemical screenings are currently addressing the need for these types of modulators and should render usable molecules in a timely fashion [135].
Acknowledgements
We thank Ms. Samantha J. Orenstein and Dr. Susmita Kaushik for critically reviewing the manuscript. Work in our laboratories is supported by grants from the National Institutes of Health/National Institute on Aging AG021904 (to A.M.C.) and AG031782 (to A.M.C. and F.M.) and National Institute of Allergy and Infectious Diseases AI059738 (to F.M.). AMC is also supported by a Hirschl/Weill-Caulier Career Scientist Award.
Footnotes
Conflict of interest: Authors declare no conflict of interest
References
- 1.De Duve C, Wattiaux R. Functions of lysosomes. [Review] Ann Rev Physiol. 1966;28:435–492. doi: 10.1146/annurev.ph.28.030166.002251. [DOI] [PubMed] [Google Scholar]
- 2.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nature cell biology. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizushima N, Levine B, Cuervo AM, et al. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell metabolism. 2011;13:495–504. doi: 10.1016/j.cmet.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizushima N. The pleiotropic role of autophagy: from protein metabolism to bactericide. Cell death and differentiation. 2005;12(Suppl 2):1535–1541. doi: 10.1038/sj.cdd.4401728. [DOI] [PubMed] [Google Scholar]
- 6.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cadwell K, Liu JY, Brown SL, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cuervo AM. Chaperone-mediated autophagy: selectivity pays off. Trends Endocrin Met. 2010;21:142–150. doi: 10.1016/j.tem.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirisako T, Baba M, Ishihara N, et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. Journal of Cell Biology. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanida I. Autophagosome formation and molecular mechanism of autophagy. Antioxidants & redox signaling. 2011;14:2201–2214. doi: 10.1089/ars.2010.3482. [DOI] [PubMed] [Google Scholar]
- 11.Mortimore GE, Lardeux BR, Adams CE. Regulation of microautophagy and basal protein turnover in rat liver. Effects of short-term starvation. The Journal of biological chemistry. 1988;263:2506–2512. [PubMed] [Google Scholar]
- 12.Mayer A. Cell-free reconstitution of microautophagy in yeast. Methods in enzymology. 2008;451:151–162. doi: 10.1016/S0076-6879(08)03211-4. [DOI] [PubMed] [Google Scholar]
- 13.Klionsky DJ, Cregg JM, Dunn WA, Jr., et al. A unified nomenclature for yeast autophagy-related genes. Developmental cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 14.Axe EL, Walker SA, Manifava M, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hailey DW, Rambold AS, Satpute-Krishnan P, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ravikumar B, Moreau K, Jahreiss L, et al. Plasma membrane contributes to the formation of preautophagosomal structures. Nat Cell Biol. 2010;12:747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang R, Zeh HJ, Lotze MT, et al. The Beclin 1 network regulates autophagy and apoptosis. Cell death and differentiation. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizushima N, Yamamoto A, Hatano M, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. The Journal of cell biology. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. Journal of Biological Chemistry. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 20.Hosokawa N, Hara T, Kaizuka T, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganley IG, Lam du H, Wang J, et al. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Settembre C, Di Malta C, Polito VA, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011 doi: 10.1126/science.1204592. DOI: 10.1126/science.1204592 Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Developmental cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 24.Melendez A, Talloczy Z, Seaman M, et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 25.Otto GP, Wu MY, Kazgan N, et al. Macroautophagy is required for multicellular development of the social amoeba Dictyostelium discoideum. The Journal of biological chemistry. 2003;278:17636–17645. doi: 10.1074/jbc.M212467200. [DOI] [PubMed] [Google Scholar]
- 26.Yoshimoto K, Hanaoka H, Sato S, et al. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. The Plant cell. 2004;16:2967–2983. doi: 10.1105/tpc.104.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alvarez VE, Kosec G, Sant’Anna C, et al. Autophagy is involved in nutritional stress response and differentiation in Trypanosoma cruzi. The Journal of biological chemistry. 2008;283:3454–3464. doi: 10.1074/jbc.M708474200. [DOI] [PubMed] [Google Scholar]
- 28.Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764–776. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mijaljica D, Prescott M, Devenish RJ. Microautophagy in mammalian cells: Revisiting a 40-year-old conundrum. Autophagy. 2011;7:673–682. doi: 10.4161/auto.7.7.14733. [DOI] [PubMed] [Google Scholar]
- 30.Tuttle DL, Lewin AS, Dunn WA., Jr Selective autophagy of peroxisomes in methylotrophic yeasts. European Journal of Cell Biology. 1993;60:283–290. [PubMed] [Google Scholar]
- 31.Sahu R, Kaushik S, Cannizzo E, et al. Microautohagy of cytosolic proteins by late endosomes. Develop Cell. 2011;20:131–139. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldman SJ, Taylor R, Zhang Y, et al. Autophagy and the degradation of mitochondria. Mitochondrion. 2010;10:309–315. doi: 10.1016/j.mito.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tolkovsky AM. Mitophagy. Biochimica et biophysica acta. 2009;1793:1508–1515. doi: 10.1016/j.bbamcr.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 34.Farre JC, Krick R, Subramani S, et al. Turnover of organelles by autophagy in yeast. Current opinion in cell biology. 2009;21:522–530. doi: 10.1016/j.ceb.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veenhuis M, Salomons FA, Van Der Klei IJ. Peroxisome biogenesis and degradation in yeast: a structure/function analysis. Microscopy research and technique. 2000;51:584–600. doi: 10.1002/1097-0029(20001215)51:6<584::AID-JEMT8>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 36.Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS biology. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamasaki M, Noda T, Baba M, et al. Starvation triggers the delivery of the endoplasmic reticulum to the vacuole via autophagy in yeast. Traffic. 2005;6:56–65. doi: 10.1111/j.1600-0854.2004.00245.x. [DOI] [PubMed] [Google Scholar]
- 38.Singh R, Kaushik S, Wang Y, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kraft C, Deplazes A, Sohrmann M, et al. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 40.Yamamoto A, Simonsen A. The elimination of accumulated and aggregated proteins: a role for aggrephagy in neurodegeneration. Neurobiology of disease. 2011;43:17–28. doi: 10.1016/j.nbd.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krick R, Muhe Y, Prick T, et al. Piecemeal microautophagy of the nucleus: genetic and morphological traits. Autophagy. 2009;5:270–272. doi: 10.4161/auto.5.2.7639. [DOI] [PubMed] [Google Scholar]
- 42.Roberts P, Moshitch-Moshkovitz S, Kvam E, et al. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Molecular biology of the cell. 2003;14:129–141. doi: 10.1091/mbc.E02-08-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kotoulas OB, Kalamidas SA, Kondomerkos DJ. Glycogen autophagy in glucose homeostasis. Pathology, research and practice. 2006;202:631–638. doi: 10.1016/j.prp.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 44.Lamark T, Kirkin V, Dikic I, et al. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell cycle. 2009;8:1986–1990. doi: 10.4161/cc.8.13.8892. [DOI] [PubMed] [Google Scholar]
- 45.Kirkin V, Lamark T, Johansen T, et al. NBR1 cooperates with p62 in selective autophagy of ubiquitinated targets. Autophagy. 2009;5:732–733. doi: 10.4161/auto.5.5.8566. [DOI] [PubMed] [Google Scholar]
- 46.Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. The Journal of biological chemistry. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 47.Arndt V, Dick N, Tawo R, et al. Chaperone-assisted selective autophagy is essential for muscle maintenance. Current biology : CB. 2010;20:143–148. doi: 10.1016/j.cub.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 48.Kaushik S, Bandyopadhyay U, Sridhar S, et al. Chaperone-mediated autophagy at a glance. Journal of cell science. 2011;124:495–499. doi: 10.1242/jcs.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Agarraberes F, Terlecky S, Dice J. An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. J Cell Biol. 1997;137:825–834. doi: 10.1083/jcb.137.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bandyopadhyay U, Kaushik S, Varticovski L, et al. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol. 2008;28:5747–5763. doi: 10.1128/MCB.02070-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bandyopadhyay U, Sridhar S, Kaushik S, et al. Identification of regulators of chaperone-nediated autophagy. Molecular cell. 2010;39:535–547. doi: 10.1016/j.molcel.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 53.Orenstein SJ, Cuervo AM. Chaperone-mediated autophagy: molecular mechanisms and physiological relevance. Seminars in cell & developmental biology. 2010;21:719–726. doi: 10.1016/j.semcdb.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mizushima N, Yamamoto A, Matsui M, et al. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Molecular biology of the cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harris RA, Goodwin GW, Paxton R, et al. Nutritional and hormonal regulation of the activity state of hepatic branched-chain alpha-keto acid dehydrogenase complex. Annals of the New York Academy of Sciences. 1989;573:306–313. doi: 10.1111/j.1749-6632.1989.tb15007.x. [DOI] [PubMed] [Google Scholar]
- 56.Pursiheimo JP, Rantanen K, Heikkinen PT, et al. Hypoxia-activated autophagy accelerates degradation of SQSTM1/p62. Oncogene. 2009;28:334–344. doi: 10.1038/onc.2008.392. [DOI] [PubMed] [Google Scholar]
- 57.Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annual review of nutrition. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 58.Moriwaki H, Miwa Y, Tajika M, et al. Branched-chain amino acids as a protein- and energy-source in liver cirrhosis. Biochemical and biophysical research communications. 2004;313:405–409. doi: 10.1016/j.bbrc.2003.07.016. [DOI] [PubMed] [Google Scholar]
- 59.Komatsu M, Waguri S, Ueno T, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. The Journal of cell biology. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuma A, Hatano M, Matsui M, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 61.Lum JJ, Bauer DE, Kong M, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 62.Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nature reviews Molecular cell biology. 2010;11:777–788. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
- 63.Douglas PM, Summers DW, Cyr DM. Molecular chaperones antagonize proteotoxicity by differentially modulating protein aggregation pathways. Prion. 2009;3:51–58. doi: 10.4161/pri.3.2.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes & development. 2008;22:1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci. 2010;13:805–811. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Menzies FM, Moreau K, Rubinsztein DC. Protein misfolding disorders and macroautophagy. Current opinion in cell biology. 2011;23:190–197. doi: 10.1016/j.ceb.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hara T, Nakamura K, Matsui M, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 68.Komatsu M, Waguri S, Chiba T, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 69.Schutt F, Bergmann M, Holz FG, et al. Proteins modified by malondialdehyde, 4-hydroxynonenal, or advanced glycation end products in lipofuscin of human retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2003;44:3663–3668. doi: 10.1167/iovs.03-0172. [DOI] [PubMed] [Google Scholar]
- 70.Dohm GL, Tapscott EB, Kasperek GJ. Protein degradation during endurance exercise and recovery. Medicine & Science in Sports & Exercise. 1987;19:S166–171. [PubMed] [Google Scholar]
- 71.Dewaele M, Maes H, Agostinis P. ROS-mediated mechanisms of autophagy stimulation and their relevance in cancer therapy. Autophagy. 2010;6:838–854. doi: 10.4161/auto.6.7.12113. [DOI] [PubMed] [Google Scholar]
- 72.Kiffin R, Christian C, Knecht E, et al. Activation of chaperone-mediated autophagy during oxidative stress. Molecular biology of the cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Human molecular genetics. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 74.Sarkar S, Ravikumar B, Rubinsztein DC. Autophagic clearance of aggregate-prone proteins associated with neurodegeneration. Methods in enzymology. 2009;453:83–110. doi: 10.1016/S0076-6879(08)04005-6. [DOI] [PubMed] [Google Scholar]
- 75.Moreau K, Luo S, Rubinsztein DC. Cytoprotective roles for autophagy. Current opinion in cell biology. 2010;22:206–211. doi: 10.1016/j.ceb.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arrasate M, Mitra S, Schweitzer ES, et al. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 77.Carew JS, Nawrocki ST, Kahue CN, et al. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood. 2007;110:313–322. doi: 10.1182/blood-2006-10-050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tolkovsky AM, Xue L, Fletcher GC, et al. Mitochondrial disappearance from cells: a clue to the role of autophagy in programmed cell death and disease? Biochimie. 2002;84:233–240. doi: 10.1016/s0300-9084(02)01371-8. [DOI] [PubMed] [Google Scholar]
- 79.Iwata J, Ezaki J, Komatsu M, et al. Excess peroxisomes are degraded by autophagic machinery in mammals. The Journal of biological chemistry. 2006;281:4035–4041. doi: 10.1074/jbc.M512283200. [DOI] [PubMed] [Google Scholar]
- 80.Mukaiyama H, Kajiwara S, Hosomi A, et al. Autophagy-deficient Schizosaccharomyces pombe mutants undergo partial sporulation during nitrogen starvation. Microbiology. 2009;155:3816–3826. doi: 10.1099/mic.0.034389-0. [DOI] [PubMed] [Google Scholar]
- 81.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tsukamoto S, Kuma A, Murakami M, et al. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117–120. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- 83.Mortensen M, Ferguson DJ, Edelmann M, et al. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:832–837. doi: 10.1073/pnas.0913170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sandoval H, Thiagarajan P, Dasgupta SK, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Takano-Ohmuro H, Mukaida M, Kominami E, et al. Autophagy in embryonic erythroid cells: its role in maturation. European journal of cell biology. 2000;79:759–764. doi: 10.1078/0171-9335-00096. [DOI] [PubMed] [Google Scholar]
- 86.Pua HH, Guo J, Komatsu M, et al. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. Journal of immunology. 2009;182:4046–4055. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- 87.Pua HH, He YW. Mitophagy in the little lymphocytes: an essential role for autophagy in mitochondrial clearance in T lymphocytes. Autophagy. 2009;5:745–746. doi: 10.4161/auto.5.5.8702. [DOI] [PubMed] [Google Scholar]
- 88.Singh R, Xiang Y, Wang Y, et al. Autophagy regulates adipose mass and differentiation in mice. The Journal of clinical investigation. 2009;119:3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang Y, Goldman S, Baerga R, et al. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Denton D, Shravage B, Simin R, et al. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Current biology : CB. 2009;19:1741–1746. doi: 10.1016/j.cub.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Grishchuk Y, Ginet V, Truttmann AC, et al. Beclin 1-independent autophagy contributes to apoptosis in cortical neurons. Autophagy. 2011;7 doi: 10.4161/auto.7.10.16608. [DOI] [PubMed] [Google Scholar]
- 92.Nishiyama J, Matsuda K, Kakegawa W, et al. Reevaluation of neurodegeneration in lurcher mice: constitutive ion fluxes cause cell death with, not by, autophagy. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:2177–2187. doi: 10.1523/JNEUROSCI.6030-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Amaravadi RK, Yu D, Lum JJ, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. The Journal of clinical investigation. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Debnath J, Baehrecke EH, Kroemer G. Does autophagy contribute to cell death? Autophagy. 2005;1:66–74. doi: 10.4161/auto.1.2.1738. [DOI] [PubMed] [Google Scholar]
- 95.Uchiyama Y. Autophagic cell death and its execution by lysosomal cathepsins. Archives of histology and cytology. 2001;64:233–246. doi: 10.1679/aohc.64.233. [DOI] [PubMed] [Google Scholar]
- 96.Gorski SM, Chittaranjan S, Pleasance ED, et al. A SAGE approach to discovery of genes involved in autophagic cell death. Current biology : CB. 2003;13:358–363. doi: 10.1016/s0960-9822(03)00082-4. [DOI] [PubMed] [Google Scholar]
- 97.Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600–606. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 99.Qu X, Yu J, Bhagat G, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. The Journal of clinical investigation. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yue Z, Jin S, Yang C, et al. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Takahashi Y, Coppola D, Matsushita N, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nature cell biology. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liang C, Feng P, Ku B, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nature cell biology. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 103.Marino G, Salvador-Montoliu N, Fueyo A, et al. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. The Journal of biological chemistry. 2007;282:18573–18583. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- 104.Cheng HC, Kim SR, Oo TF, et al. Akt suppresses retrograde degeneration of dopaminergic axons by inhibition of macroautophagy. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:2125–2135. doi: 10.1523/JNEUROSCI.5519-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Scott RC, Juhasz G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Current biology : CB. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Koneri K, Goi T, Hirono Y, et al. Beclin 1 gene inhibits tumor growth in colon cancer cell lines. Anticancer research. 2007;27:1453–1457. [PubMed] [Google Scholar]
- 107.Degenhardt K, Mathew R, Beaudoin B, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Balkwill F, Charles KA, Mantovani A. Smoldering and polarized inflammation in the initiation and promotion of malignant disease. Cancer cell. 2005;7:211–217. doi: 10.1016/j.ccr.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 109.Mantovani A, Allavena P, Sica A, et al. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 110.Folkman J. Angiogenesis and apoptosis. Seminars in cancer biology. 2003;13:159–167. doi: 10.1016/s1044-579x(02)00133-5. [DOI] [PubMed] [Google Scholar]
- 111.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 112.Jin S, White E. Role of autophagy in cancer: management of metabolic stress. Autophagy. 2007;3:28–31. doi: 10.4161/auto.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nature reviews Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 114.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 115.Thompson CB. Metabolic enzymes as oncogenes or tumor suppressors. The New England journal of medicine. 2009;360:813–815. doi: 10.1056/NEJMe0810213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Selak MA, Armour SM, MacKenzie ED, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 117.Nelson DA, Tan TT, Rabson AB, et al. Hypoxia and defective apoptosis drive genomic instability and tumorigenesis. Genes & development. 2004;18:2095–2107. doi: 10.1101/gad.1204904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim JH, Kim HY, Lee YK, et al. Involvement of mitophagy in oncogenic K-Ras-induced transformation: Overcoming a cellular energy deficit from glucose deficiency. Autophagy. 2011;7 doi: 10.4161/auto.7.10.16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Komatsu M, Kurokawa H, Waguri S, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nature cell biology. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 120.Mathew R, Karp CM, Beaudoin B, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lv L, Li D, Zhao D, et al. Acetylation Targets the M2 Isoform of Pyruvate Kinase for Degradation through Chaperone-Mediated Autophagy and Promotes Tumor Growth. Molecular cell. 2011;42:719–730. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cuervo AM, Dice JF. Lysosomes, a meeting point of proteins, chaperones, and proteases. Journal of molecular medicine. 1998;76:6–12. doi: 10.1007/s001090050185. [DOI] [PubMed] [Google Scholar]
- 123.Alirezaei M, Kemball CC, Flynn CT, et al. Short-term fasting induces profound neuronal autophagy. Autophagy. 2010;6:702–710. doi: 10.4161/auto.6.6.12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Boland B, Kumar A, Lee S, et al. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Young JE, Martinez RA, La Spada AR. Nutrient deprivation induces neuronal autophagy and implicates reduced insulin signaling in neuroprotective autophagy activation. J Biol Chem. 2009;284:2363–2373. doi: 10.1074/jbc.M806088200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rodriguez-Muela N, Germain F, Marino G, et al. Autophagy promotes survival of retinal ganglion cells after optic nerve axotomy in mice. Cell death and differentiation. 2011 doi: 10.1038/cdd.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Midorikawa R, Yamamoto-Hino M, Awano W, et al. Autophagy-dependent rhodopsin degradation prevents retinal degeneration in Drosophila. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:10703–10719. doi: 10.1523/JNEUROSCI.2061-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Piras A, Gianetto D, Conte D, et al. Activation of Autophagy in a Rat Model of Retinal Ischemia following High Intraocular Pressure. PloS one. 2011;6:e22514. doi: 10.1371/journal.pone.0022514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu C, Gao Y, Barrett J, et al. Autophagy and protein aggregation after brain ischemia. Journal of neurochemistry. 2010;115:68–78. doi: 10.1111/j.1471-4159.2010.06905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Russo R, Berliocchi L, Adornetto A, et al. Calpain-mediated cleavage of Beclin-1 and autophagy deregulation following retinal ischemic injury in vivo. Cell death & disease. 2011;2:e144. doi: 10.1038/cddis.2011.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kubota C, Torii S, Hou N, et al. Constitutive reactive oxygen species generation from autophagosome/lysosome in neuronal oxidative toxicity. J Biol Chem. 2010;285:667–674. doi: 10.1074/jbc.M109.053058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ravikumar B, Vacher C, Berger Z, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nature genetics. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 133.Nascimento-Ferreira I, Santos-Ferreira T, Sousa-Ferreira L, et al. Overexpression of the autophagic beclin-1 protein clears mutant ataxin-3 and alleviates Machado-Joseph disease. Brain : a journal of neurology. 2011;134:1400–1415. doi: 10.1093/brain/awr047. [DOI] [PubMed] [Google Scholar]
- 134.Nassif M, Hetz C. Targeting autophagy in ALS: a complex mission. Autophagy. 2011;7:450–453. doi: 10.4161/auto.7.4.14700. [DOI] [PubMed] [Google Scholar]
- 135.Fleming A, Noda T, Yoshimori T, et al. Chemical modulators of autophagy as biological probes and potential therapeutics. Nature chemical biology. 2011;7:9–17. doi: 10.1038/nchembio.500. [DOI] [PubMed] [Google Scholar]
- 136.Wong ES, Tan JM, Soong WE, et al. Autophagy-mediated clearance of aggresomes is not a universal phenomenon. Hum Mol Genet. 2008;17:2570–2582. doi: 10.1093/hmg/ddn157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lim J, Kim HW, Youdim MB, et al. Binding preference of p62 towards LC3-ll during dopaminergic neurotoxin-induced impairment of autophagic flux. Autophagy. 2011;7:51–60. doi: 10.4161/auto.7.1.13909. [DOI] [PubMed] [Google Scholar]
- 138.Winslow AR, Chen CW, Corrochano S, et al. alpha-Synuclein impairs macroautophagy: implications for Parkinson’s disease. J Cell Biol. 2010;190:1023–1037. doi: 10.1083/jcb.201003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Martinez-Vicente M, Talloczy Z, Wong E, et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s Disease. Nat Neurosci. 2010 doi: 10.1038/nn.2528. E-pub ahead of printing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ravikumar B, Acevedo-Arozena A, Imarisio S, et al. Dynein mutations impair autophagic clearance of aggregate-prone proteins. Nature genetics. 2005;37:771–776. doi: 10.1038/ng1591. [DOI] [PubMed] [Google Scholar]
- 141.Lee JY, Koga H, Kawaguchi Y, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. The EMBO journal. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Settembre C, Fraldi A, Jahreiss L, et al. A block of autophagy in lysosomal storage disorders. Hum Mol Genet. 2008;17:119–129. doi: 10.1093/hmg/ddm289. [DOI] [PubMed] [Google Scholar]
- 143.Lee J-H, Yu W, Kumar A, et al. PS1 mutations in Alzheimer’s Disease disrupt lysosomal proteolysis and autophagy. Cell. 2010;7:1146–1158. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Koga H, Cuervo AM. Chaperone-mediated autophagy dysfunction in the pathogenesis of neurodegeneration. Neurobiology of disease. 2011;43:29–37. doi: 10.1016/j.nbd.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cuervo AM, Stefanis L, Fredenburg R, et al. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 146.Martinez-Vicente M, Talloczy Z, Kaushik S, et al. Dopamine-modified alpha-synuclein blocks chaperone-mediated autophagy. J Clin Invest. 2008;118:777–788. doi: 10.1172/JCI32806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kabuta T, Wada K. Aberrant interaction between Parkinson disease-associated mutant UCH-L1 and the lysosomal receptor for chaperone-mediated autophagy. J Biol Chem. 2008;283:23731–22373. doi: 10.1074/jbc.M801918200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang Y, Martinez-Vicente M, Kruger U, et al. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Human molecular genetics. 2009;18:4153–4170. doi: 10.1093/hmg/ddp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cao DJ, Gillette TG, Hill JA. Cardiomyocyte autophagy: remodeling, repairing, and reconstructing the heart. Current hypertension reports. 2009;11:406–411. doi: 10.1007/s11906-009-0070-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hill JA, Olson EN. Cardiac plasticity. The New England journal of medicine. 2008;358:1370–1380. doi: 10.1056/NEJMra072139. [DOI] [PubMed] [Google Scholar]
- 151.Saftig P, Tanaka Y, Lullmann-Rauch R, et al. Disease model: LAMP-2 enlightens Danon disease. Trends in molecular medicine. 2001;7:37–39. doi: 10.1016/s1471-4914(00)01868-2. [DOI] [PubMed] [Google Scholar]
- 152.Tanaka Y, Guhde G, Suter A, et al. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406:902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 153.Lacoste-Collin L, Garcia V, Uro-Coste E, et al. Danon’s disease (X-linked vacuolar cardiomyopathy and myopathy): a case with a novel Lamp-2 gene mutation. Neuromuscular disorders : NMD. 2002;12:882–885. doi: 10.1016/s0960-8966(02)00179-7. [DOI] [PubMed] [Google Scholar]
- 154.Nakai A, Yamaguchi O, Takeda T, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nature medicine. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 155.Taneike M, Yamaguchi O, Nakai A, et al. Inhibition of autophagy in the heart induces age-related cardiomyopathy. Autophagy. 2010;6 doi: 10.4161/auto.6.5.11947. [DOI] [PubMed] [Google Scholar]
- 156.Wang X, Osinska H, Klevitsky R, et al. Expression of R120G-alphaB-crystallin causes aberrant desmin and alphaB-crystallin aggregation and cardiomyopathy in mice. Circulation research. 2001;89:84–91. doi: 10.1161/hh1301.092688. [DOI] [PubMed] [Google Scholar]
- 157.Tannous P, Zhu H, Johnstone JL, et al. Autophagy is an adaptive response in desmin-related cardiomyopathy. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9745–9750. doi: 10.1073/pnas.0706802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gurusamy N, Lekli I, Gorbunov NV, et al. Cardioprotection by adaptation to ischaemia augments autophagy in association with BAG-1 protein. Journal of cellular and molecular medicine. 2009;13:373–387. doi: 10.1111/j.1582-4934.2008.00495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yan L, Vatner DE, Kim SJ, et al. Autophagy in chronically ischemic myocardium. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13807–13812. doi: 10.1073/pnas.0506843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hamacher-Brady A, Brady NR, Gottlieb RA. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. J Biol Chem. 2006;281:29776–29787. doi: 10.1074/jbc.M603783200. [DOI] [PubMed] [Google Scholar]
- 161.Huang C, Yitzhaki S, Perry CN, et al. Autophagy induced by ischemic preconditioning is essential for cardioprotection. Journal of cardiovascular translational research. 2010;3:365–373. doi: 10.1007/s12265-010-9189-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Valentim L, Laurence KM, Townsend PA, et al. Urocortin inhibits Beclin1-mediated autophagic cell death in cardiac myocytes exposed to ischaemia/reperfusion injury. Journal of molecular and cellular cardiology. 2006;40:846–852. doi: 10.1016/j.yjmcc.2006.03.428. [DOI] [PubMed] [Google Scholar]
- 163.Matsui Y, Takagi H, Qu X, et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circulation research. 2007;100:914–922. doi: 10.1161/01.RES.0000261924.76669.36. [DOI] [PubMed] [Google Scholar]
- 164.Zhu H, Tannous P, Johnstone JL, et al. Cardiac autophagy is a maladaptive response to hemodynamic stress. The Journal of clinical investigation. 2007;117:1782–1793. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Morselli E, Galluzzi L, Kepp O, et al. Autophagy mediates pharmacological lifespan extension by spermidine and resveratrol. Aging. 2009;1:961–970. doi: 10.18632/aging.100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Das M, Das DK. Resveratrol and cardiovascular health. Molecular aspects of medicine. 2010;31:503–512. doi: 10.1016/j.mam.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 167.Lekli I, Ray D, Mukherjee S, et al. Co-ordinated autophagy with resveratrol and gamma-tocotrienol confers synergetic cardioprotection. Journal of cellular and molecular medicine. 2010;14:2506–2518. doi: 10.1111/j.1582-4934.2009.00921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wing SS, Chiang HL, Goldberg AL, et al. Proteins containing peptide sequences related to Lys-Phe-Glu-Arg-Gln are selectively depleted in liver and heart, but not skeletal muscle, of fasted rats. The Biochemical journal. 1991;275(Pt 1):165–169. doi: 10.1042/bj2750165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Eskelinen EL, Cuervo AM, Taylor MR, et al. Unifying Nomenclature for the Isoforms of the Lysosomal Membrane Protein LAMP-2. Traffic. 2005;6:1058–1061. doi: 10.1111/j.1600-0854.2005.00337.x. [DOI] [PubMed] [Google Scholar]
- 170.Massey AC, Kaushik S, Sovak G, et al. Consequences of the selective blockage of chaperone-mediated autophagy. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:5805–5810. doi: 10.1073/pnas.0507436103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Deretic V. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunological reviews. 2011;240:92–104. doi: 10.1111/j.1600-065X.2010.00995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Nakagawa I, Amano A, Mizushima N, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- 173.Blanchet FP, Moris A, Nikolic DS, et al. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity. 2010;32:654–669. doi: 10.1016/j.immuni.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Shelly S, Lukinova N, Bambina S, et al. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30:588–598. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Orvedahl A, Alexander D, Talloczy Z, et al. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 176.Rich KA, Burkett C, Webster P. Cytoplasmic bacteria can be targets for autophagy. Cell Microbiol. 2003;5:455–468. doi: 10.1046/j.1462-5822.2003.00292.x. [DOI] [PubMed] [Google Scholar]
- 177.Gutierrez MG, Master SS, Singh SB, et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 178.Dorn BR, Dunn WA, Jr., Progulske-Fox A. Bacterial interactions with the autophagic pathway. Cell Microbiol. 2002;4:1–10. doi: 10.1046/j.1462-5822.2002.00164.x. [DOI] [PubMed] [Google Scholar]
- 179.Colombo MI. Autophagy: a pathogen driven process. IUBMB Life. 2007;59:238–242. doi: 10.1080/15216540701230503. [DOI] [PubMed] [Google Scholar]
- 180.Gutierrez MG, Vazquez CL, Munafo DB, et al. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cell Microbiol. 2005;7:981–993. doi: 10.1111/j.1462-5822.2005.00527.x. [DOI] [PubMed] [Google Scholar]
- 181.Dorn BR, Dunn WA., Jr. Progulske-Fox A. Porphyromonas gingivalis traffics to autophagosomes in human coronary artery endothelial cells. Infect Immun. 2001;69:5698–5708. doi: 10.1128/IAI.69.9.5698-5708.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mestre MB, Fader CM, Sola C, et al. Alpha-hemolysin is required for the activation of the autophagic pathway in Staphylococcus aureus-infected cells. Autophagy. 2010;6:110–125. doi: 10.4161/auto.6.1.10698. [DOI] [PubMed] [Google Scholar]
- 183.Ogawa M, Yoshimori T, Suzuki T, et al. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- 184.Kirkegaard K, Taylor MP, Jackson WT. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat Rev Microbiol. 2004;2:301–314. doi: 10.1038/nrmicro865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Munz C. Autophagy and antigen presentation. Cell Microbiol. 2006;8:891–898. doi: 10.1111/j.1462-5822.2006.00714.x. [DOI] [PubMed] [Google Scholar]
- 186.Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Crotzer VL, Blum JS. Autophagy and its role in MHC-mediated antigen presentation. J Immunol. 2009;182:3335–3341. doi: 10.4049/jimmunol.0803458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Friede T, Gnau V, Jung G, et al. Natural ligand motifs of closely related HLA-DR4 molecules predict features of rheumatoid arthritis associated peptides. Biochim Biophys Acta. 1996;1316:85–101. doi: 10.1016/0925-4439(96)00010-5. [DOI] [PubMed] [Google Scholar]
- 189.Dongre AR, Kovats S, deRoos P, et al. In vivo MHC class II presentation of cytosolic proteins revealed by rapid automated tandem mass spectrometry and functional analyses. Eur J Immunol. 2001;31:1485–1494. doi: 10.1002/1521-4141(200105)31:5<1485::AID-IMMU1485>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 190.Nimmerjahn F, Milosevic S, Behrends U, et al. Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy. Eur J Immunol. 2003;33:1250–1259. doi: 10.1002/eji.200323730. [DOI] [PubMed] [Google Scholar]
- 191.Dorfel D, Appel S, Grunebach F, et al. Processing and presentation of HLA class I and II epitopes by dendritic cells after transfection with in vitro-transcribed MUC1 RNA. Blood. 2005;105:3199–3205. doi: 10.1182/blood-2004-09-3556. [DOI] [PubMed] [Google Scholar]
- 192.Brazil MI, Weiss S, Stockinger B. Excessive degradation of intracellular protein in macrophages prevents presentation in the context of major histocompatibility complex class II molecules. Eur J Immunol. 1997;27:1506–1514. doi: 10.1002/eji.1830270629. [DOI] [PubMed] [Google Scholar]
- 193.Dengjel J, Schoor O, Fischer R, et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc Natl Acad Sci U S A. 2005;102:7922–7927. doi: 10.1073/pnas.0501190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Paludan C, Schmid D, Landthaler M, et al. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307:593–596. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- 195.Lee HK, Mattei LM, Steinberg BE, et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 2010;32:227–239. doi: 10.1016/j.immuni.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zhou D, Li P, Lin Y, et al. Lamp-2a facilitates MHC class II presentation of cytoplasmic antigens. Immunity. 2005;22:571–581. doi: 10.1016/j.immuni.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 197.Munz C. Antigen processing via autophagy--not only for MHC class II presentation anymore? Curr Opin Immunol. 2010;22:89–93. doi: 10.1016/j.coi.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.English L, Chemali M, Duron J, et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol. 2009;10:480–487. doi: 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- 200.Nedjic J, Aichinger M, Emmerich J, et al. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008;455:396–400. doi: 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- 201.Schmid D, Pypaert M, Munz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Rioux JD, Xavier RJ, Taylor KD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nature genetics. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Prescott NJ, Fisher SA, Franke A, et al. A nonsynonymous SNP in ATG16L1 predisposes to ileal Crohn’s disease and is independent of CARD15 and IBD5. Gastroenterology. 2007;132:1665–1671. doi: 10.1053/j.gastro.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 204.Lunemann JD, Munz C. Autophagy in CD4+ T-cell immunity and tolerance. Cell death and differentiation. 2009;16:79–86. doi: 10.1038/cdd.2008.113. [DOI] [PubMed] [Google Scholar]
- 205.Qu X, Zou Z, Sun Q, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 206.Bratton DL, Henson PM. Autoimmunity and apoptosis: refusing to go quietly. Nat Med. 2005;11:26–27. doi: 10.1038/nm0105-26. [DOI] [PubMed] [Google Scholar]
- 207.Jia W, Pua HH, Li QJ, et al. Autophagy regulates endoplasmic reticulum homeostasis and calcium mobilization in T lymphocytes. J Immunol. 2011;186:1564–1574. doi: 10.4049/jimmunol.1001822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Pua HH, Dzhagalov I, Chuck M, et al. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Li C, Capan E, Zhao Y, et al. Autophagy is induced in CD4+ T cells and important for the growth factor-withdrawal cell death. J Immunol. 2006;177:5163–5168. doi: 10.4049/jimmunol.177.8.5163. [DOI] [PubMed] [Google Scholar]
- 210.Hubbard VM, Valdor R, Patel B, et al. Macroautophagy Regulates Energy Metabolism during Effector T Cell Activation. J Immunol. 2010;185:7349–7357. doi: 10.4049/jimmunol.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 212.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell host & microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]