Abstract
Introduction
In a North American, HIV-positive, highly active antiretroviral therapy (HAART)-treated, adherent cohort of self-identified white and black patients, we previously observed that chemokine (C-C motif) receptor 5 (CCR5) –2459G>A genotype had a strong association with time to achieve virologic success (TVLS) in black but not in white patients.
Methods
Using 128 genome-wide ancestry informative markers, we performed a quantitative assessment of ancestry in these patients (n = 310) to determine (1) whether CCR5 –2459G>A genotype is still associated with TVLS of HAART when ancestry, not self-identified race, is considered and (2) whether this association is influenced by varying African ancestry.
Results
We found that the interaction between CCR5 –2459G>A genotype and African ancestry (≤0.125 vs. ≥0.425 and <0.71 vs. ≥0.71) was significantly associated with TVLS (GG compared with AA, P = 0.044 and 0.018, respectively). Furthermore, the association between CCR5 –2459G>A genotype and TVLS was stronger in patients with African ancestry ≥0.71 than in patients with African ancestry ≥0.452, in both Kaplan-Meier (log-rank P = 0.039 and 0.057, respectively, for AA, GA, and GG) and Cox proportional hazards regression (relative hazard for GG compared with AA 2.59 [95% CI, 1.27–5.22; P = 0.01] and 2.26 [95% CI, 1.18–4.32; P = 0.01], respectively) analyses.
Conclusions
We observed that the association between CCR5 –2459G>A genotype and TVLS of HAART increased with stronger African ancestry. Understanding the genomic mechanisms by which African ancestry influences this association is critical, and requires further studies.
Keywords: African American, ancestry informative markers, CCR5, HAART, virologic success
INTRODUCTION
In patients on highly active antiretroviral therapy (HAART), time to virologic suppression is one of the important factors shown to influence CD4+ T-cell recovery and clinical outcomes.1 Minimizing the time to virologic suppression and attaining durable suppression, together with the early and overall patterns of viral load decay among virologic suppressed patients, lead to better immunologic and clinical responses to HAART.1 Recently, in a North American, HIV-positive, HAART-treated, adherent cohort of self-identified white (Caucasian) and black (African American) patients, who were followed for ≥6 months after initiation of HAART, we observed that the chemokine (C-C motif) receptor 5 (CCR5) –2459G>A genotype (rs1799987) had a strong association with time to achieve virologic success (TVLS) in black but not in white patients.2 The CCR5 –2459G/A allelic variants are also known as 59029G/A (based on GenBank accession no. U95626) and 303G/A,3 and the A allele-containing most prevalent haplotype is called HHE.3 In our study, black patients carrying the –2459G allele achieved virologic success significantly earlier (AA, GA, and GG, 131, 98, and 76 days, respectively [log-rank P = 0.04]), and this association remained significant in a Cox proportional hazards model that included a total of 10 covariates.2 None of the other genetic polymorphisms, including a 32 base-pair deletion in the open reading frame of CCR5 (Δ32, rs333), CCR2 190G>A (Val64Ile, rs1799864), and a total of 5 SNPs in 2 drug-metabolizing enzyme and one transporter genes, was significantly associated with TVLS in either racial group of patients. These observations suggested that the association between CCR5 –2459G>A genotype and TVLS of HAART was race-specific in this cohort.2
Admixture is a type of gene flow that occurs when individuals from 2 or more parental populations that have been isolated for several generations form a new population.4 Admixed populations are common in North America, as the history of North America has been marked by the encounter of populations from different continents. Recent genetic association analyses in admixed populations, such as African Americans, have highlighted the complexities due to confounding by ancestry.5 In African Americans, systematic differences among individuals occur in the proportion of European ancestry,6 which can be accurately inferred using genome-wide ancestry informative markers (AIMs).7, 8 Several recent epidemiological studies in African Americans have taken advantage of this, revealing that gene expression patterns and many disease-related phenotypes vary with the proportion of European ancestry.9–14 Herein, we applied this idea to measure the effects of varying African ancestry on CCR5 –2459G>A genotype-associated TVLS of HAART in a North American cohort.2 The major aims of the study were to determine (1) whether CCR5 –2459G>A genotype is still associated with TVLS of HAART when ancestry, not self-identified race, is considered and (2) whether the association between CCR5 –2459G>A genotype and TVLS of HAART is influenced by varying African ancestry.
MATERIALS AND METHODS
Study Cohort
All subjects (n = 393) were adults with confirmed HIV infection, receiving care at the Special Immunology Unit of Case Western Reserve University/University Hospitals Case Medical Center, Cleveland, OH. The characteristics of this cohort (self-identified white patients, n = 175; black patients, n = 218), including HAART regimens, adherence, and prior antiretroviral exposure, as well as distribution of various allele frequencies have been reported previously.2 All patients provided written informed consent for de-identified clinical data and specimen collection, storage, and usage in genetic and non-genetic studies. The data and specimen collection protocol was approved by the Institutional Review Board of University Hospitals Case Medical Center.
AIMs Genotyping
We used a set of 128 genome-wide AIMs (Supplemental Table 1), which have been characterized and validated for determining continental origin and admixture proportions in common populations in North America.7, 8 TaqMan® single nucleotide polymorphism (SNP) genotyping assays for these AIMs are commercially available (Life Technologies Corporation [Applied Biosystems], Carlsbad, CA). Assays were performed on the TaqMan® OpenArray® Genotyping System (Applied Biosystems), following the manufacturer’s protocol. Each array contained several water blanks as negative controls, and 4 DNA samples (3 European and 1 African American) from the Coriell Cell Repositories (Coriell Institute for Medical Research, Camden, NJ) as positive controls.
The TaqMan® Genotyper software v1.0.1 (Applied Biosystems) was used to analyze the primary data. The assay results were validated by re-running randomly-selected 20 of the 393 (5%) samples. All SNPs with >10% missing data (6 SNPs excluded, 122 SNPs remaining), and all samples with >20% missing SNP data (67 samples excluded, 326 samples remaining) were removed.
Ancestry Analysis
Ancestry proportions in these 326 subjects were determined using the Bayesian clustering algorithms implemented in the program STRUCTURE v2.3.2.1,15, 16 with the following run parameters: ancestral populations (k) = 3; burn-in iterations = 50,000; and subsequent iterations = 50,000. To have a better assessment of ancestry in these subjects, we spiked our dataset with 502 HapMap phase III samples of known ancestry (Northern European, n = 165; Yoruban, n = 167; Japanese + Chinese, n = 170). Among the 326 subjects, those with ≥10% Southeast Asian ancestry were removed (16 samples excluded, 310 remaining).
Statistical Analysis
Virologic success was defined as a viral load <400 copies/mL within 6 months of HAART initiation.2 Patients who had not achieved virologic success within this time were censored at 6 months. To describe the probability of achieving virologic success according to CCR5 –2459G>A genotype, Kaplan-Meier curves were plotted. The association between CCR5 –2459G>A genotype and TVLS was examined using Cox proportional hazards regression (Cox model).17 There was no significant interaction between follow-up time and CCR5 –2459G>A genotype in the primary Cox model, suggesting that the proportional hazards assumption was appropriate. Covariates included in the analysis were age, gender, baseline CD4+ T-cell count, baseline viral load, and CCR2 190G>A genotype.2 All analyses were performed using SAS software (version 9.3). A two-sided P < 0.05 was considered statistically significant.
RESULTS
The characteristics of the whole cohort (n = 310) and various African ancestry proportion groups are presented in Table 1.
TABLE 1.
Characteristics of All Patients and Various African Ancestry Proportion Groups
| Characteristic | All patients (n = 310) | African Ancestry
|
||
|---|---|---|---|---|
| ≤0.125 (n = 135) | ≥0.425 (n = 175) | ≥0.71 (n = 155) | ||
| Age (yrs), Mean (SD) | 37.9 (8.9) | 38.9 (8.8) | 37.1 (9) | 37.3 (9.3) |
| Race† | ||||
| White, n (%) | 137 (44.2) | 135 (100) | 2 (1.1) | 1 (0.6) |
| Black, n (%) | 173 (55.8) | 0 (0.0) | 173 (98.9) | 154 (99.4) |
| Gender | ||||
| Male, n (%) | 246 (79.4) | 125 (92.6) | 121 (69.1) | 106 (68.4) |
| Female, n (%) | 64 (20.6) | 10 (7.4) | 54 (30.9) | 49 (31.6) |
| Baseline CD4+ T-cell count (cells/mm3), Mean (SD) | 213 (215)a | 215 (227) | 211 (205) | 217 (208) |
| Baseline viral load (×1000 copies/mL), Mean (SD) | 159 (294)b | 168 (230) | 152 (335) | 119 (206) |
SD, standard deviation.
Self-identified.
Available in a total of 283 patients (127 White, 156 Black).
Available in a total of 261 patients (113 White, 148 Black).
Ancestry Proportion Estimates in HapMap Samples and Study Cohort
The STRUCTURE program was run on all samples 3 times independently, and the European and African ancestry proportion estimates were highly concordant across runs (Pearson’s correlation coefficient, r > 0.999 in all pairwise comparisons). Similar to previous studies that have applied these 128 genome-wide AIMs,7, 8 we observed very high proportion of corresponding ancestry in the HapMap samples (Supplemental Table 2).
The distribution of ancestry in our final samples (n = 310) is shown in the supplemental Figure 1. In these samples, the proportion of European ancestry ranged from 0.003–0.996, the proportion of African ancestry ranged from 0.002–0.993, and the proportion of Southeast Asian ancestry ranged from 0.002–0.096. No subject was characterized by a proportion of African ancestry between 0.125 and 0.452. All subjects with African ancestry ≤0.125 (n = 135) self-identified as white (Caucasian), and all but 2 with African ancestry ≥0.452 (n = 175) self-identified as black (African American) (Table 1). In the subjects with African ancestry ≥0.452, the mean and median proportions of African ancestry were 0.845 and 0.859, respectively; these values are consistent with previous AIMs-based estimates of African ancestry in African American communities in nearby areas of the Northeast and Midwest regions of the United States.6
Effect of Ancestry on TVLS
We tested for association of ancestry alone with TVLS. Ancestry was not associated with TVLS (≤0.125 vs. ≥0.452, log-rank P = 0.72). We conducted a second set of analysis dividing the entire sample at the median proportion of African ancestry (<0.71 and ≥0.71, n = 155 each). Because it splits the sample evenly, it maximizes power to detect differences between the two groups. Here also ancestry was not associated with TVLS (<0.71 vs. ≥0.71, log-rank P = 0.90).
Interaction Effect of CCR5 –2459 Genotype and African Ancestry on TVLS
In our previous study, CCR5 –2459G>A genotype had race-specific association with TVLS.2 In order to examine this issue more thoroughly at the genomic level, we tested for effect modification by including a CCR5 –2459G>A genotype × African ancestry interaction term in a Cox model. When we modeled an interaction between CCR5 –2459G>A genotype and African ancestry ≤0.125 vs. ≥0.452, the effect of the GG genotype on TVLS, compared with the AA genotype (reference), was significantly different between the 2 ancestry proportion groups (P = 0.044). In contrast, the effect of the GA genotype on TVLS, compared with the AA genotype (reference), was not significantly different (P = 0.081).
In a Cox model including an interaction term between CCR5 –2459G>A genotype and African ancestry <0.71 vs. ≥0.71, the effect of the GG genotype on TVLS was stronger (P = 0.018), whereas the effect of the GA genotype was again not significantly different between the 2 ancestry proportion groups (P = 0.094). That the effect of the GA genotype, compared with the AA genotype, was not significantly different between 2 ancestry proportion groups suggests a dose effect of the –2459G allele on TVLS.
CCR5 –2459 Genotype-Associated TVLS within African Ancestry Proportion Groups
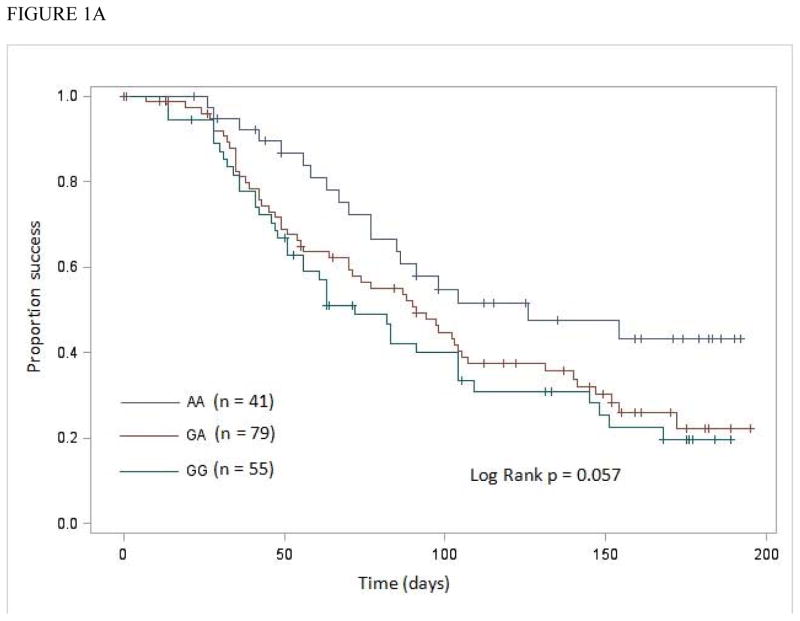
Since we identified a significant interaction between CCR5 –2459G>A genotype and African ancestry, we conducted stratified Kaplan-Meier and Cox model analyses to estimate CCR5 –2459G>A genotype effects within each group of proportions of African ancestry. Kaplan-Meier analysis showed no significant difference in TVLS among CCR5 –2459G>A genotypes in patients with African ancestry ≤0.125 (log-rank P = 0.72). Marginally significant difference was observed in patients with African ancestry ≥0.452 (log-rank P = 0.057) (Figure 1A). In these patients, the association between the –2459G allele and TVLS was more significant after adjusting for age, gender, baseline CD4+ T-cell count, baseline viral load, and CCR2 190G>A genotype (Table 2).
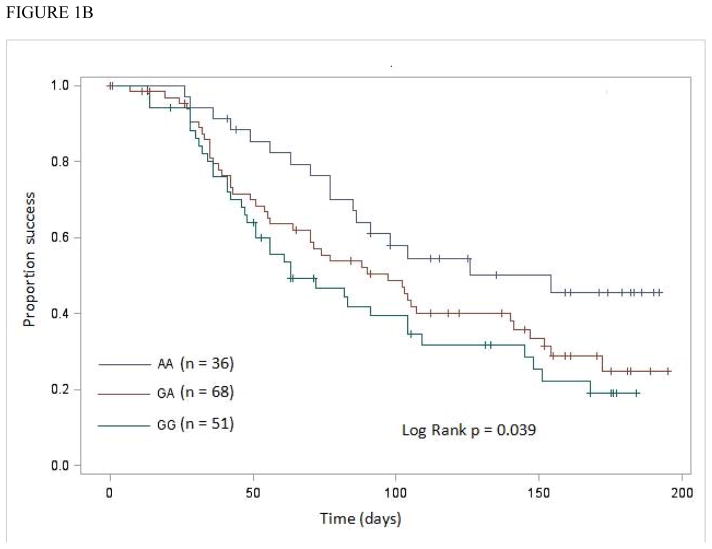
Figure 1.
Kaplan-Meier analysis of influence of CCR5 –2459G>A genotypes on time to achieve virologic success (TimeVLSucc; measured in days)
A, in patients with proportions of African ancestry ≥0.452 and
B, in patients with proportions of African ancestry ≥0.71.
TABLE 2.
CCR5 -2459G>A Genotype Effect Estimates within Low and High African Ancestry Proportion Groups
| Genotype | African ancestry ≤0.125
|
African ancestry ≥0.452
|
||||||
|---|---|---|---|---|---|---|---|---|
| n | TVLSa | ARH(95% CI)b,c | P | n | TVLSa | ARH(95% CI)b,c | P | |
| AA | 39 | 111 | - | 41 | 126 | - | ||
| GA | 77 | 93 | 0.83 (0.48–1.43) | 0.50 | 79 | 91 | 1.72 (0.96–3.10) | 0.07 |
| GG | 19 | 112 | 0.68 (0.31–1.51) | 0.35 | 55 | 72 | 2.26 (1.18–4.32) | 0.01 |
Time to virologic success, median number of days.
Adjusted relative hazard (95% confidence interval). Covariates included in the Cox models were age, gender, baseline CD4+ T-cell count, baseline viral load, and CCR2 190G>A genotype.
ARH are for GA vs. AA and GG vs. AA.
- AA is the reference.
No significant difference was observed in TVLS among CCR5 –2459G>A genotypes in patients with African ancestry <0.71 (log-rank P = 0.67). However, a significant difference was observed in patients with African ancestry ≥0.71 (log-rank P = 0.039) (Figure 1B). Again, the significance of the association between the –2459G allele and TVLS was enhanced with adjustment for age, gender, baseline CD4+ T-cell count, baseline viral load, and CCR2 190G>A genotype (Table 3).
TABLE 3.
CCR5 -2459G>A Genotype Effect Estimates within African Ancestry Proportion Groups Divided at the Median Value
| Genotype | African ancestry <0.71
|
African ancestry ≥0.71
|
||||||
|---|---|---|---|---|---|---|---|---|
| n | TVLSa | ARH(95% CI)b,c | P | n | TVLSa | ARH(95% CI)b,c | P | |
| AA | 44 | 111 | - | 36 | 154 | - | ||
| GA | 88 | 92 | 0.84 (0.50–1.41) | 0.51 | 68 | 97 | 1.83 (0.97–3.44) | 0.06 |
| GG | 23 | 104 | 0.65 (0.32–1.32) | 0.23 | 51 | 63 | 2.59 (1.27–5.22) | 0.01 |
Time to virologic success, median number of days.
Adjusted relative hazard (95% confidence interval). Covariates included in the Cox models were age, gender, baseline CD4+ T-cell count, baseline viral load, and CCR2 190G>A genotype.
ARH are for GA vs. AA and GG vs. AA.
- AA is the reference.
Both Kaplan-Meier and Cox model analyses showed a stronger significant association between CCR5 –2459G>A genotype and TVLS in patients with African ancestry ≥0.71 (Figure 1B; Table 3) than in patients with African ancestry ≥0.452 (Figure 1A; Table 2).
African Ancestry Proportions among CCR5 –2459 Genotypes
Although the allele frequencies of CCR5 –2459G and –2459A do not differ greatly among continental groups,2, 18 we verified in our samples that the genotypes at this locus were not confounded with African ancestry. The mean proportions of African ancestry within a group, determined by a threshold of 0.452 or 0.71, did not differ significantly among CCR5 –2459G>A genotypes (Student’s t test, P > 0.1 for all comparisons).
DISCUSSION
In the present study, using 128 genome-wide AIMs, we found that the interaction between CCR5 –2459G>A genotype and African ancestry, but not ancestry alone, was significantly associated with TVLS. Furthermore, when we performed analyses stratified by proportions of African ancestry, we found that the association between CCR5 –2459G>A genotype and TVLS was stronger in patients with African ancestry ≥0.71 than in patients with African ancestry ≥0.452. This result was intriguing, mainly because the sample size was reduced by 20 (≥0.452, n = 175; ≥0.71, n = 155). Among these 20 subjects, 19 were self-identified blacks, whereas one was self-identified white. The African ancestry proportions were 0.452, 0.49, and 0.534 for 3 of the 20 subjects, and >0.6 but <0.71 for the remaining 17 subjects. Conversely, the European ancestry proportions were 0.539, 0.5, and 0.429, respectively, for the 3 subjects, and ranged from 0.265–0.369 for the 17 subjects; these estimates are largely consistent with previous reports.6, 19 Thus, these 20 subjects are more heterogeneous than the remaining 155 subjects in terms of their African ancestry proportions, and removing them creates a more homogeneous group with African ancestry proportions ≥0.71, showing a stronger association between CCR5 –2459G>A genotype and TVLS.
Studies investigating the association between genetic markers and HIV/AIDS outcomes have heavily relied upon self-identified race classification. To our knowledge, only recently have researchers begun to consider genetic ancestry into their analyses. In a multiethnic HIV-infected women cohort, Frasco et al.20 showed evidence of substantial confounding of the association between cytochrome P450 2B6 enzyme polymorphisms and virologic response to efavirenz and nevirapine by genetic ancestry. In a multiethnic HIV-infected men cohort, Nicholaou et al.21 showed that the effect of HIV+/HAART status on lipid levels, relative to HIV− status, varies by genetic ancestry (African/European admixed ancestry vs. European ancestry). However, in these studies, there was a poor concordance between the self-identified race and genetic ancestry, and they did not elaborate on whether varying genetic ancestry among their subjects had any effect on the phenotypes investigated. Although self-identified race may serve as a good surrogate for genetic ancestry in the clinical setting, our results suggest the importance of considering quantitative assessment of ancestry in order to determine whether HIV/AIDS outcomes vary among African Americans with varying ancestry.
Our results draw support from several recent studies showing a significant relationship between African ancestry and a variety of disease-related phenotypes.9–11, 13, 14, 22–24 In many of these studies, a specific genetic variant10, 22 or multiple common SNPs in a single gene11 were found to be significantly associated with phenotype. Of particular interest, a quantitative assessment of ancestry revealed that the phenotypic effect increased significantly with stronger ancestry.11, 13, 22, 24 Among male subjects, the relative rate of a severe asthma exacerbation was increased more than 4-fold for every 20% increase in the proportion of African ancestry.13 The risk of developing estrogen receptor negative breast cancer, particularly triple-negative breast cancer, increased 3 times in women in the highest quintile of percent African ancestry relative to those in the lowest quintile.22 Odds of uterine leiomyomata were 34% lower among women in the highest quartile of percent European ancestry relative to those in the lowest quartile.24
An effect of CCR5 –2459G>A genotype on TVLS influenced by African ancestry implies the existence of one or more modifier genetic factors within the African ancestral genome. Although CCR5 is a major determinant, other chemokine receptor genes located on chromosome 3 near CCR5, and chemokine genes located elsewhere are also potentially important in HIV/AIDS pathogenesis.25–28 In addition, outside the chemokine receptor-ligand nexus, host genetic factors that are associated with viral load control have been identified by recent genome-wide association studies.29 Thus, the effect of CCR5 –2459G>A on virologic response to HAART might be due to strong linkage disequilibrium with SNP(s) in other gene(s) on the same chromosome,18 and/or epistatic interaction(s) with SNP(s) elsewhere in the genome. Also, in addition to commonly occurring polymorphisms in CCR5,26 a number of other polymorphisms, mostly non-synonymous, have been discovered in Caucasians and African Americans.30–32 Although these additional polymorphisms occur at low frequencies (<5%), most of them are functionally significant, showing profound effects on both HIV-1 co-receptor and specific ligand-induced functions.33, 34 Therefore, by re-sequencing the CCR5 locus, the possibility should be explored that additional polymorphisms, in conjunction with –2459G>A, could offer insight regarding the race-specific difference in virologic response to HAART.
We acknowledge that our study has some limitations. For certain genotypic associations in African Americans, the effect of ancestry proximal to the gene of interest (local or cis genetic ancestry) is more significant than the effect of ancestry elsewhere in the genome (global or trans genetic ancestry).10–12 However, we were unable to perform a meaningful analysis of the local ancestry in our study, because we have only 9 markers located on chromosome 3, and the marker (rs9809104, C/T) closest to –2459G>A (rs1799987) is 7.25 Mb away (Supplemental Table 1). This marker cannot distinguish local African vs. European ancestry with high accuracy (allele T frequency in 1000 Genomes project African samples = 0.14, and in European samples = 0.79). For such analysis, a high-density African SNP panel providing higher resolution of global as well as local ancestry35 is needed, whereby the effect of ancestry proximal to CCR5 on virologic response to HAART can be evaluated, and can be compared with the effect of global ancestry. Regarding our results pertaining to African ancestry proportions ≥0.452 and ≥0.71, specifically the 20 subjects with more heterogeneous ancestry proportions, we cannot exclude the fact that residual confounding may exist due to unmeasured ethnic factors (environmental, social, cultural, or behavioral). A number of studies have reported correlation between these factors and adherence vis-à-vis response to HAART.36, 37 However, a residual correlation of individual admixture with social or other environmental factors is less likely.38 Furthermore, using a subjective but clinically validated adherence assessment tool, we intentionally selected a stringently high level of adherence (≥90%) as an inclusion criterion for this cohort.2
In conclusion, we observed that stronger African ancestry influenced CCR5 –2459G>A genotype-associated TVLS of HAART in a North American cohort. Further studies to validate this observation in HAART-treated patients in other settings, ideally in a larger and more diverse cohort, and to explore the genomic mechanisms vs. ethnic factors contributing to this influence are important and highly relevant. Furthermore, given that HIV/AIDS continues to disproportionately affect African Americans,39 identifying and better understanding the genetic predictors of antiretroviral treatment response in this population is critical.
Supplementary Material
Acknowledgments
Supported by a development award from the Center for AIDS Research University Hospitals Case Medical Center (National Institutes of Health [NIH] grant AI36219) (R. K. M.); an Infectious Diseases research support from STERIS Corporation (R. K. M. and B. R.); a large pilot grant from Case Western Reserve University/Cleveland Clinic CTSA grant number UL1RR024989 (National Center for Research Resources) (R. K. M.); and the National Institute of Dental and Craniofacial Research (NIH grant 1P01DE019759, Project 4 to R. J. J.).
We sincerely thank Dr. Sudha Iyengar, Dr. Michael Lederman, Dave McNamara, Dr. Carolyn Myers, and Alex Barron for helpful discussions and critical evaluation of this manuscript. We are thankful to Dr. Martina Veigl and Debora Poruban (Gene Expression & Genotyping Core Facility) for performing the ancestry informative marker genotyping.
Footnotes
Meeting at which parts of the data were presented is as follows: XIX International AIDS Conference, July 2012, held in Washington D.C., U.S.A. (Abstract no. TUPE009).
Conflict of interest: none declared.
References
- 1.Marconi VC, Grandits G, Okulicz JF, et al. Cumulative viral load and virologic decay patterns after antiretroviral therapy in HIV-infected subjects influence CD4 recovery and AIDS. PLoS One. 2011;6:e17956. doi: 10.1371/journal.pone.0017956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mehlotra RK, Cheruvu VK, Blood Zikursh MJ, et al. Chemokine (C-C motif) receptor 5–2459 genotype in patients receiving highly active antiretroviral therapy: race-specific influence on virologic success. J Infect Dis. 2011;204:291–298. doi: 10.1093/infdis/jir262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gonzalez E, Bamshad M, Sato N, et al. Race-specific HIV-1 disease-modifying effects associated with CCR5 haplotypes. Proc Natl Acad Sci U S A. 1999;96:12004–12009. doi: 10.1073/pnas.96.21.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakraborty R. Gene admixture in human populations: models and predictions. Am J Phys Anthropol. 1986;29(S7):1–43. [Google Scholar]
- 5.Deo RC, Reich D, Tandon A, et al. Genetic differences between the determinants of lipid profile phenotypes in African and European Americans: the Jackson Heart Study. PLoS Genet. 2009;5:e1000342. doi: 10.1371/journal.pgen.1000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parra EJ. Admixture in North America. In: Suarez-Kurtz G, editor. Pharmacogenomics in Admixed Populations. Austin: Landes Bioscience; 2007. pp. 28–46. [Google Scholar]
- 7.Kidd JR, Friedlaender FR, Speed WC, et al. Analyses of a set of 128 ancestry informative single-nucleotide polymorphisms in a global set of 119 population samples. Investig Genet. 2011;2:1. doi: 10.1186/2041-2223-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kosoy R, Nassir R, Tian C, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat. 2009;30:69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng CY, Reich D, Haiman CA, et al. African ancestry and its correlation to type 2 diabetes in African Americans: a genetic admixture analysis in three U.S. population cohorts. PLoS One. 2012;7:e32840. doi: 10.1371/journal.pone.0032840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deo RC, Wilson JG, Xing C, et al. Single-nucleotide polymorphisms in LPA explain most of the ancestry-specific variation in Lp(a) levels in African Americans. PLoS One. 2011;6:e14581. doi: 10.1371/journal.pone.0014581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kao WH, Klag MJ, Meoni LA, et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Genet. 2008;40:1185–1192. doi: 10.1038/ng.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price AL, Patterson N, Hancks DC, et al. Effects of cis and trans genetic ancestry on gene expression in African Americans. PLoS Genet. 2008;4:e1000294. doi: 10.1371/journal.pgen.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rumpel JA, Ahmedani BK, Peterson EL, et al. Genetic ancestry and its association with asthma exacerbations among African American subjects with asthma. J Allergy Clin Immunol. 2012;130:1302–1306. doi: 10.1016/j.jaci.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlesinger D, Grinberg LT, Alba JG, et al. African ancestry protects against Alzheimer’s disease-related neuropathology. Mol Psychiatry. 2011;18:79–85. doi: 10.1038/mp.2011.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox DR. Regression models and life tables. J R Stat Soc [B] 1972;34:187–220. [Google Scholar]
- 18.Clark VJ, Dean M. Haplotype structure and linkage disequilibrium in chemokine and chemokine receptor genes. Hum Genomics. 2004;1:255–273. doi: 10.1186/1479-7364-1-4-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zakharia F, Basu A, Absher D, et al. Characterizing the admixed African ancestry of African Americans. Genome Biol. 2009;10:R141. doi: 10.1186/gb-2009-10-12-r141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frasco MA, Mack WJ, Van Den Berg D, et al. Underlying genetic structure impacts the association between CYP2B6 polymorphisms and response to efavirenz and nevirapine. AIDS. 2012;26:2097–2106. doi: 10.1097/QAD.0b013e3283593602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholaou MJ, Martinson JJ, Abraham AG, et al. HAART-associated dyslipidemia varies by biogeographical ancestry in the Multicenter AIDS Cohort Study. AIDS Research and Human Retroviruses. 2013;29:871–879. doi: 10.1089/aid.2012.0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmer JR, Ruiz-Narvaez EA, Rotimi CN, et al. Genetic susceptibility loci for subtypes of breast cancer in an African American population. Cancer Epidemiol Biomarkers Prev. 2013;22:127–134. doi: 10.1158/1055-9965.EPI-12-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi L, Nassir R, Kosoy R, et al. Relationship between hysterectomy and admixture in African American women. Am J Obstet Gynecol. 2013;208:279.e271–279.e277. doi: 10.1016/j.ajog.2013.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wise LA, Ruiz-Narvaez EA, Palmer JR, et al. African ancestry and genetic risk for uterine leiomyomata. Am J Epidemiol. 2012;176:1159–1168. doi: 10.1093/aje/kws276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.An P, Li R, Wang JM, et al. Role of exonic variation in chemokine receptor genes on AIDS: CCRL2 F167Y association with pneumocystis pneumonia. PLoS Genet. 2011;7:e1002328. doi: 10.1371/journal.pgen.1002328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzalez E, Dhanda R, Bamshad M, et al. Global survey of genetic variation in CCR5, RANTES, and MIP-1alpha: impact on the epidemiology of the HIV-1 pandemic. Proc Natl Acad Sci USA. 2001;98:5199–5204. doi: 10.1073/pnas.091056898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonzalez E, Kulkarni H, Bolivar H, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 28.McDermott DH, Beecroft MJ, Kleeberger CA, et al. Chemokine RANTES promoter polymorphism affects risk of both HIV infection and disease progression in the Multicenter AIDS Cohort Study. AIDS. 2000;14:2671–2678. doi: 10.1097/00002030-200012010-00006. [DOI] [PubMed] [Google Scholar]
- 29.van Manen D, van ‘t Wout AB, Schuitemaker H. Genome-wide association studies on HIV susceptibility, pathogenesis and pharmacogenomics. Retrovirology. 2012;9:70. doi: 10.1186/1742-4690-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ansari-Lari MA, Liu XM, Metzker ML, et al. The extent of genetic variation in the CCR5 gene. Nat Genet. 1997;16:221–222. doi: 10.1038/ng0797-221. [DOI] [PubMed] [Google Scholar]
- 31.Carrington M, Dean M, Martin MP, et al. Genetics of HIV-1 infection: chemokine receptor CCR5 polymorphism and its consequences. Hum Mol Genet. 1999;8:1939–1945. doi: 10.1093/hmg/8.10.1939. [DOI] [PubMed] [Google Scholar]
- 32.Carrington M, Kissner T, Gerrard B, et al. Novel alleles of the chemokine-receptor gene CCR5. Am J Hum Genet. 1997;61:1261–1267. doi: 10.1086/301645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blanpain C, Lee B, Tackoen M, et al. Multiple nonfunctional alleles of CCR5 are frequent in various human populations. Blood. 2000;96:1638–1645. [PubMed] [Google Scholar]
- 34.Howard OM, Shirakawa AK, Turpin JA, et al. Naturally occurring CCR5 extracellular and transmembrane domain variants affect HIV-1 co-receptor and ligand binding function. J Biol Chem. 1999;274:16228–16234. doi: 10.1074/jbc.274.23.16228. [DOI] [PubMed] [Google Scholar]
- 35.Tandon A, Patterson N, Reich D. Ancestry informative marker panels for African Americans based on subsets of commercially available SNP arrays. Genet Epidemiol. 2011;35:80–83. doi: 10.1002/gepi.20550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Connor JL, Gardner EM, Mannheimer SB, et al. Factors associated with adherence amongst 5295 people receiving antiretroviral therapy as part of an international trial. J Infect Dis. 2013;208:40–49. doi: 10.1093/infdis/jis731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oh DL, Sarafian F, Silvestre A, et al. Evaluation of adherence and factors affecting adherence to combination antiretroviral therapy among White, Hispanic, and Black men in the MACS Cohort. J Acquir Immune Defic Syndr. 2009;52:290–293. doi: 10.1097/QAI.0b013e3181ab6d48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mountain JL, Risch N. Assessing genetic contributions to phenotypic differences among ‘racial’ and ‘ethnic’ groups. Nat Genet. 2004;36:S48–53. doi: 10.1038/ng1456. [DOI] [PubMed] [Google Scholar]
- 39.Prejean J, Song R, Hernandez A, et al. Estimated HIV incidence in the United States, 2006–2009. PLoS One. 2011;6:e17502. doi: 10.1371/journal.pone.0017502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.