Abstract
Substrates that selectively encourage the growth of specific cell types are valuable for the engineering of complex tissues. Some cell-selective peptides have been identified from extracellular matrix proteins; these peptides have proven useful for biomaterials-based approaches to tissue repair or regeneration. However, there are very few examples of synthetic materials that display selectivity in supporting cell growth. We describe nylon-3 polymers that support in vitro culture of endothelial cells, but do not support the culture of smooth muscle cells or fibroblasts. These materials may be promising for vascular biomaterials applications.
Synthetic biomaterials have been widely studied for biomedical applications, including regenerative medicine.1–15 Some limitations intrinsic to the use of naturally-derived materials, such as susceptibility to biodegradation, high cost of production and possible contamination, can be overcome with synthetic biomaterials.16–28 However, unnatural materials lack specific cell-binding motifs, and cellular interactions with these materials often depend upon adsorbed proteins.29–31 Efforts to control cell adhesion to synthetic materials typically involve modification with specific proteins or molecular motifs derived from extracellular matrix (ECM) components, such as the arginine-glycine-aspartic acid (RGD) segment from fibronectin.18,19,23,32–38 Peptides that are selectively adhesive toward a specific cell type can be advantageous in this context. Such peptides might enable the segregation of different cell types into defined areas, in order to mimic a natural structure (e.g., a blood vessel), or to inhibit undesired cell adhesion on a construct implanted in vivo (e.g., blocking smooth muscle cell population of a vascular graft bearing a pre-established endothelial cell layer). However, to date only a small handful of cell-selective peptides have been identified,39–43 and all are derived from native ECM components. These peptides must be tethered to the biomaterial surface and are susceptible to cleavage or degradation under cell culture conditions.44 Moreover, both the activity and selectivity of such peptides can depend on details of the tethering process, such as the length and chemical nature of the tether and the density of surface functionalization.45 Approaches that do not rely on naturally cell-selective motifs are rare.46–48
Our approach to finding cell-selective synthetic materials focuses on nylon-3 polymers (poly-β-peptides), which have previously been shown to display intriguing biological properties,49–52 including potential utility in tissue engineering applications.30,53,54 The nylon-3 backbone contains closely spaced secondary amide units and is therefore chemically similar to the backbone of proteins. This similarity is expected to promote biocompatibility. The nylon-3 backbone differs sufficiently from proteins, however, that these materials resist protease action. Synthesis involves anionic ring-opening polymerization (AROP) of β-lactams.55 Nylon-3 homo-polymers or co-polymers are far easier to synthesize than are sequence-specific peptides.
At the outset of the present work, we wondered whether we could identify a nylon-3 polymer that would promote the adhesion and growth of endothelial cells (ECs) without the need to append specific peptide motifs. Such a material might ultimately be useful as a coating that would promote the formation of an EC layer on the surface of a vascular graft or stent before implantation. The coating material would be particularly valuable if it discouraged the growth of fibroblasts or smooth muscle cells (SMCs), because invasion of the EC layer on an implant by these types of cells can lead to occlusion of blood vessels and failure of a graft or stent.56–59
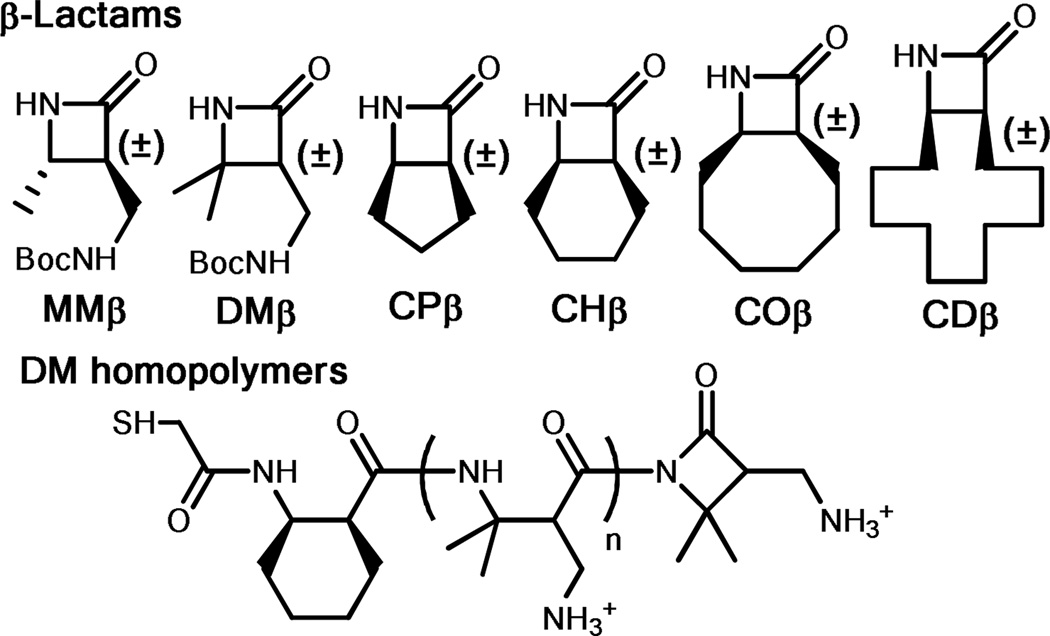
For initial cell adhesion screening, we selected several nylon-3 co-polymers (Figures 1 and S1) from a previously reported set.54 Each sample was generated by co-polymerization of two β-lactams, one that leads to a hydrophobic nylon-3 subunit (CPβ, CHβ, COβ or CDβ) and another that leads to a cationic nylon-3 subunit after side chain deprotection (MMβ or DMβ).54 In addition, we examined cationic homopolymers generated from MMβ or DMβ. These polymers all contain an N-terminal sulfhydryl group, which enables covalent attachment to a glass surface that bears maleimide groups (Figure S2). Previous adhesion evaluation with NIH 3T3 fibroblasts yielded a range of behaviors, with some nylon-3 polymers supporting excellent adhesion and cell morphology and others showing poor adhesion and morphology.30 Poly-DM seemed to be particularly poor in terms of promoting fibroblast growth (Figure S3), which led us to focus on this nylon-3 polymer in our search for EC selectivity.
Figure 1.
β-Lactams and DM homopolymers (poly-DM) used in this study. All β-lactams are racemic and all polymers are heterochiral.
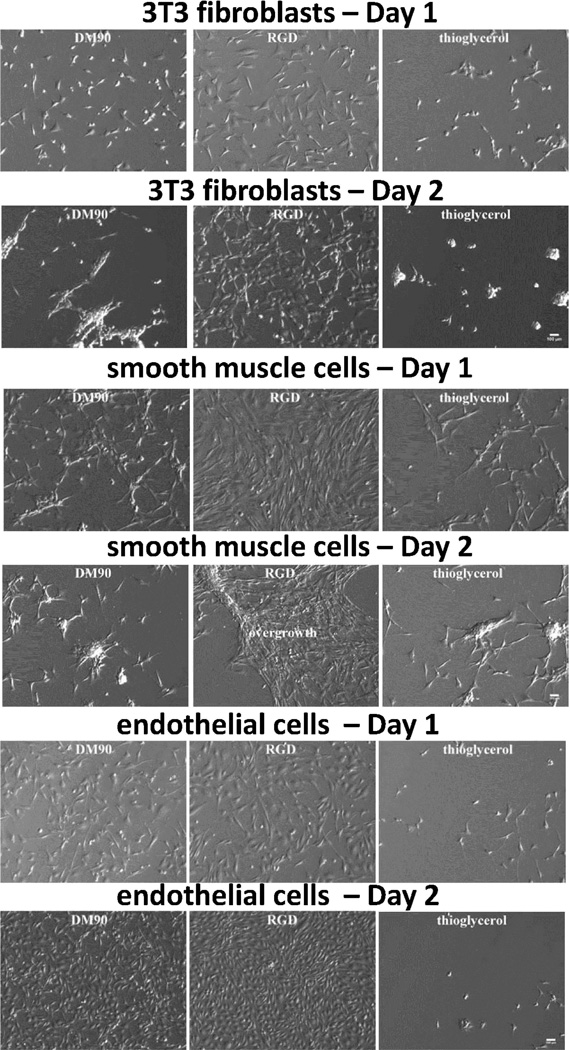
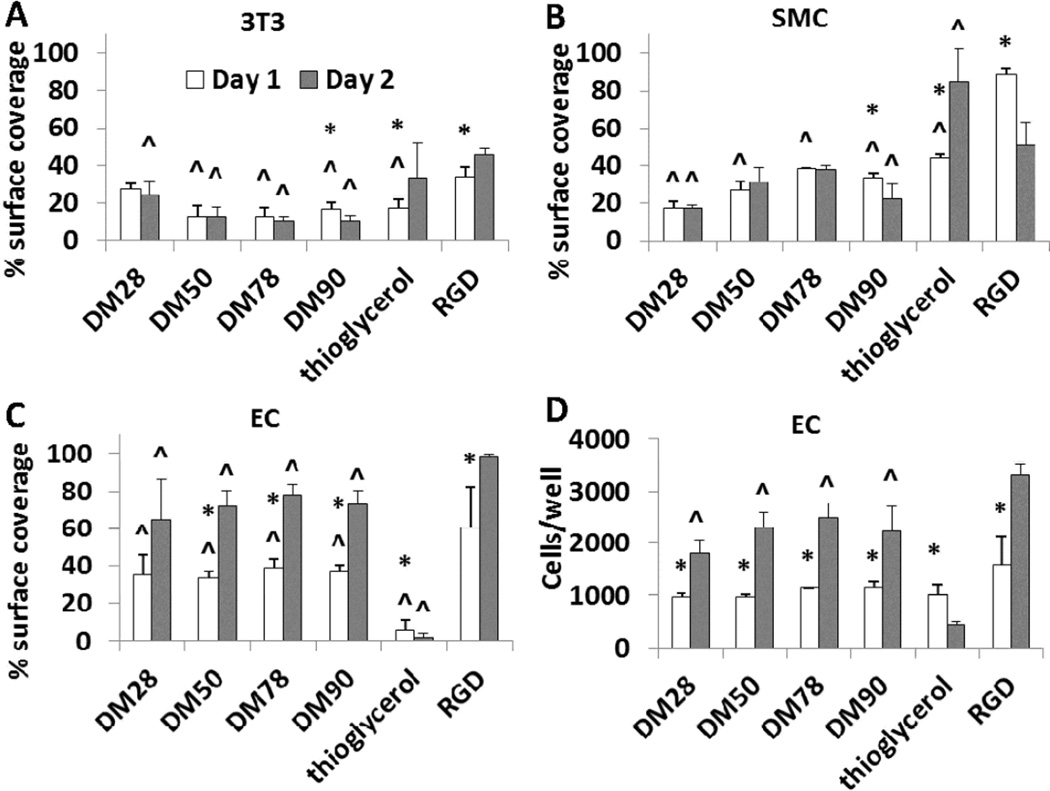
Nylon-3 chain length can influence interactions between cells and surfaces bearing immobilized polymers;54 therefore, poly-DM samples with four different average lengths were examined for cell growth (28-, 50-, 78-, and 90-mer averages, designated DM28, DM50, DM78, and DM90, respectively). A surface modified with the peptide RGDSPC60,61 was used as a positive control, and a thioglycerol-modified surface was used as a negative control. Microscopic evaluation of 3T3 fibroblasts cultured on the four different poly-DM-modified substrates confirmed that all chain lengths were unsupportive of fibroblast growth (DM90 result in Figure 2; remaining data in Figure S4). Initial fibroblast adhesion on poly-DM surfaces was accompanied by normal cell morphology and higher cell density than the negative control, but by Day 2 the fibroblasts on poly-DM had aggregated into clumps and resembled those cultured on the thioglycerol negative control surface. Fibroblasts on the RGD surface remained healthy in appearance at Day 2. Because of fibroblast clumping on poly-DM and negative control surfaces on Day 2, cell number determination via DNA quantification was unreliable. We therefore evaluated cell growth by quantification of surface coverage, based on analysis of microscopic images. A lower extent of surface coverage was observed for poly-DM surfaces relative to the RGD surface on both Day 1 and Day 2 (Figure 3a). In addition, there was no increase in the extent of fibroblast surface coverage on poly-DM surfaces from Day 1 to Day 2.
Figure 2.
Photomicrographs of 3T3 fibroblasts, SMCs and ECs cultured on representative DM90 modified surface and control surfaces. RGD and thioglycerol served as positive and negative controls, respectively. Scale bar = 100 µm.
Figure 3.
Quantification of cells on surfaces, as measured by percentage of surface covered by cells (A, B, C) or by a DNA assay followed by conversion to cell number using a calibration curve (D). (*) p < 0.05 compared to the same surface on Day 2. (^) p < 0.05 compared to positive control (RGD) on Day 2. Scale bar = 100 µm. The apparent reduction in SMC coverage at Day 2 relative to Day 1 (part B) is due to the overgrowth of SMCs (see SMC data in Figure 2).
We next examined the responses of SMCs and ECs to poly-DM-bearing surfaces (DM90 results in Figure 2; remaining data in Figure S5). Microscopic evaluation indicated SMC adhesion to the DM90 surface on Day 1 to be significantly inferior to Day 1 adhesion to the RGD surface. SMCs on the poly-DM surfaces were clumped and exhibited altered cell morphology relative to SMCs on the RGD surface. By Day 2, these trends had intensified, making it impossible to determine accurately adherent cell number via DNA quantification. Moreover, the SMC aggregates on poly-DM surfaces were weakly attached at Day 2 and easily detached upon gentle handling of the culture dish. In contrast, SMCs on the RGD surface possessed appropriate morphology, were firmly attached and proliferated rapidly, leading to overgrowth of the culture by Day 2. There was no increase of surface coverage by SMCs from Day 1 to Day 2 for any of the poly-DM modified surfaces (Figure 3b). The peptide CGREDV, which contains the REDV motif found in fibronectin and has been shown to display EC-selective properties,34 was used to generate a control surface by covalent attachment via the terminal Cys. The resulting surface supported some SMC adhesion on Day 1, but no increase in cell number by Day 2 (Figure S6).
Surfaces bearing DM homopolymers were very effective for culture of ECs, in contrast to the inhospitable environment they provided to fibroblasts and SMCs. On all poly-DM-modified surfaces, ECs cultured in EC-specific medium exhibited a healthy morphology, similar to that obtained on the RGD-bearing surface, on both Days 1 and 2 post-seeding (DM90 results in Figure 2; remaining data in Figure S7). The extent of surface coverage by ECs almost doubled from Day 1 to Day 2, according to image analysis (Figure 3c). EC number was quantified independently via DNA quantification, which indicated that EC density grew ≥2-fold on all poly-DM-modified surfaces between Days 1 and 2, paralleling behavior on the RGD surface (Figure 3d). The number of ECs adherent to the REDV-modified surface on Day 1 was lower than on the polymer- or RGD-modified surfaces, but ECs on the REDV surface had doubled by Day 2 (Figure S6). Although the negative control surface (functionalized with thioglycerol) initially supported some EC adhesion and spreading, this surface did not support EC growth; a net cell loss occurred by Day 2 (Figures 2 and 3). ECs seeded onto a surface bearing DM90 in DMEM, the medium used for culturing 3T3 fibroblasts and SMCs on the nylon-3 surfaces, retained excellent growth characteristics; these ECs formed a confluent monolayer on the DM90 surface on Day 2 post-seeding (Figure S8). This observation indicates that the difference in culture medium formulation across the cell types is not responsible for the poor support of SMC and fibroblast culture on poly-DM-modified surfaces. The ECs cultured in DMEM tended to lose their characteristic cobblestone morphology, which is a common consequence of using a non-EC-specific culture medium.
The results presented above show that surfaces bearing poly-DM support the growth of ECs in preference to fibroblasts or SMCs. Many efforts have been directed toward the generation of surfaces with these properties, for applications in vascular tissue engineering and vascular graft development, but these precedents have relied on incorporation of biomolecules or biomoleculederived units (e.g., specific peptides) into the surface.18,23,33,34,62,63 Our discovery that attachment of a fully synthetic and easily-prepared polymer can render a surface preferentially favorable for EC growth may create new avenues for tissue engineering.
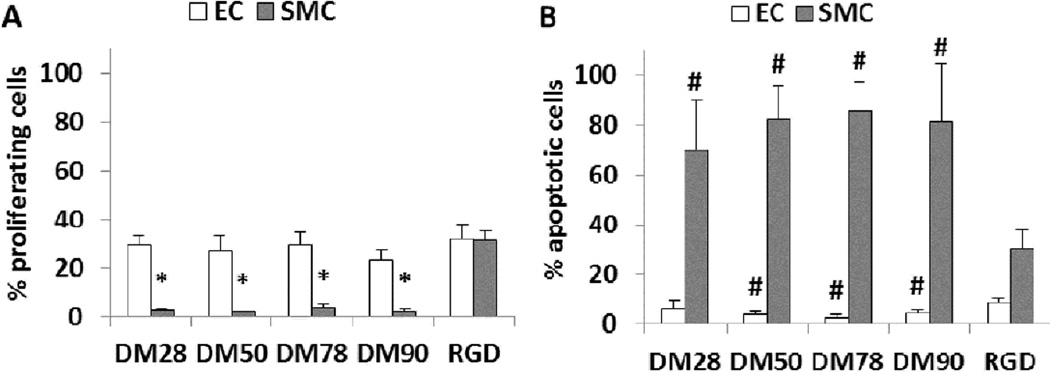
The cell adhesion data (Figures 3) indicate that the preferential support for EC growth relative to SMC or fibroblast growth manifested by poly-DM surfaces tended to emerge after Day 1. Thus, the preference seems to arise not from initial cell adhesion, but rather from effects of the modified surface on cell proliferation and survival. This hypothesis was explored by quantifying EC and SMC proliferation and apoptosis on poly-DM- and RGD-bearing substrates at 30 hours post-seeding. ECs exhibited excellent proliferation (~30%) and low levels of apoptosis (<7%) on surfaces bearing poly-DM at this time point (Figure 4). In contrast, SMC proliferation on poly-DM-modified surfaces was poor (<4%), and apoptosis was very high (70–85%). Staining for apoptosis markers (fragmented DNA) demonstrated that the SMC aggregates formed on poly-DM-modified surfaces (Figure 2) were composed primarily of apoptotic cells (Figure S9). These proliferation and apoptosis data are consistent with the data in Figure 3 in suggesting that poly-DM-modified surfaces provide selective support for culture of ECs relative to other cell types.
Figure 4.
Endothelial cell and smooth muscle cell proliferation (A) and apoptosis (B) on poly-DM modified surfaces. (*) p < 0.0001 compared to RGD surface. (#) p < 0.03 compared to RGD surface.
DM homopolymers presumably lack a molecular substructure that can interact directly with cell-surface adhesion receptors; therefore, the favorability of poly-DM-modified surfaces for EC culture may be mediated by attached proteins.30,31,53,64 We evaluated the adsorption of total protein and of three individual proteins, fibronectin (FN), collagen (Coll), and vitronectin (VN), to nylon-3 modified surfaces (Figure S10), in order to probe for initial adsorption differences that might explain differences in cell culture selectivity. FN, Coll, and VN are the most common cell adhesion proteins found in serum-based cell culture media. However, the results of these studies suggested that there is little or no correlation between total protein or specific protein adsorption patterns and adhesion of any of the three cell types at Day 1.
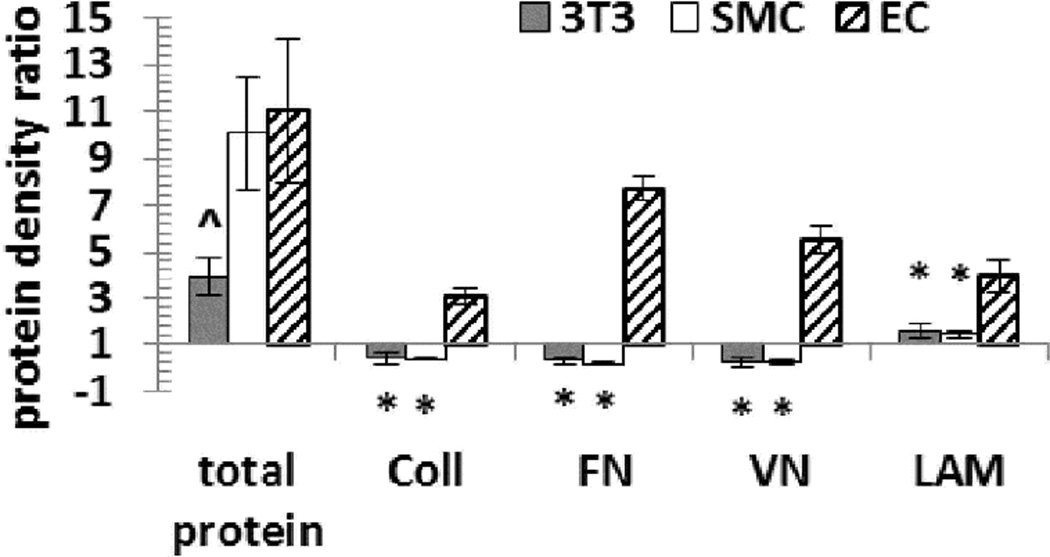
In a further effort to understand the selective promotion of EC culture by poly-DM-modified surfaces, we compared ECM deposition by adherent fibroblasts, SMCs and ECs on functionalized surfaces.65 Protein deposition profiles were quantified on Day 2 after cell seeding and compared with protein adsorption profiles in the absence of cells (i.e., protein adsorption from cell culture medium, Figure S10). The results in Figures 5 and S11 reveal increased surface density of Coll, FN, VN, and laminin (LAM) on EC-cultured poly-DM surfaces, which contrasts with decreased surface density of Coll, FN and VN on fibroblast- and SMC-cultured poly-DM surfaces. Therefore, the inability of the poly-DM-bearing surfaces to support Coll, FN and VN deposition by SMCs and fibroblasts appears to be correlated with the unfavorable culture of these two cell types on poly-DM surfaces.
Figure 5.
Changes in density of adsorbed protein on a DM90- modified surface after 2 days of cell culture, relative to the density adsorbed from culture medium in the absence of cells. Y axis represents the ratio between protein density after 2 days of cell culture and protein density in the absence of cells. A ratio > 1 means increased surface protein density after cell culture. Total adsorbed protein was measured via a NanoOrange assay, while individual proteins (collagen = Coll; fibronectin = FN; vitronectin = VN; laminin = LAM) were measured via immunofluorescent staining. (*) p<0.0001, (^) p<0.002 compared to EC-cultured DM90 surface.
Our findings reveal a new strategy for creating surfaces that selectively favor proliferation of one cell type relative to others, based on a system pertinent to vascular tissue engineering. The use of a completely synthetic material,46–48 lacking evident molecular-level signals that could be recognized by cell-surface receptors, differs significantly from previously described approaches, which rely upon specific ligand-receptor interactions to enable the adhesion of a targeted cell type in preference to other cells.39–42 The class of synthetic materials we have examined, nylon-3 polymers, is advantageous relative to the sequence-specific peptides that have been used in other studies because the polymers are readily prepared on a large scale and resist enzymatic proteolysis. Our initial studies show that the cationic poly-DM homopolymer is superior to a small set of cationic-hydrophobic nylon-3 co-polymers, but it seems possible that other nylon-3 co-polymer compositions could be even more effective. The ease with which nylon-3 co-polymer parameters can be varied will facilitate a broader exploration of structure-activity relationships. In addition, the synthetic accessibility and versatility of this polymer type will enable further studies to elucidate the mechanism by which poly-DM preferentially encourages endothelial cell proliferation.
Supplementary Material
ACKNOWLEDGMENT
This research was supported by the NIH (R21EB013259, to K.S.M and S.H.G.). X. C. was supported in part by the Nanoscale Science and Engineering Center at UW-Madison (DMR-0425880).
Footnotes
ASSOCIATED CONTENT
Supporting Information Available.
Experimental details for monomer and polymer synthesis, substrate surface modification, cell culture, surface cell density and protein density quantification, quantification of cell proliferation and apoptosis. This information is available free of charge via the Internet at http://pubs.acs.org/.
REFERENCES
- 1.Kiick KL. Science. 2007;317:1182. doi: 10.1126/science.1145951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang CY, Song BB, Ao Y, Nowak AP, Abelowitz RB, Korsak RA, Havton LA, Deming TJ, Sofroniew MV. Biomaterials. 2009;30:2881. doi: 10.1016/j.biomaterials.2009.01.056. [DOI] [PubMed] [Google Scholar]
- 3.Martina M, Hutmacher DW. Polym Int. 2007;56:145. [Google Scholar]
- 4.Chan G, Mooney DJ. Trends Biotechnol. 2008;26:382. doi: 10.1016/j.tibtech.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 5.Liao SW, Yu TB, Guan ZB. J Am Chem Soc. 2009;131:17638. doi: 10.1021/ja907097t. [DOI] [PubMed] [Google Scholar]
- 6.Pashuck ET, Stevens MM. Sci Transl Med. 2012;4:160sr4. doi: 10.1126/scitranslmed.3002717. [DOI] [PubMed] [Google Scholar]
- 7.Ryan DM, Nilsson BL. Polym Chem-Uk. 2012;3:18. [Google Scholar]
- 8.Ma PX. Adv Drug Deliver Rev. 2008;60:184. doi: 10.1016/j.addr.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Branco MC, Schneider JP. Acta Biomater. 2009;5:817. doi: 10.1016/j.actbio.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langer R, Tirrell DA. Nature. 2004;428:487. doi: 10.1038/nature02388. [DOI] [PubMed] [Google Scholar]
- 11.Fisher OZ, Khademhosseini A, Langer R, Peppas NA. Accounts Chem Res. 2010;43:419. doi: 10.1021/ar900226q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palermo EF, Kuroda K. Appl Microbiol Biot. 2010;87:1605. doi: 10.1007/s00253-010-2687-z. [DOI] [PubMed] [Google Scholar]
- 13.Cui HG, Webber MJ, Stupp SI. Biopolymers. 2010;94:1. doi: 10.1002/bip.21328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hauser CAE, Zhang SG. Chem Soc Rev. 2010;39:2780. doi: 10.1039/b921448h. [DOI] [PubMed] [Google Scholar]
- 15.Pires MM, Chmielewski J. J Am Chem Soc. 2009;131:2706. doi: 10.1021/ja8088845. [DOI] [PubMed] [Google Scholar]
- 16.Rosso F, Giordano A, Barbarisi M, Barbarisi A. J Cell Physiol. 2004;199:174. doi: 10.1002/jcp.10471. [DOI] [PubMed] [Google Scholar]
- 17.Tibbitt MW, Anseth KS. Biotechnol Bioeng. 2009;103:655. doi: 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fioretta ES, Fledderus JO, Burakowska-Meise EA, Baaijens FPT, Verhaar MC, Bouten CVC. Macromol Biosci. 2012;12:577. doi: 10.1002/mabi.201100315. [DOI] [PubMed] [Google Scholar]
- 19.Liu XH, Holzwarth JM, Ma PX. Macromol Biosci. 2012;12:911. doi: 10.1002/mabi.201100466. [DOI] [PubMed] [Google Scholar]
- 20.Robinson KG, Nie T, Baldwin AD, Yang EC, Kiick KL, Akins RE. J Biomed Mater Res A. 2012;100A:1356. doi: 10.1002/jbm.a.34075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tessmar JK, Gopferich AM. Macromol Biosci. 2007;7:23. doi: 10.1002/mabi.200600096. [DOI] [PubMed] [Google Scholar]
- 22.Wischerhoff E, Uhlig K, Lankenau A, Borner HG, Laschewsky A, Duschl C, Lutz JF. Angew Chem Int Edit. 2008;47:5666. doi: 10.1002/anie.200801202. [DOI] [PubMed] [Google Scholar]
- 23.Jun HW, West JL. Tissue Eng. 2005;11:1133. doi: 10.1089/ten.2005.11.1133. [DOI] [PubMed] [Google Scholar]
- 24.Chuang TW, Masters KS. Biomaterials. 2009;30:5341. doi: 10.1016/j.biomaterials.2009.06.029. [DOI] [PubMed] [Google Scholar]
- 25.Xu F, Nacker JC, Crone WC, Masters KS. Biomaterials. 2008;29:150. doi: 10.1016/j.biomaterials.2007.09.028. [DOI] [PubMed] [Google Scholar]
- 26.Xu FM, Flanagan CE, Ruiz A, Crone WC, Masters KS. Macromol Biosci. 2011;11:257. doi: 10.1002/mabi.201000313. [DOI] [PubMed] [Google Scholar]
- 27.Altmann B, Steinberg T, Giselbrecht S, Gottwald E, Tomakidi P, Bachle-Haas M, Kohal RJ. Biomaterials. 2011;32:8947. doi: 10.1016/j.biomaterials.2011.08.023. [DOI] [PubMed] [Google Scholar]
- 28.Perez CMR, Panitch A, Chmielewski J. Macromol Biosci. 2011;11:1426. doi: 10.1002/mabi.201100230. [DOI] [PubMed] [Google Scholar]
- 29.Grinnell F, Feld MK. Cell. 1979;17:117. doi: 10.1016/0092-8674(79)90300-3. [DOI] [PubMed] [Google Scholar]
- 30.Lee MR, Stahl SS, Gellman SH, Masters KS. J Am Chem Soc. 2009;131:16779. doi: 10.1021/ja9050636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steele JG, Johnson G, Underwood PA. J Biomed Mater Res. 1992;26:861. doi: 10.1002/jbm.820260704. [DOI] [PubMed] [Google Scholar]
- 32.Meinhart JG, Schense JC, Schima H, Gorlitzer M, Hubbell JA, Deutsch M, Zilla P. Tissue Eng. 2005;11:887. doi: 10.1089/ten.2005.11.887. [DOI] [PubMed] [Google Scholar]
- 33.Fittkau MH, Zilla P, Bezuidenhout D, Lutolf M, Human P, Hubbell JA, Davies N. Biomaterials. 2005;26:167. doi: 10.1016/j.biomaterials.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 34.Hubbell JA, Massia SP, Desai NP, Drumheller PD. Bio-Technol. 1991;9:568. doi: 10.1038/nbt0691-568. [DOI] [PubMed] [Google Scholar]
- 35.Liu JC, Heilshorn SC, Tirrell DA. Biomacromolecules. 2004;5:497. doi: 10.1021/bm034340z. [DOI] [PubMed] [Google Scholar]
- 36.Rodin S, Domogatskaya A, Strom S, Hansson EM, Chien KR, Inzunza J, Hovatta O, Tryggvason K. Nat Biotechnol. 2010;28:611. doi: 10.1038/nbt.1620. [DOI] [PubMed] [Google Scholar]
- 37.Willcox PJ, Reinhart-King CA, Lahr SJ, DeGrado WF, Hammer DA. Biomaterials. 2005;26:4757. doi: 10.1016/j.biomaterials.2004.11.060. [DOI] [PubMed] [Google Scholar]
- 38.Galler KM, Aulisa L, Regan KR, D'Souza RN, Hartgerink JD. J Am Chem Soc. 2010;132:3217. doi: 10.1021/ja910481t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Humphries MJ, Akiyama SK, Komoriya A, Olden K, Yamada KM. J Cell Biol. 1986;103:2637. doi: 10.1083/jcb.103.6.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Veleva AN, Heath DE, Cooper SL, Patterson C. Biomaterials. 2008;29:3656. doi: 10.1016/j.biomaterials.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 41.Dee KC, Andersen TT, Bizios R. J Biomed Mater Res. 1998;40:371. doi: 10.1002/(sici)1097-4636(19980605)40:3<371::aid-jbm5>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 42.Gobin AS, West JL. J Biomed Mater Res A. 2003;67:255. doi: 10.1002/jbm.a.10110. [DOI] [PubMed] [Google Scholar]
- 43.Kanie K, Narita Y, Zhao Y, Kuwabara F, Satake M, Honda S, Kaneko H, Yoshioka T, Okochi M, Honda H, Kato R. Biotechnol Bioeng. 2012;109:1808. doi: 10.1002/bit.24459. [DOI] [PubMed] [Google Scholar]
- 44.Bogdanowich-Knipp SJ, Chakrabarti S, Williams TD, Dillman RK, Siahaan TJ. J Pept Res. 1999;53:530. doi: 10.1034/j.1399-3011.1999.00052.x. [DOI] [PubMed] [Google Scholar]
- 45.Wei Y, Ji Y, Xiao L, Lin Q, Ji J. Colloids Surf B Biointerfaces. 2011;84:369. doi: 10.1016/j.colsurfb.2011.01.028. [DOI] [PubMed] [Google Scholar]
- 46.Boese G, Trimpert C, Albrecht W, Malsch G, Groth T, Lendlein A. Tissue Eng. 2007;13:2995. doi: 10.1089/ten.2006.0442. [DOI] [PubMed] [Google Scholar]
- 47.Csaderova L, Martines E, Seunarine K, Gadegaard N, Wilkinson CDW, Riehle MO. Small. 2010;6:2755. doi: 10.1002/smll.201000193. [DOI] [PubMed] [Google Scholar]
- 48.Kanie K, Kato R, Zhao YZ, Narita Y, Okochi M, Honda H. J Pept Sci. 2011;17:479. doi: 10.1002/psc.1355. [DOI] [PubMed] [Google Scholar]
- 49.Mowery BP, Lee SE, Kissounko DA, Epand RF, Epand RM, Weisblum B, Stahl SS, Gellman SH. J Am Chem Soc. 2007;129:15474. doi: 10.1021/ja077288d. [DOI] [PubMed] [Google Scholar]
- 50.Dohm MT, Mowery BP, Czyzewski AM, Stahl SS, Gellman SH, Barron AE. J Am Chem Soc. 2010;132:7957. doi: 10.1021/ja909734n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dane EL, Grinstaff MW. J Am Chem Soc. 2012;134:16255. doi: 10.1021/ja305900r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu R, Chen X, Hayouka Z, Chakraborty S, Falk SP, Weisblum B, Masters KS, Gellman SH. J Am Chem Soc. 2013;135:5270. doi: 10.1021/ja4006404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu R, Vang KZ, Kreeger PK, Gellman SH, Masters KS. J Biomed Mater Res A. 2012;100:2750. doi: 10.1002/jbm.a.34211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu RH, Masters KS, Gellman SH. Biomacromolecules. 2012;13:1100. doi: 10.1021/bm201847n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hashimoto K. Prog Polym Sci. 2000;25:1411. [Google Scholar]
- 56.Chlupac J, Filova E, Bacakova L. Physiol Res. 2009;58:S119. doi: 10.33549/physiolres.931918. [DOI] [PubMed] [Google Scholar]
- 57.Li JH, Ding MM, Fu Q, Tan H, Xie XY, Zhong YP. J Mater Sci-Mater M. 2008;19:2595. doi: 10.1007/s10856-007-3354-5. [DOI] [PubMed] [Google Scholar]
- 58.Parizek M, Novotna K, Bacakova L. Physiol Res. 2011;60:419. doi: 10.33549/physiolres.932038. [DOI] [PubMed] [Google Scholar]
- 59.Garanich JS, Mathura RA, Shi ZD, Tarbell JM. Am J Physiol-Heart C. 2007;292:H3128. doi: 10.1152/ajpheart.00578.2006. [DOI] [PubMed] [Google Scholar]
- 60.Ruoslahti E, Pierschbacher MD. Science. 1987;238:491. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 61.Massia SP, Hubbell JA. Journal of Cell Biology. 1991;114:1089. doi: 10.1083/jcb.114.5.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee JM, Choe W, Kim BK, Seo WW, Lim WH, Kang CK, Kyeong S, Eom KD, Cho HJ, Kim YC, Hur J, Yang HM, Lee YS, Kim HS. Biomaterials. 2012;33:8917. doi: 10.1016/j.biomaterials.2012.08.066. [DOI] [PubMed] [Google Scholar]
- 63.de Mel A, Jell G, Stevens MM, Seifalian AM. Biomacromolecules. 2008;9:2969. doi: 10.1021/bm800681k. [DOI] [PubMed] [Google Scholar]
- 64.Kowalczynska HM, Nowak-Wyrzykowska M, Kolos R, Dobkowski J, Kaminski J. J Biomed Mater Res A. 2008;87A:944. doi: 10.1002/jbm.a.31868. [DOI] [PubMed] [Google Scholar]
- 65.Underwood PA, Bennett FA. J Cell Sci. 1989;93(Pt 4):641. doi: 10.1242/jcs.93.4.641. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.